

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC133139.5 + phase: 0
(1086 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
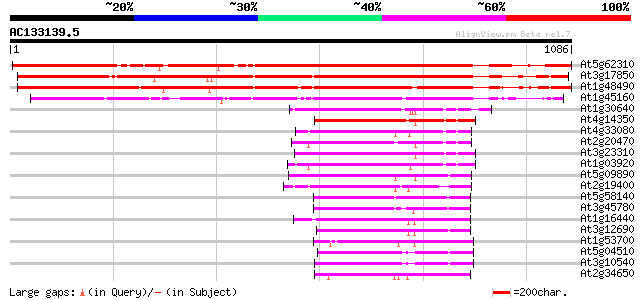
Score E
Sequences producing significant alignments: (bits) Value
At5g62310 IRE (root hair elongation) 1186 0.0
At3g17850 protein kinase like protein 962 0.0
At1g48490 IRE - like protein kinase 920 0.0
At1g45160 similar to IRE homolog 1 dbj|BAA89784.1 607 e-174
At1g30640 putative protein kinase 260 3e-69
At4g14350 protein kinase 247 3e-65
At4g33080 protein kinase like protein 246 4e-65
At2g20470 putative protein kinase 246 5e-65
At3g23310 protein kinase like protein 245 1e-64
At1g03920 putative protein kinase 244 2e-64
At5g09890 protein kinase 243 3e-64
At2g19400 putative protein kinase 227 2e-59
At5g58140 non phototropic hypocotyl 1-like (NPL1) 223 6e-58
At3g45780 nonphototropic hypocotyl 1 (NPH1) 209 5e-54
At1g16440 putative protein kinase 206 6e-53
At3g12690 protein kinase, putative 203 5e-52
At1g53700 auxin-induced protein kinase like protein 184 3e-46
At5g04510 3-phosphoinositide-dependent protein kinase-1 (PDK1) 182 7e-46
At3g10540 3-phosphoinositide-dependent protein kinase-1 like 181 2e-45
At2g34650 protein kinase PINOID 178 2e-44
>At5g62310 IRE (root hair elongation)
Length = 1168
Score = 1186 bits (3067), Expect = 0.0
Identities = 658/1113 (59%), Positives = 792/1113 (71%), Gaps = 145/1113 (13%)
Query: 6 VQGNHAAYAKDFQSPRYQEILRLTSGKKRKNRPDIKSFSHELNSKGVRPFPVWKNRAFG- 64
V+ H AK+ QSPR+Q ILR+TSG+K+K DIKSFSHELNSKGVRPFPVW++RA G
Sbjct: 163 VESTHVGLAKETQSPRFQAILRVTSGRKKKAH-DIKSFSHELNSKGVRPFPVWRSRAVGH 221
Query: 65 -QEIMEEIRAKFEKLKEEVDSDLGGFAGDLVGTLEKIPGSHPEWKEGLEDLLVVARQCAK 123
+EIM IR KF+K KE+VD+DLG FAG LV TLE P S+ E + GLEDLLV ARQCA
Sbjct: 222 MEEIMAAIRTKFDKQKEDVDADLGVFAGYLVTTLESTPESNKELRVGLEDLLVEARQCAT 281
Query: 124 MTAAEFWINCETIVQKLDDKRQDIPVGILKQAHTRLLFILSRCTRLVQFQKESVKEQDHI 183
M A+EFW+ CE IVQKLDDKRQ++P+G LKQAH RLLFIL+RC RLVQF+KES ++HI
Sbjct: 282 MPASEFWLKCEGIVQKLDDKRQELPMGGLKQAHNRLLFILTRCNRLVQFRKESGYVEEHI 341
Query: 184 LGLHQLSDLGVYSEQIMKAEESCGFPPSDHEMAEKLIKKSHGKEQDKPITKQSQADQHAS 243
LG+HQLSDLGVY EQ+++ + EK I+K + K+ Q DQ+++
Sbjct: 342 LGMHQLSDLGVYPEQMVEISRQQDL------LREKEIQKINEKQN----LAGKQDDQNSN 391
Query: 244 VVIDNVEVTTA-STDSTPGSSYKMASWRKLPSAAEKNRVGQDAVK-------------DE 289
D VEV TA STDST S+++M+SW+KLPSAAEKNR + K DE
Sbjct: 392 SGADGVEVNTARSTDST-SSNFRMSSWKKLPSAAEKNRSLNNTPKAKGESKIQPKVYGDE 450
Query: 290 NAENWDTLSCHPDQHSQPSSRTRRPSWGYWGDQQNLLHDDSMICRICEVEIPILHVEEHS 349
NAEN + S QP+S R WG+W D Q + +D+SMICRICEVEIP++HVEEHS
Sbjct: 451 NAENLHSPS------GQPASADRSALWGFWADHQCVTYDNSMICRICEVEIPVVHVEEHS 504
Query: 350 RICTIADKCDLKGLTVNERLERVAETIEMLLDSLTPTSSLHEEFNELSLERNN------- 402
RICTIAD+CDLKG+ VN RLERVAE++E +L+S TP SS+ S +N
Sbjct: 505 RICTIADRCDLKGINVNLRLERVAESLEKILESWTPKSSVTPRAVADSARLSNSSRQEDL 564
Query: 403 --MSSRCSEDMLDLAP--DNTFVADDLNLSREISCEAHSLKPDHGAKISSPESLTPRSPL 458
+S RCS+DMLD P NTF D+LN+ E+S +G K SS SLTP SP
Sbjct: 565 DEISQRCSDDMLDCVPRSQNTFSLDELNILNEMSMT-------NGTKDSSAGSLTPPSPA 617
Query: 459 ITPRTSQIEMLLSVSGRRPISELESYDQINKLVEIARAVANANSCDESAFQDIVDCVEDL 518
TPR SQ+++LLS GR+ ISELE+Y QINKL++IAR+VAN N C S+ +++ +++L
Sbjct: 618 -TPRNSQVDLLLS--GRKTISELENYQQINKLLDIARSVANVNVCGYSSLDFMIEQLDEL 674
Query: 519 RCVIQNRKEDALIVDTFGRRIEKLLQEKYLTLCEQIHDERAESSNSMADEESSVDDDTIR 578
+ VIQ+RK DAL+V+TFGRRIEKLLQEKY+ LC I DE+ +SSN+M DEESS D+DT+R
Sbjct: 675 KYVIQDRKADALVVETFGRRIEKLLQEKYIELCGLIDDEKVDSSNAMPDEESSADEDTVR 734
Query: 579 SLRASPINGFSKDRTSIEDFEIIKPISRGAFGRVFLAQKRSTGDLFAIKVLKKADMIRKN 638
SLRASP+N +KDRTSIEDFEIIKPISRGAFGRVFLA+KR+TGDLFAIKVLKKADMIRKN
Sbjct: 735 SLRASPLNPRAKDRTSIEDFEIIKPISRGAFGRVFLAKKRATGDLFAIKVLKKADMIRKN 794
Query: 639 AVEGILAERDILISVRNPFVVRFYYSFTCKENLYLVMEYLNGGDLYSMLRNLGCLDEDMA 698
AVE ILAER+ILISVRNPFVVRF+YSFTC+ENLYLVMEYLNGGDL+S+LRNLGCLDEDMA
Sbjct: 795 AVESILAERNILISVRNPFVVRFFYSFTCRENLYLVMEYLNGGDLFSLLRNLGCLDEDMA 854
Query: 699 RVYIAEVVLALEYLHSQSIVHRDLKPDNLLIGQDGHIKLTDFGLSKVGLINSTEDLSAPA 758
R+YIAEVVLALEYLHS +I+HRDLKPDNLLI QDGHIKLTDFGLSKVGLINST+DLS +
Sbjct: 855 RIYIAEVVLALEYLHSVNIIHRDLKPDNLLINQDGHIKLTDFGLSKVGLINSTDDLSGES 914
Query: 759 SFTN-GFLVDDEPKPRHVSKREARQQQSIVGTPDYLAPEILLGMGHGTTADWWSVGVILY 817
S N GF +D K +H +++R++ ++VGTPDYLAPEILLGMGHG TADWWSVGVIL+
Sbjct: 915 SLGNSGFFAEDGSKAQHSQGKDSRKKHAVVGTPDYLAPEILLGMGHGKTADWWSVGVILF 974
Query: 818 ELLVGIPPFNADHAQQIFDNIINRDIQWPKHPEEISFEAYDLMNKLLIENPVQRLGVTGA 877
E+LVGIPPFNA+ QQIF+NIINRDI WP PEEIS+EA+DL+NKLL ENPVQRLG TGA
Sbjct: 975 EVLVGIPPFNAETPQQIFENIINRDIPWPNVPEEISYEAHDLINKLLTENPVQRLGATGA 1034
Query: 878 TEVKRHAFFKDVNWDTLARQKVVILILFEFDWFAPYLLISHTWDPTNFLQAMFIPSAEAH 937
EVK+H FFKD+NWDTLARQK AMF+PSAE
Sbjct: 1035 GEVKQHHFFKDINWDTLARQK-----------------------------AMFVPSAEPQ 1065
Query: 938 DTSYFMSRYIWNVEDDEHCAGGSDFYDHSETSSSGSGSDSLDEDAMFIPSAEAHDTSYFM 997
DTSYFMSRYIWN E DE+ GGSDF D ++T SS S
Sbjct: 1066 DTSYFMSRYIWNPE-DENVHGGSDFDDLTDTCSSSS------------------------ 1100
Query: 998 SRYIWNVEDDEHCAGGSDFYDHSETSSSGSGSDSLDEDGDECASLTEFGNSA-LGVQYSF 1056
+N +++ DGDEC SL EFGN L V+YSF
Sbjct: 1101 ----FNTQEE---------------------------DGDECGSLAEFGNGPNLAVKYSF 1129
Query: 1057 SNFSFKNISQLVSIN---MMHISKETPDDSNPS 1086
SNFSFKN+SQL SIN ++ +KE+ + SN S
Sbjct: 1130 SNFSFKNLSQLASINYDLVLKNAKESVEASNQS 1162
>At3g17850 protein kinase like protein
Length = 1296
Score = 962 bits (2486), Expect = 0.0
Identities = 549/1112 (49%), Positives = 696/1112 (62%), Gaps = 136/1112 (12%)
Query: 15 KDFQSPRYQEILRLTSGKKRKNRPDIKSFSHELNSKGVRPFPVWKNRAFG--QEIMEEIR 72
K+ +SPRYQ +LR+TS +++ DIKSFSHELNSKGVRPFP+WK R +E++ IR
Sbjct: 269 KESESPRYQALLRMTSAPRKRFPGDIKSFSHELNSKGVRPFPLWKPRRSNNVEEVLNLIR 328
Query: 73 AKFEKLKEEVDSDLGGFAGDLVGTLEKIPGSHPEWKEGLEDLLVVARQCAKMTAAEFWIN 132
AKFEK KEEV+SDL FA DLVG LEK SHPEW+E EDLL++AR CA T +FW+
Sbjct: 329 AKFEKAKEEVNSDLAVFAADLVGVLEKNAESHPEWEETFEDLLILARSCAMTTPGDFWLQ 388
Query: 133 CETIVQKLDDKRQDIPVGILKQAHTRLLFILSRCTRLVQFQKESVKEQDHILGLHQLSDL 192
CE IVQ LDD+RQ++P G+LKQ HTR+LFIL+RCTRL+QF KES E++ ++ L Q L
Sbjct: 389 CEGIVQDLDDRRQELPPGVLKQLHTRMLFILTRCTRLLQFHKESWGEEEQVVQLRQSRVL 448
Query: 193 GVYSEQIMKAEESCGFPPSDHEMAEKLIKKSHGKEQDKPITKQSQADQHASVVIDNVEVT 252
I K S G S KK++ +EQ K+ + +
Sbjct: 449 ----HSIEKIPPS-GAGRSYSAAKVPSTKKAYSQEQHGLDWKEDAVVRSVPPLAPPENYA 503
Query: 253 TASTDSTPGSSYKMASWRKLPSAAEK-------------------NRVGQDAVKDENAEN 293
++S P + +M+SW+KLPS A K N VG +D+ A
Sbjct: 504 IKESES-PANIDRMSSWKKLPSPALKTVKEAPASEEQNDSKVEPPNIVGSRQGRDDAAVA 562
Query: 294 WDTLSCHPDQHSQPSSRTRRPSWGYWGDQQNLLHDDSMICRICEVEIPILHVEEHSRICT 353
D H S SWGYWG+Q + + S++CRICE E+P HVE+HSR+CT
Sbjct: 563 ILNFPPAKDSHEHSSKHRHNISWGYWGEQPLISEESSIMCRICEEEVPTTHVEDHSRVCT 622
Query: 354 IADKCDLKGLTVNERLERVAETIEMLL------DSLTPTSS----------LHEEFNELS 397
+ADK D KGL+V+ERL VA T++ + DSL S L EE + LS
Sbjct: 623 LADKYDQKGLSVDERLMAVAGTLDKIAETFRHKDSLAAAESPDGMKVSNSHLTEESDVLS 682
Query: 398 LERNNMSSRCSEDMLDLAP--DNTFVADDLNLSREISCEAH-SLKPDHGAKISSPESLTP 454
++ S + SEDMLD P DN+ DDL +SC K D G SS S+TP
Sbjct: 683 PRLSDWSRKGSEDMLDCFPEADNSIFMDDLRGLPLMSCRTRFGPKSDQGMTTSSASSMTP 742
Query: 455 RSPLITPRTSQIEMLLSVSGRRPISELESYDQINKLVEIARAVANANSCDESAFQDIVDC 514
RSP+ TPR IE +L G+ + + Q+++L +IA+ A+A D+ + ++ C
Sbjct: 743 RSPIPTPRPDPIEQILG--GKGTFHDQDDIPQMSELADIAKCAADAIPGDDQSIPFLLSC 800
Query: 515 VEDLRCVIQNRKEDALIVDTFGRRIEKLLQEKYLTLCEQIHDERAESSNSMADEESSVDD 574
+EDLR VI RK DAL V+TFG RIEKL++EKY+ +CE + DE+ + +++ DE++ ++D
Sbjct: 801 LEDLRVVIDRRKFDALTVETFGTRIEKLIREKYVHMCELMDDEKVDLLSTVIDEDAPLED 860
Query: 575 DTIRSLRASPINGFSKDRTSIEDFEIIKPISRGAFGRVFLAQKRSTGDLFAIKVLKKADM 634
D +RSLR SP++ +DRTSI+DFEIIKPISRGAFGRVFLA+KR+TGDLFAIKVLKKADM
Sbjct: 861 DVVRSLRTSPVH--PRDRTSIDDFEIIKPISRGAFGRVFLAKKRTTGDLFAIKVLKKADM 918
Query: 635 IRKNAVEGILAERDILISVRNPFVVRFYYSFTCKENLYLVMEYLNGGDLYSMLRNLGCLD 694
IRKNAVE ILAERDILI+VRNPFVVRF+YSFTC++NLYLVMEYLNGGDLYS+LRNLGCL+
Sbjct: 919 IRKNAVESILAERDILINVRNPFVVRFFYSFTCRDNLYLVMEYLNGGDLYSLLRNLGCLE 978
Query: 695 EDMARVYIAEVVLALEYLHSQSIVHRDLKPDNLLIGQDGHIKLTDFGLSKVGLINSTEDL 754
ED+ RVYIAEVVLALEYLHS+ +VHRDLKPDNLLI DGHIKLTDFGLSKVGLINST+DL
Sbjct: 979 EDIVRVYIAEVVLALEYLHSEGVVHRDLKPDNLLIAHDGHIKLTDFGLSKVGLINSTDDL 1038
Query: 755 SAPASFTNGFLVDDEPK-PRHVSKREARQQQSIVGTPDYLAPEILLGMGHGTTADWWSVG 813
+ PA L ++E + + E R+++S VGTPDYLAPEILLG GHG TADWWSVG
Sbjct: 1039 AGPAVSGTSLLDEEESRLAASEEQLERRKKRSAVGTPDYLAPEILLGTGHGATADWWSVG 1098
Query: 814 VILYELLVGIPPFNADHAQQIFDNIINRDIQWPKHPEEISFEAYDLMNKLLIENPVQRLG 873
+IL+EL+VGIPPFNA+H QQIFDNI+NR I WP PEE+S EA+D++++ L E+P QRLG
Sbjct: 1099 IILFELIVGIPPFNAEHPQQIFDNILNRKIPWPHVPEEMSAEAHDIIDRFLTEDPHQRLG 1158
Query: 874 VTGATEVKRHAFFKDVNWDTLARQKVVILILFEFDWFAPYLLISHTWDPTNFLQAMFIPS 933
GA EVK+H FFKD+NWDTLARQK A F+P+
Sbjct: 1159 ARGAAEVKQHIFFKDINWDTLARQK-----------------------------AAFVPA 1189
Query: 934 AE-AHDTSYFMSRYIWNVEDDEHCAGGSDFYDHSETSSSGSGSDSLDEDAMFIPSAEAHD 992
+E A DTSYF SRY WN D++ F PS E D
Sbjct: 1190 SESAIDTSYFRSRYSWNTSDEQ-----------------------------FFPSGEVPD 1220
Query: 993 TSYFMSRYIWNVEDDEHCAGGSDFYDHSETSSSGSGSDSLDEDG--DECASLTEFGNSAL 1050
S D ++S S + E+G +EC EF S +
Sbjct: 1221 YS-----------------------DADSMTNSSGCSSNHHEEGEAEECEGHAEF-ESGI 1256
Query: 1051 GVQYSFSNFSFKNISQLVSINMMHISKETPDD 1082
V YSFSNFSFKN+SQL SIN +SK D+
Sbjct: 1257 PVDYSFSNFSFKNLSQLASINYDLLSKGWKDE 1288
>At1g48490 IRE - like protein kinase
Length = 1235
Score = 920 bits (2379), Expect = 0.0
Identities = 538/1117 (48%), Positives = 681/1117 (60%), Gaps = 142/1117 (12%)
Query: 15 KDFQSPRYQEILRLTSGKKRKNRPDIKSFSHELNSKGVRPFPVWKNRAFG--QEIMEEIR 72
K+ SPRYQ +LR+TS +++ DIKSFSHELNSKGVRPFP+WK R ++I+ IR
Sbjct: 212 KESDSPRYQALLRMTSAPRKRFPGDIKSFSHELNSKGVRPFPLWKPRRLNNLEDILNLIR 271
Query: 73 AKFEKLKEEVDSDLGGFAGDLVGTLEKIPGSHPEWKEGLEDLLVVARQCAKMTAAEFWIN 132
KF+K KEEV+SDL F GDL+ +K SHPE +EDLLV+A+ CAK T+ EFW+
Sbjct: 272 TKFDKAKEEVNSDLFAFGGDLLDIYDKNKESHPELLVTIEDLLVLAKTCAKTTSKEFWLQ 331
Query: 133 CETIVQKLDDKRQDIPVGILKQAHTRLLFILSRCTRLVQFQKESVKEQDHILGLHQLSDL 192
CE IVQ LDD+RQ++P G+LKQ HTR+LFIL+RCTRL+QF KES +++ + L Q L
Sbjct: 332 CEGIVQDLDDRRQELPPGVLKQLHTRMLFILTRCTRLLQFHKESWGQEEDAVQLRQSGVL 391
Query: 193 GVYSEQIMKAEESCGFPPSD-HEMAEKLIKKSHGKEQDKPITKQSQADQHASVVIDNVEV 251
++ E G S + + KK++ +EQ + + A + E
Sbjct: 392 HSADKRDPTGEVRDGKGSSTANALKVPSTKKAYSQEQRGLNWIEGFFVRPAPLSSPYNE- 450
Query: 252 TTASTDSTPGSSYKMASWRKLPSAAEKNRVGQDAVKDENAENWDT--------------- 296
T+ +P + KM+SW++LPS A K K++N +
Sbjct: 451 -TSKDSESPANIDKMSSWKRLPSPASKGVQEAAVSKEQNDRKVEPPQVVKKLVAISDDMA 509
Query: 297 ------LSCHPDQHSQPSSRTRRPSWGYWGDQQNLLHDDSMICRICEVEIPILHVEEHSR 350
+S S SWGYWG Q + + S+ICRICE EIP HVE+HSR
Sbjct: 510 VAKLPEVSSAKASQEHMSKNRHNISWGYWGHQSCISEESSIICRICEEEIPTTHVEDHSR 569
Query: 351 ICTIADKCDLKGLTVNERLERVAETIEMLLDSLTP----------------TSSLHEEFN 394
IC +ADK D KG+ V+ERL VA T+E + D++ +SL EE +
Sbjct: 570 ICALADKYDQKGVGVDERLMAVAVTLEKITDNVIQKDSLAAVESPEGMKISNASLTEELD 629
Query: 395 ELSLERNNMSSRCSEDMLDLAP--DNTFVADDLNLSREISCEAH-SLKPDHGAKISSPES 451
LS + ++ S R SEDMLD P DN+ DD+ +SC K D G SS S
Sbjct: 630 VLSPKLSDWSRRGSEDMLDCFPETDNSVFMDDMGCLPSMSCRTRFGPKSDQGMATSSAGS 689
Query: 452 LTPRSPLITPRTSQIEMLLSVSGRRPISELESYDQINKLVEIARAVANANSCDESAFQDI 511
+TPRSP+ TPR IE+LL G+ + + + Q+++L +IAR ANA D+ + Q +
Sbjct: 690 MTPRSPIPTPRPDPIELLLE--GKGTFHDQDDFPQMSELADIARCAANAIPVDDQSIQLL 747
Query: 512 VDCVEDLRCVIQNRKEDALIVDTFGRRIEKLLQEKYLTLCEQIHDERAESSNSMADEESS 571
+ C+EDLR VI RK DALIV+TFG RIEKL+QEKYL LCE + DE+ ++ DE++
Sbjct: 748 LSCLEDLRVVIDRRKFDALIVETFGTRIEKLIQEKYLQLCELMDDEKG----TIIDEDAP 803
Query: 572 VDDDTIRSLRASPINGFSKDRTSIEDFEIIKPISRGAFGRVFLAQKRSTGDLFAIKVLKK 631
++DD +RSLR SP++ +DR SI+DFE++K ISRGAFG V LA+K +TGDLFAIKVL+K
Sbjct: 804 LEDDVVRSLRTSPVH--LRDRISIDDFEVMKSISRGAFGHVILARKNTTGDLFAIKVLRK 861
Query: 632 ADMIRKNAVEGILAERDILISVRNPFVVRFYYSFTCKENLYLVMEYLNGGDLYSMLRNLG 691
ADMIRKNAVE ILAERDILI+ RNPFVVRF+YSFTC ENLYLVMEYLNGGD YSMLR +G
Sbjct: 862 ADMIRKNAVESILAERDILINARNPFVVRFFYSFTCSENLYLVMEYLNGGDFYSMLRKIG 921
Query: 692 CLDEDMARVYIAEVVLALEYLHSQSIVHRDLKPDNLLIGQDGHIKLTDFGLSKVGLINST 751
CLDE ARVYIAEVVLALEYLHS+ +VHRDLKPDNLLI DGH+KLTDFGLSKVGLIN+T
Sbjct: 922 CLDEANARVYIAEVVLALEYLHSEGVVHRDLKPDNLLIAHDGHVKLTDFGLSKVGLINNT 981
Query: 752 EDLSAPASFTNGFLVDDEPK-PRHVSKREARQQQSIVGTPDYLAPEILLGMGHGTTADWW 810
+DLS P S LV+++PK P KR A VGTPDYLAPEILLG GHG TADWW
Sbjct: 982 DDLSGPVSSATSLLVEEKPKLPTLDHKRSA------VGTPDYLAPEILLGTGHGATADWW 1035
Query: 811 SVGVILYELLVGIPPFNADHAQQIFDNIINRDIQWPKHPEEISFEAYDLMNKLLIENPVQ 870
SVG+ILYE LVGIPPFNADH QQIFDNI+NR+IQWP PE++S EA DL+++LL E+P Q
Sbjct: 1036 SVGIILYEFLVGIPPFNADHPQQIFDNILNRNIQWPPVPEDMSHEARDLIDRLLTEDPHQ 1095
Query: 871 RLGVTGATEVKRHAFFKDVNWDTLARQKVVILILFEFDWFAPYLLISHTWDPTNFLQAMF 930
RLG GA EVK+H+FFKD++W+TLA+QK A F
Sbjct: 1096 RLGARGAAEVKQHSFFKDIDWNTLAQQK-----------------------------AAF 1126
Query: 931 IPSAE-AHDTSYFMSRYIWNVEDDEHCAGGSDFYDHSETSSSGSGSDSLDEDAMFIPSAE 989
+P +E A DTSYF SRY WN E C P+ E
Sbjct: 1127 VPDSENAFDTSYFQSRYSWNY-SGERC----------------------------FPTNE 1157
Query: 990 AHDTSYFMSRYIWNVEDDEHCAGGSDFYDHSETSSSGSGSDSLDEDGDECASLTEFGNSA 1049
D+S E D C SSG S+ DE D EF ++
Sbjct: 1158 NEDSS----------EGDSLC------------GSSGRLSNHHDEGVDIPCGPAEF-ETS 1194
Query: 1050 LGVQYSFSNFSFKNISQLVSINMMHISKETPDDSNPS 1086
+ Y F NFSFKN+SQL IN +SK D++ PS
Sbjct: 1195 VSENYPFDNFSFKNLSQLAYINYNLMSKGHKDETQPS 1231
>At1g45160 similar to IRE homolog 1 dbj|BAA89784.1
Length = 1067
Score = 607 bits (1566), Expect = e-174
Identities = 418/1058 (39%), Positives = 562/1058 (52%), Gaps = 173/1058 (16%)
Query: 40 IKSFSHELNSKGVRPFPVWKNRAFG--QEIMEEIRAKFEKLKEEVDSDLGGFAGDLVGTL 97
IKSFSHEL +G P + ++ +E++ + ++F+ KE VD L F D+ +
Sbjct: 134 IKSFSHELGPRGGVQTPYPRPHSYNDLKELLGSLHSRFDVAKETVDKKLDVFVRDVKEAM 193
Query: 98 EKIPGSHPEWKEGLEDLLVVARQCAKMTAAEFWINCETIVQKLDDKRQDIPVGILKQAHT 157
EK+ S PE +E E LL VAR C +MT+A+ CE+IVQ L KR+ G++K +
Sbjct: 194 EKMDPSCPEDREMAEQLLDVARACMEMTSAQLRATCESIVQDLTRKRKQCQAGLVKWLFS 253
Query: 158 RLLFILSRCTRLVQFQKESVKEQDHILGLHQLSDLGVYSEQIMKAEESCGFPP--SDHEM 215
+LLFIL+ CTR+V FQKE+ + E+I E G P D
Sbjct: 254 QLLFILTHCTRVVMFQKETEPIDES-----SFRKFKECLERIPALETDWGSTPRVDDSGS 308
Query: 216 AEKLIKKSHGKEQDKPITKQSQADQHA-SVVIDNVEVTTASTDSTPGSSYKMASWRKLPS 274
+++ ++ K K+S + A V+ N A+ + + + S P
Sbjct: 309 GYPEYQRNEAGQKFKRRDKESLESETALDYVVPNDHGNNAAREGYAAAKQEFPSHE--PQ 366
Query: 275 AAEKNRVGQDAVKDENAENWDTLSCHPDQHSQPSSRTRRPSWGYWGDQQNLLHDDSMICR 334
K + + DE D +S P + S D +ICR
Sbjct: 367 FDSKVVEQRFYLSDEYE---DKMSNEPGKELGGS--------------------DYVICR 403
Query: 335 ICEVEIPILHVEEHSRICTIADKCDLKGLTVNERLERVAETIEMLLDSLTPTSSLHEEFN 394
ICE E+P+ H+E HS IC ADKC++ + V+ERL ++ E +E ++DS + S
Sbjct: 404 ICEEEVPLFHLEPHSYICAYADKCEINCVDVDERLLKLEEILEQIIDSRSLNSFTQAGGL 463
Query: 395 ELSLERNN--MSSRCS---------------EDMLDLAPDNTFVADDLNLSREISCEAH- 436
E S+ R + S CS ED+ ++ D F+ D + + I ++H
Sbjct: 464 ENSVLRKSGVASEGCSPKINEWRNKGLEGMFEDLHEM--DTAFI--DESYTYPIHLKSHV 519
Query: 437 SLKPDHGAKISSPESLTPRSPLITPRTSQIEMLLSVSGRRPISELESYDQINKLVEIARA 496
K H A SS S+T S TPRTS + S R E E + L +IAR
Sbjct: 520 GAKFCHHATSSSTGSITSVSSTNTPRTSHFD---SYWLERHCPEQEDLRLMMDLSDIARC 576
Query: 497 VANANSCDESAFQDIVDCVEDLRCVIQNRKEDALIVDTFGRRIEKLLQEKYLTLCEQIHD 556
A+ + E + I+ C++D++ V++ K AL++DTFG RIEKLL EKYL E D
Sbjct: 577 GASTDFSKEGSCDYIMACMQDIQAVLKQGKLKALVIDTFGGRIEKLLCEKYLHARELTAD 636
Query: 557 ERAESSNSMADEESSVDDDTIRSLRASPINGFSKDRTSIEDFEIIKPISRGAFGRVFLAQ 616
+ +S+ + + S +D + A+P KDR SI+DFEIIKPISRGAFG+VFLA+
Sbjct: 637 K-----SSVGNIKES--EDVLEHASATP-QLLLKDRISIDDFEIIKPISRGAFGKVFLAR 688
Query: 617 KRSTGDLFAIKVLKKADMIRKNAVEGILAERDILISVRNPFVVRFYYSFTCKENLYLVME 676
KR+TGD FAIKVLKK DMIRKN +E IL ER+ILI+VR PF+VRF+YSFTC++NLYLVME
Sbjct: 689 KRTTGDFFAIKVLKKLDMIRKNDIERILQERNILITVRYPFLVRFFYSFTCRDNLYLVME 748
Query: 677 YLNGGDLYSMLRNLGCLDEDMARVYIAEVVLALEYLHSQSIVHRDLKPDNLLIGQDGHIK 736
YLNGGDLYS+L+ +GCLDE++AR+YIAE+VLALEYLHS IVHRDLKPDNLLI +GHIK
Sbjct: 749 YLNGGDLYSLLQKVGCLDEEIARIYIAELVLALEYLHSLKIVHRDLKPDNLLIAYNGHIK 808
Query: 737 LTDFGLSKVGLINSTEDLSAPASFTNGFLVDDEPKPRHVSK--REARQQQSIVGTPDYLA 794
LTDFGLSK+GLIN+T DLS S V H K E R + S VGTPDYLA
Sbjct: 809 LTDFGLSKIGLINNTIDLSGHESD-----VSPRTNSHHFQKNQEEERIRHSAVGTPDYLA 863
Query: 795 PEILLGMGHGTTADWWSVGVILYELLVGIPPFNADHAQQIFDNIINRDIQWPKHPEEISF 854
PEILLG HG ADWWS G++L+ELL GIPPF A ++IFDNI+N + WP P E+S+
Sbjct: 864 PEILLGTEHGYAADWWSAGIVLFELLTGIPPFTASRPEKIFDNILNGKMPWPDVPGEMSY 923
Query: 855 EAYDLMNKLLIENPVQRLGVTGATEVKRHAFFKDVNWDTLARQKVVILILFEFDWFAPYL 914
EA DL+N+LL+ P +RLG GA EVK H FF+ V+W+ LA QK
Sbjct: 924 EAQDLINRLLVHEPEKRLGANGAAEVKSHPFFQGVDWENLALQK---------------- 967
Query: 915 LISHTWDPTNFLQAMFIPSAEA-HDTSYFMSRYIWNVEDDEHCAGGSDFYDHSETSSSGS 973
A F+P E+ +DTSYF+SR+ + C+ D ++SGS
Sbjct: 968 -------------AAFVPQPESINDTSYFVSRF-----SESSCS------DTETGNNSGS 1003
Query: 974 GSDSLDEDAMFIPSAEAHDTSYFMSRYIWNVEDDEHCAGGSDFYDHSETSSSGSGSDSLD 1033
DS DE LD
Sbjct: 1004 NPDSGDE---------------------------------------------------LD 1012
Query: 1034 EDGDECASLTEFGNSALGVQYSFSNFSFKNISQLVSIN 1071
E C +L +F + S NFSFKN+SQL SIN
Sbjct: 1013 E----CTNLEKFDSPP--YYLSLINFSFKNLSQLASIN 1044
>At1g30640 putative protein kinase
Length = 562
Score = 260 bits (664), Expect = 3e-69
Identities = 159/423 (37%), Positives = 240/423 (56%), Gaps = 59/423 (13%)
Query: 543 LQEKYLTLCEQIHDERAESSNSMADEESSVDD--DTIRSLRASPIN--GFSKDRTSIEDF 598
+Q+K L ++ + R+ ++AD + +V+D D +++ + + + ++DF
Sbjct: 64 IQKKSL---QERKERRSILEQNLADADVTVEDKMDILKNFEKKEMEYMRLQRQKMGVDDF 120
Query: 599 EIIKPISRGAFGRVFLAQKRSTGDLFAIKVLKKADMIRKNAVEGILAERDILISVRNPFV 658
E++ I RGAFG V + +++STG ++A+K LKK++M+R+ VE + AER++L V +PF+
Sbjct: 121 ELLSIIGRGAFGEVRICKEKSTGSVYAMKKLKKSEMLRRGQVEHVKAERNVLAEVDSPFI 180
Query: 659 VRFYYSFTCKENLYLVMEYLNGGDLYSMLRNLGCLDEDMARVYIAEVVLALEYLHSQSIV 718
V+ YSF E+LYL+MEYL GGD+ ++L L ED R Y+A+ +LA+E +H + V
Sbjct: 181 VKLCYSFQDDEHLYLIMEYLPGGDMMTLLMRKDTLREDETRFYVAQTILAIESIHKHNYV 240
Query: 719 HRDLKPDNLLIGQDGHIKLTDFGLSKVGLINSTEDLSAPASFTNGFLVDDEPKPR----H 774
HRD+KPDNLLI ++GHIKL+DFGLSK + D A LVD KP
Sbjct: 241 HRDIKPDNLLITRNGHIKLSDFGLSKSLESKNFPDFKAE-------LVDRSTKPAAEHDR 293
Query: 775 VSK-----REARQQQ-------------SIVGTPDYLAPEILLGMGHGTTADWWSVGVIL 816
+SK R +Q+Q S VGTPDY+APE+LL G+G DWWS+G I+
Sbjct: 294 LSKPPSAPRRTQQEQLLHWQQNRRTLAFSTVGTPDYIAPEVLLKKGYGMECDWWSLGAIM 353
Query: 817 YELLVGIPPFNADHAQQIFDNIINRDIQWP---KHPEE--ISFEAYDLMNKLLIENPVQR 871
+E+LVG PPF ++ I+N W K P+E +S E DL+ +LL N QR
Sbjct: 354 FEMLVGFPPFYSEEPLATCRKIVN----WKTCLKFPDEAKLSIEVKDLIRRLLC-NVEQR 408
Query: 872 LGVTGATEVKRHAFFKDVNWDTLARQKVVILILFEFDWFAPYL-LISHTWDPTNFLQAMF 930
LG G E+K H +F+ V W+ L ++ APY+ + H D NF +
Sbjct: 409 LGTKGVHEIKAHPWFRGVEWERL------------YESNAPYIPQVKHELDTQNFEKFDE 456
Query: 931 IPS 933
+PS
Sbjct: 457 VPS 459
>At4g14350 protein kinase
Length = 551
Score = 247 bits (630), Expect = 3e-65
Identities = 139/329 (42%), Positives = 199/329 (60%), Gaps = 21/329 (6%)
Query: 590 KDRTSIEDFEIIKPISRGAFGRVFLAQKRSTGDLFAIKVLKKADMIRKNAVEGILAERDI 649
+ + +DFE + I +GAFG V + +++ TG+++A+K LKK++M+R+ VE + AER++
Sbjct: 111 RHKMGADDFEPLTMIGKGAFGEVRICREKGTGNVYAMKKLKKSEMLRRGQVEHVKAERNL 170
Query: 650 LISVRNPFVVRFYYSFTCKENLYLVMEYLNGGDLYSMLRNLGCLDEDMARVYIAEVVLAL 709
L V + +V+ Y SF +E LYL+MEYL GGD+ ++L L ED AR YI E VLA+
Sbjct: 171 LAEVDSNCIVKLYCSFQDEEYLYLIMEYLPGGDMMTLLMRKDTLTEDEARFYIGETVLAI 230
Query: 710 EYLHSQSIVHRDLKPDNLLIGQDGHIKLTDFGLSK-VGLINSTEDLSAPASFTNGFLVDD 768
E +H + +HRD+KPDNLL+ +DGH+KL+DFGL K + N E A +G L D
Sbjct: 231 ESIHKHNYIHRDIKPDNLLLDKDGHMKLSDFGLCKPLDCSNLQEKDFTVARNVSGALQSD 290
Query: 769 EPKPRHVSKREARQQQ-------------SIVGTPDYLAPEILLGMGHGTTADWWSVGVI 815
R V+ R +Q+Q S VGTPDY+APE+LL G+G DWWS+G I
Sbjct: 291 ---GRPVATRRTQQEQLLNWQRNRRMLAYSTVGTPDYIAPEVLLKKGYGMECDWWSLGAI 347
Query: 816 LYELLVGIPPFNADHAQQIFDNIINRDIQWPKHPEEI--SFEAYDLMNKLLIENPVQRLG 873
+YE+LVG PPF +D I+N + K P+E+ S EA DL+ +LL N QRLG
Sbjct: 348 MYEMLVGFPPFYSDDPMTTCRKIVNWR-NYLKFPDEVRLSPEAKDLICRLLC-NVEQRLG 405
Query: 874 VTGATEVKRHAFFKDVNWDTLARQKVVIL 902
GA E+K H +F+ W L + K +
Sbjct: 406 TKGADEIKGHPWFRGTEWGKLYQMKAAFI 434
>At4g33080 protein kinase like protein
Length = 519
Score = 246 bits (629), Expect = 4e-65
Identities = 144/366 (39%), Positives = 211/366 (57%), Gaps = 32/366 (8%)
Query: 554 IHDERAESSNSMADEESSVDDDTIRSLRASPINGFSKDRTSIEDFEIIKPISRGAFGRVF 613
I + + SS +E+ ++ D R + + +++ S++DFE++ I RGAFG V
Sbjct: 52 ILERKLASSGVPKEEQINMIKDLER--KETEFMRLKRNKISVDDFELLTIIGRGAFGEVR 109
Query: 614 LAQKRSTGDLFAIKVLKKADMIRKNAVEGILAERDILISVRNPFVVRFYYSFTCKENLYL 673
L ++R +G+++A+K LKK++M+ + VE + AER++L V + ++V+ YYSF E LYL
Sbjct: 110 LCRERKSGNIYAMKKLKKSEMVMRGQVEHVRAERNLLAEVESHYIVKLYYSFQDPEYLYL 169
Query: 674 VMEYLNGGDLYSMLRNLGCLDEDMARVYIAEVVLALEYLHSQSIVHRDLKPDNLLIGQDG 733
+MEYL GGD+ ++L L ED+AR YIA+ VLA+E +H + +HRD+KPDNLL+ +DG
Sbjct: 170 IMEYLPGGDMMTLLMREDTLREDVARFYIAQSVLAIESIHRYNYIHRDIKPDNLLLDKDG 229
Query: 734 HIKLTDFGLSK-----------VGLINSTEDLSAPASFTNGFLVDDEPKP--------RH 774
H+KL+DFGL K E +S P F D + +H
Sbjct: 230 HMKLSDFGLCKPLDCRNLPSIQENRATDDETMSEPMDVDRCFPDTDNKRSWRSPQEQLQH 289
Query: 775 VSKREARQQQSIVGTPDYLAPEILLGMGHGTTADWWSVGVILYELLVGIPPFNADHAQQI 834
+ S VGTPDY+APE+LL G+G DWWS+G I+YE+LVG PPF AD
Sbjct: 290 WQMNRRKLAFSTVGTPDYIAPEVLLKKGYGMECDWWSLGAIMYEMLVGYPPFYADDPIST 349
Query: 835 FDNIINRDIQWPKH---PEEISF--EAYDLMNKLLIENPVQRLGV-TGATEVKRHAFFKD 888
I++ W H PE+ F EA DL+ +LL N RLG GA ++K H +FKD
Sbjct: 350 CRKIVH----WRNHLKFPEDAKFSSEAKDLICRLLC-NVDHRLGTGGGAQQIKDHPWFKD 404
Query: 889 VNWDTL 894
V W+ L
Sbjct: 405 VVWEKL 410
>At2g20470 putative protein kinase
Length = 596
Score = 246 bits (628), Expect = 5e-65
Identities = 139/374 (37%), Positives = 218/374 (58%), Gaps = 35/374 (9%)
Query: 545 EKYLTLCEQIHDERAESSNSMADEESSVDDDT----IRSLRASPINGFSKDRTSIEDFEI 600
++ + + ++ + R+ +AD + S +D + + + + + DF++
Sbjct: 67 KEQMKILQERKERRSMLEQKLADADVSEEDQNNLLKFLEKKETEYMRLQRHKLGVADFDL 126
Query: 601 IKPISRGAFGRVFLAQKRSTGDLFAIKVLKKADMIRKNAVEGILAERDILISVRNPFVVR 660
+ I +GAFG V + ++++TG ++A+K LKKA+M+R+ VE + AER++L V + ++V+
Sbjct: 127 LTMIGKGAFGEVRVCREKTTGQVYAMKKLKKAEMLRRGQVEHVRAERNLLAEVDSNYIVK 186
Query: 661 FYYSFTCKENLYLVMEYLNGGDLYSMLRNLGCLDEDMARVYIAEVVLALEYLHSQSIVHR 720
Y SF ++LYLVMEYL GGD+ ++L L E+ A+ Y+AE VLA+E +H + +HR
Sbjct: 187 LYCSFQDDDHLYLVMEYLPGGDMMTLLMRKDTLTEEEAKFYVAETVLAIESIHRHNYIHR 246
Query: 721 DLKPDNLLIGQDGHIKLTDFGLSKVGLINSTEDLSA--PASFTNGFLVDDEPKPRHVSKR 778
D+KPDNLL+ + GH++L+DFGL K D SA F+N E + + +
Sbjct: 247 DIKPDNLLLDRYGHLRLSDFGLCK------PLDCSAIGENDFSNNSNGSTEQEAGSTAPK 300
Query: 779 EARQQQ-------------SIVGTPDYLAPEILLGMGHGTTADWWSVGVILYELLVGIPP 825
+Q+Q S VGTPDY+APE+LL G+G DWWS+G I+YE+LVG PP
Sbjct: 301 RTQQEQLEHWQRNRRTLAYSTVGTPDYIAPEVLLKKGYGMECDWWSLGAIMYEMLVGYPP 360
Query: 826 FNADHAQQIFDNIINRDIQWPKH---PEE--ISFEAYDLMNKLLIENPVQRLGVTGATEV 880
F +D I+N W H PEE +S EA DL+N LL + +RLG GA E+
Sbjct: 361 FYSDDPMSTCRKIVN----WKSHLKFPEEAILSREAKDLINSLLC-SVRRRLGSKGADEL 415
Query: 881 KRHAFFKDVNWDTL 894
K H +F+ V+WDT+
Sbjct: 416 KAHTWFETVDWDTI 429
>At3g23310 protein kinase like protein
Length = 568
Score = 245 bits (625), Expect = 1e-64
Identities = 140/370 (37%), Positives = 212/370 (56%), Gaps = 21/370 (5%)
Query: 552 EQIHDERAESSNSMADEESSVDD--DTIRSLRASPINGFSKDRTSI--EDFEIIKPISRG 607
+Q + R N +A E S ++ + ++ L + R + +DFE + I +G
Sbjct: 70 QQRKERRDMLENKLAAAEVSEEEQKNLLKDLEKKETEYMRRQRHKMGTDDFEPLTMIGKG 129
Query: 608 AFGRVFLAQKRSTGDLFAIKVLKKADMIRKNAVEGILAERDILISVRNPFVVRFYYSFTC 667
AFG V + ++++TG+++A+K LKK++M+R+ VE + AER++L V + +V+ Y SF
Sbjct: 130 AFGEVRICREKTTGNVYAMKKLKKSEMLRRGQVEHVKAERNLLAEVDSNCIVKLYCSFQD 189
Query: 668 KENLYLVMEYLNGGDLYSMLRNLGCLDEDMARVYIAEVVLALEYLHSQSIVHRDLKPDNL 727
+E LYL+MEYL GGD+ ++L L ED AR Y+ E VLA+E +H + +HRD+KPDNL
Sbjct: 190 EEYLYLIMEYLPGGDMMTLLMRKDTLTEDEARFYVGETVLAIESIHKHNYIHRDIKPDNL 249
Query: 728 LIGQDGHIKLTDFGLSKV--GLINSTEDLSAPASFTNGFLVDDEPKPRHVSKREARQQQ- 784
L+ + GH+KL+DFGL K I +D + + D P ++ + Q Q
Sbjct: 250 LLDRSGHMKLSDFGLCKPLDCSILQEKDFVVAHNLSGALQSDGRPVAPRRTRSQMEQLQN 309
Query: 785 ----------SIVGTPDYLAPEILLGMGHGTTADWWSVGVILYELLVGIPPFNADHAQQI 834
S VGTPDY+APE+LL G+G DWWS+G I+YE+LVG PPF +D
Sbjct: 310 WQRNRRMLAYSTVGTPDYIAPEVLLKKGYGMECDWWSLGAIMYEMLVGFPPFYSDEPMTT 369
Query: 835 FDNIINRDIQWPKHPEEI--SFEAYDLMNKLLIENPVQRLGVTGATEVKRHAFFKDVNWD 892
I+N + K P+E+ S EA DL+ +LL N QR+G GA E+K H +F V W+
Sbjct: 370 CRKIVNWK-NYLKFPDEVRLSPEAKDLICRLLC-NVEQRIGTKGANEIKEHPWFSGVEWE 427
Query: 893 TLARQKVVIL 902
L + K +
Sbjct: 428 KLYQMKAAFI 437
>At1g03920 putative protein kinase
Length = 569
Score = 244 bits (623), Expect = 2e-64
Identities = 145/384 (37%), Positives = 219/384 (56%), Gaps = 27/384 (7%)
Query: 539 IEKLLQEKYLTLCEQIHDERAESSNSMADEESSVDDDT----IRSLRASPINGFSKDRTS 594
IE +E+ L E+ + R +AD + +D T + + + +
Sbjct: 75 IENHYKEQMKNLNER-KERRTTLEKKLADADVCEEDQTNLMKFLEKKETEYMRLQRHKMG 133
Query: 595 IEDFEIIKPISRGAFGRVFLAQKRSTGDLFAIKVLKKADMIRKNAVEGILAERDILISVR 654
+DFE++ I +GAFG V + ++ +TG +FA+K LKK++M+R+ VE + AER++L V
Sbjct: 134 ADDFELLTMIGKGAFGEVRVVREINTGHVFAMKKLKKSEMLRRGQVEHVRAERNLLAEVD 193
Query: 655 NPFVVRFYYSFTCKENLYLVMEYLNGGDLYSMLRNLGCLDEDMARVYIAEVVLALEYLHS 714
+ +V+ Y SF E LYL+MEYL GGD+ ++L L ED A+ YIAE VLA+E +H+
Sbjct: 194 SNCIVKLYCSFQDNEYLYLIMEYLPGGDMMTLLMRKDTLSEDEAKFYIAESVLAIESIHN 253
Query: 715 QSIVHRDLKPDNLLIGQDGHIKLTDFGLSK---VGLINSTEDLSAPASFTNGFLVDDEPK 771
++ +HRD+KPDNLL+ + GH++L+DFGL K +I+ ED + + + G
Sbjct: 254 RNYIHRDIKPDNLLLDRYGHLRLSDFGLCKPLDCSVIDG-EDFTVGNAGSGGGSESVSTT 312
Query: 772 PR--------HVSKREARQQQSIVGTPDYLAPEILLGMGHGTTADWWSVGVILYELLVGI 823
P+ H K S VGTPDY+APE+LL G+G DWWS+G I+YE+LVG
Sbjct: 313 PKRSQQEQLEHWQKNRRMLAYSTVGTPDYIAPEVLLKKGYGMECDWWSLGAIMYEMLVGY 372
Query: 824 PPFNADHAQQIFDNIINRDIQWPKH---PEE--ISFEAYDLMNKLLIENPVQRLGVTGAT 878
PPF AD I+N W H PEE +S A DL+ KLL + QRLG TGA+
Sbjct: 373 PPFYADDPMSTCRKIVN----WKTHLKFPEESRLSRGARDLIGKLLC-SVNQRLGSTGAS 427
Query: 879 EVKRHAFFKDVNWDTLARQKVVIL 902
++K H +F+ V W+ + + + +
Sbjct: 428 QIKAHPWFEGVQWEKIYQMEAAFI 451
>At5g09890 protein kinase
Length = 515
Score = 243 bits (621), Expect = 3e-64
Identities = 141/378 (37%), Positives = 220/378 (57%), Gaps = 25/378 (6%)
Query: 540 EKLLQEKYLTLCEQIHDE---RAESSNSMADEESSVD--DDTIRSL--RASPINGFSKDR 592
++ ++ Y + +H+ R E + + + V+ D+ +R+L R + + +
Sbjct: 37 KQFIENHYKNYLQGLHERMERRREFQRKVQEAQLPVEEQDEMMRNLARRETEYMRLQRRK 96
Query: 593 TSIEDFEIIKPISRGAFGRVFLAQKRSTGDLFAIKVLKKADMIRKNAVEGILAERDILIS 652
I+DFE++ I +GAFG V L + RST +++A+K LKK +M+ + VE + +ER++L
Sbjct: 97 IGIDDFELLTVIGKGAFGEVRLCRLRSTSEVYAMKKLKKTEMLSRGQVEHVRSERNLLAE 156
Query: 653 VRNPFVVRFYYSFTCKENLYLVMEYLNGGDLYSMLRNLGCLDEDMARVYIAEVVLALEYL 712
V + ++V+ +YSF E LYL+MEYL GGD+ ++L L ED+AR YIAE +LA+ +
Sbjct: 157 VDSRYIVKLFYSFQDSECLYLIMEYLPGGDIMTLLMREDILSEDVARFYIAESILAIHSI 216
Query: 713 HSQSIVHRDLKPDNLLIGQDGHIKLTDFGLSK------VGLINSTEDLSAPASFTNGFLV 766
H + VHRD+KPDNL++ + GH+KL+DFGL K L+ +++ + S
Sbjct: 217 HQHNYVHRDIKPDNLILDKSGHLKLSDFGLCKPLDDKYSSLLLEDDEMLSQDSENQSGKS 276
Query: 767 DDEPKPRHVSKREARQQQ--------SIVGTPDYLAPEILLGMGHGTTADWWSVGVILYE 818
D + P + K + Q + S VGT DY+APE+LL G+G DWWS+G ILYE
Sbjct: 277 DADKAPWQMPKEQLLQWKRNRRALAYSTVGTLDYMAPEVLLKKGYGMECDWWSLGAILYE 336
Query: 819 LLVGIPPFNADHAQQIFDNIINRDI--QWPKHPEEISFEAYDLMNKLLIENPVQRLGVTG 876
+LVG PPF +D + IIN + ++P+ P +IS EA DL+ +LL + RLG G
Sbjct: 337 MLVGYPPFCSDDPRITCRKIINWRVCLKFPEEP-KISDEARDLICRLLCDVD-SRLGTRG 394
Query: 877 ATEVKRHAFFKDVNWDTL 894
E+K H +FK WD L
Sbjct: 395 VEEIKSHPWFKGTPWDKL 412
>At2g19400 putative protein kinase
Length = 509
Score = 227 bits (579), Expect = 2e-59
Identities = 138/387 (35%), Positives = 215/387 (54%), Gaps = 48/387 (12%)
Query: 531 IVDTFGRRIEKLLQEKYLTLCEQIHDERAESSNSMADEESSVDDDTIRSLRASPINGFSK 590
I + + RR+ + Q K + +++ S + E+ + +D R + + +
Sbjct: 43 IENHYNRRMRHIQQRKER---RWVLEQKIASLDVSEKEQLELLEDLQR--KETEYTRLMR 97
Query: 591 DRTSIEDFEIIKPISRGAFGRVFLAQKRSTGDLFAIKVLKKADMIRKNAVEGILAERDIL 650
+R ++DF+++ I RGAFG V L +++ TG+++A+K LKK++M+ + VE + AER++L
Sbjct: 98 NRLCVDDFDLLSIIGRGAFGEVRLCREKKTGNIYAMKKLKKSEMLSRGQVEHVRAERNLL 157
Query: 651 ISVRNPFVVRFYYSFTCKENLYLVMEYLNGGDLYSMLRNLGCLDEDMARVYIAEVVLALE 710
V + +V+ YYSF E LYL+MEYL+GGD+ ++L L E +AR YIA+ VLA+E
Sbjct: 158 AEVASDCIVKLYYSFQDPEYLYLIMEYLSGGDVMTLLMREETLTETVARFYIAQSVLAIE 217
Query: 711 YLHSQSIVHRDLKPDNLLIGQDGHIKLTDFGLSK------VGLINSTEDLSAPASFTNGF 764
+H + VHRD+KPDNLL+ + GH+KL+DFGL K + +N E L+ N
Sbjct: 218 SIHKHNYVHRDIKPDNLLLDKYGHMKLSDFGLCKPLDCRNISAMNVNEPLN--DENINES 275
Query: 765 LVDDE---------------PKPRHVSKREARQQQSIVGTPDYLAPEILLGMGHGTTADW 809
+ DE + +H + S VGTPDY+APE+LL G+G DW
Sbjct: 276 IDGDENCSIGRRGRRWKSPLEQLQHWQINRRKLAYSTVGTPDYIAPEVLLKKGYGVECDW 335
Query: 810 WSVGVILYELLVGIPPFNADHAQQIFDNIINRDIQWPKHPEEISFEAYDLMNKLLIENPV 869
WS+G I+YE+LVG PPF +D ++ EA DL+ +LL ++
Sbjct: 336 WSLGAIMYEMLVGYPPFYSDDP-----------------GARLTPEARDLICRLLCDSE- 377
Query: 870 QRLGV--TGATEVKRHAFFKDVNWDTL 894
RLG GA ++K H +FKDV W+ L
Sbjct: 378 HRLGSHGAGAEQIKAHTWFKDVEWEKL 404
>At5g58140 non phototropic hypocotyl 1-like (NPL1)
Length = 915
Score = 223 bits (567), Expect = 6e-58
Identities = 119/306 (38%), Positives = 184/306 (59%), Gaps = 7/306 (2%)
Query: 589 SKDRTSIEDFEIIKPISRGAFGRVFLAQKRSTGDLFAIKVLKKADMIRKNAVEGILAERD 648
S + + F+ IKP+ G G V L + + TG+L+A+K ++K M+ +N ER+
Sbjct: 568 SGETVGLHHFKPIKPLGSGDTGSVHLVELKGTGELYAMKAMEKTMMLNRNKAHRACIERE 627
Query: 649 ILISVRNPFVVRFYYSFTCKENLYLVMEYLNGGDLYSML--RNLGCLDEDMARVYIAEVV 706
I+ + +PF+ Y SF ++ L+ ++ GG+L+++L + + L ED AR Y AEVV
Sbjct: 628 IISLLDHPFLPTLYASFQTSTHVCLITDFCPGGELFALLDRQPMKILTEDSARFYAAEVV 687
Query: 707 LALEYLHSQSIVHRDLKPDNLLIGQDGHIKLTDFGLSKVGLINSTEDLSAPASFTNGFLV 766
+ LEYLH IV+RDLKP+N+L+ +DGHI L DF LS + T L PA+ +
Sbjct: 688 IGLEYLHCLGIVYRDLKPENILLKKDGHIVLADFDLSF--MTTCTPQLIIPAAPSKRRRS 745
Query: 767 DDEPKPRHVSKREARQQQSIVGTPDYLAPEILLGMGHGTTADWWSVGVILYELLVGIPPF 826
+P P V++ + Q S VGT +Y+APEI+ G GH + DWW++G++LYE+L G PF
Sbjct: 746 KSQPLPTFVAE-PSTQSNSFVGTEEYIAPEIITGAGHTSAIDWWALGILLYEMLYGRTPF 804
Query: 827 NADHAQQIFDNIINRDIQWPKHPEEISFEAYDLMNKLLIENPVQRLGVT-GATEVKRHAF 885
+ Q+ F NI+++D+ +P +S L+N LL +P RLG GA E+K+HAF
Sbjct: 805 RGKNRQKTFANILHKDLTFPS-SIPVSLVGRQLINTLLNRDPSSRLGSKGGANEIKQHAF 863
Query: 886 FKDVNW 891
F+ +NW
Sbjct: 864 FRGINW 869
>At3g45780 nonphototropic hypocotyl 1 (NPH1)
Length = 996
Score = 209 bits (533), Expect = 5e-54
Identities = 118/314 (37%), Positives = 182/314 (57%), Gaps = 21/314 (6%)
Query: 589 SKDRTSIEDFEIIKPISRGAFGRVFLAQKRSTGDLFAIKVLKKADMIRKNAVEGILAERD 648
S + ++ F+ +KP+ G G V L + T LFA+K + KA M+ +N V AER+
Sbjct: 654 SGEPIGLKHFKPVKPLGSGDTGSVHLVELVGTDQLFAMKAMDKAVMLNRNKVHRARAERE 713
Query: 649 ILISVRNPFVVRFYYSFTCKENLYLVMEYLNGGDLYSML--RNLGCLDEDMARVYIAEVV 706
IL + +PF+ Y SF K ++ L+ +Y GG+L+ +L + L ED R Y A+VV
Sbjct: 714 ILDLLDHPFLPALYASFQTKTHICLITDYYPGGELFMLLDRQPRKVLKEDAVRFYAAQVV 773
Query: 707 LALEYLHSQSIVHRDLKPDNLLIGQDGHIKLTDFGLSKVGLINSTEDLSAPASFTNGFLV 766
+ALEYLH Q I++RDLKP+N+LI +G I L+DF LS L + L P+ +
Sbjct: 774 VALEYLHCQGIIYRDLKPENVLIQGNGDISLSDFDLSC--LTSCKPQLLIPS-------I 824
Query: 767 DDEPKPRHVSKREA--------RQQQSIVGTPDYLAPEILLGMGHGTTADWWSVGVILYE 818
D++ K + ++ R S VGT +Y+APEI+ G GH + DWW++G+++YE
Sbjct: 825 DEKKKKKQQKSQQTPIFMAEPMRASNSFVGTEEYIAPEIISGAGHTSAVDWWALGILMYE 884
Query: 819 LLVGIPPFNADHAQQIFDNIINRDIQWPKHPEEISFEAYDLMNKLLIENPVQRLG-VTGA 877
+L G PF Q+ F N++ +D+++P S + L+ +LL +P +RLG GA
Sbjct: 885 MLYGYTPFRGKTRQKTFTNVLQKDLKFPA-SIPASLQVKQLIFRLLQRDPKKRLGCFEGA 943
Query: 878 TEVKRHAFFKDVNW 891
EVK+H+FFK +NW
Sbjct: 944 NEVKQHSFFKGINW 957
>At1g16440 putative protein kinase
Length = 431
Score = 206 bits (524), Expect = 6e-53
Identities = 127/380 (33%), Positives = 199/380 (51%), Gaps = 45/380 (11%)
Query: 550 LCEQIHDERAESSNSMADEESSVDD--DTIRSLRASPINGFSKDRTSIEDFEIIKPISRG 607
+ Q ++ + S+N+ + + D D + SL++ I + I DF ++K + G
Sbjct: 1 MSSQSNNSESTSTNNSSKPHTGGDIRWDAVNSLKSRGI------KLGISDFRVLKRLGYG 54
Query: 608 AFGRVFLAQKRSTGDL--FAIKVLKKADMIRKNAVEGILAERDILISVRNPFVVRFYYSF 665
G V+L + + FA+KV+ KA ++ +N + ER+IL + +PF+ Y F
Sbjct: 55 DIGSVYLVELKGANPTTYFAMKVMDKASLVSRNKLLRAQTEREILSQLDHPFLPTLYSHF 114
Query: 666 TCKENLYLVMEYLNGGDLYSMLRNLG--CLDEDMARVYIAEVVLALEYLHSQSIVHRDLK 723
+ LVME+ +GG+LYS+ + C ED AR + +EV+LALEYLH IV+RDLK
Sbjct: 115 ETDKFYCLVMEFCSGGNLYSLRQKQPNKCFTEDAARFFASEVLLALEYLHMLGIVYRDLK 174
Query: 724 PDNLLIGQDGHIKLTDFGLSKVGLINSTEDLSAPASFTNGFLVDDE------------PK 771
P+N+L+ DGHI L+DF LS +N T S T G + D+ P+
Sbjct: 175 PENVLVRDDGHIMLSDFDLSLRCSVNPTLVKSFNGGGTTGIIDDNAAVQGCYQPSAFFPR 234
Query: 772 PRHVSKREAR-------------------QQQSIVGTPDYLAPEILLGMGHGTTADWWSV 812
SK+ + + S VGT +YLAPEI+ GHG+ DWW+
Sbjct: 235 MLQSSKKNRKSKSDFDGSLPELMAEPTNVKSMSFVGTHEYLAPEIIKNEGHGSAVDWWTF 294
Query: 813 GVILYELLVGIPPFNADHAQQIFDNIINRDIQWPKHPEEISFEAYDLMNKLLIENPVQRL 872
G+ +YELL G PF + N+I + +++P++ ++S A DL+ LL++ P R+
Sbjct: 295 GIFIYELLHGATPFKGQGNKATLYNVIGQPLRFPEY-SQVSSTAKDLIKGLLVKEPQNRI 353
Query: 873 GV-TGATEVKRHAFFKDVNW 891
GATE+K+H FF+ VNW
Sbjct: 354 AYKRGATEIKQHPFFEGVNW 373
>At3g12690 protein kinase, putative
Length = 577
Score = 203 bits (516), Expect = 5e-52
Identities = 119/334 (35%), Positives = 184/334 (54%), Gaps = 38/334 (11%)
Query: 595 IEDFEIIKPISRGAFGRVFLAQKRSTGDLFAIKVLKKADMIRKNAVEGILAERDILISVR 654
+++F ++K + G G V+LA R T +FA+KV+ KA + +N + ER+IL +
Sbjct: 182 LDNFRLLKRLGYGDIGSVYLADLRGTNAVFAMKVMDKASLASRNKLLRAQTEREILSLLD 241
Query: 655 NPFVVRFYYSFTCKENLYLVMEYLNGGDLYSMLRNLGC--LDEDMARVYIAEVVLALEYL 712
+PF+ Y F + LVME+ +GG+L+S+ + E+ AR Y +EV+LALEYL
Sbjct: 242 HPFLPTLYSYFETDKFYCLVMEFCSGGNLHSLRQKQPSRRFTEEAARFYASEVLLALEYL 301
Query: 713 HSQSIVHRDLKPDNLLIGQDGHIKLTDFGLSKVGLINSTEDLSAPASFTNGFLVDDE--- 769
H +V+RDLKP+N+L+ +GHI L+DF LS N T S+ G ++++E
Sbjct: 302 HMLGVVYRDLKPENILVRDEGHIMLSDFDLSLRCTFNPTLVKSSSVCSGGGAILNEEFAV 361
Query: 770 -----PK---PRHVSKREAR-----------------------QQQSIVGTPDYLAPEIL 798
P PR + ++ R + S VGT +YLAPEI+
Sbjct: 362 NGCMHPSAFLPRLLPSKKTRKAKSDSGLGGLSMPELMAEPTDVRSMSFVGTHEYLAPEII 421
Query: 799 LGMGHGTTADWWSVGVILYELLVGIPPFNADHAQQIFDNIINRDIQWPKHPEEISFEAYD 858
G GHG+ DWW+ G+ LYELL G PF + N++ + +++P P +S A D
Sbjct: 422 RGEGHGSAVDWWTFGIFLYELLHGTTPFKGQGNRATLHNVVGQPLKFPDTP-HVSSAARD 480
Query: 859 LMNKLLIENPVQRLGVT-GATEVKRHAFFKDVNW 891
L+ LL+++P +R+ T GATE+K+H FF+ VNW
Sbjct: 481 LIRGLLVKDPHRRIAYTRGATEIKQHPFFEGVNW 514
>At1g53700 auxin-induced protein kinase like protein
Length = 476
Score = 184 bits (466), Expect = 3e-46
Identities = 117/332 (35%), Positives = 175/332 (52%), Gaps = 25/332 (7%)
Query: 589 SKDRTSIEDFEIIKPISRGAFGRVFLAQKRS----TGDLFAIKVLKKADMIRKNAVEGIL 644
S R + F++++ + G GRVFL R TG FA+KV+ + D++ + +
Sbjct: 84 SDGRLHLRHFKLVRHLGTGNLGRVFLCHLRDCPNPTG--FALKVIDR-DVLTAKKISHVE 140
Query: 645 AERDILISVRNPFVVRFYYSFTCKENLYLVMEYLNGGDLYSMLRNL--GCLDEDMARVYI 702
E +IL + +PF+ Y L+++Y GDL+S+LR L R +
Sbjct: 141 TEAEILSLLDHPFLPTLYARIDASHYTCLLIDYCPNGDLHSLLRKQPNNRLPISPVRFFA 200
Query: 703 AEVVLALEYLHSQSIVHRDLKPDNLLIGQDGHIKLTDFGLSKVGLINST-------EDLS 755
AEV++ALEYLH+ IV+RDLKP+N+LI +DGHI L+DF L + T S
Sbjct: 201 AEVLVALEYLHALGIVYRDLKPENILIREDGHIMLSDFDLCFKADVVPTFRSRRFRRTSS 260
Query: 756 APASFTNG---FLVDDEPKPRHVSKREARQ-----QQSIVGTPDYLAPEILLGMGHGTTA 807
+P G F + E + + A + +S VGT +YLAPE++ G GHG+
Sbjct: 261 SPRKTRRGGGCFSTEVEYEREEIVAEFAAEPVTAFSKSCVGTHEYLAPELVAGNGHGSGV 320
Query: 808 DWWSVGVILYELLVGIPPFNADHAQQIFDNIINRDIQWPKHPEEISFEAYDLMNKLLIEN 867
DWW+ G+ LYE+L G PF +Q NI++ D EE EA DL+ KLL+++
Sbjct: 321 DWWAFGIFLYEMLYGTTPFKGGTKEQTLRNIVSNDDVAFTLEEEGMVEAKDLIEKLLVKD 380
Query: 868 PVQRLG-VTGATEVKRHAFFKDVNWDTLARQK 898
P +RLG GA ++KRH FF+ + W + K
Sbjct: 381 PRKRLGCARGAQDIKRHEFFEGIKWPLIRNYK 412
>At5g04510 3-phosphoinositide-dependent protein kinase-1 (PDK1)
Length = 491
Score = 182 bits (463), Expect = 7e-46
Identities = 109/303 (35%), Positives = 161/303 (52%), Gaps = 25/303 (8%)
Query: 597 DFEIIKPISRGAFGRVFLAQKRSTGDLFAIKVLKKADMIRKNAVEGILAERDILISVRNP 656
DFE K G++ +V A+K+ TG ++A+K++ K + ++N + ER +L + +P
Sbjct: 43 DFEFGKIYGVGSYSKVVRAKKKETGTVYALKIMDKKFITKENKTAYVKLERIVLDQLEHP 102
Query: 657 FVVRFYYSFTCKENLYLVMEYLNGGDLYSMLRNLGCLDEDMARVYIAEVVLALEYLHSQS 716
+++ Y++F +LY+ +E GG+L+ + G L ED AR Y AEVV ALEY+HS
Sbjct: 103 GIIKLYFTFQDTSSLYMALESCEGGELFDQITRKGRLSEDEARFYTAEVVDALEYIHSMG 162
Query: 717 IVHRDLKPDNLLIGQDGHIKLTDFGLSKVGLINSTEDLSAPASFTNGFLVDDEPKPRHVS 776
++HRD+KP+NLL+ DGHIK+ DFG K + L AS DD+
Sbjct: 163 LIHRDIKPENLLLTSDGHIKIADFGSVKPMQDSQITVLPNAAS-------DDKAC----- 210
Query: 777 KREARQQQSIVGTPDYLAPEILLGMGHGTTADWWSVGVILYELLVGIPPFNADHAQQIFD 836
+ VGT Y+ PE+L D W++G LY++L G PF IF
Sbjct: 211 --------TFVGTAAYVPPEVLNSSPATFGNDLWALGCTLYQMLSGTSPFKDASEWLIFQ 262
Query: 837 NIINRDIQWPKHPEEISFEAYDLMNKLLIENPVQR--LGVTGATEVKRHAFFKDVNWDTL 894
II RDI++P H S A DL+++LL P +R G G +KRH FF V+W L
Sbjct: 263 RIIARDIKFPNH---FSEAARDLIDRLLDTEPSRRPGAGSEGYVALKRHPFFNGVDWKNL 319
Query: 895 ARQ 897
Q
Sbjct: 320 RSQ 322
>At3g10540 3-phosphoinositide-dependent protein kinase-1 like
Length = 486
Score = 181 bits (460), Expect = 2e-45
Identities = 108/310 (34%), Positives = 167/310 (53%), Gaps = 25/310 (8%)
Query: 590 KDRTSIEDFEIIKPISRGAFGRVFLAQKRSTGDLFAIKVLKKADMIRKNAVEGILAERDI 649
++ + DFE+ K G++ +V A+K+ G ++A+K++ K + ++N + ER +
Sbjct: 37 QENFTYHDFELGKIYGVGSYSKVVRAKKKDNGTVYALKIMDKKFITKENKTAYVKLERIV 96
Query: 650 LISVRNPFVVRFYYSFTCKENLYLVMEYLNGGDLYSMLRNLGCLDEDMARVYIAEVVLAL 709
L + +P +V+ +++F ++LY+ +E GG+L+ + G L ED AR Y AEVV AL
Sbjct: 97 LDQLEHPGIVKLFFTFQDTQSLYMALESCEGGELFDQITRKGRLSEDEARFYSAEVVDAL 156
Query: 710 EYLHSQSIVHRDLKPDNLLIGQDGHIKLTDFGLSKVGLINSTEDLSAPASFTNGFLVDDE 769
EY+H+ ++HRD+KP+NLL+ DGHIK+ DFG K + L AS DD+
Sbjct: 157 EYIHNMGLIHRDIKPENLLLTLDGHIKIADFGSVKPMQDSQITVLPNAAS-------DDK 209
Query: 770 PKPRHVSKREARQQQSIVGTPDYLAPEILLGMGHGTTADWWSVGVILYELLVGIPPFNAD 829
+ VGT Y+ PE+L D W++G LY++L G PF
Sbjct: 210 AC-------------TFVGTAAYVPPEVLNSSPATFGNDLWALGCTLYQMLSGTSPFKDA 256
Query: 830 HAQQIFDNIINRDIQWPKHPEEISFEAYDLMNKLLIENPVQR--LGVTGATEVKRHAFFK 887
IF II RDI++P H S A DL+++LL +P +R G G +KRH FFK
Sbjct: 257 SEWLIFQRIIARDIKFPNH---FSEAARDLIDRLLDTDPSRRPGAGSEGYDSLKRHPFFK 313
Query: 888 DVNWDTLARQ 897
V+W L Q
Sbjct: 314 GVDWKNLRSQ 323
>At2g34650 protein kinase PINOID
Length = 438
Score = 178 bits (451), Expect = 2e-44
Identities = 116/333 (34%), Positives = 169/333 (49%), Gaps = 31/333 (9%)
Query: 590 KDRTSIEDFEIIKPISRGAFGRVFLA-----QKRSTGDLFAIKVLKKADMIRKNAVEGIL 644
K + DF +++ I G G V+L ++ S FA+KV+ K + K +
Sbjct: 67 KQGLTFRDFRLMRRIGAGDIGTVYLCRLAGDEEESRSSYFAMKVVDKEALALKKKMHRAE 126
Query: 645 AERDILISVRNPFVVRFYYSFTCKENLYLVMEYLNGGDLYSMLRNLGC--LDEDMARVYI 702
E+ IL + +PF+ Y F +VMEY +GGDL+S+ AR Y
Sbjct: 127 MEKTILKMLDHPFLPTLYAEFEASHFSCIVMEYCSGGDLHSLRHRQPHRRFSLSSARFYA 186
Query: 703 AEVVLALEYLHSQSIVHRDLKPDNLLIGQDGHIKLTDFGLS----KVGLINST------E 752
AEV++ALEYLH I++RDLKP+N+L+ DGHI L+DF LS + + S+ +
Sbjct: 187 AEVLVALEYLHMLGIIYRDLKPENILVRSDGHIMLSDFDLSLCSDSIAAVESSSSSPENQ 246
Query: 753 DLSAPASFTNGFLVDD-----------EPKPRHVSKREARQQQSIVGTPDYLAPEILLGM 801
L +P FT + EP V++ + S VGT +Y+APE+ G
Sbjct: 247 QLRSPRRFTRLARLFQRVLRSKKVQTLEPTRLFVAEPVTARSGSFVGTHEYVAPEVASGG 306
Query: 802 GHGTTADWWSVGVILYELLVGIPPFNADHAQQIFDNIINRDIQWPKHPEEISFE--AYDL 859
HG DWW+ GV LYE++ G PF A I NI+ R + +P FE A +L
Sbjct: 307 SHGNAVDWWAFGVFLYEMIYGKTPFVAPTNDVILRNIVKRQLSFPTDSPATMFELHARNL 366
Query: 860 MNKLLIENPVQRLGV-TGATEVKRHAFFKDVNW 891
++ LL ++P +RLG GA EVK H FFK +N+
Sbjct: 367 ISGLLNKDPTKRLGSRRGAAEVKVHPFFKGLNF 399
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.317 0.134 0.392
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 25,468,618
Number of Sequences: 26719
Number of extensions: 1160097
Number of successful extensions: 6221
Number of sequences better than 10.0: 990
Number of HSP's better than 10.0 without gapping: 865
Number of HSP's successfully gapped in prelim test: 125
Number of HSP's that attempted gapping in prelim test: 3693
Number of HSP's gapped (non-prelim): 1694
length of query: 1086
length of database: 11,318,596
effective HSP length: 110
effective length of query: 976
effective length of database: 8,379,506
effective search space: 8178397856
effective search space used: 8178397856
T: 11
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.6 bits)
S2: 65 (29.6 bits)
Medicago: description of AC133139.5


