

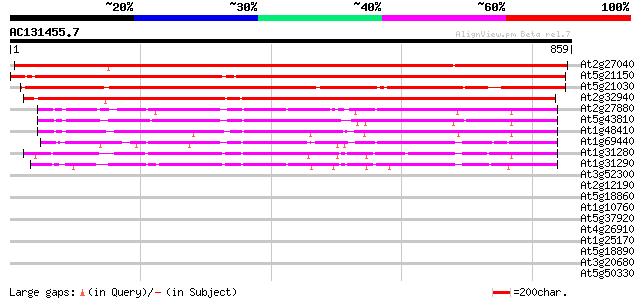
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC131455.7 + phase: 0 /pseudo
(859 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
At2g27040 Argonaute (AGO1)-like protein 1194 0.0
At5g21150 zwille/pinhead-like protein 1106 0.0
At5g21030 zwille/pinhead-like protein 920 0.0
At2g32940 Argonaute (AGO1)-like protein 885 0.0
At2g27880 Argonaute (AGO1)-like protein 458 e-129
At5g43810 PINHEAD (gb|AAD40098.1); translation initiation factor 440 e-123
At1g48410 Argonaute protein (AGO1) 430 e-120
At1g69440 hypothetical protein 330 2e-90
At1g31280 unknown protein 304 2e-82
At1g31290 hypothetical protein 302 6e-82
At3g52300 unknown protein 33 0.52
At2g12190 pseudogene 32 2.0
At5g18860 unknown protein 31 2.6
At1g10760 unknown protein 31 3.4
At5g37920 putative protein 30 4.4
At4g26910 putative dihydrolipoamide succinyltransferase 30 4.4
At1g25170 30 4.4
At5g18890 putative protein 30 5.8
At3g20680 unknown protein 30 5.8
At5g50330 putative protein 29 9.8
>At2g27040 Argonaute (AGO1)-like protein
Length = 924
Score = 1194 bits (3088), Expect = 0.0
Identities = 587/863 (68%), Positives = 709/863 (82%), Gaps = 19/863 (2%)
Query: 8 ECLPPPPPIVPPNVKPIKIEQEHLKKKLPTKAPMARRGLGTKGAKLPLLTNHFEVNVANT 67
E LPPPPP++PPNV+P++++ E +KK P + PMAR+G GT+G K+PLLTNHF+V+VAN
Sbjct: 24 EALPPPPPVIPPNVEPVRVKTELAEKKGPVRVPMARKGFGTRGQKIPLLTNHFKVDVANL 83
Query: 68 NRVFFQYSVALFYEDGRPVEGKGAGRKIIDKVQETYDSELNGKDLAYDGE-TLFTIGSLA 126
FF YSVALFY+DGRPVE KG GRKI+DKV +TY S+L+GK+ AYDGE TLFT G+L
Sbjct: 84 QGHFFHYSVALFYDDGRPVEQKGVGRKILDKVHQTYHSDLDGKEFAYDGEKTLFTYGALP 143
Query: 127 QKKLEFIVVVEDVASNRNNANTSP---------DKKRIRKSYRSKTYKVEINFAKEIPLQ 177
K++F VV+E+V++ R N N SP D+KR+R+ RSK ++VEI++A +IPLQ
Sbjct: 144 SNKMDFSVVLEEVSATRANGNGSPNGNESPSDGDRKRLRRPNRSKNFRVEISYAAKIPLQ 203
Query: 178 AIANALKGHEAENYQEAIRVLDIILRQHSAKQGCLLVRQNFFHNDPNNLNDVGGGVLSCK 237
A+ANA++G E+EN QEAIRVLDIILRQH+A+QGCLLVRQ+FFHNDP N VGG +L C+
Sbjct: 204 ALANAMRGQESENSQEAIRVLDIILRQHAARQGCLLVRQSFFHNDPTNCEPVGGNILGCR 263
Query: 238 GLHSSFRTTQSGLSLNIDVSTTMIVRPGPVVDFLIENQNVRDPFSLDWNKAKRTLKNLRI 297
G HSSFRTTQ G+SLN+DV+TTMI++PGPVVDFLI NQN RDP+S+DW+KAKRTLKNLR+
Sbjct: 264 GFHSSFRTTQGGMSLNMDVTTTMIIKPGPVVDFLIANQNARDPYSIDWSKAKRTLKNLRV 323
Query: 298 TAKPSNQEYKITGLSELSCKDQLFTMKKRGAVA-GEDDTEEITVYDYFVHRRKIDLQYSA 356
PS QE+KITGLS+ C++Q F +KKR GE +T E+TV DYF R IDLQYSA
Sbjct: 324 KVSPSGQEFKITGLSDKPCREQTFELKKRNPNENGEFETTEVTVADYFRDTRHIDLQYSA 383
Query: 357 GLPCINVGKPKRPTYIPIELCSLISLQRYTKALSTSQRSSLVEKSRQKPVERMRVLSNAL 416
LPCINVGKPKRPTYIP+ELC+L+ LQRYTKAL+T QRS+LVEKSRQKP ERM VLS AL
Sbjct: 384 DLPCINVGKPKRPTYIPLELCALVPLQRYTKALTTFQRSALVEKSRQKPQERMTVLSKAL 443
Query: 417 KASNYGSEPMLRNCGISITSEFTQVDGRVLQAPRLKFG--NEDFNPRNGRWNFNNKKFVE 474
K SNY +EP+LR+CGISI+S FTQV+GRVL AP+LK G +E F PRNGRWNFNNK+FVE
Sbjct: 444 KVSNYDAEPLLRSCGISISSNFTQVEGRVLPAPKLKMGCGSETF-PRNGRWNFNNKEFVE 502
Query: 475 PVSLGNWSVVNFSARCDVRGLVRDLIKCGGMKGILVEQPKDVIEENRQFKGEPPVFRVEK 534
P + W VVNFSARC+VR +V DLIK GG KGI + P V EE QF+ PP+ RVE
Sbjct: 503 PTKIQRWVVVNFSARCNVRQVVDDLIKIGGSKGIEIASPFQVFEEGNQFRRAPPMIRVEN 562
Query: 535 MFADVL-KLSKRPSFLLCLLPERKNSDLYGPWKKKNLAEFGIVTQCIAPTRV-NDQYLTN 592
MF D+ KL P F+LC+LP++KNSDLYGPWKKKNL EFGIVTQC+APTR NDQYLTN
Sbjct: 563 MFKDIQSKLPGVPQFILCVLPDKKNSDLYGPWKKKNLTEFGIVTQCMAPTRQPNDQYLTN 622
Query: 593 VLLKINAKLGGMNSWLGVEHSRSIPIVSKVPTLILGMDVSHGSPGQPDIPSIAAVVSSRK 652
+LLKINAKLGG+NS L VE + + ++SKVPT+ILGMDVSHGSPGQ D+PSIAAVVSSR+
Sbjct: 623 LLLKINAKLGGLNSMLSVERTPAFTVISKVPTIILGMDVSHGSPGQSDVPSIAAVVSSRE 682
Query: 653 WPLISKYRACVRTQGSKVEMIDNLFKPVSDKEDEGIIRELLLDFFHSSEERRPENIIIFR 712
WPLISKYRA VRTQ SK EMI++L K + ED+GII+ELL+DF+ SS +R+PE+IIIFR
Sbjct: 683 WPLISKYRASVRTQPSKAEMIESLVKK-NGTEDDGIIKELLVDFYTSSNKRKPEHIIIFR 741
Query: 713 DGVSESQFNEVLNVELSQIIEACKFLDENWNPKFMVIVAQKNHHTKFFQPRSPDNVPPGT 772
DGVSESQFN+VLN+EL QIIEACK LD NWNPKF+++VAQKNHHTKFFQP SP+NVPPGT
Sbjct: 742 DGVSESQFNQVLNIELDQIIEACKLLDANWNPKFLLLVAQKNHHTKFFQPTSPENVPPGT 801
Query: 773 VVDSKICHPRNYDFYMCAHAGMIGTSRPTHYHVLLDEIGFSPDDLQELVHSLSYVYQRST 832
++D+KICHP+N DFY+CAHAGMIGT+RPTHYHVL DEIGFS D+LQELVHSLSYVYQRST
Sbjct: 802 IIDNKICHPKNNDFYLCAHAGMIGTTRPTHYHVLYDEIGFSADELQELVHSLSYVYQRST 861
Query: 833 TAISVVK--TYMQLLQSAMHTWL 853
+AISVV Y L + + T++
Sbjct: 862 SAISVVAPICYAHLAAAQLGTFM 884
>At5g21150 zwille/pinhead-like protein
Length = 892
Score = 1106 bits (2861), Expect = 0.0
Identities = 562/860 (65%), Positives = 674/860 (78%), Gaps = 22/860 (2%)
Query: 2 SSQENKECLPPPPPIVPPNVKPIKIEQEHLKKKLPTKAPMAR-RGLGTKGAKLPLLTNHF 60
S + N LPPPPP VP N+ P E E +KK + PMAR RG G+KG K+PLLTNHF
Sbjct: 3 SDEPNGSGLPPPPPFVPANLVP---EVEPVKKNI--LLPMARPRGSGSKGQKIPLLTNHF 57
Query: 61 EVNVANTNRVFFQYSVALFYEDGRPVEGKGAGRKIIDKVQETYDSELNGKDLAYDGE-TL 119
V + FF YSVA+ YEDGRPVE KG GRKI+DKVQETY S+L K AYDGE TL
Sbjct: 58 GVKFNKPSGYFFHYSVAINYEDGRPVEAKGIGRKILDKVQETYQSDLGAKYFAYDGEKTL 117
Query: 120 FTIGSLAQKKLEFIVVVEDVASNRNNANTSP---DKKRIRKSYRSKTYKVEINFAKEIPL 176
FT+G+L KL+F VV+E++ S+RN+A D+KR R+ ++K + VEI++A +IP+
Sbjct: 118 FTVGALPSNKLDFSVVLEEIPSSRNHAGNDTNDADRKRSRRPNQTKKFMVEISYAAKIPM 177
Query: 177 QAIANALKGHEAENYQEAIRVLDIILRQHSAKQGCLLVRQNFFHNDPNNLNDVGGGVLSC 236
QAIA+AL+G E EN Q+A+RVLDIILRQ +A+QGCLLVRQ+FFHND N +GGGV C
Sbjct: 178 QAIASALQGKETENLQDALRVLDIILRQSAARQGCLLVRQSFFHNDVKNFVPIGGGVSGC 237
Query: 237 KGLHSSFRTTQSGLSLNIDVSTTMIVRPGPVVDFLIENQNVRDPFSLDWNKAKRTLKNLR 296
+G HSSFRTTQ GLSLNID STTMIV+PGPVVDFL+ NQN +DP+ +DWNKA+R LKNLR
Sbjct: 238 RGFHSSFRTTQGGLSLNIDTSTTMIVQPGPVVDFLLANQNKKDPYGMDWNKARRVLKNLR 297
Query: 297 ITAKPSNQEYKITGLSELSCKDQLFTMKKRGAVAGEDDTEEITVYDYFVHRRKIDLQYSA 356
+ SN+EYKI+GLSE SCKDQL K GE + EITV +Y+ R I+++YS
Sbjct: 298 VQITLSNREYKISGLSEHSCKDQLKPNDK-----GEFEEVEITVLNYY-KERNIEVRYSG 351
Query: 357 GLPCINVGKPKRPTYIPIELCSLISLQRYTKALSTSQRSSLVEKSRQKPVERMRVLSNAL 416
PCINVGKPKRPTY PIE C+L+SLQRYTK+L+ QR++LVEKSRQKP ERM L+ L
Sbjct: 352 DFPCINVGKPKRPTYFPIEFCNLVSLQRYTKSLTNFQRAALVEKSRQKPPERMASLTKGL 411
Query: 417 KASNYGSEPMLRNCGISITSEFTQVDGRVLQAPRLKFGN-EDFNPRNGRWNFNNKKFVEP 475
K SNY ++P+L++ G+SI + FTQV+GR+L P LK G E+ +P G+WNF K EP
Sbjct: 412 KDSNYNADPVLQDSGVSIITNFTQVEGRILPTPMLKVGKGENLSPIKGKWNFMRKTLAEP 471
Query: 476 VSLGNWSVVNFSARCDVRGLVRDLIKCGGMKGILVEQP-KDVIEENRQFKGEPPVFRVEK 534
++ W+VVNFSARCD L+RDLIKCG KGI VE P KDVI EN QF+ P RVE
Sbjct: 472 TTVTRWAVVNFSARCDTNTLIRDLIKCGREKGINVEPPFKDVINENPQFRNAPATVRVEN 531
Query: 535 MFADVL-KLSKRPSFLLCLLPERKNSDLYGPWKKKNLAEFGIVTQCIAPTRVNDQYLTNV 593
MF + KL K P FLLC+L ERKNSD+YGPWKKKNL + GIVTQCIAPTR+NDQYLTNV
Sbjct: 532 MFEQIKSKLPKPPLFLLCILAERKNSDVYGPWKKKNLVDLGIVTQCIAPTRLNDQYLTNV 591
Query: 594 LLKINAKLGGMNSWLGVEHSRSIPIVSKVPTLILGMDVSHGSPGQPDIPSIAAVVSSRKW 653
LLKINAKLGG+NS L +E S ++P V++VPT+I+GMDVSHGSPGQ DIPSIAAVVSSR+W
Sbjct: 592 LLKINAKLGGLNSLLAMERSPAMPKVTQVPTIIVGMDVSHGSPGQSDIPSIAAVVSSRQW 651
Query: 654 PLISKYRACVRTQGSKVEMIDNLFKPVSDKEDEGIIRELLLDFFHSSEERRPENIIIFRD 713
PLISKY+ACVRTQ K+EMIDNLFKPV+ K DEG+ RELLLDF++SSE R+PE+IIIFRD
Sbjct: 652 PLISKYKACVRTQSRKMEMIDNLFKPVNGK-DEGMFRELLLDFYYSSENRKPEHIIIFRD 710
Query: 714 GVSESQFNEVLNVELSQIIEACKFLDENWNPKFMVIVAQKNHHTKFFQPRSPDNVPPGTV 773
GVSESQFN+VLN+EL Q+++ACKFLD+ W+PKF VIVAQKNHHTKFFQ R PDNVPPGT+
Sbjct: 711 GVSESQFNQVLNIELDQMMQACKFLDDTWHPKFTVIVAQKNHHTKFFQSRGPDNVPPGTI 770
Query: 774 VDSKICHPRNYDFYMCAHAGMIGTSRPTHYHVLLDEIGFSPDDLQELVHSLSYVYQRSTT 833
+DS+ICHPRN+DFY+CAHAGMIGT+RPTHYHVL DEIGF+ DDLQELVHSLSYVYQRSTT
Sbjct: 771 IDSQICHPRNFDFYLCAHAGMIGTTRPTHYHVLYDEIGFATDDLQELVHSLSYVYQRSTT 830
Query: 834 AISVVK--TYMQLLQSAMHT 851
AISVV Y L + M T
Sbjct: 831 AISVVAPVCYAHLAAAQMGT 850
>At5g21030 zwille/pinhead-like protein
Length = 850
Score = 920 bits (2379), Expect = 0.0
Identities = 501/846 (59%), Positives = 612/846 (72%), Gaps = 53/846 (6%)
Query: 17 VPPNVKPIKIEQEHLKKKLPTKAPMARRGLGTKGAKLPLLTNHFEVNVANTN-RVFFQYS 75
+PP P +E+E LK K + PM RRG G+KG K+ LLTNHF VN N FF YS
Sbjct: 5 LPP---PQHMEREPLKSK-SSLLPMTRRGNGSKGQKILLLTNHFRVNFRKPNSHNFFHYS 60
Query: 76 VALFYEDGRPVEGKGAGRKIIDKVQETYDSELNGKDLAYDGE-TLFTIGSLAQKKLEFIV 134
V + YEDG P+ KG GRKI++KVQ+T ++L K AYDG+ L+T+G L + L+F V
Sbjct: 61 VTITYEDGSPLLAKGFGRKILEKVQQTCQADLGCKHFAYDGDKNLYTVGPLPRSSLDFSV 120
Query: 135 VVEDVASNRNNANTSPDKKRIRKSYRSKTYKVEINFAK-EIPLQAIANALKGHEAENYQE 193
V+E S RN KR++ ++SK + V I FA EIP++AIANAL+G + ++ +
Sbjct: 121 VLETAPSRRNA------DKRLKLPHQSKKFNVAILFAPPEIPMEAIANALQGKKTKHLLD 174
Query: 194 AIRVLDIILRQHSAKQGCLLVRQNFFHNDPNNLNDVGGGVLSCKGLHSSFRTTQSGLSLN 253
AIRV+D IL Q++A+QGCLLVRQ+FFHND ++G GV CKG HSSFRTTQ GLSLN
Sbjct: 175 AIRVMDCILSQNAARQGCLLVRQSFFHNDAKYFANIGEGVDCCKGFHSSFRTTQGGLSLN 234
Query: 254 IDVSTTMIVRPGPVVDFLIENQNVRDPFSLDWNKAKRTLKNLRITAKPSNQEYKITGLSE 313
IDVST MIV+PGPVVDFLI NQ V DPFS++W KAK TLKNLR+ PSNQEYKITGLS
Sbjct: 235 IDVSTAMIVKPGPVVDFLIANQGVNDPFSINWKKAKNTLKNLRVKVLPSNQEYKITGLSG 294
Query: 314 LSCKDQLFTMKKRGAVAGEDDTEEITVYDYFVHRRKIDLQYSAGLPCINVGKPKRPTYIP 373
L CKDQ FT KKR E + EITV DYF R+I+L+YS GLPCINVGKP RPTY P
Sbjct: 295 LHCKDQTFTWKKRNQ-NREFEEVEITVSDYFTRIREIELRYSGGLPCINVGKPNRPTYFP 353
Query: 374 IELCSLISLQRYTKALSTSQRSSLVEKSRQKPVERMRVLSNALKASNYGSEPMLRNCGIS 433
IELC L+SLQRYTKAL+ QRS+L+++SRQ P +R+ VL+ ALK SNY +PML+ CG+
Sbjct: 354 IELCELVSLQRYTKALTKFQRSNLIKESRQNPQQRIGVLTRALKTSNYNDDPMLQECGVR 413
Query: 434 ITSEFTQVDGRVLQAPRLKFGNE-DFNPRNGRWNFNNKKFVEPVSLGNWSVVNFSARCDV 492
I S+FTQV+GRVL P+LK G E D P NG WNF NK P ++ W+VVNFSARCD
Sbjct: 414 IGSDFTQVEGRVLPTPKLKAGKEQDIYPINGSWNFKNK----PATVTRWAVVNFSARCDP 469
Query: 493 RGLVRDLIKCGGMKGILVEQPKDVI-EENRQFKGEPPVFRVEKMFADVLKL--SKRPSFL 549
+ ++ DL +CG MKGI V+ P V+ EEN QFK RV+KMF + + P FL
Sbjct: 470 QKIIDDLTRCGKMKGINVDSPYHVVFEENPQFKDATGSVRVDKMFQHLQSILGEVPPKFL 529
Query: 550 LCLLPERKNSDLYGPWKKKNLAEFGIVTQCIAPTR-VNDQYLTNVLLKINAKLGGMNSWL 608
LC+L E+KNSD+Y +K+ + + +CI P + +NDQYLTN+LLKINAKLGG+NS L
Sbjct: 530 LCIL-EKKNSDVY----EKSCSMWN--CECIVPPQNLNDQYLTNLLLKINAKLGGLNSVL 582
Query: 609 GVEHSRSIPIVSKVPTLILGMDVSHGSPGQPD-IPSIAAVVSSRKWPLISKYRACVRTQG 667
+E S ++P+V +VPT+I+GMDVSHGSPGQ D IPSIAAVVSSR+WPLISKYRACVRTQ
Sbjct: 583 DMELSGTMPLVMRVPTIIIGMDVSHGSPGQSDHIPSIAAVVSSREWPLISKYRACVRTQS 642
Query: 668 SKVEMIDNLFKPVSDKEDEGIIRELLLDFFHSSEERRPENIIIFRDGVSESQFNEVLNVE 727
KVEMID+LFKPVSDK+D+GI+RELLLD FHSS ++P +IIIFRDGVSESQFN+VLN+E
Sbjct: 643 PKVEMIDSLFKPVSDKDDQGIMRELLLD-FHSSSGKKPNHIIIFRDGVSESQFNQVLNIE 701
Query: 728 LSQIIEACKFLDENWNPKFMVIVAQKNHHTKFFQPRSPDNVPPGTVVDSKICHPRNYDFY 787
L Q++ Q NHHTKFFQ SP+NV PGT++DS ICH N DFY
Sbjct: 702 LDQMM-------------------QINHHTKFFQTESPNNVLPGTIIDSNICHQHNNDFY 742
Query: 788 MCAHAGMIGTSRPTHYHVLLDEIGFSPDDLQELVHSLSYVYQRSTTAISVVK--TYMQLL 845
+CAHAG IGT+RPTHYHVL DEIGF D LQELVHSLSYVYQRSTTAIS+V Y L
Sbjct: 743 LCAHAGKIGTTRPTHYHVLYDEIGFDTDQLQELVHSLSYVYQRSTTAISLVAPICYAHLA 802
Query: 846 QSAMHT 851
+ M T
Sbjct: 803 AAQMAT 808
>At2g32940 Argonaute (AGO1)-like protein
Length = 887
Score = 885 bits (2287), Expect = 0.0
Identities = 459/832 (55%), Positives = 600/832 (71%), Gaps = 24/832 (2%)
Query: 21 VKPIKIEQE---HLKKKLPTKAPMARRGLGTKGAKLPLLTNHFEVNVANTNRVFFQYSVA 77
+ PI IE E H + T RRG+GT G + L TNHF V+V + VF+QY+V+
Sbjct: 9 LSPISIEPEQPSHRDYDITT-----RRGVGTTGNPIELCTNHFNVSVRQPDVVFYQYTVS 63
Query: 78 LFYEDGRPVEGKGAGRKIIDKVQETYDSELNGKDLAYDGE-TLFTIGSLAQKKLEFIVVV 136
+ E+G V+G G RK++D++ +TY S+L+GK LAYDGE TL+T+G L Q + +F+V+V
Sbjct: 64 ITTENGDAVDGTGISRKLMDQLFKTYSSDLDGKRLAYDGEKTLYTVGPLPQNEFDFLVIV 123
Query: 137 EDVASNRN-----NANTSPDKKRIRKSYRSKTYKVEINFAKEIPLQAIANALKGHEA--E 189
E S R+ ++S KR ++S+ ++YKV+I++A EIPL+ + +G +
Sbjct: 124 EGSFSKRDCGVSDGGSSSGTCKRSKRSFLPRSYKVQIHYAAEIPLKTVLGTQRGAYTPDK 183
Query: 190 NYQEAIRVLDIILRQHSAKQGCLLVRQNFFHNDPNNLNDVGGGVLSCKGLHSSFRTTQSG 249
+ Q+A+RVLDI+LRQ +A++GCLLVRQ FFH+D + + VGGGV+ +GLHSSFR T G
Sbjct: 184 SAQDALRVLDIVLRQQAAERGCLLVRQAFFHSDGHPMK-VGGGVIGIRGLHSSFRPTHGG 242
Query: 250 LSLNIDVSTTMIVRPGPVVDFLIENQNVRDPFSLDWNK-AKRTLKNLRITAKPSNQEYKI 308
LSLNIDVSTTMI+ PGPV++FL NQ+V P +DW K A + LK++R+ A N E+KI
Sbjct: 243 LSLNIDVSTTMILEPGPVIEFLKANQSVETPRQIDWIKVAAKMLKHMRVKATHRNMEFKI 302
Query: 309 TGLSELSCKDQLFTMKKRGAVAGEDDTEEITVYDYFVHRRKIDLQYSAGLPCINVGKPKR 368
GLS C QLF+MK + E EITVYDYF + SA PC++VGKP R
Sbjct: 303 IGLSSKPCNQQLFSMKIKDGER-EVPIREITVYDYFKQTYTEPIS-SAYFPCLDVGKPDR 360
Query: 369 PTYIPIELCSLISLQRYTKALSTSQRSSLVEKSRQKPVERMRVLSNALKASNYGSEPMLR 428
P Y+P+E C+L+SLQRYTK LS QR LVE SRQKP+ER++ L++A+ Y +P L
Sbjct: 361 PNYLPLEFCNLVSLQRYTKPLSGRQRVLLVESSRQKPLERIKTLNDAMHTYCYDKDPFLA 420
Query: 429 NCGISITSEFTQVDGRVLQAPRLKFG-NEDFNPRNGRWNFNNKKFVEPVSLGNWSVVNFS 487
CGISI E TQV+GRVL+ P LKFG NEDF P NGRWNFNNK +EP ++ +W++VNFS
Sbjct: 421 GCGISIEKEMTQVEGRVLKPPMLKFGKNEDFQPCNGRWNFNNKMLLEPRAIKSWAIVNFS 480
Query: 488 ARCDVRGLVRDLIKCGGMKGILVEQPKDVIEENRQFKGEPPVFRVEKMFADV-LKLSKRP 546
CD + R+LI CG KGI +++P ++EE+ Q+K PV RVEKM A + LK P
Sbjct: 481 FPCDSSHISRELISCGMRKGIEIDRPFALVEEDPQYKKAGPVERVEKMIATMKLKFPDPP 540
Query: 547 SFLLCLLPERKNSDLYGPWKKKNLAEFGIVTQCIAPTRVNDQYLTNVLLKINAKLGGMNS 606
F+LC+LPERK SD+YGPWKK L E GI TQCI P +++DQYLTNVLLKIN+KLGG+NS
Sbjct: 541 HFILCILPERKTSDIYGPWKKICLTEEGIHTQCICPIKISDQYLTNVLLKINSKLGGINS 600
Query: 607 WLGVEHSRSIPIVSKVPTLILGMDVSHGSPGQPDIPSIAAVVSSRKWPLISKYRACVRTQ 666
LG+E+S +IP+++K+PTLILGMDVSHG PG+ D+PS+AAVV S+ WPLIS+YRA VRTQ
Sbjct: 601 LLGIEYSYNIPLINKIPTLILGMDVSHGPPGRADVPSVAAVVGSKCWPLISRYRAAVRTQ 660
Query: 667 GSKVEMIDNLFKPV--SDKEDEGIIRELLLDFFHSSEERRPENIIIFRDGVSESQFNEVL 724
++EMID+LF+P+ ++K D GI+ EL ++F+ +S R+P+ IIIFRDGVSESQF +VL
Sbjct: 661 SPRLEMIDSLFQPIENTEKGDNGIMNELFVEFYRTSRARKPKQIIIFRDGVSESQFEQVL 720
Query: 725 NVELSQIIEACKFLDENWNPKFMVIVAQKNHHTKFFQPRSPDNVPPGTVVDSKICHPRNY 784
+E+ QII+A + L E+ PKF VIVAQKNHHTK FQ + P+NVP GTVVD+KI HP NY
Sbjct: 721 KIEVDQIIKAYQRLGESDVPKFTVIVAQKNHHTKLFQAKGPENVPAGTVVDTKIVHPTNY 780
Query: 785 DFYMCAHAGMIGTSRPTHYHVLLDEIGFSPDDLQELVHSLSYVYQRSTTAIS 836
DFYMCAHAG IGTSRP HYHVLLDEIGFSPDDLQ L+HSLSY S +S
Sbjct: 781 DFYMCAHAGKIGTSRPAHYHVLLDEIGFSPDDLQNLIHSLSYKLLNSIFNVS 832
>At2g27880 Argonaute (AGO1)-like protein
Length = 997
Score = 458 bits (1179), Expect = e-129
Identities = 302/835 (36%), Positives = 451/835 (53%), Gaps = 83/835 (9%)
Query: 43 RRGLGTKGAKLPLLTNHFEVNVANTNRVFFQYSVALFYEDGRPVEGKGAGRKIIDKVQET 102
R G GT G K+ + NHF V VA+ R + Y V++ E V K R ++ + +
Sbjct: 150 RPGRGTLGKKVMVRANHFLVQVAD--RDLYHYDVSINPE----VISKTVNRNVMKLLVKN 203
Query: 103 Y-DSELNGKDLAYDG-ETLFTIGSLAQKKLEFIVVVEDVASNRNNANTSPDKKRIRKSYR 160
Y DS L GK AYDG ++L+T G L EF+V ++A R + ++ D+
Sbjct: 204 YKDSHLGGKSPAYDGRKSLYTAGPLPFDSKEFVV---NLAEKRADGSSGKDRP------- 253
Query: 161 SKTYKVEINFAKEIPLQAIANALKGHEAENYQEAIRVLDIILRQHSAKQGCLLVRQNFFH 220
+KV + L + L + E + I+VLD++LR + + V ++FFH
Sbjct: 254 ---FKVAVKNVTSTDLYQLQQFLDRKQREAPYDTIQVLDVVLRDKPSND-YVSVGRSFFH 309
Query: 221 ND-----PNNLNDVGGGVLSCKGLHSSFRTTQSGLSLNIDVSTTMIVRPGPVVDFLIENQ 275
+ ++G G+ +G S R TQ GLSLNIDVS P V DF+ +
Sbjct: 310 TSLGKDARDGRGELGDGIEYWRGYFQSLRLTQMGLSLNIDVSARSFYEPIVVTDFISKFL 369
Query: 276 NVRD---PF-SLDWNKAKRTLKNLRITAKPSN--QEYKITGLSELSCKDQLFTMKKRGAV 329
N+RD P D K K+ L+ L++ N + KI+G+S L ++ FT+
Sbjct: 370 NIRDLNRPLRDSDRLKVKKVLRTLKVKLLHWNCTKSAKISGISSLPIRELRFTL------ 423
Query: 330 AGEDDTEEITVYDYFVHRRKIDLQYSAGLPCINVGKPKRPTYIPIELCSLISLQRYTKAL 389
+D E TV YF + ++Y A LP I G RP Y+P+ELC + QRYTK L
Sbjct: 424 ---EDKSEKTVVQYFAEKYNYRVKYQA-LPAIQTGSDTRPVYLPMELCQIDEGQRYTKRL 479
Query: 390 STSQRSSLVEKSRQKPVERMRVLSNALKASNYGSEPMLRNCGISITSEFTQVDGRVLQAP 449
+ Q ++L++ + Q+P +R + N + +NY + + + G+S+T++ ++ RVL P
Sbjct: 480 NEKQVTALLKATCQRPPDRENSIKNLVVKNNYNDD-LSKEFGMSVTTQLASIEARVLPPP 538
Query: 450 RLKF---GNEDF-NPRNGRWNFNNKKFVEPVSLGNWSVVNFSARCDVRGLVRDLIKCGGM 505
LK+ G E NPR G+WN +KK V + +W+ V+FS R D RGL ++ C +
Sbjct: 539 MLKYHDSGKEKMVNPRLGQWNMIDKKMVNGAKVTSWTCVSFSTRID-RGLPQEF--CKQL 595
Query: 506 KGILVEQPKDVIEENRQFKGEP-------PVFRVEKMFADVLKLSKRPSFLLCLLPERKN 558
G+ V + +FK +P P +E+ D+ K + L+ +LP+
Sbjct: 596 IGMCVSK-------GMEFKPQPAIPFISCPPEHIEEALLDIHKRAPGLQLLIVILPDVTG 648
Query: 559 SDLYGPWKKKNLAEFGIVTQCIAPTRVND---QYLTNVLLKINAKLGGMNSWLGVEHSRS 615
S YG K+ E GIV+QC P +VN QY+ NV LKIN K GG N+ L R+
Sbjct: 649 S--YGKIKRICETELGIVSQCCQPRQVNKLNKQYMENVALKINVKTGGRNTVLNDAIRRN 706
Query: 616 IPIVSKVPTLILGMDVSHGSPGQPDIPSIAAVVSSRKWPLISKYRACVRTQGSKVEMIDN 675
IP+++ PT+I+G DV+H PG+ PSIAAVV+S WP I+KYR V Q + E+I +
Sbjct: 707 IPLITDRPTIIMGADVTHPQPGEDSSPSIAAVVASMDWPEINKYRGLVSAQAHREEIIQD 766
Query: 676 LFKPVSDKE----DEGIIRELLLDFFHSSEERRPENIIIFRDGVSESQFNEVLNVELSQI 731
L+K V D + G+IRE + F ++ + P+ II +RDGVSE QF++VL E++ I
Sbjct: 767 LYKLVQDPQRGLVHSGLIREHFIAFRRATGQI-PQRIIFYRDGVSEGQFSQVLLHEMTAI 825
Query: 732 IEACKFLDENWNPKFMVIVAQKNHHTKFFQPRSPD--------NVPPGTVVDSKICHPRN 783
+AC L EN+ P+ ++ QK HHT+ F + + N+ PGTVVD+KICHP
Sbjct: 826 RKACNSLQENYVPRVTFVIVQKRHHTRLFPEQHGNRDMTDKSGNIQPGTVVDTKICHPNE 885
Query: 784 YDFYMCAHAGMIGTSRPTHYHVLLDEIGFSPDDLQELVHSLSYVYQRSTTAISVV 838
+DFY+ +HAG+ GTSRP HYHVLLDE GF+ D LQ L ++L Y Y R T ++S+V
Sbjct: 886 FDFYLNSHAGIQGTSRPAHYHVLLDENGFTADQLQMLTNNLCYTYARCTKSVSIV 940
>At5g43810 PINHEAD (gb|AAD40098.1); translation initiation factor
Length = 988
Score = 440 bits (1131), Expect = e-123
Identities = 292/840 (34%), Positives = 444/840 (52%), Gaps = 86/840 (10%)
Query: 43 RRGLGTKGAKLPLLTNHFEVNVANTNRVFFQYSVALFYEDGRPVEGKGAGRKIIDKVQET 102
R G GT G K + NHF ++ + QY V + E V K R II ++
Sbjct: 131 RPGFGTLGTKCIVKANHFLADLPTKD--LNQYDVTITPE----VSSKSVNRAIIAELVRL 184
Query: 103 Y-DSELNGKDLAYDG-ETLFTIGSLAQKKLEFIVVVEDVASNRNNANTSPDKKRIRKSYR 160
Y +S+L + AYDG ++L+T G L EF V + D D I R
Sbjct: 185 YKESDLGRRLPAYDGRKSLYTAGELPFTWKEFSVKIVD-----------EDDGIINGPKR 233
Query: 161 SKTYKVEINFAKEIPLQAIANALKGHEAENYQEAIRVLDIILRQHSAKQGCLLVRQNFFH 220
++YKV I F + + L G A+ QEA+++LDI+LR+ S K+ C + R +FF
Sbjct: 234 ERSYKVAIKFVARANMHHLGEFLAGKRADCPQEAVQILDIVLRELSVKRFCPVGR-SFFS 292
Query: 221 NDPNNLNDVGGGVLSCKGLHSSFRTTQSGLSLNIDVSTTMIVRPGPVVDF---LIENQNV 277
D +G G+ S G + S R TQ GLSLNID+++ + P PV++F L+ +
Sbjct: 293 PDIKTPQRLGEGLESWCGFYQSIRPTQMGLSLNIDMASAAFIEPLPVIEFVAQLLGKDVL 352
Query: 278 RDPFS-LDWNKAKRTLKNLRITAKPS---NQEYKITGLSELSCKDQLFTMKKRGAVAGED 333
P S D K K+ L+ +++ ++Y++ GL+ ++ +F + ++
Sbjct: 353 SKPLSDSDRVKIKKGLRGVKVEVTHRANVRRKYRVAGLTTQPTRELMFPV--------DE 404
Query: 334 DTEEITVYDYFVHRRKIDLQYSAGLPCINVGKPKRPTYIPIELCSLISLQRYTKALSTSQ 393
+ +V +YF +Q++ LPC+ VG K+ +Y+P+E C ++ QRYTK L+ Q
Sbjct: 405 NCTMKSVIEYFQEMYGFTIQHTH-LPCLQVGNQKKASYLPMEACKIVEGQRYTKRLNEKQ 463
Query: 394 RSSLVEKSRQKPVERMRVLSNALKASNYGSEPMLRNCGISITSEFTQVDGRVLQAPRLKF 453
++L++ + Q+P +R + ++ + Y +P + G++I+ + V+ R+L AP LK+
Sbjct: 464 ITALLKVTCQRPRDRENDILRTVQHNAYDQDPYAKEFGMNISEKLASVEARILPAPWLKY 523
Query: 454 ---GNE-DFNPRNGRWNFNNKKFVEPVSLGNWSVVNFSARCD---VRGLVRDLIKCGGMK 506
G E D P+ G+WN NKK + +++ W+ VNFS RG +L + +
Sbjct: 524 HENGKEKDCLPQVGQWNMMNKKMINGMTVSRWACVNFSRSVQENVARGFCNELGQMCEVS 583
Query: 507 GILVEQPKDVIEENRQFKGEP--PVF-----RVEKMFADVLKLS------KRPSFLLCLL 553
G+ +F EP P++ +VEK V S K LL +L
Sbjct: 584 GM-------------EFNPEPVIPIYSARPDQVEKALKHVYHTSMNKTKGKELELLLAIL 630
Query: 554 PERKNSDLYGPWKKKNLAEFGIVTQCIAPT---RVNDQYLTNVLLKINAKLGGMNSWLGV 610
P+ N LYG K+ E G+++QC +++ QYL NV LKIN K+GG N+ L
Sbjct: 631 PDN-NGSLYGDLKRICETELGLISQCCLTKHVFKISKQYLANVSLKINVKMGGRNTVLVD 689
Query: 611 EHSRSIPIVSKVPTLILGMDVSHGSPGQPDIPSIAAVVSSRKWPLISKYRACVRTQGSKV 670
S IP+VS +PT+I G DV+H G+ PSIAAVV+S+ WP ++KY V Q +
Sbjct: 690 AISCRIPLVSDIPTIIFGADVTHPENGEESSPSIAAVVASQDWPEVTKYAGLVCAQAHRQ 749
Query: 671 EMIDNLFK----PVSDKEDEGIIRELLLDFFHSSEERRPENIIIFRDGVSESQFNEVLNV 726
E+I +L+K PV G+IR+LL+ F + ++P II +RDGVSE QF +VL
Sbjct: 750 ELIQDLYKTWQDPVRGTVSGGMIRDLLISF-RKATGQKPLRIIFYRDGVSEGQFYQVLLY 808
Query: 727 ELSQIIEACKFLDENWNPKFMVIVAQKNHHTKFFQPRSPD--------NVPPGTVVDSKI 778
EL I +AC L+ N+ P IV QK HHT+ F D N+ PGTVVD+KI
Sbjct: 809 ELDAIRKACASLEPNYQPPVTFIVVQKRHHTRLFANNHRDKNSTDRSGNILPGTVVDTKI 868
Query: 779 CHPRNYDFYMCAHAGMIGTSRPTHYHVLLDEIGFSPDDLQELVHSLSYVYQRSTTAISVV 838
CHP +DFY+C+HAG+ GTSRP HYHVL DE F+ D +Q L ++L Y Y R T ++S+V
Sbjct: 869 CHPTEFDFYLCSHAGIQGTSRPAHYHVLWDENNFTADGIQSLTNNLCYTYARCTRSVSIV 928
>At1g48410 Argonaute protein (AGO1)
Length = 870
Score = 430 bits (1105), Expect = e-120
Identities = 284/840 (33%), Positives = 439/840 (51%), Gaps = 84/840 (10%)
Query: 43 RRGLGTKGAKLPLLTNHFEVNVANTNRVFFQYSVALFYEDGRPVEGKGAGRKIIDKVQET 102
R G G G + + NHF + + + Y V + E V +G R ++ ++ +
Sbjct: 2 RPGKGQSGKRCIVKANHFFAELPDKD--LHHYDVTITPE----VTSRGVNRAVMKQLVDN 55
Query: 103 Y-DSELNGKDLAYDG-ETLFTIGSLAQKKLEFIVVVEDVASNRNNANTSPDKKRIRKSYR 160
Y DS L + AYDG ++L+T G L EF + + D R
Sbjct: 56 YRDSHLGSRLPAYDGRKSLYTAGPLPFNSKEFRINLLDEEVGAGGQR------------R 103
Query: 161 SKTYKVEINFAKEIPLQAIANALKGHEAENYQEAIRVLDIILRQ-HSAKQGCLLVRQNFF 219
+ +KV I L + L+G +++ QEA++VLDI+LR+ +++ + V ++F+
Sbjct: 104 EREFKVVIKLVARADLHHLGMFLEGKQSDAPQEALQVLDIVLRELPTSRIRYIPVGRSFY 163
Query: 220 HNDPNNLNDVGGGVLSCKGLHSSFRTTQSGLSLNIDVSTTMIVRPGPVVDFLIENQNVRD 279
D +G G+ S +G + S R TQ GLSLNID+S+T + PV+ F+ + N RD
Sbjct: 164 SPDIGKKQSLGDGLESWRGFYQSIRPTQMGLSLNIDMSSTAFIEANPVIQFVCDLLN-RD 222
Query: 280 ----PFS-LDWNKAKRTLKNLRITAKPSN---QEYKITGLSELSCKDQLFTMKKRGAVAG 331
P S D K K+ L+ +++ ++Y+I+GL+ ++ ++ F + +R
Sbjct: 223 ISSRPLSDADRVKIKKALRGVKVEVTHRGNMRRKYRISGLTAVATRELTFPVDERNT--- 279
Query: 332 EDDTEEITVYDYFVHRRKIDLQYSAGLPCINVGKPKRPTYIPIELCSLISLQRYTKALST 391
+ +V +YF +Q++ LPC+ VG RP Y+P+E+C ++ QRY+K L+
Sbjct: 280 -----QKSVVEYFHETYGFRIQHTQ-LPCLQVGNSNRPNYLPMEVCKIVEGQRYSKRLNE 333
Query: 392 SQRSSLVEKSRQKPVERMRVLSNALKASNYGSEPMLRNCGISITSEFTQVDGRVLQAPRL 451
Q ++L++ + Q+P++R + + ++ ++Y + + GI I++ V+ R+L P L
Sbjct: 334 RQITALLKVTCQRPIDREKDILQTVQLNDYAKDNYAQEFGIKISTSLASVEARILPPPWL 393
Query: 452 KFGNEDFN----PRNGRWNFNNKKFVEPVSLGNWSVVNFSARCD---VRGLVRDLIKCGG 504
K+ P+ G+WN NKK + ++ NW +NFS + R ++L +
Sbjct: 394 KYHESGREGTCLPQVGQWNMMNKKMINGGTVNNWICINFSRQVQDNLARTFCQELAQMCY 453
Query: 505 MKGILVEQPKDVIEENRQFKGEPPVFRVEKMFADVLKL-----------SKRPSFLLCLL 553
+ G+ P+ V+ PPV + VLK K L+ +L
Sbjct: 454 VSGMAFN-PEPVL---------PPVSARPEQVEKVLKTRYHDATSKLSQGKEIDLLIVIL 503
Query: 554 PERKNSDLYGPWKKKNLAEFGIVTQCIAPTRV---NDQYLTNVLLKINAKLGGMNSWLGV 610
P+ N LYG K+ E GIV+QC V + QY+ NV LKIN K+GG N+ L
Sbjct: 504 PDN-NGSLYGDLKRICETELGIVSQCCLTKHVFKMSKQYMANVALKINVKVGGRNTVLVD 562
Query: 611 EHSRSIPIVSKVPTLILGMDVSHGSPGQPDIPSIAAVVSSRKWPLISKYRACVRTQGSKV 670
SR IP+VS PT+I G DV+H PG+ PSIAAVV+S+ WP I+KY V Q +
Sbjct: 563 ALSRRIPLVSDRPTIIFGADVTHPHPGEDSSPSIAAVVASQDWPEITKYAGLVCAQAHRQ 622
Query: 671 EMIDNLFKPVSDKEDE----GIIRELLLDFFHSSEERRPENIIIFRDGVSESQFNEVLNV 726
E+I +LFK D + G+I+ELL+ F S+ + P II +RDGVSE QF +VL
Sbjct: 623 ELIQDLFKEWKDPQKGVVTGGMIKELLIAFRRSTGHK-PLRIIFYRDGVSEGQFYQVLLY 681
Query: 727 ELSQIIEACKFLDENWNPKFMVIVAQKNHHTKFFQPRSPD--------NVPPGTVVDSKI 778
EL I +AC L+ + P +V QK HHT+ F D N+ PGTVVDSKI
Sbjct: 682 ELDAIRKACASLEAGYQPPVTFVVVQKRHHTRLFAQNHNDRHSVDRSGNILPGTVVDSKI 741
Query: 779 CHPRNYDFYMCAHAGMIGTSRPTHYHVLLDEIGFSPDDLQELVHSLSYVYQRSTTAISVV 838
CHP +DFY+C+HAG+ GTSRP HYHVL DE F+ D LQ L ++L Y Y R T ++S+V
Sbjct: 742 CHPTEFDFYLCSHAGIQGTSRPAHYHVLWDENNFTADGLQSLTNNLCYTYARCTRSVSIV 801
>At1g69440 hypothetical protein
Length = 990
Score = 330 bits (847), Expect = 2e-90
Identities = 248/834 (29%), Positives = 410/834 (48%), Gaps = 101/834 (12%)
Query: 47 GTKGAKLPLLTNHFEVNVANTNRVFFQYSVALFYEDGRPVEGKGAGRKIIDKVQETYDSE 106
G G+ + LL NHF V ++ R++ Y+V + P K R I K+ ET +
Sbjct: 158 GQDGSVIYLLANHFLVKFDSSQRIY-HYNVEI-----SPQPSKEIARMIKQKLVETDRNS 211
Query: 107 LNGKDLAYDG-ETLFTIGSLAQKKLEFIVVVE----DVASNRNNANTSPDKKRIRKSYRS 161
+G A+DG + +++ +LEF V + N + +K+I K +R
Sbjct: 212 FSGVVPAFDGRQNIYSPVEFQGDRLEFFVNLPIPSCKAVMNYGDLREKQPQKKIEKLFRV 271
Query: 162 KTYKVEINFAKEIPLQAIANALKGHEAENYQ----EAIRVLDIILRQHSAKQGCLLVRQN 217
V KE + E E++ E I LD+ILR++ ++ C + ++
Sbjct: 272 NMKLVSKFDGKE----------QRKEGEDWAPLPPEYIHALDVILRENPMEK-CTSIGRS 320
Query: 218 FFHNDPNNLNDVGGGVLSCKGLHSSFRTTQSGLSLNIDVSTTMIVRPGPVVDFLIEN--- 274
F+ + ++GGG + +G S R TQ GL+LN+D+S T V+ +L +
Sbjct: 321 FYSSSMGGSKEIGGGAVGLRGFFQSLRHTQQGLALNMDLSITAFHESIGVIAYLQKRLEF 380
Query: 275 -----QNVRDPFSLDWNK-AKRTLKNLRITA--KPSNQEYKITGLSELSCKDQLFTMKKR 326
+N SL+ + ++ LKN+R+ + + Q Y++ GL+E ++ F
Sbjct: 381 LTDLPRNKGRELSLEEKREVEKALKNIRVFVCHRETVQRYRVYGLTEEITENIWFP---- 436
Query: 327 GAVAGEDDTEEITVYDYFVHRRKIDLQYSAGLPCINVGKPKRPTYIPIELCSLISLQRYT 386
+ + + + + YF ++Q+ LPC+ + + RP Y+P+ELC + Q++
Sbjct: 437 -----DREGKYLRLMSYFKDHYGYEIQFK-NLPCLQISRA-RPCYLPMELCMICEGQKFL 489
Query: 387 KALSTSQRSSLVEKSRQKPVERMRVLSNALKAS-NYGSEPMLRNCGISITSEFTQVDGRV 445
LS Q + +++ QKP ER ++ + S S R + ++ E T + GR+
Sbjct: 490 GKLSDDQAAKIMKMGCQKPNERKAIIDKVMTGSVGPSSGNQTREFNLEVSREMTLLKGRI 549
Query: 446 LQAPRLKFGNEDFNPRNGRWNFNNKKFVEPVSLGNWSVVNFSARCDVRGLVRDLI----- 500
LQ P+LK PRN K + + W++++ D + + I
Sbjct: 550 LQPPKLKLDR----PRN----LKESKVFKGTRIERWALMSIGGSSDQKSTIPKFINELTQ 601
Query: 501 KCGGMKGILVE--------QPKDVIEENRQFKGEPPVFRVEKMFADVLKLSKRPSFLLCL 552
KC + L + +P ++ + +E ++ + + L+
Sbjct: 602 KCEHLGVFLSKNTLSSTFFEPSHILNN---------ISLLESKLKEIQRAASNNLQLIIC 652
Query: 553 LPERKNSDLYGPWKKKNLAEFGIVTQC-IAP--TRVNDQYLTNVLLKINAKLGGMNSWLG 609
+ E+K+ YG K+ + G+VTQC + P T+++ Q+++N+ LKINAK+GG + L
Sbjct: 653 VMEKKHKG-YGDLKRISETRIGVVTQCCLYPNITKLSSQFVSNLALKINAKIGGSMTELY 711
Query: 610 VEHSRSIPIVSKV--PTLILGMDVSHGSPGQPDIPSIAAVVSSRKWPLISKYRACVRTQG 667
IP + + P + +G DV+H P PS+AAVV S WP ++Y + +R+Q
Sbjct: 712 NSIPSHIPRLLRPDEPVIFMGADVTHPHPFDDCSPSVAAVVGSINWPEANRYVSRMRSQT 771
Query: 668 SKVEMIDNLFKPVSDKEDEGIIRELLLDFFHSSEERRPENIIIFRDGVSESQFNEVLNVE 727
+ E+I +L V + LLD F+ + ++ P II FRDGVSE+QF +VL E
Sbjct: 772 HRQEIIQDLDLMVKE----------LLDDFYKAVKKLPNRIIFFRDGVSETQFKKVLQEE 821
Query: 728 LSQIIEAC-KFLDENWNPKFMVIVAQKNHHTKFFQPRSPD--NVPPGTVVDSKICHPRNY 784
L I AC KF D +NP V QK HHT+ F+ PD N+PPGTVVD+ I HP+ +
Sbjct: 822 LQSIKTACSKFQD--YNPSITFAVVQKRHHTRLFRC-DPDHENIPPGTVVDTVITHPKEF 878
Query: 785 DFYMCAHAGMIGTSRPTHYHVLLDEIGFSPDDLQELVHSLSYVYQRSTTAISVV 838
DFY+C+H G+ GTSRPTHYH+L DE F+ D+LQ LV++L Y + R T IS+V
Sbjct: 879 DFYLCSHLGVKGTSRPTHYHILWDENEFTSDELQRLVYNLCYTFVRCTKPISIV 932
>At1g31280 unknown protein
Length = 1013
Score = 304 bits (778), Expect = 2e-82
Identities = 247/859 (28%), Positives = 402/859 (46%), Gaps = 91/859 (10%)
Query: 22 KPIKIEQEHLKKKLPT-----KAPMAR--RGLGTKGAKLPLLTNHFEVNVANTNRVFFQY 74
+P++ E +LK + K PM R RG ++ L NH++VN N V Y
Sbjct: 137 EPVRAEVMNLKPSVQVATSDRKEPMKRPDRGGVVAVRRVNLYVNHYKVNF-NPESVIRHY 195
Query: 75 SVALFYEDGRPVEGKGAGRKIIDKVQETYDSELNGKDLAYDGE-TLFTIGSL--AQKKLE 131
V + E + + DKV E AYDG+ +F+ L K+E
Sbjct: 196 DVEIKGEIPTKKVSRFELAMVRDKVFTDNPDEFPLAMTAYDGQKNIFSAVELPTGSYKVE 255
Query: 132 FIVVVEDVASNRNNANTSPDKKRIRKSYRSKTYKVEINFAKEIPLQAIANALKGHEAENY 191
+ E R ++Y I + L + + G + N
Sbjct: 256 YPKTEE---------------------MRGRSYTFTIKQVNVLKLGDLKEYMTGRSSFNP 294
Query: 192 QEAIRVLDIILRQHSAKQGCLL-VRQNFFHNDPNNLNDVGGGVLSCKGLHSSFRTTQSGL 250
++ ++ +D+++++H +K C++ V ++FF + D GV++ KG + + T GL
Sbjct: 295 RDVLQGMDVVMKEHPSK--CMITVGKSFFTRETEPDEDFRFGVIAAKGYRHTLKPTAQGL 352
Query: 251 SLNIDVSTTMIVRPGPVVDFLIENQNVRDPFSLDWNKAKRTLKNLRITA--KPSNQEYKI 308
SL +D S + V+++L N D + L L++T + + Q+ I
Sbjct: 353 SLCLDYSVLAFRKAMSVIEYLKLYFNWSDMRQFRRRDVEEELIGLKVTVNHRKNKQKLTI 412
Query: 309 TGLSELSCKDQLFTMKKRGAVAGEDDTEEITVYDYFVHRRKIDLQYSAGLPCINVGKPKR 368
GLS + KD F + + G + + ++ +YF + + + +PC+++GK R
Sbjct: 413 VGLSMQNTKDIKFDLIDQ---EGNEPPRKTSIVEYFRIKYGRHIVHK-DIPCLDLGKNGR 468
Query: 369 PTYIPIELCSLISLQRYTKA-LSTSQRSSLVEKSRQKPVERMRVLSNALKASNYGSE-PM 426
++P+E C L+ Q Y K L L + S P +R R + +KA N S +
Sbjct: 469 QNFVPMEFCDLVEGQIYPKDNLDKDSALWLKKLSLVNPQQRQRNIDKMIKARNGPSGGEI 528
Query: 427 LRNCGISITSEFTQVDGRVLQAPRLKFGN------EDFNPR-NGRWNFNNKKFVEPVSLG 479
+ N G+ + + T V+GRVL+AP LK E+ NPR N +WN K +
Sbjct: 529 IGNFGLKVDTNMTPVEGRVLKAPSLKLAERGRVVREEPNPRQNNQWNLMKKGVTRGSIVK 588
Query: 480 NWSVVNFSARCDVRGLVRDLI-----KCGGMKGILVEQPKDVIEENRQFKGEPPVFRVEK 534
+W+V++F+A + D + +C + G+ +E P ++ + + + +E+
Sbjct: 589 HWAVLDFTASERFNKMPNDFVDNLIDRCWRL-GMQMEAP--IVYKTSRMETLSNGNAIEE 645
Query: 535 MFADVLKLSKR------PSFLLCLLPERKNSDLYGPWKKKNLAEFGIVTQCIAP---TRV 585
+ V+ + R P+ +LC + + D Y K + G+VTQC T+
Sbjct: 646 LLRSVIDEASRKHGGARPTLVLCAMSRK--DDGYKTLKWIAETKLGLVTQCFLTGPATKG 703
Query: 586 NDQYLTNVLLKINAKLGGMNSWLGVEHSRSIPIVSKVP-TLILGMDVSHGSPGQPDIPSI 644
DQY N+ LK+NAK+GG N VE + K + +G DV+H + PSI
Sbjct: 704 GDQYRANLALKMNAKVGGSN----VELMDTFSFFKKEDEVMFIGADVNHPAARDKMSPSI 759
Query: 645 AAVVSSRKWPLISKYRACVRTQGSKVEMIDNLFKPVSDKEDEGIIRELLLDFFHSSEERR 704
AVV + WP ++Y A V Q + E I + L+ + +R
Sbjct: 760 VAVVGTLNWPEANRYAARVIAQPHRKEEIQGFGDACLE----------LVKAHVQATGKR 809
Query: 705 PENIIIFRDGVSESQFNEVLNVELSQIIEACKFLDENWNPKFMVIVAQKNHHTKFFQPRS 764
P I+IFRDGVS++QF+ VLNVEL + F +NPK VIVAQK H T+FF +
Sbjct: 810 PNKIVIFRDGVSDAQFDMVLNVELLDV--KLTFEKNGYNPKITVIVAQKRHQTRFFPATN 867
Query: 765 PD-----NVPPGTVVDSKICHPRNYDFYMCAHAGMIGTSRPTHYHVLLDEIGFSPDDLQE 819
D NVP GTVVD+K+ HP YDFY+C+H G IGTS+PTHY+ L DE+GF+ D +Q+
Sbjct: 868 NDGSDKGNVPSGTVVDTKVIHPYEYDFYLCSHHGGIGTSKPTHYYTLWDELGFTSDQVQK 927
Query: 820 LVHSLSYVYQRSTTAISVV 838
L+ + + + R T +S+V
Sbjct: 928 LIFEMCFTFTRCTKPVSLV 946
>At1g31290 hypothetical protein
Length = 1194
Score = 302 bits (773), Expect = 6e-82
Identities = 248/853 (29%), Positives = 412/853 (48%), Gaps = 102/853 (11%)
Query: 33 KKLPTKAPMARRGLGTKGAKLPLLTNHFEVNVANTNRVFFQYSVALFYEDGRPVEGKGAG 92
KK P K P + KG + L NHF V+ + T V Y V ++G+ +
Sbjct: 330 KKEPVKRPDKGGNIKVKGV-INLSVNHFRVSFS-TESVIRHYDV--------DIKGENSS 379
Query: 93 RKI----IDKVQETYDSELN---GKDLAYDGE-TLFTIGSLAQKKLEFIVVVEDVASNRN 144
+KI + V+E + N AYDG+ +F+ L +
Sbjct: 380 KKISRFELAMVKEKLFKDNNDFPNAMTAYDGQKNIFSAVELPTGSFKV------------ 427
Query: 145 NANTSPDKKRIRKSYRSKTYKVEINFAKEIPLQAIANALKGHEAENYQEAIRVLDIILRQ 204
D + R ++Y I KE+ L + + G ++ ++ +D+++++
Sbjct: 428 ------DFSETEEIMRGRSYTFIIKQVKELKLLDLQAYIDGRSTFIPRDVLQGMDVVMKE 481
Query: 205 HSAKQGCLLVRQNFFHNDPNNLNDVGGGVLSCKGLHSSFRTTQSGLSLNIDVSTTMIVRP 264
H +K+ + V + FF D G GV + KG H + + T GLSL ++ S +
Sbjct: 482 HPSKR-MITVGKRFFSTRLEI--DFGYGVGAAKGFHHTLKPTVQGLSLCLNSSLLAFRKA 538
Query: 265 GPVVDFL---IENQNVRDPFSLDWNKAKRTLKNLRITA--KPSNQEYKITGLSELSCKDQ 319
V+++L +N+R + + + L L++T + + Q++ I GLS+ KD
Sbjct: 539 ISVIEYLKLYFGWRNIRQFKNCRPDDVVQELIGLKVTVDHRKTKQKFIIMGLSKDDTKDI 598
Query: 320 LFTMKKRGAVAGEDDTEEITVYDYFVHRRKIDLQYSAGLPCINVGKPKRPTYIPIELCSL 379
F AG +I++ +YF + D+ + +PC+N+GK R ++P+E C+L
Sbjct: 599 KFDFIDH---AGNQPPRKISIVEYFKEKYGRDIDHK-DIPCLNLGKKGRENFVPMEFCNL 654
Query: 380 ISLQRYTKA-LSTSQRSSLVEKSRQKPVERMRVLSNALKASN--YGSEPMLRNCGISITS 436
+ Q + K L + L E S P +R+ ++ +K+S+ G + ++ N G+ +
Sbjct: 655 VEGQIFPKEKLYRDSAAWLKELSLVTPQQRLENINKMIKSSDGPRGGD-IIGNFGLRVDP 713
Query: 437 EFTQVDGRVLQAPRLKFGNEDFNP-------RNGRWNFNNKKFVEPVSLGNWSVVNFSAR 489
T V+GRVL+AP LK + NP + +WN K + + +W+V++F+A
Sbjct: 714 NMTTVEGRVLEAPTLKLTDRRGNPIHEKLMSESNQWNLTTKGVTKGSIIKHWAVLDFTAS 773
Query: 490 CDVRG-----LVRDLI-KCGGMKGILVEQPKDVIEENRQFKGEPPVFRVEKMFADVLKLS 543
++ V LI +C G+ G+ +E P ++ + + +E++ V+ +
Sbjct: 774 ESLKKKMPGYFVNKLIERCKGL-GMQMEAP--IVCKTSSMETLYDGNALEELLRSVIDEA 830
Query: 544 KR------PSFLLCLLPERKNSDLYGPWKKKNLAEFGIVTQC------IAPTRVNDQYLT 591
P+ +LC + + D Y K + G+VTQC I V+DQYL
Sbjct: 831 SHNHGGACPTLVLCAMTGKH--DGYKTLKWIAETKLGLVTQCFLTISAIKGETVSDQYLA 888
Query: 592 NVLLKINAKLGGMNSWLGVEHSRSIPIVSKVPTLILGMDVSHGSPGQPDIPSIAAVVSSR 651
N+ LKINAK+GG N L V++ S + + +G DV+H + PSI AVV +
Sbjct: 889 NLALKINAKVGGTNVEL-VDNIFSF-FKKEDKVMFIGADVNHPAAHDNMSPSIVAVVGTL 946
Query: 652 KWPLISKYRACVRTQGSKVEMIDNLFKPVSDKEDEGIIRELLLDFFHSSEERRPENIIIF 711
WP ++Y A V+ Q + E I + + L++ + E+RP I+IF
Sbjct: 947 NWPEANRYAARVKAQSHRKEEIQGFGETCWE----------LIEAHSQAPEKRPNKIVIF 996
Query: 712 RDGVSESQFNEVLNVELSQIIEACKFLDENWNPKFMVIVAQKNHHTKFFQP------RSP 765
RDGVS+ QF+ VLNVEL + + F +NP+ VIVAQK H T+FF R+
Sbjct: 997 RDGVSDGQFDMVLNVELQNVKDV--FAKVGYNPQITVIVAQKRHQTRFFPATTSKDGRAK 1054
Query: 766 DNVPPGTVVDSKICHPRNYDFYMCAHAGMIGTSRPTHYHVLLDEIGFSPDDLQELVHSLS 825
NVP GTVVD+ I HP YDFY+C+ G IGTS+PTHY+VL DEIGF+ + +Q+L+ L
Sbjct: 1055 GNVPSGTVVDTTIIHPFEYDFYLCSQHGAIGTSKPTHYYVLSDEIGFNSNQIQKLIFDLC 1114
Query: 826 YVYQRSTTAISVV 838
+ + R T +++V
Sbjct: 1115 FTFTRCTKPVALV 1127
>At3g52300 unknown protein
Length = 168
Score = 33.5 bits (75), Expect = 0.52
Identities = 24/80 (30%), Positives = 37/80 (46%), Gaps = 7/80 (8%)
Query: 33 KKLPTKAPMARRGLGTKGAKLPLLTNHFEVNVANTNRVFFQYSVAL---FYEDGRPVEG- 88
KK+ A A R + G L+T+ +N R F + + L F ++ P++
Sbjct: 6 KKIADVAFKASRTIDWDGMAKVLVTDEARREFSNLRRAFDEVNTQLQTKFSQEPEPIDWD 65
Query: 89 ---KGAGRKIIDKVQETYDS 105
KG G I+DK +E YDS
Sbjct: 66 YYRKGIGAGIVDKYKEAYDS 85
>At2g12190 pseudogene
Length = 512
Score = 31.6 bits (70), Expect = 2.0
Identities = 44/192 (22%), Positives = 77/192 (39%), Gaps = 30/192 (15%)
Query: 601 LGGMNSWLGVEHSRSIPIV-----SKVPTLILGMDVSHGS--------PGQPDIPSIAAV 647
LGG+N++L H R PI+ S+ + ++H + +P I+ +
Sbjct: 52 LGGLNNYLRSVHHRLGPIITLRITSRPAIFVADRSLAHQALVLNGAVFADRPPAAPISKI 111
Query: 648 VSSRKWPLISK-YRACVR-TQGSKVEMIDNLFKPVSDKEDEGIIRELLLDFFHSSEERRP 705
+SS + + S Y A R + + I + + S + E+L D F S P
Sbjct: 112 ISSNQHNISSSLYGATWRLLRRNLTSEILHPSRVRSYSHARRWVLEILFDRFGKSRGEEP 171
Query: 706 ENII--------------IFRDGVSESQFNEVLNVELSQIIEACKFLDENWNPKFMVIVA 751
++ F D + E Q +V V+ Q++ +F N PKF ++
Sbjct: 172 IVVVDHLHYAMFALLVLMCFGDKLDEKQIKQVEYVQRRQLLGFSRFNILNLWPKFTKLIL 231
Query: 752 QKNHHTKFFQPR 763
+K +FFQ R
Sbjct: 232 RKRWE-EFFQMR 242
>At5g18860 unknown protein
Length = 890
Score = 31.2 bits (69), Expect = 2.6
Identities = 25/101 (24%), Positives = 47/101 (45%), Gaps = 10/101 (9%)
Query: 631 VSHGSPGQPDIPSIAAVVSSRKWPLISK-----YRACVRTQGSKVEMIDNLFKPV-SDKE 684
V+HG+P D P + ++ W ++K + V T G + NL K + SDK+
Sbjct: 631 VTHGAPRDTDRPELRQPLAIEVWQNLTKSGNGVSKITVLTNGP----LTNLAKIISSDKK 686
Query: 685 DEGIIRELLLDFFHSSEERRPENIIIFRDGVSESQFNEVLN 725
+I+E+ + H + E+ + I + ++FN L+
Sbjct: 687 SSSLIKEVYIVGGHINREKSDKGNIFTIPSNAYAEFNMFLD 727
>At1g10760 unknown protein
Length = 1399
Score = 30.8 bits (68), Expect = 3.4
Identities = 26/109 (23%), Positives = 50/109 (45%), Gaps = 14/109 (12%)
Query: 3 SQENKECLPPPPPIVPPNVKPIK--IEQEHLKKKLPTKAPMARRGLGTKGAK---LPLLT 57
SQ+ E L PP I+PPN P++ ++ + +P+ L +G +P +
Sbjct: 399 SQKGGEWLDPPSDILPPNSLPVRGAVDTKLTITSTDLPSPVQTFELEIEGDSYKGMPFVL 458
Query: 58 NHFEVNVANTNRVFFQYSVALFYEDGRPV-----EGKGAGRKIIDKVQE 101
N E + N + F+ F ++ + V +GKG + ++DK+ +
Sbjct: 459 NAGERWIKNNDSDFY----VDFAKEEKHVQKDYGDGKGTAKHLLDKIAD 503
>At5g37920 putative protein
Length = 609
Score = 30.4 bits (67), Expect = 4.4
Identities = 15/45 (33%), Positives = 24/45 (53%)
Query: 708 IIIFRDGVSESQFNEVLNVELSQIIEACKFLDENWNPKFMVIVAQ 752
I F G+ + ++L +L I+E+ K L +N N F+V V Q
Sbjct: 213 IQFFNTGMKQDVIKKILRFQLVNIVESAKKLVDNGNEMFLVRVLQ 257
>At4g26910 putative dihydrolipoamide succinyltransferase
Length = 464
Score = 30.4 bits (67), Expect = 4.4
Identities = 20/79 (25%), Positives = 36/79 (45%), Gaps = 13/79 (16%)
Query: 11 PPPP------PIVPPNVKPIKIEQEHLKKKLPTKAPMARRGLGTKGAKLPLLTNHFEVNV 64
PPPP P +PP + ++ L+K++ T+ ++ LLT EV++
Sbjct: 217 PPPPKQSAKEPQLPPKERERRVPMTRLRKRVATRLKDSQNTFA-------LLTTFNEVDM 269
Query: 65 ANTNRVFFQYSVALFYEDG 83
N ++ QY A + + G
Sbjct: 270 TNLMKLRSQYKDAFYEKHG 288
>At1g25170
Length = 469
Score = 30.4 bits (67), Expect = 4.4
Identities = 17/77 (22%), Positives = 36/77 (46%)
Query: 660 RACVRTQGSKVEMIDNLFKPVSDKEDEGIIRELLLDFFHSSEERRPENIIIFRDGVSESQ 719
R VR +G + +L+KPV D++ + + ++ D EE+ + +I R + +
Sbjct: 128 RKTVRNRGHCGTRLSHLYKPVFDRQFKMLKIHMMEDMIKDGEEKSLDTFVISRQLTTFTD 187
Query: 720 FNEVLNVELSQIIEACK 736
L+ + I++ K
Sbjct: 188 LTVCLSTSATYILDRFK 204
>At5g18890 putative protein
Length = 550
Score = 30.0 bits (66), Expect = 5.8
Identities = 26/101 (25%), Positives = 45/101 (43%), Gaps = 10/101 (9%)
Query: 631 VSHGSPGQPDIPSIAAVVSSRKWPLISK-----YRACVRTQGSKVEMIDNLFKPV-SDKE 684
V+HG+P D P + ++ W ++K + V T G + +L K + SDK
Sbjct: 282 VAHGAPSDTDRPELRQPLALEVWQNLTKSVDEVSKITVLTNGP----LTSLAKIISSDKN 337
Query: 685 DEGIIRELLLDFFHSSEERRPENIIIFRDGVSESQFNEVLN 725
II+E+ + H S + + I S ++FN L+
Sbjct: 338 SSSIIKEVYIVGGHISRGKSDKGNIFTVPSNSYAEFNMFLD 378
>At3g20680 unknown protein
Length = 338
Score = 30.0 bits (66), Expect = 5.8
Identities = 29/135 (21%), Positives = 56/135 (41%), Gaps = 24/135 (17%)
Query: 433 SITSEFTQVDGRVLQAPR------LKFGNEDFNPR-----NGRWNFNNKKFVEPVSLGNW 481
S+ + D + AP +K +E F+ + N RW F +K +G++
Sbjct: 183 SLKTALRSADVAIFMAPEKSQLEDVKTASEAFSTKPVVMINPRWLFEEEKSFGDGFIGSF 242
Query: 482 SVVNFSARCDVRGL-----------VRDLIKCGGMKGILVEQPK--DVIEENRQFKGEPP 528
V+ +VRG+ VRD + G +LVE+ + ++ QF P
Sbjct: 243 EVIYAFTGLEVRGVFSKRKGVIFKCVRDGVVSGERWNVLVEEEEGNGTLKVVSQFNSRPS 302
Query: 529 VFRVEKMFADVLKLS 543
+ VE + +++ ++
Sbjct: 303 IEEVELVLYNLMAMN 317
>At5g50330 putative protein
Length = 423
Score = 29.3 bits (64), Expect = 9.8
Identities = 15/42 (35%), Positives = 22/42 (51%), Gaps = 5/42 (11%)
Query: 201 ILRQHSAKQGCLLVRQNFFHNDPNNLNDVGGGVLSCKGLHSS 242
IL S G ++++ FFH DP+ G +L CKG +S
Sbjct: 270 ILNSLSRAYGQMILKSGFFHADPH-----PGNILICKGQEAS 306
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.318 0.136 0.399
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 20,060,220
Number of Sequences: 26719
Number of extensions: 915291
Number of successful extensions: 3231
Number of sequences better than 10.0: 23
Number of HSP's better than 10.0 without gapping: 13
Number of HSP's successfully gapped in prelim test: 10
Number of HSP's that attempted gapping in prelim test: 3108
Number of HSP's gapped (non-prelim): 37
length of query: 859
length of database: 11,318,596
effective HSP length: 108
effective length of query: 751
effective length of database: 8,432,944
effective search space: 6333140944
effective search space used: 6333140944
T: 11
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 64 (29.3 bits)
Medicago: description of AC131455.7


