

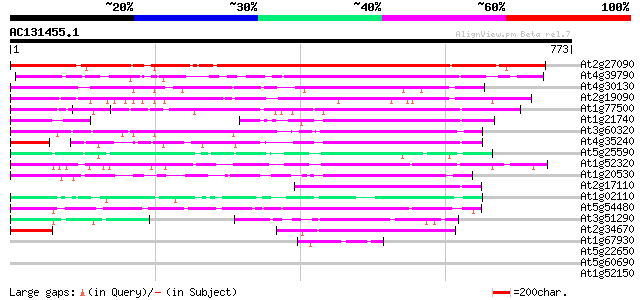
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC131455.1 + phase: 0
(773 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
At2g27090 unknown protein 808 0.0
At4g39790 putative protein 341 1e-93
At4g30130 putative protein 268 1e-71
At2g19090 hypothetical protein 246 5e-65
At1g77500 unknown protein 181 2e-45
At1g21740 putative bzip-like transcription factor 177 3e-44
At3g60320 bZIP like protein 174 2e-43
At4g35240 putative protein 168 9e-42
At5g25590 unknown protein (At5g25590) 164 2e-40
At1g52320 bZIP protein, putative 158 1e-38
At1g20530 hypothetical protein 154 2e-37
At2g17110 unknown protein 144 2e-34
At1g02110 bZIP-like protein 141 1e-33
At5g54480 unknown protein 138 1e-32
At3g51290 putative protein 104 2e-22
At2g34670 unknown protein 101 1e-21
At1g67930 putative golgi transport complex protein 43 8e-04
At5g22650 putative histone deacetylase (HD2B) 40 0.004
At5g60690 REVOLUTA or interfascicular fiberless 1 38 0.019
At1g52150 unknown protein 38 0.019
>At2g27090 unknown protein
Length = 743
Score = 808 bits (2086), Expect = 0.0
Identities = 443/758 (58%), Positives = 542/758 (71%), Gaps = 44/758 (5%)
Query: 1 MGASSSKMEEDKALQLCRERKKFVRQALDGRCSLAAAHISYVQSLRIVGTALSKFTEPEP 60
MGAS+S+++EDKALQLCRERKKFV+QALDGRC LAAAH+SYVQSL+ GTAL KF+E E
Sbjct: 1 MGASTSRIDEDKALQLCRERKKFVQQALDGRCLLAAAHVSYVQSLKSTGTALRKFSETEV 60
Query: 61 HSESSLYTN--ATPEA-LALTDKAMSQSSPSMSQQIDAGETHSFYTT----PSPPSSSKF 113
ESSLYT+ ATPE LAL +K++S S S +HS + T PSPPS+S F
Sbjct: 61 PVESSLYTSTSATPEQPLALIEKSVSHLSYSPPPA-----SHSHHDTYSPPPSPPSTSPF 115
Query: 114 HANHMKFGSFSSKKVEEKPPVPVIATVTSSGIAQNGS--RMSETEAFEDSSVPDGTPQWD 171
NHMKF FSSKKVEEKPPV +IATVTSS I + S +M T E SS+P P WD
Sbjct: 116 QVNHMKFKGFSSKKVEEKPPVSIIATVTSSSIPPSRSIEKMESTPFEESSSMPPEAP-WD 174
Query: 172 FFGLFNPVDHQFSFQEPKGMPHHIGNA-------EEGGIPDLEDDEEKASSCGSEGSRDS 224
+FGL +P+D+QFS H+GN EE G P+ EDD E S E SRDS
Sbjct: 175 YFGLSHPIDNQFSSS-------HVGNGHVSRSVKEEDGTPEEEDDGEDFSFQEREESRDS 227
Query: 225 EDEFDEEPTAETLVQRFENLNRANNHVQENVLPVKGDSVSEVELVSEKGNFPNSSPSKKL 284
+D+ +EPT++TLV+ FEN NR + LP + + V +EK P SP
Sbjct: 228 DDDEFDEPTSDTLVRSFENFNRVRR--DHSTLPQR-EGVESDSSDAEKSKTPELSP---- 280
Query: 285 PMAALLPPEANKSTEKENHSENKVTPKNFFSSMKDIEVLFNRASDSGKEVPRMLEANKFH 344
P+ L+ NK+ K +H+ENK+ P++F SSMK+IE+LF +AS++GKEVPRMLEANK H
Sbjct: 281 PVTPLVATPVNKTPNKGDHTENKLPPRDFLSSMKEIELLFVKASETGKEVPRMLEANKLH 340
Query: 345 FRPIFQGKKDGSLASFICKACFSCGEDPSQVPEEPAQNSVKYLTWHRTASSRSSSSKNPL 404
FRPI K+ GS AS + K C SCGEDP VPEEPAQNSVKYLTWHRT SSRSSSS+NPL
Sbjct: 341 FRPIVPSKESGSGASSLFKTCLSCGEDPKDVPEEPAQNSVKYLTWHRTESSRSSSSRNPL 400
Query: 405 DANSRENVEDLTNNLFDNSCMNAGSHASTLDRLHAWERKLYDEVKASEIVRKEYDAKCKI 464
+ ++VE+L +NLF+N CM AGSHASTLDRL+AWERKLYDEVK S+ VR+EYD KC+I
Sbjct: 401 GGMNSDDVEELNSNLFENICMIAGSHASTLDRLYAWERKLYDEVKGSQTVRREYDEKCRI 460
Query: 465 LRNLESKAEKTSTVDKTRAAVKDLHSRIRVAIHRIDSISKRIEELRDKELQPQLEELIEG 524
LR LES+ + + +DKTRA VKDLHSRIRVAIHRIDSIS+RIEELRD ELQPQLEELIEG
Sbjct: 461 LRELESEGKGSQRIDKTRAVVKDLHSRIRVAIHRIDSISRRIEELRDNELQPQLEELIEG 520
Query: 525 LNRMWEVMHECHKLQFQIMSASYNNSHARITMHSELRRQITAYLENELQFLSSSFTKWIE 584
L+RMWEVM ECHK+QFQ++ A Y + ++ M SEL RQ+T++LE+EL L+SSFTKWI
Sbjct: 521 LSRMWEVMLECHKVQFQLIKACYRGGNIKLNMQSELHRQVTSHLEDELCALASSFTKWIT 580
Query: 585 AQKSYLQAINGWIHKCVPLQQKSVKRKRRPQSELLIQYGPPIYATCDVWLKKLGTLPVKD 644
QKSY+QAIN W+ KCV L Q+S KRKRR L YGPPIYATC +WL+KL LP K+
Sbjct: 581 GQKSYIQAINEWLVKCVALPQRS-KRKRRAPQPSLRNYGPPIYATCGIWLEKLEVLPTKE 639
Query: 645 VVDSIKSLAADTARFLPYQDKNQGKEPHSHIGGESADGL----LRDDISEDWISGFDRFR 700
V SIK+LA+D ARFLP Q+KN+ K+ H GE+ + L L+D+ ED GFDRFR
Sbjct: 640 VSGSIKALASDVARFLPRQEKNRTKK---HRSGENKNDLTAHMLQDETLEDCGPGFDRFR 696
Query: 701 ANLIRFLGQLNSLSGSSVKMYRELRQAIQEAKNNYHRL 738
+L F+GQLN + SSVKMY EL++ I AKNNY +L
Sbjct: 697 TSLEGFVGQLNQFAESSVKMYEELKEGIHGAKNNYEQL 734
>At4g39790 putative protein
Length = 631
Score = 341 bits (874), Expect = 1e-93
Identities = 255/742 (34%), Positives = 365/742 (48%), Gaps = 131/742 (17%)
Query: 9 EEDKALQLCRERKKFVRQALDGRCSLAAAHISYVQSLRIVGTALSKFTEPEPHSESSLYT 68
++++ L LC+ERK+FV+QA+D RC+LAAAH+SY++SLR +G L ++ E E ESS
Sbjct: 3 KKNEPLHLCKERKRFVKQAMDSRCALAAAHVSYIRSLRNIGACLRQYAEAETAHESSPSL 62
Query: 69 NATPEALALTDKAMSQSSPSMSQQIDAGETHSFYTTPSPPSSSKFHANHMKFGSFSSKKV 128
AT +K+ S +S +D+ +H+ + P+P F+ ++MK + SS
Sbjct: 63 TATE-----PEKSPSHNSSYPDDSVDSPLSHN--SNPNPNPKPLFNLSYMKTETASS--- 112
Query: 129 EEKPPVPVIATVTSSGIAQNGSRMSET-EAFEDSSVP----DGTPQWDFFGLFNPVDHQF 183
T T + ++ + T AF P T WD+F + D F
Sbjct: 113 --------TVTFTINPLSDGDDDLEVTMPAFSPPPPPRPRRPETSSWDYFDTCDDFD-SF 163
Query: 184 SFQEPKGMPHHIGNAEEGGIPDLEDDE-----EKASSCGSEGSRDSEDEFDEEPTAETLV 238
F +G +E+ I D E D EK +S G+ SE D +
Sbjct: 164 RF---------VGLSEQTEI-DSECDAAVIGLEKITSQGNVAKSGSETLQDSSFKTKQRK 213
Query: 239 QRFENLNRANNHVQENVLPVKGDSVSEVELVSEKGNFPNSSPSKKLPMAALLPPEANKST 298
Q E+ N PS+ + A
Sbjct: 214 QSCED------------------------------NDEREDPSEFITHRA---------- 233
Query: 299 EKENHSENKVTPKNFFSSMKDIEVLFNRASDSGKEVPRMLEANKFH--FRPIFQGKKDGS 356
K+ S K FF RAS+SG+EV RMLE NK F + +GK
Sbjct: 234 -KDFVSSMKDIEHKFF-----------RASESGREVSRMLEVNKIRVGFADMTEGK---- 277
Query: 357 LASFICKACFSCGEDPSQVPEEPAQNSV-KYLTWHRTASSRSSSSKNPLDANSRENVEDL 415
+ C E V EP + V K + W RT+SSRSS+S+NPL S+E+ +D
Sbjct: 278 ----VIHQFLKCSELNFNV--EPLSHQVTKVIVWKRTSSSRSSTSRNPLIQTSKEDHDDE 331
Query: 416 TNNLF-DNSCMNAGSHASTLDRLHAWERKLYDEVKASEIVRKEYDAKCKILRNLESKAEK 474
+ + F + CM +GSH+S+LDRL+AWERKLYDEVKASE++RKEYD KC+ LRN +K
Sbjct: 332 SGSDFIEEFCMISGSHSSSLDRLYAWERKLYDEVKASEMIRKEYDRKCEQLRNQFAKDHS 391
Query: 475 TSTVDKTRAAVKDLHSRIRVAIHRIDSISKRIEELRDKELQPQLEELIEGLNRMWEVMHE 534
++DKTRAA KDLHSRIRVAI ++SISKRIE +RD EL PQL E ++GL RMW+ M E
Sbjct: 392 AKSMDKTRAAAKDLHSRIRVAIQSVESISKRIERIRDDELHPQLLEFLQGLIRMWKAMLE 451
Query: 535 CHKLQFQIMSASYNNSHARITMH-SELRRQITAYLENELQFLSSSFTKWIEAQKSYLQAI 593
CH Q+ +S +Y+ H+ T H S L+R+I A L E + SF + + SY++A+
Sbjct: 452 CHHTQYITISLAYHCRHSSKTAHESVLKRRILAELLEETECFGLSFVDLVHSMASYVEAL 511
Query: 594 NGWIHKCVPLQQKSVKRKRRPQSELLIQYGPPIYATCDVWLKKLGTLPVKDVVDSIKSLA 653
NGW+H CV L Q+ R RRP S + PPI+ C W + TLP ++ SIK +
Sbjct: 512 NGWLHNCVLLPQERSTRNRRPWSPRRV-LAPPIFVLCRDWSAGIKTLPSDELSGSIKGFS 570
Query: 654 ADTARFLPYQDKNQGKEPHSHIGGESADGLLRDDISEDWISGFDRFRANLIRFLGQLNSL 713
D +G E LL D+S ++L + L +L
Sbjct: 571 LDM----------------EMLGEEKGGSLLVSDLSS--------VHSSLAKLLERLKKF 606
Query: 714 SGSSVKMYRELRQAIQEAKNNY 735
S +S+KMY +++ + A+ Y
Sbjct: 607 SEASLKMYEDVKVKSEAARVAY 628
>At4g30130 putative protein
Length = 725
Score = 268 bits (684), Expect = 1e-71
Identities = 210/687 (30%), Positives = 332/687 (47%), Gaps = 91/687 (13%)
Query: 1 MGASSSKMEEDKALQLCRERKKFVRQALDGRCSLAAAHISYVQSLRIVGTALSKFTEPEP 60
MG S SK ++D+A+Q+C++RK+F++QA++ R A+ HI+Y+QSLR V AL ++ E +
Sbjct: 1 MGCSHSKFDDDEAVQICKDRKRFIKQAVEHRTGFASGHIAYIQSLRKVSDALREYIEGDE 60
Query: 61 HSESSLYTNATP-EALALTDKAMSQSSPSMSQQIDAGETHSFYTTPSPPSSSKFHANHMK 119
E L T TP + ++ + + S PS Q +A SK + N
Sbjct: 61 PHEFMLDTCVTPVKRMSSSGGFIEISPPSKMVQSEA--------------ESKLNVNSYL 106
Query: 120 FGSFSSK-KVEEKPP-VPVIATVTSSGIAQN-GSRMS-ETEAFEDSSVPDGTPQ---WDF 172
S S +VEEKPP P V + G G M+ + ++P +PQ WDF
Sbjct: 107 MASGSRPVRVEEKPPRSPETFQVETYGADSFFGMNMNMNSPGLGSHNIPPPSPQNSQWDF 166
Query: 173 F-GLFNPVD-HQFSFQEPKGMPHHIGNA----EEGGIPDLEDDEEKASSCGSEGSRDSED 226
F F+ +D + +S+ GM + EE GIPDLE+DE ED
Sbjct: 167 FWNPFSALDQYGYSYDNQNGMDDDMRRLRRVREEEGIPDLEEDEYVKF----------ED 216
Query: 227 EFDEEPTAET---LVQRFENLNRANNHVQENVLPVKGDSVSEVELVSEKGNFPNSSPSKK 283
+ + T + + + + + N ++ ++ +S E+ + S
Sbjct: 217 HHNMKATEDFNGGKMYQEDKVEHVNEEFTDSGCKIENESDKNCNGTQERRSLEVSRGGTT 276
Query: 284 LPMAALLPPEANKSTEKENHSENKVTPKNFFSSMKDIEVLFNRASDSGKEVPRMLEANKF 343
+ + + T N+ P + +KD+E F +GKEV +LEA++
Sbjct: 277 GHVVGVTSDDGKGETPGFTVYLNR-RPTSMAEVIKDLEDQFAIICTAGKEVSGLLEASRV 335
Query: 344 HFRPIFQGKKDGSLASFICKACFSCGEDPSQVPEEPAQNSVKYLTWHRTASSRSSSSKNP 403
+ + ++ A F G SSRSSSS
Sbjct: 336 QYT---SSNELSAMTMLNPVALFRSG-----------------------GSSRSSSSSRF 369
Query: 404 L---DANSRENVEDLTNNLFDNSCMNAGSHASTLDRLHAWERKLYDEVKASEIVRKEYDA 460
L SR + + ++ + SCM +GSH STLDRL+AWE+KLYDEVK+ + +R Y+
Sbjct: 370 LISSSGGSRASEFESSSEFSEESCMLSGSHQSTLDRLYAWEKKLYDEVKSGDRIRIAYEK 429
Query: 461 KCKILRNLESKAEKTSTVDKTRAAVKDLHSRIRVAIHRIDSISKRIEELRDKELQPQLEE 520
KC +LRN + K +S VDKTRA ++DLH++I+V+IH I+SIS+RIE LRD+EL PQL E
Sbjct: 430 KCLVLRNQDVKGADSSAVDKTRATIRDLHTQIKVSIHSIESISERIETLRDQELLPQLLE 489
Query: 521 LIEGLNRMWEVMHECHKLQFQ-------IMSASYNNSHARITMHS--ELRRQITA----Y 567
L++GL +MW+VM ECH++Q + +++ + +N H + S E+ Q A +
Sbjct: 490 LVQGLAQMWKVMAECHQIQKRTLDEAKLLLATTPSNRHKKQQQTSLPEINSQRLARSALH 549
Query: 568 LENELQFLSSSFTKWIEAQKSYLQAINGWIHKCVPLQQKSVKRKRRPQSELLIQYGPPIY 627
L +L+ + F WI +Q+SY+ ++ GW+ +C P+ L PIY
Sbjct: 550 LVVQLRNWRACFQAWITSQRSYILSLTGWLLRCFRCDPD-------PEKVTLTSCPHPIY 602
Query: 628 ATCDVWLKKLGTLPVKDVVDSIKSLAA 654
C W + L L K V+D + A+
Sbjct: 603 EVCIQWSRLLNGLNEKPVLDKLDFFAS 629
>At2g19090 hypothetical protein
Length = 844
Score = 246 bits (627), Expect = 5e-65
Identities = 230/837 (27%), Positives = 374/837 (44%), Gaps = 146/837 (17%)
Query: 1 MGASSSKMEEDKALQLCRERKKFVRQALDGRCSLAAAHISYVQSLRIVGTALSKFTEPEP 60
MG S SK+++++A+Q+C++RK+F++QA++ R A+ HI+Y+ SLR V AL F
Sbjct: 1 MGCSHSKLDDEEAVQICKDRKRFIKQAIEHRTKFASGHIAYIHSLRKVSDALHDFILQGD 60
Query: 61 HSESSLYTNATPEALALTDKAMSQSSPSMSQQIDAGETHSFYTTPSPPSS------SKFH 114
++ + T ++ K M Q S+ ++GE S + PP S
Sbjct: 61 NNNEFVPT-LCQDSFVTPVKRMPQRRRRSSRS-NSGEFISISPSSIPPKMIQGRPRSNVK 118
Query: 115 ANHMKFGSFSSKKVEEKPP----VPVIATVTSS----------GIAQNGSRMSETEA--- 157
AN++ +VE++ P V + +SS G+ N + + T +
Sbjct: 119 ANYLMANRSRPVRVEQRSPETFRVETFSPPSSSNQYGESDGFFGMNMNMNTSASTSSSFW 178
Query: 158 ---------FEDSSVPDGTPQ---WDFF-GLFNPVDH-------QFSFQEPKGMPHHIGN 197
++P +PQ WDFF F+ +D+ + S G+ I
Sbjct: 179 NPLSSPEQRLSSHNIPPPSPQNSQWDFFWNPFSSLDYYGYNSYDRGSVDSRSGIDDEIRG 238
Query: 198 A----EEGGIPDLEDDEEK-----------------------------ASSCGSEGSRDS 224
EE GIPDLE+D+E S C E +
Sbjct: 239 LRRVREEEGIPDLEEDDEPHKPEPVVNVRFQNHNPNPKATEENRGKVDKSCCNEEVKVED 298
Query: 225 EDEFDEEPTAETLVQRFENLNRANNHVQENVL--PVKGDSVSEVELVSEK-GNFPNSSPS 281
DE ++E E + ++ N E + P + +EV E GN
Sbjct: 299 VDEDEDEDGDEDDDEFTDSGCETENEGDEKCVAPPTQEQRKAEVCRGGEATGNVVGVGKV 358
Query: 282 KKLPMAALLPPEANKSTEKENHSENKVTPKNFFSSMKDIEVLFNRASDSGKEVPRMLEAN 341
+++ + +A K+T + + P + +KD+E F D+ KEV +LEA
Sbjct: 359 QEMKNVVGVRDDA-KTTGFTVYVNRR--PTSMAEVIKDLEDQFTTICDAAKEVSGLLEAG 415
Query: 342 KFHFRPIFQGKKDGSLASFICKACFSCGEDPSQVPEEPAQNSVKYLTWHRTASSRSSSSK 401
+ + F GE + V ++ + + R+ SSRSSSS+
Sbjct: 416 RAQYTSSFNDHS---------------GEFMNNVAIFLSRKMLNPVALFRSGSSRSSSSR 460
Query: 402 NPLDAN--SRENVEDLTNNLFDNSCMNAGSHASTLDRLHAWERKLYDEVKAS-------- 451
+ ++ SRE+ + +++ D SCM +GSH +TLDRL AWE+KLYDEV+ S
Sbjct: 461 FLITSSGGSRESGSESRSDVSDESCMISGSHQTTLDRLFAWEKKLYDEVRVSNKRYKQLS 520
Query: 452 -EIVRKEYDAKCKILRNLESKAEKTSTVDKTRAAVKDLHSRIRVAIHRIDSISKRIEELR 510
E VR+ Y+ KC LRN + K + VDKTRA ++DL ++I+V+IH I+SISKRIE LR
Sbjct: 521 GERVRRAYEKKCMQLRNQDVKGDDPLAVDKTRATIRDLDTQIKVSIHSIESISKRIETLR 580
Query: 511 DKELQPQLEELIEG----------LNRMWEVMHECHKLQFQIMSAS--------YNNSHA 552
D+EL PQL EL+EG L RMW+VM E H++Q + + + + H
Sbjct: 581 DQELLPQLLELVEGLTNGNHDHNRLTRMWKVMAESHQIQKRTLDEAKLLLAGTPVSKRHK 640
Query: 553 R-------ITMHSELRRQITAYLENELQFLSSSFTKWIEAQKSYLQAINGWIHKCVPLQQ 605
+ ++S+ Q LE +L+ + F WI +Q+SY++A++GW+ +C
Sbjct: 641 KRQPPIMPEAINSQRLAQSALNLEAQLRNWRTCFEFWITSQRSYMKALSGWLLRCFRCDP 700
Query: 606 KSVKRKRRPQSELLIQYGPPIYATCDVWLKKLGTLPVKDVVDSIKSLAADTARFLPYQ-- 663
P+ L PIY C W + L +L K V+D ++ A+ Q
Sbjct: 701 D-------PEKVRLSSCLHPIYRVCIQWSRLLNSLNEKPVLDKLEFFASGMGSIYARQVR 753
Query: 664 -DKN-QGKEPHSHIGGESADGLLRDDISEDWISGFDRFRANLIRFLGQLNSLSGSSV 718
D N G + G ES + ++ D ED + ++ ++ L S++ SS+
Sbjct: 754 EDPNWSGSGSRRYSGSESMELVVADKGEEDVVMTAEKLAEVAVKVLCHGMSVAVSSL 810
>At1g77500 unknown protein
Length = 879
Score = 181 bits (458), Expect = 2e-45
Identities = 167/658 (25%), Positives = 283/658 (42%), Gaps = 69/658 (10%)
Query: 87 PSMSQQIDAGETHSFYTTPSPPSSSKF---HANHMKFGSFSSKKVEEKPPVPVIATVTSS 143
P+ + + G +Y S P S F NH +VEE P + S
Sbjct: 187 PNYTGENPYGNRGMYYMKKSAPQSRPFIFQPENH---------RVEENAQWP-----SDS 232
Query: 144 GIAQNGSRMSETEAFEDSSVPDGTPQWDFFGLFNPVDHQFSFQEPKGMPHHIGNAEEGGI 203
G G + S P WDF +F+ D+ + G + +G A
Sbjct: 233 GFRNTGVQRRSPSPLPPPSPPT-VSTWDFLNVFDTYDYSNARSRASGY-YPMGMASISSS 290
Query: 204 PDLEDDEEKASSCGSEGSRDSEDEFDEEPTAETLVQ-RFENLNRANNHVQE---NVLPVK 259
PD ++ E+ EG + E+ ++E + + + L + H E NV P +
Sbjct: 291 PDSKEVRER------EGIPELEEVTEQEVIKQVYRRPKRPGLEKVKEHRDEHKHNVFPER 344
Query: 260 GDSVSEVELVSEKGNFPNSSPSKKLPMAALLPPEAN--KSTEKENHSENKVTPKNFFSSM 317
+ EV + + S + + + E + E ++ S + ++ S
Sbjct: 345 NINKREVPMPEQVTESSLDSETISSFSGSDVESEFHYVNGGEGKSSSISSISHGAGTKSS 404
Query: 318 KDIEVLFNRASDSGKEVPRMLEANKFHFRPIFQGKKDGSLASFICKA------------- 364
+++E + R E+ + F + K SL+S A
Sbjct: 405 REVEEQYGRKKGVSFELEETTSTSSFDV----ESSKISSLSSLSIHATRDLREVVKEIKS 460
Query: 365 ----CFSCGEDPS------QVPEEPAQNSVKYLT----WHRTASSRSSSSKNPLDANSRE 410
SCG++ + ++P + N VK + + S+RSS S+ L
Sbjct: 461 EFEIASSCGKEVALLLEVGKLPYQHKNNGVKVILSRIMYLVAPSTRSSHSQPRLSIRLTS 520
Query: 411 NVEDLTNNLFDNSCMNAGSH---ASTLDRLHAWERKLYDEVKASEIVRKEYDAKCKILRN 467
+ + ++ +N G + +STL++L+AWE+KLY EVK E +R Y+ KC+ L+
Sbjct: 521 RTRKMAKS-YNGQDVNGGFNGNLSSTLEKLYAWEKKLYKEVKDEEKLRAIYEEKCRRLKK 579
Query: 468 LESKAEKTSTVDKTRAAVKDLHSRIRVAIHRIDSISKRIEELRDKELQPQLEELIEGLNR 527
++S ++ +D TRAA++ L ++I V I +DSIS RI +LRD+ELQPQL +LI GL R
Sbjct: 580 MDSHGAESIKIDATRAAIRKLLTKIDVCIRSVDSISSRIHKLRDEELQPQLIQLIHGLIR 639
Query: 528 MWEVMHECHKLQFQ-IMSASYNNSHARITMHSELRRQITAYLENELQFLSSSFTKWIEAQ 586
MW M CH+ QFQ I + + A T+ ++ LE EL+ SF W+ Q
Sbjct: 640 MWRSMLRCHQKQFQAIRESKVRSLKANTTLQNDSGSTAILDLEIELREWCISFNNWVNTQ 699
Query: 587 KSYLQAINGWIHKCVPLQQKSVKRKRRPQSELLIQYGPPIYATCDVWLKKLGTLPVKDVV 646
KSY+Q ++GW+ KC+ + ++ P S I PPI+ C W + + + ++V
Sbjct: 700 KSYVQFLSGWLTKCLHYEPEATDDGIAPFSPSQIG-APPIFIICKDWQEAMCRISGENVT 758
Query: 647 DSIKSLAADTARFLPYQDKNQG-KEPHSHIGGESADGLLRDDISEDWISGFDRFRANL 703
++++ A+ Q++ Q K ES ++ SE IS D + +L
Sbjct: 759 NAMQGFASSLHELWEKQEEEQRVKAQSEQRDAESERSVVSKGRSESGISALDDLKVDL 816
Score = 59.3 bits (142), Expect = 8e-09
Identities = 40/140 (28%), Positives = 71/140 (50%), Gaps = 4/140 (2%)
Query: 1 MGASSSKMEEDKALQLCRERKKFVRQALDGRCSLAAAHISYVQSLRIVGTALSKFTEPEP 60
MG SK++ + LCRERK+ ++ A R +LA AH++Y QSL VG A+ +F + E
Sbjct: 1 MGCGGSKVDNQPIVILCRERKELLKAASYHRSALAVAHLTYFQSLSDVGEAIQRFVDDEV 60
Query: 61 HSESSLYTNATPEALALTDKAMSQSSPSMSQQIDAGETHSF-YTTPSPPSSSKFHANHMK 119
+ S ++++P++ LT + P+ ++I T S ++ + H+
Sbjct: 61 VAGFS--SSSSPDSPVLT-LPSDEGKPTKHKRISPSSTTSISHSVIEEEDTDDDSHLHLS 117
Query: 120 FGSFSSKKVEEKPPVPVIAT 139
GS S +V K + + +T
Sbjct: 118 SGSESESEVGSKDHIQITST 137
>At1g21740 putative bzip-like transcription factor
Length = 953
Score = 177 bits (448), Expect = 3e-44
Identities = 118/362 (32%), Positives = 183/362 (49%), Gaps = 48/362 (13%)
Query: 317 MKDIEVLFNRASDSGKEVPRMLEANKFHFRPIFQGKKDGSLASFICKACFSCGEDPSQVP 376
+K+I+ F AS GKEV +LE +K + Q K G K FS
Sbjct: 503 VKEIKSEFEVASSHGKEVAVLLEVSKLPY----QQKSSG------LKVIFS--------- 543
Query: 377 EEPAQNSVKYLTWHRTASSRSSSS----------KNPLDANSRENVEDLTNNLFDNSCMN 426
+ YL T SSRS K N ++ E LT NL
Sbjct: 544 ------RIMYLVAPSTVSSRSQPQPSIRLTSRILKIAKSYNGQDVREGLTGNL------- 590
Query: 427 AGSHASTLDRLHAWERKLYDEVKASEIVRKEYDAKCKILRNLESKAEKTSTVDKTRAAVK 486
++TL++L+AWE+KLY EVK E +R Y+ KC+ L+ L+S ++S +D TRAA++
Sbjct: 591 ----SATLEQLYAWEKKLYKEVKDEEKLRVVYEEKCRTLKKLDSLGAESSKIDTTRAAIR 646
Query: 487 DLHSRIRVAIHRIDSISKRIEELRDKELQPQLEELIEGLNRMWEVMHECHKLQFQ-IMSA 545
L +++ V I +DSIS RI +LRD+ELQPQL +LI GL RMW M +CH+ QFQ IM +
Sbjct: 647 KLLTKLDVCIRSVDSISSRIHKLRDEELQPQLTQLIHGLIRMWRSMLKCHQKQFQAIMES 706
Query: 546 SYNNSHARITMHSELRRQITAYLENELQFLSSSFTKWIEAQKSYLQAINGWIHKCVPLQQ 605
+ A + + + LE EL+ SF W+ QKSY++++NGW+ +C+ +
Sbjct: 707 KVRSLRANTGLQRDSGLKAILDLEMELREWCISFNDWVNTQKSYVESLNGWLSRCLHYEP 766
Query: 606 KSVKRKRRPQSELLIQYGPPIYATCDVWLKKLGTLPVKDVVDSIKSLAADTARFLPYQDK 665
+S + P S + P ++ C W + + + ++V ++++ A+ QD+
Sbjct: 767 ESTEDGIAPFSPSRVG-APQVFVICKDWQEAMARISGENVSNAMQGFASSLHELWERQDE 825
Query: 666 NQ 667
Q
Sbjct: 826 EQ 827
Score = 63.5 bits (153), Expect = 4e-10
Identities = 38/111 (34%), Positives = 56/111 (50%), Gaps = 13/111 (11%)
Query: 1 MGASSSKMEEDKALQLCRERKKFVRQALDGRCSLAAAHISYVQSLRIVGTALSKFTEPEP 60
MG SK+++ + LCRERK+ ++ A RC+LAAAH+SY QSL VG ++ +F + E
Sbjct: 1 MGCGGSKVDDQPLVILCRERKQLIKAASHHRCALAAAHLSYFQSLCDVGDSIKRFVDEE- 59
Query: 61 HSESSLYTNATPEALALTDKAMSQSSPSMSQQIDAGETHSFYTTPSPPSSS 111
L T + S SP ++ D G+ H + S S S
Sbjct: 60 ------------LVLVGTSSSSSPDSPVLTLPSDEGKPHKHKISSSSTSVS 98
>At3g60320 bZIP like protein
Length = 796
Score = 174 bits (441), Expect = 2e-43
Identities = 176/702 (25%), Positives = 301/702 (42%), Gaps = 97/702 (13%)
Query: 1 MGASSSKMEEDKALQLCRERKKFVRQALDGRCSLAAAHISYVQSLRIVGTALSKFTEPEP 60
MG ++SK++ + A++ C++R++ +++A+ R LAAAH Y +SLRI G+ALS F EP
Sbjct: 1 MGCAASKLDNEDAVRRCKDRRRLMKEAVYARHHLAAAHADYCRSLRITGSALSSFASGEP 60
Query: 61 HSES----SLYTNATPEALALTDKAMSQSSPSMSQQIDAGETHSFYTTPSPPSSSKFHAN 116
S S +++ + P L+ A P S + T+PS SS + +
Sbjct: 61 LSVSDQTPAVFLHTPPPPLSEQSPAKFVP-PRFSPSPAPSSVYPPSTSPSVASSKQ--PS 117
Query: 117 HMKFGSFSSKKVEEKPPVPVIATVTSSGIAQNGSRMS-----ETEAFEDSSVPDGTPQ-- 169
M S +K + KP +P I + +S + R + A+++S+ TP
Sbjct: 118 VMSTSSNRRRKQQPKPRLPHILSESSPSSSPRSERSNFMPNLYPSAYQNSTY-SATPSHA 176
Query: 170 -----WDFFGLFNPVDHQFSFQEPKGMPHHIGN-----------AEEGGIPDLEDDEEKA 213
W+ F +P D +F ++ + H+ N +E + +++
Sbjct: 177 SSVWNWENFYPPSPPDSEFFNRKAQEKKHNSDNRFNDEDTETVRSEYDFFDTRKQKQKQF 236
Query: 214 SSCGSEGSRDSEDEFDEEPTAETLVQRFENLNRANNHVQENVLPVKGDSVSEVELVSEKG 273
S ++ ++E E +E +E + +++ +E +S+SEV SE G
Sbjct: 237 ESMRNQVEEETETEREEVQCSEWEDHDHYSTTSSSDAAEEEEEDDDRESISEVGTRSEFG 296
Query: 274 NFPNSSPSKKLPMAALLPPEANKSTEKENHSENK---------------------VTPKN 312
+ S+ ++ P+ E+ + + V ++
Sbjct: 297 STVRSNSMRRHHQQPSPMPQVYGGAEQSKYDKADDATISSGSYRGGGDIADMKMVVRHRD 356
Query: 313 FFSSMKDIEVLFNRASDSGKEVPRMLEANKFHFRPIFQGKKDGSLASFICKACFSCGEDP 372
+ I+ F++A+ SG++V +MLE + F K + S + S
Sbjct: 357 LKEIIDAIKENFDKAAASGEQVSQMLELGRAELDRSFSQLKKTVIHSSSLLSNLSS---- 412
Query: 373 SQVPEEPAQNSVKYLTWHRTASSRSSSSKNPLDANSRENVEDLTNNLFDNSCMNAGSHAS 432
TW +SK PL R + L NS + S S
Sbjct: 413 ---------------TW---------TSKPPLAVKYRIDTTALDQ---PNS---SKSLCS 442
Query: 433 TLDRLHAWERKLYDEVKASEIVRKEYDAKCKILRNLESKAEKTSTVDKTRAAVKDLHSRI 492
TLDRL AWE+KLY+E+KA E + E++ K L++ E K E + +DKT+A++ L S I
Sbjct: 443 TLDRLLAWEKKLYEEIKAREGFKIEHEKKLSQLQSQEYKGEDEAKLDKTKASITRLQSLI 502
Query: 493 RVAIHRIDSISKRIEELRDKELQPQLEELIEGLNRMWEVMHECHKLQFQIMS--ASYNNS 550
V + + S I LRD +L PQL EL G MW+ MH+ H+ Q I+ N
Sbjct: 503 IVTSQAVTTTSTAIIRLRDTDLVPQLVELCHGFMYMWKSMHQYHETQNSIVEQVRGLINR 562
Query: 551 HARITMHSELRRQITAYLENELQFLSSSFTKWIEAQKSYLQAINGWIH-KCVPLQQKSVK 609
+ SEL RQ T LE+ + SSF+ I+ Q+ ++ +++ W +P+ Q+
Sbjct: 563 SGKGESTSELHRQATRDLESAVSSWHSSFSSLIKFQRDFIHSVHAWFKLTLLPVCQEDAA 622
Query: 610 RKRRPQSELLIQYGPPIYATCDVWLKKLGTLPVKDVVDSIKS 651
+ + YA CD W L +P ++IKS
Sbjct: 623 NHHKEPLD--------AYAFCDEWKLALDRIPDTVASEAIKS 656
>At4g35240 putative protein
Length = 828
Score = 168 bits (426), Expect = 9e-42
Identities = 156/592 (26%), Positives = 249/592 (41%), Gaps = 118/592 (19%)
Query: 84 QSSPSMSQQIDAGETHSFYTTPSPPSSSKFHANHMKFG---------SFSSKKVEEKPPV 134
+ P+ Q++ GE+ S Y P PP +S F ++ G S S+ KPP
Sbjct: 210 EQRPTSPQRVYIGESSSSY--PYPPQNSYFGYSNPVPGPGPGYYGSSSASTTAAATKPPP 267
Query: 135 PVIATVTSSGIAQNGSRMSETEAFEDSSVPDGTPQWDFFGLFNPVDHQFSFQEPKGMPHH 194
P + S+G WDF NP D + P
Sbjct: 268 PPPSPPRSNG-------------------------WDFL---NPFDTYYPPYTPSRDSRE 299
Query: 195 IGNAEEGGIPDLEDDE---EKASSCGSEGSRDSEDEFDEEPTAETLVQRFENLNRANNHV 251
+ EE GIPDLEDD+ E + + P A +++
Sbjct: 300 L--REEEGIPDLEDDDSHYEVVKEVYGKPKFAAGGGHQPNPAAVHMMRE---------ES 348
Query: 252 QENVLPVKGDSVS---EVELVSEKGNFPNSSPSKKLPMAALLPPEANKSTEKENHSENKV 308
L G S S +V S + P+ S K+ + E ++E S
Sbjct: 349 PSPPLDKSGASTSGGGDVGDASAYQSRPSVSVEKEGMEYEVHVVEKKVVEDEERRSNATA 408
Query: 309 T--------PKNFFSSMKDIEVLFNRASDSGKEVPRMLEANKFHFRPIFQGKKDGSLASF 360
T P+ K+IE F +A++SG E+ ++LE K + G+K
Sbjct: 409 TRGGGGGGGPRAVPEVAKEIENQFVKAAESGSEIAKLLEVGKHPY-----GRK------- 456
Query: 361 ICKACFSCGEDPSQVPEEPAQNSVKYLTWHRTASSRSSSSKNPLDANSRENVEDLTNNLF 420
H T+SS +++ P A+ E + + NL
Sbjct: 457 -----------------------------HGTSSSAAAAVVPPTYADIEEELASRSRNL- 486
Query: 421 DNSCMNAGSHASTLDRLHAWERKLYDEVKASEIVRKEYDAKCKILRNLESKAEKTSTVDK 480
+STL +LH WE+KLY EVKA E +R ++ K + L+ L+ + + VDK
Sbjct: 487 ----------SSTLHKLHLWEKKLYHEVKAEEKLRLAHEKKLRKLKRLDQRGAEAIKVDK 536
Query: 481 TRAAVKDLHSRIRVAIHRIDSISKRIEELRDKELQPQLEELIEGLNRMWEVMHECHKLQF 540
TR V+D+ ++IR+AI +D IS I ++RD++L PQL LI+GL RMW+ M ECH+ Q
Sbjct: 537 TRKLVRDMSTKIRIAIQVVDKISVTINKIRDEDLWPQLNALIQGLTRMWKTMLECHQSQC 596
Query: 541 QIMSASYNNSHARITMH-SELRRQITAYLENELQFLSSSFTKWIEAQKSYLQAINGWIHK 599
Q + + R + + + T+ L +EL F+ W+ AQK Y++ +N W+ K
Sbjct: 597 QAIREAQGLGPIRASKKLGDEHLEATSLLGHELINWILGFSSWVSAQKGYVKELNKWLMK 656
Query: 600 CVPLQQKSVKRKRRPQSELLIQYGPPIYATCDVWLKKLGTLPVKDVVDSIKS 651
C+ + + P S I PPI+ C+ W + L + K+V+++++S
Sbjct: 657 CLLYEPEETPDGIVPFSPGRIG-APPIFVICNQWSQALDRISEKEVIEAMRS 707
Score = 50.4 bits (119), Expect = 4e-06
Identities = 24/55 (43%), Positives = 37/55 (66%)
Query: 1 MGASSSKMEEDKALQLCRERKKFVRQALDGRCSLAAAHISYVQSLRIVGTALSKF 55
MG +SSK+++ A+ LCRER F+ A+ R +LA +H++Y SLR +G +L F
Sbjct: 1 MGCTSSKLDDLPAVALCRERCAFLEAAIHQRYALAESHVAYTHSLREIGHSLHLF 55
>At5g25590 unknown protein (At5g25590)
Length = 775
Score = 164 bits (415), Expect = 2e-40
Identities = 161/712 (22%), Positives = 285/712 (39%), Gaps = 115/712 (16%)
Query: 1 MGASSSKMEEDKALQLCRERKKFVRQALDGRCSLAAAHISYVQSLRIVGTALSKFTEPEP 60
MG + S+++ ++A+ C+ER+ +++A+ + AA H +Y +L+ G ALS + E
Sbjct: 1 MGCAQSRVDNEEAVARCKERRNVIKEAVSASKAFAAGHFAYAIALKNTGAALSDYGHGE- 59
Query: 61 HSESSLYTNATPEALALTDKAMSQSSPSMSQQIDAGETHSFYTTPSPPSSSKFHANHMKF 120
S+ + + ++ + P+ Q + P PP KF + +K
Sbjct: 60 -SDQKALDDVLLDQQHYEKQSRNNVDPASPQPPPPPPIENL--PPPPPPLPKFSPSPIKR 116
Query: 121 ------------------------------------GSFSSKKVEEKPPVPVIATVTSSG 144
GS EE+ P T + G
Sbjct: 117 AISLPSMAVRGRKVQTLDGMAIEEEEEDEEEEEEVKGSGRDTAQEEEEP----RTPENVG 172
Query: 145 IAQNGSRMSETEAFEDSSVPDGTPQWDFFGLFN--PVDHQFSFQEPKGMPHHIGNAEEGG 202
+ R+ +T S+ P + WD+F + P + + G + + +
Sbjct: 173 KSNGRKRLEKTTPEIVSASPANSMAWDYFFMVENMPGPNLDDREVRNGYENQSSHFQFNE 232
Query: 203 IPDLEDDEEKASSCGSEGSRDSEDEFDEEPTAETLVQRFENLNRANNHVQENVLPVKGDS 262
D E++EE+ S + S + + EP V+ E + + +E
Sbjct: 233 EDDEEEEEEERSGIYRKKSGSGKVVEEMEPKTPEKVEEEEEEDEEEDEEEEEE------- 285
Query: 263 VSEVELVSEKGNFPNSSPSKKLPMAALLPPEANKSTEKENHSENKVTPKNFFSSMKDIEV 322
E E+V E K+ ++ PPE ++ K + + + N + +I+
Sbjct: 286 -EEEEVVVEVKK--KKKGKAKIEHSSTAPPEFRRAVAKTSAAASSSV--NLMKILDEIDD 340
Query: 323 LFNRASDSGKEVPRMLEANKFHFRPIFQGKKDGSLASFICKACFSCGEDPSQVPEEPAQN 382
F +AS+ +EV +MLEA + H+ F + ++ + +
Sbjct: 341 RFLKASECAQEVSKMLEATRLHYHSNFADNR-----GYV----------------DHSAR 379
Query: 383 SVKYLTWHRTASSRSSSSKNPLDANSRENVEDLTNNLFDNSCMNAGSHASTLDRLHAWER 442
++ +TW+++ S+ D S E+ +HA+ LD+L AWE+
Sbjct: 380 VMRVITWNKSLRGISNGEGGKDDQESDEHE----------------THATVLDKLLAWEK 423
Query: 443 KLYDEVKASEIVRKEYDAKCKILRNLESKAEKTSTVDKTRAAVKDLHSRIRVAIHRIDSI 502
KLYDEVK E+++ EY K +L + + TV+KT+AAV LH+R V + +DS
Sbjct: 424 KLYDEVKQGELMKIEYQKKVSLLNRHKKRGASAETVEKTKAAVSHLHTRYIVDMQSMDST 483
Query: 503 SKRIEELRDKELQPQLEELIEGLNRMWEVMHECHKLQF----QIMSASYNNSHARIT-MH 557
+ LRD +L P+L L+EG+ +MW M H Q ++ + + S T H
Sbjct: 484 VSEVNRLRDDQLYPRLVALVEGMAKMWTNMCIHHDTQLGIVGELKALEISTSLKETTKQH 543
Query: 558 SELRRQITAYLENELQFLSSSFTKWIEAQKSYLQAINGWIH-KCVPLQ---QKSVKRKRR 613
RQ LE F + QK Y+ ++N W+ +P++ ++ V R
Sbjct: 544 HHQTRQFCTVLEE----WHVQFDTLVTHQKQYINSLNNWLKLNLIPIESSLKEKVSSPPR 599
Query: 614 PQSELLIQYGPPIYATCDVWLKKLGTLPVKDVVDSIKSLAADTARFLPYQDK 665
PQ PPI A W +L LP + +I S AA L +Q++
Sbjct: 600 PQR-------PPIQALLHSWHDRLEKLPDEVAKSAISSFAAVIKTILLHQEE 644
>At1g52320 bZIP protein, putative
Length = 798
Score = 158 bits (399), Expect = 1e-38
Identities = 181/822 (22%), Positives = 337/822 (40%), Gaps = 161/822 (19%)
Query: 1 MGASSSKMEEDKALQLCRERKKFVRQALDGRCSLAAAHISYVQSLRIVGTALSKFTEPE- 59
MG + SK+E ++A+ C+ERK+ ++ A+ R + AAAH +Y +L+ G ALS ++ E
Sbjct: 1 MGCAQSKIENEEAVTRCKERKQLMKDAVTARNAFAAAHSAYAMALKNTGAALSDYSHGEF 60
Query: 60 ---PHSESSLY-----TNATPEALA---------LTDKAMSQSSPSMSQQIDAGETHSFY 102
HS SS T++ P A++ +++ S SS ++ Q I
Sbjct: 61 LVSNHSSSSAAAAIASTSSLPTAISPPLPSSTAPVSNSTASSSSAAVPQPIPD------- 113
Query: 103 TTPSPP---------SSSKFHANHMKFGSFSSKK---VEEKPPVP--------------- 135
T P PP +++ N G + +EE +
Sbjct: 114 TLPPPPPPPPLPLQRAATMPEMNGRSGGGHAGSGLNGIEEDGALDNDDDDDDDDDDSEME 173
Query: 136 ----VIATVTSSGIAQNGSRMS-ETEAFEDSSVPDGTPQWDFFGLFNPVDHQFSFQEPKG 190
+I S G + G+R + E ++ P P + + P HQ Q+ +
Sbjct: 174 NRDRLIRKSRSRGGSTRGNRTTIEDHHLQEEKAPPPPPLANSRPIPPPRQHQHQHQQQQQ 233
Query: 191 MPHH---IGNAEEGGIPDLEDDEEKA-------------SSCGSEGSRDSEDEFDEEPTA 234
P + N E LED + S +E D E+E +EE
Sbjct: 234 QPFYDYFFPNVENMPGTTLEDTPPQPQPQPTRPVPPQPHSPVVTEDDEDEEEEEEEEEEE 293
Query: 235 E-TLVQRFENLNRANNHVQENVLPVKGDSVSEVE-LVSEKG-NFPNSSPSKKLPMAALLP 291
E T+++R + V+E + ++ + V+ + + KG P ++P
Sbjct: 294 EETVIERKPLVEERPKRVEE--VTIELEKVTNLRGMKKSKGIGIPGERRGMRMP------ 345
Query: 292 PEANKSTEKENHSENKVTPKNFFSSMKDIEVLFNRASDSGKEVPRMLEANKFHFRPIFQG 351
VT + + +++ F +AS+S +V +MLEA + H+ F
Sbjct: 346 ----------------VTATHLANVFIELDDNFLKASESAHDVSKMLEATRLHYHSNFAD 389
Query: 352 KKDGSLASFICKACFSCGEDPSQVPEEPAQNSVKYLTWHRTASSRSSSSKNPLDANSREN 411
+ G + + + ++ +TW+R+ ++ D + EN
Sbjct: 390 NR-GHI--------------------DHSARVMRVITWNRSFRGIPNADDGKDDVDLEEN 428
Query: 412 VEDLTNNLFDNSCMNAGSHASTLDRLHAWERKLYDEVKASEIVRKEYDAKCKILRNLESK 471
+HA+ LD+L AWE+KLYDEVKA E+++ EY K L ++ +
Sbjct: 429 E----------------THATVLDKLLAWEKKLYDEVKAGELMKIEYQKKVAHLNRVKKR 472
Query: 472 AEKTSTVDKTRAAVKDLHSRIRVAIHRIDSISKRIEELRDKELQPQLEELIEGLNRMWEV 531
+ ++++ +AAV LH+R V + +DS I LRD++L +L L+E + +MWE+
Sbjct: 473 GGHSDSLERAKAAVSHLHTRYIVDMQSMDSTVSEINRLRDEQLYLKLVHLVEAMGKMWEM 532
Query: 532 MHECHKLQFQIMSA--SYNNSHARITMHSELRRQITAYLENELQFLSSSFTKWIEAQKSY 589
M H+ Q +I S + S A + ++ + T L +Q + F + I+ QK Y
Sbjct: 533 MQIHHQRQAEISKVLRSLDVSQA-VKETNDHHHERTIQLLAVVQEWHTQFCRMIDHQKEY 591
Query: 590 LQAINGWIH-KCVPLQQKSVKRKRRPQSELLIQYGPPIYATCDVWLKKLGTLPVKDVVDS 648
++A+ GW+ +P++ ++ P P I W +L +P + +
Sbjct: 592 IKALGGWLKLNLIPIESTLKEKVSSPPR----VPNPAIQKLLHAWYDRLDKIPDEMAKSA 647
Query: 649 IKSLAADTARFLPYQD-----KNQGKEPHSHIGGESADGLLRDDISEDWISGFDRFRANL 703
I + AA + + Q+ +N+ +E +G + EDW + + R
Sbjct: 648 IINFAAVVSTIMQQQEDEISLRNKCEETRKELGRKIRQ-------FEDWYHKYIQKRGPE 700
Query: 704 IRFLGQLNSLSGSSVKM----YRELRQAIQEAKNNYHRLNSQ 741
+ ++ V + ++++ ++E + YHR + Q
Sbjct: 701 GMNPDEADNDHNDEVAVRQFNVEQIKKRLEEEEEAYHRQSHQ 742
>At1g20530 hypothetical protein
Length = 614
Score = 154 bits (389), Expect = 2e-37
Identities = 157/649 (24%), Positives = 262/649 (40%), Gaps = 153/649 (23%)
Query: 1 MGASSSKMEEDKALQLCRERKKFVRQALDGRCSLAAAHISYVQSLRIVGTALSKFTEPEP 60
MG S SK++ A+ LCR+R + L +LA AH +Y+ SL VG AL +F +
Sbjct: 1 MGCSPSKLDGLPAVSLCRDRCSSLEDTLRRSYALADAHSAYLLSLNTVGPALHRFFQHAV 60
Query: 61 HSESSLYTNA-------TPEALALTDKAMSQSS---PSMSQQIDAGETHSFYTTPSPPSS 110
S + + A +PE+ + S S P + GE T S
Sbjct: 61 ESPPDVESEADESPETYSPESCSSAHSVTSSDSDLPPKFDSDCEEGEG----TNCDLLSG 116
Query: 111 SKFHANHMKFGSFSSKKVEEKPPVPVIATVTSSGIAQNGSRMSETEAFEDSSVPDGTPQW 170
SK + ++ SF S++ E P S A W
Sbjct: 117 SKPESLNLNHDSFYSRRYESGTITPPPPPPAPSNYA-----------------------W 153
Query: 171 DFFGLFNPVDHQFSFQEPKGMPHHIGNAEEGGIPDLEDDEEKASSCGSEGSRDSEDEFDE 230
DF F + ++ + DL+D E S E
Sbjct: 154 DFINFFESYELPYTTD----------------VNDLKDRETTPSH--------------E 183
Query: 231 EPTAETLVQRFENLNRANNHVQENVLPVKGDSVSEVELVSEKGNFPNSSPSKKLPMAALL 290
E + L + V +N +K + + +SEK
Sbjct: 184 E----------DKLKKKKITVSQNDEKIKVEEEKKTLRISEKNR---------------- 217
Query: 291 PPEANKSTEKENHSENKVTPKNFFSSMKDIEVLFNRASDSGKEVPRMLEANKFHFRPIFQ 350
K KE S+++ + K ++ +F +AS+SG +V +M + ++F +
Sbjct: 218 -----KRAPKE--SKDQKVSSDLSEVTKQLQEMFKKASESGNDVSKMFDTSRFRY----- 265
Query: 351 GKKDGSLASFICKACFSCGEDPSQVPEEPAQNSVKYLTWHRTASSRSSSSKNPLDANSRE 410
+ + + C +P + K +T P D E
Sbjct: 266 ---------YQKSSVYQCNVR-ILLPSSNILYTKKVMT--------------PFDPKPVE 301
Query: 411 NVEDLTNNLFDNSCMNAGSHASTLDRLHAWERKLYDEVKASEIVRKEYDAKCKILRNLES 470
E NNL +STL +L WE+KLY EVKA E +R + K+LR LE+
Sbjct: 302 --ESNFNNL-----------SSTLKKLFMWEKKLYQEVKAEEKLRTSHMKNYKLLRRLEA 348
Query: 471 KAEKTSTVDKTRAAVKDLHSRIRVAIHRIDSISKRIEELRDKELQPQLEELIEGLNRMWE 530
K+ S ++ R++++ L +R+RV+IH+I++I I +LRD+EL Q++ELI L+ MW
Sbjct: 349 KSADLSKIEAIRSSIQCLSTRMRVSIHKINNICLTINKLRDEELWFQMKELIHRLSEMWN 408
Query: 531 VMHECHKLQFQIMSASYNNSHARITMHSELRR-QITAYLENELQFLSSSFTKWIEAQKSY 589
M ECH Q ++++ + I + +L + ++ L+ EL+ S S + WI+AQ Y
Sbjct: 409 SMLECHSRQSKVIAEAKKLDKMTIKENLDLSQLELAMELKLELRNWSLSMSNWIDAQAQY 468
Query: 590 LQAINGWIHKCVPLQQKSVKRKRRPQSELL-IQYGPPIYATCDVWLKKL 637
++A+N W+ +C+ K+ PQ + PP++ + W + L
Sbjct: 469 VKALNNWLMRCL---------KQEPQEPTPDLSEEPPLFGAINTWSQNL 508
>At2g17110 unknown protein
Length = 399
Score = 144 bits (363), Expect = 2e-34
Identities = 83/259 (32%), Positives = 142/259 (54%), Gaps = 2/259 (0%)
Query: 393 ASSRSSSSKNPLDANSRENVEDLTNNLFDNSCMNAGSHASTLDRLHAWERKLYDEVKASE 452
+S++SS+SK S ++ + + + +STL +LH WE+KLYDEVKA E
Sbjct: 30 SSAQSSTSKKAKAEASSSVTAPTYADIEAELALKSRNLSSTLHKLHLWEKKLYDEVKAEE 89
Query: 453 IVRKEYDAKCKILRNLESKAEKTSTVDKTRAAVKDLHSRIRVAIHRIDSISKRIEELRDK 512
+R ++ K + L+ ++ + + VD TR V+ L ++IR+AI +D IS I ++RD+
Sbjct: 90 KMRVNHEKKLRKLKRMDERGAENQKVDSTRKLVRSLSTKIRIAIQVVDKISVTINKIRDE 149
Query: 513 ELQPQLEELIEGLNRMWEVMHECHKLQFQIMSASYNNSHARITMH-SELRRQITAYLENE 571
EL QL ELI+GL++MW+ M ECHK Q + + + R + + ++T L E
Sbjct: 150 ELWLQLNELIQGLSKMWKSMLECHKSQCEAIKEARGLGPIRASKNFGGEHLEVTRTLGYE 209
Query: 572 LQFLSSSFTKWIEAQKSYLQAINGWIHKCVPLQQKSVKRKRRPQSELLIQYGPPIYATCD 631
L F+ W+ AQK +++ +N W+ KC+ + + P S I P I+ C+
Sbjct: 210 LINWIVGFSSWVSAQKGFVRELNSWLMKCLFYEPEETPDGIVPFSPGRIG-APMIFVICN 268
Query: 632 VWLKKLGTLPVKDVVDSIK 650
W + L + K+V+++I+
Sbjct: 269 QWEQALDRISEKEVIEAIR 287
>At1g02110 bZIP-like protein
Length = 679
Score = 141 bits (356), Expect = 1e-33
Identities = 164/664 (24%), Positives = 267/664 (39%), Gaps = 132/664 (19%)
Query: 1 MGASSSKMEEDKALQLCRERKKFVRQALDGRCSLAAAHISYVQSLRIVGTALSKFTEPEP 60
MG ++SK++ + A++ C+ER++ ++ A+ R LAAAH Y +SLR+ G+ALS F EP
Sbjct: 1 MGCTASKLDSEDAVRRCKERRRLMKDAVYARHHLAAAHSDYCRSLRLTGSALSSFAAGEP 60
Query: 61 HSESSLYTNATPEALALTDKAMSQSSPSMSQQIDAGETHSFYTTPSPPSSSKFHANHMKF 120
S S TP PS SQ DA S ++ PP +
Sbjct: 61 LSVSE----NTPAVFL---------RPSSSQ--DAPRVPSSHSPEPPPPPIR-------- 97
Query: 121 GSFSSKKVEEKP---PVPVIATVTSSGIAQNGSRMSETEAFEDSSVPDGTPQWDFFGLFN 177
SK +P P + + SS A + + + S + W++ +
Sbjct: 98 ----SKPKPTRPRRLPHILSDSSPSSSPATSFYPTAHQNSTYSRSPSQASSVWNWENFYP 153
Query: 178 PVDHQFSFQEPKGMPHHIGNAEEGGIPDLEDDEEKASSCGSEGSRDSEDE-----FDEEP 232
P + E K +H D + + E++ RD+ +E + ++
Sbjct: 154 PSPPDSEYFERKARQNHKHRPPS----DYDAETERSDHDYCHSRRDAAEEVHCSEWGDDH 209
Query: 233 TAETLVQRFENLNRANNHVQENVLPVKGDSVSEVELVSEKGNFPNSSPSKKLPMAALLPP 292
T + HV + + E E V + P+ P+ K
Sbjct: 210 DRFTATSSSDGDGEVETHVSRSGI--------EEEPVKQ----PHQDPNGK--------E 249
Query: 293 EANKSTEKENHSENK--VTPKNFFSSMKDIEVLFNRASDSGKEVPRMLEANKFHFRPIFQ 350
++ T + + K V KN + ++ F++A+ +G +V MLE I +
Sbjct: 250 HSDHVTTSSDCYKTKLVVRHKNLKEILDAVQDYFDKAASAGDQVSAMLE--------IGR 301
Query: 351 GKKDGSLASFICKACFSCGEDPSQVPEEPAQNSVKYLTWHRTASSRSSSSKNPLDANSRE 410
+ D S + + +V + + + S S +SK PL +
Sbjct: 302 AELDRSFSKL--------------------RKTVYHSSSVFSNLSASWTSKPPLAVKYKL 341
Query: 411 NVEDLTNNLFDNSCMNAGSHASTLDRLHAWERKLYDEVKASEIVRKEYDAKCKILRNLES 470
+ L + S STLDRL AWE+KLY++VKA E V+ E++ K L++ E
Sbjct: 342 DASTLNDEQG-----GLKSLCSTLDRLLAWEKKLYEDVKAREGVKIEHEKKLSALQSQEY 396
Query: 471 KAEKTSTVDKTRAAVKDLHSRIRVAIHRIDSISKRIEELRDKELQPQLEELIEGLNRMWE 530
K E T S I LRD +L PQL EL GL MW+
Sbjct: 397 KGEAVLTT------------------------SNAILRLRDTDLVPQLVELCHGLMYMWK 432
Query: 531 VMHECHKLQFQIMS--ASYNNSHARITMHSELRRQITAYLENELQFLSSSFTKWIEAQKS 588
MHE H++Q I+ N R SE+ RQ+T LE+ + SSF + I+ Q+
Sbjct: 433 SMHEYHEIQNNIVQQVRGLINQTERGESTSEVHRQVTRDLESAVSLWHSSFCRIIKFQRE 492
Query: 589 YLQAINGWIH-KCVPLQQKSVKRKRRPQSELLIQYGPPIYATCDVWLKKLGTLPVKDVVD 647
++ +++ W VPL K++R P +A C+ W + L +P +
Sbjct: 493 FICSLHAWFKLSLVPLSNGDPKKQR-----------PDSFALCEEWKQSLERVPDTVASE 541
Query: 648 SIKS 651
+IKS
Sbjct: 542 AIKS 545
>At5g54480 unknown protein
Length = 720
Score = 138 bits (347), Expect = 1e-32
Identities = 145/663 (21%), Positives = 281/663 (41%), Gaps = 69/663 (10%)
Query: 1 MGASSSKMEEDKAL-QLCRERKKFVRQALDGRCSLAAAHISYVQSLRIVGTALSKFTEPE 59
MG +SK + + L LC+ERK+ ++ A D R LA +H+ Y +SL AL +F +
Sbjct: 1 MGIVASKGDSNTPLLNLCKERKELIKAARDARYHLARSHLLYFKSLLDFSKALKQFLHKD 60
Query: 60 ----PHSESSLYTNATPEALALTDKAMSQSSPSMSQQIDAGETHSFYTTPSPPSSSKFHA 115
P+S+ + + + S S S ++ S P ++
Sbjct: 61 LVVIPYSDDD----------SSSSDIICSGSDSDSDTDSDSDSDSDCIVCDQPQTAPLSN 110
Query: 116 NHMKFGSFSSKKVEEKPPVPVIATVTSSGIAQNGSRMS-ETEAFEDSSVPDGTPQWDFFG 174
N + ++ PV T +S Q G S E FE + + +
Sbjct: 111 NDDQ---------SQRKPVVGDGTCGASNNGQEGMEKSREGLGFEGTQFVSDPSEDEMKS 161
Query: 175 LFNPVDHQFSFQEPKGMPHHIGNAEEGGIPDLEDDEEKASSCGSEGSRDSEDEFDEEPTA 234
+P D G + N ++ I +L+ +++ S G R+ E D EP +
Sbjct: 162 SQHPYDIL-------GFYKSVYNMQQCEIIELDHRDDEVESDGFREIREREGIPDLEPES 214
Query: 235 E--TLVQRFENLNRANNHVQENVLPVKGDS--VSEVELVSEKGNFPNSSPSKKLPMAALL 290
+ +L+++ N+ N ++ V+ S SE++ + N N + +
Sbjct: 215 DYNSLIRK----NKKKNKKKKKKKNVRESSSVASEIDKRDVEANTCNGQVADETD----- 265
Query: 291 PPEANKSTEKENHSENKVTPKNFFSSMKDIEVLFNRASDSGKEVPRMLEANKFHFRPIFQ 350
+N++ +E + ++ + S +++EV+ + E E++ F
Sbjct: 266 --NSNEACAQETENTPEIVTEEVTRSDEEVEVVARSDEEVSDEA---YESSSSCFSES-S 319
Query: 351 GKKDGSLASFICKACFSCGEDPSQVPEEPAQNSVKYLTWHRTASSRSSSSKNPLDANSRE 410
G L + + CG+ E V + +H+ S+ + + +S
Sbjct: 320 GSGLTDLRKVVERINSICGKAAGN-SEVSELLEVSRVVYHQPLGSQFKGFASRVIGSSGT 378
Query: 411 NVEDLTNNLFDNSCMNAGSHASTLDRLHAWERKLYDEVKASEIVRKEYDAKCKILRNLES 470
DL A S + TL++L+ WE+KL+ EV A E +R Y+ KIL NL+
Sbjct: 379 TTRDLVLKRRFRLDDLAVSLSMTLEKLYMWEKKLHAEVTAEEKLRVAYEKAYKILNNLDQ 438
Query: 471 KAEKTSTVDKTRAAVKDLHSRIRVAIHRIDSISKRIEELRDKELQPQLEELIEGLNRMWE 530
++S + + VK S++ V++ ++SIS RI ++RD+EL Q+ E+I G MW
Sbjct: 439 NGAESSELYEAETLVKLHLSKVNVSVRAVESISMRIHKIRDEELSFQVIEIINGFKTMWR 498
Query: 531 VMHECHKLQFQIMSASYNNSHARITMHSELRRQITAYLENELQFLSSSFTKWIEAQKSYL 590
+ +CH QF++++ S + H I + R+ T +E +++ S +I+A + ++
Sbjct: 499 FLAKCHHKQFRVIARSKSCVH--IVENGSSSRKATQQVEKQIRRYRESLKGYIDAHRGFV 556
Query: 591 QAINGWIHKCVPLQQKSVKRKRRPQSELLIQYGPPIYATCDVWLKKL---GTLPVKDVVD 647
+ +N W+++ + ++ P I+ C WL+++ T+ V VV+
Sbjct: 557 KLLNEWLNRIIMEDDETETE------------APEIFRVCSEWLREIENVDTIKVLSVVE 604
Query: 648 SIK 650
++
Sbjct: 605 EMR 607
>At3g51290 putative protein
Length = 602
Score = 104 bits (259), Expect = 2e-22
Identities = 95/333 (28%), Positives = 151/333 (44%), Gaps = 66/333 (19%)
Query: 311 KNFFSSMKDIEVLFNRASDSGKEVPRMLEANKFHFRPIFQGKKDGSLASFICKACFSCGE 370
K+ +K+++ F +A+DSG + +LE + K G + S + + C
Sbjct: 200 KDLMEIIKEVDEYFLKAADSGAPLSSLLEIST-SITDFSGHSKSGKMYS---SSNYECNL 255
Query: 371 DPSQVPEEPAQNSVKYLTWHRT-ASSRSSSSKNPLDANSRENVEDLTNNLFDNSCMNAGS 429
+P+ W R A S+ S +N + +C+ GS
Sbjct: 256 NPTSF-------------WTRGFAPSKLSEYRN-------------AGGVIGGNCI-VGS 288
Query: 430 HASTLDRLHAWERKLYDEVKASEIVRKEYDAKCKILRNLESKAEKTSTVDKTRAAVKDLH 489
H+ST+DRL+AWE+KLY EVK +E ++ +++ K + +R LE K + +K + V+ L
Sbjct: 289 HSSTVDRLYAWEKKLYQEVKYAESIKMDHEKKVEQVRRLEMKRAEYVKTEKAKKDVEKLE 348
Query: 490 SRIRVAIHRIDSISKRIEELRDKELQPQLEELIEGLNRMWEVMHECHKLQFQIM-SASYN 548
S++ V+ I S S I +LR+ EL PQL EL++G M+E H++Q I+ Y
Sbjct: 349 SQLSVSSQAIQSASNEIIKLRETELYPQLVELVKG------SMYESHQVQTHIVQQLKYL 402
Query: 549 NSHARITMHSELRRQITAYLENELQ---FLSSSFTKWI--------------------EA 585
N+ SEL RQ T LE E + SS+ I E
Sbjct: 403 NTIPSTEPTSELHRQSTLQLELEFSKNPLVRSSYESKIYSFCEEWHLAIDRIPDKVASEG 462
Query: 586 QKSYLQAINGWIHKCVPLQQKSVKRKRRPQSEL 618
KS+L A++G V Q K+K+R +S L
Sbjct: 463 IKSFLTAVHG----IVAQQADEHKQKKRTESML 491
Score = 45.8 bits (107), Expect = 9e-05
Identities = 51/231 (22%), Positives = 84/231 (36%), Gaps = 48/231 (20%)
Query: 1 MGASSSKMEEDKALQLCRERKKFVRQALDGRCSLAAAHISYVQSLRIVGTALSKFTEPE- 59
MG S+++ + + C+ RK++++ + R +L+ +H Y++SLR VG++L F+ E
Sbjct: 1 MGCCQSRIDSKEIVSRCKARKRYLKHLVKARQTLSVSHALYLRSLRAVGSSLVHFSSKET 60
Query: 60 ---------------------------PHSESSLYTNATPEALALTDKAMSQSSPSMSQQ 92
P SE++ +T T T + P
Sbjct: 61 PLHLHHNPPSPSPPPPPPPRPPPPPLSPGSETTTWTTTT------TSSVLPPPPPPPPPP 114
Query: 93 IDAGETHSFYT--TPSPPSSSKFH-------ANHMKFGSFSSKKVEEKPPVPVIATVTSS 143
T F+ P PPSSS+ A G+ S V P AT +S
Sbjct: 115 PPPSSTWDFWDPFIPPPPSSSEEEWEEETTTATRTATGTGSDAAV---TTAPTTATPQAS 171
Query: 144 GIAQNGSR--MSETEAFEDSSVPDGTPQWDFFGLFNPVDHQFSFQEPKGMP 192
+ S+ M+ T + +V D + VD F G P
Sbjct: 172 SVVSGFSKDTMTTTTTGSELAVVVSRNGKDLMEIIKEVDEYFLKAADSGAP 222
>At2g34670 unknown protein
Length = 561
Score = 101 bits (252), Expect = 1e-21
Identities = 68/254 (26%), Positives = 126/254 (48%), Gaps = 8/254 (3%)
Query: 368 CGEDPSQVPEEPAQNSVKYLTWHRTASSRSSSSKN------PLDANSRENVEDLTNNLFD 421
C ++ + + + ++++V + T RSSS+K + S + +D T +
Sbjct: 268 CEKEIAVIVDINSRDTVDPFRYQETRRKRSSSAKVFSALSWSWSSKSLQLGKDATTSGTV 327
Query: 422 NSCMNAGSHASTLDRLHAWERKLYDEVKASEIVRKEYDAKCKILRNLESKAEKTSTVDKT 481
C G+H STL++L+ E+KLY V+ EI + E++ K +L+ + + S ++K
Sbjct: 328 EPC-RPGAHCSTLEKLYTAEKKLYQLVRNKEIAKVEHERKSALLQKQDGETYDLSKMEKA 386
Query: 482 RAAVKDLHSRIRVAIHRIDSISKRIEELRDKELQPQLEELIEGLNRMWEVMHECHKLQFQ 541
R +++ L + I+ I + + L + EL PQL L GL +MW+ M +CH++Q
Sbjct: 387 RLSLESLETEIQRLEDSITTTRSCLLNLINDELYPQLVALTSGLAQMWKTMLKCHQVQIH 446
Query: 542 I-MSASYNNSHARITMHSELRRQITAYLENELQFLSSSFTKWIEAQKSYLQAINGWIHKC 600
I ++ + I + SE +RQ LE E+ +SF K + +Q+ Y++ + WI
Sbjct: 447 ISQQLNHLPDYPSIDLSSEYKRQAVNELETEVTCWYNSFCKLVNSQREYVKTLCTWIQLT 506
Query: 601 VPLQQKSVKRKRRP 614
L + +R P
Sbjct: 507 DRLSNEDNQRSSLP 520
Score = 47.8 bits (112), Expect = 2e-05
Identities = 20/59 (33%), Positives = 40/59 (66%)
Query: 1 MGASSSKMEEDKALQLCRERKKFVRQALDGRCSLAAAHISYVQSLRIVGTALSKFTEPE 59
MG ++S+++ ++ + +CR+RK+ +++ L R A A ++Y+++LR G L +FTE E
Sbjct: 1 MGCAASRIDNEEKVLVCRQRKRLMKKLLGFRGEFADAQLAYLRALRNTGVTLRQFTESE 59
>At1g67930 putative golgi transport complex protein
Length = 832
Score = 42.7 bits (99), Expect = 8e-04
Identities = 36/126 (28%), Positives = 64/126 (50%), Gaps = 16/126 (12%)
Query: 397 SSSSKNPLDANSRENV------EDLTNNLFDNSCMNAGSHASTLDRLHAWERKLYDEVKA 450
SSSS +PLD+ + + + ++ F ++ + +GS AST +RLH R L +++
Sbjct: 40 SSSSSSPLDSFATDPILSPFLSSSFSSASFSSAALASGSPASTAERLHQAIRLLDSQLRN 99
Query: 451 SEIVRKEYDAKCKILRNLESKAEKTSTVDKTRAAVKDLHSRIRVAIHRIDS-ISKRIEEL 509
I R ++L L S + ++ R++V L S IR R+ S +S+ I+ +
Sbjct: 100 DVISRHP-----ELLAQLSSLSHADVSLSSLRSSVSSLQSSIR----RVRSDLSEPIKSI 150
Query: 510 RDKELQ 515
R K +Q
Sbjct: 151 RSKSVQ 156
>At5g22650 putative histone deacetylase (HD2B)
Length = 306
Score = 40.4 bits (93), Expect = 0.004
Identities = 41/168 (24%), Positives = 62/168 (36%), Gaps = 25/168 (14%)
Query: 187 EPKGMPHHIGNAEEGGIPDLED------------DEEKASSCGSEGSRDSEDEFDEEPTA 234
+PK P + AEE D ED D EK + S D E+E E+
Sbjct: 137 KPKAKPAEVKPAEEKPESDEEDESDDEDESEEDDDSEKGMDVDEDDSDDDEEEDSEDEEE 196
Query: 235 ETLVQRFENLNRANNHVQENVLPVKG----------DSVSEVELVSEKGNFPNSSPSKKL 284
E ++ E +N+ + + PV G + + E + G+ P+KK
Sbjct: 197 EETPKKPEPINKKRPNESVSKTPVSGKKAKPAAAPASTPQKTEEKKKGGHTATPHPAKK- 255
Query: 285 PMAALLPPEANKSTEKENHSENKVTPKNFFSSMKDIEVLFNRASDSGK 332
P AN+S + S K F+S K N+ S+ GK
Sbjct: 256 --GGKSPVNANQSPKSGGQSSGGNNNKKPFNSGKQFGGSNNKGSNKGK 301
>At5g60690 REVOLUTA or interfascicular fiberless 1
Length = 842
Score = 38.1 bits (87), Expect = 0.019
Identities = 33/119 (27%), Positives = 59/119 (48%), Gaps = 7/119 (5%)
Query: 406 ANSRENVEDLTNNLFDNSCMNAGSHAS-TLDRLHAWERKLYDEVKASEIVRKEYDAKCKI 464
AN RE D N D+S G + T +++ A ER + K S + R++ +C I
Sbjct: 6 ANHRERSSDSMNRHLDSS----GKYVRYTAEQVEALERVYAECPKPSSLRRQQLIRECSI 61
Query: 465 LRNLESKAEKTSTVD-KTRAAVKDLHSRIRVAIHRIDSISKRIEELRDKELQPQLEELI 522
L N+E K K + + R + SR++ ++ +++K + E D+ LQ Q+ +L+
Sbjct: 62 LANIEPKQIKVWFQNRRCRDKQRKEASRLQSVNRKLSAMNKLLMEENDR-LQKQVSQLV 119
>At1g52150 unknown protein
Length = 836
Score = 38.1 bits (87), Expect = 0.019
Identities = 28/101 (27%), Positives = 54/101 (52%), Gaps = 3/101 (2%)
Query: 424 CMNAGSHAS-TLDRLHAWERKLYDEVKASEIVRKEYDAKCKILRNLESKAEKTSTVD-KT 481
C++ G + T +++ A ER +D K S I R++ +C IL N+E K K + +
Sbjct: 12 CLDNGKYVRYTPEQVEALERLYHDCPKPSSIRRQQLIRECPILSNIEPKQIKVWFQNRRC 71
Query: 482 RAAVKDLHSRIRVAIHRIDSISKRIEELRDKELQPQLEELI 522
R + SR++ ++ +++K + E D+ LQ Q+ +L+
Sbjct: 72 REKQRKEASRLQAVNRKLTAMNKLLMEENDR-LQKQVSQLV 111
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.312 0.128 0.365
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 17,812,945
Number of Sequences: 26719
Number of extensions: 803727
Number of successful extensions: 3217
Number of sequences better than 10.0: 83
Number of HSP's better than 10.0 without gapping: 23
Number of HSP's successfully gapped in prelim test: 60
Number of HSP's that attempted gapping in prelim test: 3094
Number of HSP's gapped (non-prelim): 134
length of query: 773
length of database: 11,318,596
effective HSP length: 107
effective length of query: 666
effective length of database: 8,459,663
effective search space: 5634135558
effective search space used: 5634135558
T: 11
A: 40
X1: 16 ( 7.2 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 42 (21.9 bits)
S2: 64 (29.3 bits)
Medicago: description of AC131455.1


