

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC131248.8 + phase: 0
(285 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
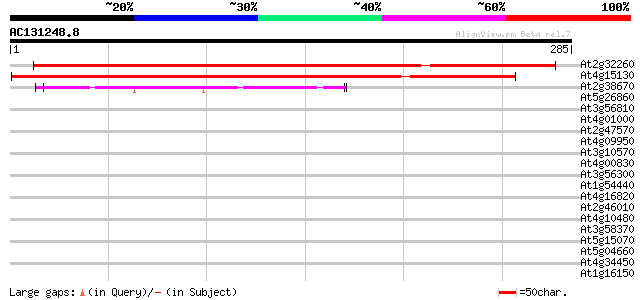
At2g32260 putative phospholipid cytidylyltransferase 432 e-121
At4g15130 CTP:phosphorylcholine cytidylyltransferase (AtCCT2) 404 e-113
At2g38670 putative phospholipid cytidylyltransferase 108 3e-24
At5g26860 LON protease homolog (LON_ARA) 33 0.23
At3g56810 unknown protein 33 0.23
At4g01000 putative protein 30 1.1
At2g47570 60S ribosomal protein L18 30 1.1
At4g09950 AIG1-like protein 30 1.9
At3g10570 putative cytochrome P450 29 2.5
At4g00830 unknown protein 29 3.3
At3g56300 cysteine-tRNA ligase-like protein 29 3.3
At1g54440 hypothetical protein 29 3.3
At4g16820 triacylglycerol lipase like protein 28 4.3
At2g46010 hypothetical protein 28 4.3
At4g10480 putative alpha NAC 28 5.6
At3g58370 putative protein 28 5.6
At5g15070 unknown protein 28 7.4
At5g04660 cytochrom P450 - like protein 28 7.4
At4g34450 Nonclathrin coat protein gamma - like protein 28 7.4
At1g16150 28 7.4
>At2g32260 putative phospholipid cytidylyltransferase
Length = 332
Score = 432 bits (1111), Expect = e-121
Identities = 209/266 (78%), Positives = 236/266 (88%), Gaps = 5/266 (1%)
Query: 13 DKPVRVYADGIYDLFHFGHARSLEQAKKSFPN-TYLLVGCCSDEITHKYKGKTVMNEQER 71
D+PVRVYADGIYDLFHFGHARSLEQAK +FPN TYLLVGCC+DE THKYKG+TVM +ER
Sbjct: 32 DRPVRVYADGIYDLFHFGHARSLEQAKLAFPNNTYLLVGCCNDETTHKYKGRTVMTAEER 91
Query: 72 YESLRHCKWVDEVIPDVPWVINQEFIDKHKIDYVAHDALPYADTSGAGNDVYEFVKAIGK 131
YESLRHCKWVDEVIPD PWV+NQEF+DKH+IDYVAHD+LPYAD+SGAG DVYEFVK +G+
Sbjct: 92 YESLRHCKWVDEVIPDAPWVVNQEFLDKHQIDYVAHDSLPYADSSGAGKDVYEFVKKVGR 151
Query: 132 FKETKRTEGISTSDIIMRIIKDYNQYVMRNLDRGYSRKELGVSYVKEKRLRMNMGLKKLQ 191
FKET+RTEGISTSDIIMRI+KDYNQYVMRNLDRGYSR++LGVS+VKEKRLR+NM LKKLQ
Sbjct: 152 FKETQRTEGISTSDIIMRIVKDYNQYVMRNLDRGYSREDLGVSFVKEKRLRVNMRLKKLQ 211
Query: 192 ERVKKQQETVGKKIGTVRRIAGMNRTEWVENADRLVAGFLEMFEEGCHKMGTAIRDRIQE 251
ERVK+QQE VG+KI TV+ M R EWVENADR VAGFLE+FEEGCHKMGTAI D IQE
Sbjct: 212 ERVKEQQERVGEKIQTVK----MLRNEWVENADRWVAGFLEIFEEGCHKMGTAIVDSIQE 267
Query: 252 QLKAQQLKSLLYDEWDDDVDDEFYED 277
+L Q+ L + DDD DD+FYE+
Sbjct: 268 RLMRQKSAERLENGQDDDTDDQFYEE 293
>At4g15130 CTP:phosphorylcholine cytidylyltransferase (AtCCT2)
Length = 300
Score = 404 bits (1039), Expect = e-113
Identities = 195/256 (76%), Positives = 221/256 (86%), Gaps = 4/256 (1%)
Query: 2 NEQDDSEVGKNDKPVRVYADGIYDLFHFGHARSLEQAKKSFPNTYLLVGCCSDEITHKYK 61
N+ + +D+PVRVYADGI+DLFHFGHAR++EQAKKSFPNTYLLVGCC+DEIT+K+K
Sbjct: 7 NKVSGGDSSSSDRPVRVYADGIFDLFHFGHARAIEQAKKSFPNTYLLVGCCNDEITNKFK 66
Query: 62 GKTVMNEQERYESLRHCKWVDEVIPDVPWVINQEFIDKHKIDYVAHDALPYADTSGAGND 121
GKTVM E ERYESLRHCKWVDEVIPD PWV+ EF+DKHKIDYVAHDALPYADTSGAGND
Sbjct: 67 GKTVMTESERYESLRHCKWVDEVIPDAPWVLTTEFLDKHKIDYVAHDALPYADTSGAGND 126
Query: 122 VYEFVKAIGKFKETKRTEGISTSDIIMRIIKDYNQYVMRNLDRGYSRKELGVSYVKEKRL 181
VYEFVK+IGKFKETKRTEGISTSDIIMRI+KDYNQYV+RN YSR+EL +EKRL
Sbjct: 127 VYEFVKSIGKFKETKRTEGISTSDIIMRIVKDYNQYVLRNPGPRYSREELSCQLEQEKRL 186
Query: 182 RMNMGLKKLQERVKKQQETVGKKIGTVRRIAGMNRTEWVENADRLVAGFLEMFEEGCHKM 241
R+NM LKKLQE+VK+QQE KI TV + AGM+ EW+ENADR VAGFLEMFEEGCHKM
Sbjct: 187 RVNMRLKKLQEKVKEQQE----KIQTVAKTAGMHHDEWLENADRWVAGFLEMFEEGCHKM 242
Query: 242 GTAIRDRIQEQLKAQQ 257
GTAIRD IQ++L Q+
Sbjct: 243 GTAIRDGIQQRLMRQE 258
>At2g38670 putative phospholipid cytidylyltransferase
Length = 421
Score = 108 bits (270), Expect = 3e-24
Identities = 62/162 (38%), Positives = 91/162 (55%), Gaps = 11/162 (6%)
Query: 14 KPVRVYADGIYDLFHFGHARSLEQAKKSFPNTYLLVGCCSDEITHKYKGKTVMNEQERYE 73
KPVRVY DG +D+ H+GH +L QA+ L+VG SDE KG V ER
Sbjct: 53 KPVRVYMDGCFDMMHYGHCNALRQARAL--GDQLVVGVVSDEEIIANKGPPVTPLHERMT 110
Query: 74 SLRHCKWVDEVIPDVPWVINQEFI----DKHKIDYVAHDALPYADTSGAGNDVYEFVKAI 129
++ KWVDEVI D P+ I ++F+ D+++IDY+ H P G D Y K
Sbjct: 111 MVKAVKWVDEVISDAPYAITEDFMKKLFDEYQIDYIIHGDDPCVLPD--GTDAYALAKKA 168
Query: 130 GKFKETKRTEGISTSDIIMRIIKDYNQYVMRNLDRGYSRKEL 171
G++K+ KRTEG+S++DI+ R++ + R++ +SR L
Sbjct: 169 GRYKQIKRTEGVSSTDIVGRMLLCVRE---RSISDTHSRSSL 207
Score = 81.6 bits (200), Expect = 4e-16
Identities = 51/156 (32%), Positives = 80/156 (50%), Gaps = 5/156 (3%)
Query: 18 VYADGIYDLFHFGHARSLEQAKKSFPNTYLLVGCCSDEITHKYKG--KTVMNEQERYESL 75
+Y DG +DLFH GH L +A++ +LLVG +D+ +G + +MN ER S+
Sbjct: 257 IYIDGAFDLFHAGHVEILRRAREL--GDFLLVGIHNDQTVSAKRGAHRPIMNLHERSLSV 314
Query: 76 RHCKWVDEVIPDVPWVINQEFIDKHKIDYVAHDALPYADTSGAGND-VYEFVKAIGKFKE 134
C++VDEVI PW ++++ I I V H + +D D Y ++G F+
Sbjct: 315 LACRYVDEVIIGAPWEVSRDTITTFDISLVVHGTVAESDDFRKEEDNPYSVPISMGIFQV 374
Query: 135 TKRTEGISTSDIIMRIIKDYNQYVMRNLDRGYSRKE 170
I+TS II RI+ ++ Y RN + S K+
Sbjct: 375 LDSPLDITTSTIIRRIVANHEAYQKRNAKKEASEKK 410
>At5g26860 LON protease homolog (LON_ARA)
Length = 985
Score = 32.7 bits (73), Expect = 0.23
Identities = 29/93 (31%), Positives = 42/93 (44%), Gaps = 13/93 (13%)
Query: 179 KRLRMNMGLKKLQERVKKQQETVGKKI-----GTVRR------IAGMNRTEWVENADR-- 225
KRLR+ + L K + + K QET+ K I G RR + + + VE D+
Sbjct: 341 KRLRLTLELMKKEMEISKIQETIAKAIEEKISGEQRRYLLNEQLKAIKKELGVETDDKSA 400
Query: 226 LVAGFLEMFEEGCHKMGTAIRDRIQEQLKAQQL 258
L A F E E K+ + I+E+L QL
Sbjct: 401 LSAKFKERIEPNKEKIPAHVLQVIEEELTKLQL 433
>At3g56810 unknown protein
Length = 332
Score = 32.7 bits (73), Expect = 0.23
Identities = 43/203 (21%), Positives = 74/203 (36%), Gaps = 35/203 (17%)
Query: 97 IDKHKIDYVAHDALPYADTSGAGNDVYEFVKAIGKF-------KETKRTEGISTSDIIMR 149
ID K Y+ H L + + N + G K TK T+ +D ++
Sbjct: 120 IDVIKTTYILHKRLEVSPPTTRENFKFSGTVLAGDVVVGTHTQKNTKTTQHEPDTDTMLS 179
Query: 150 II-------KDYNQYVMRNLDRGYSRKELGVSYVKEKRLRMNMGLKKLQERVKKQQETVG 202
++ KD Y + N++ + V KR T
Sbjct: 180 MMNHHEYCCKDNKPYKVNNINLITRPDHYSIHDVISKR---------------STSTTTT 224
Query: 203 KKIGTVRRIAGMNRTEWVENADRLVAGFLEMFEEGCHKMGTAIRDRIQEQLKAQQLKSLL 262
+ GT+ + R +E+ +++ K+G A+ D I L A+ ++ L
Sbjct: 225 ESCGTLPLVVRRVRRSLMESVNQVCDDVASGQRREVAKIGLALHDHICRDLIAETVRELS 284
Query: 263 YDEWDDDVDDEFYE---DESVEY 282
+ ++D DDEFY+ D SV Y
Sbjct: 285 FSDYD---DDEFYQSPVDSSVCY 304
>At4g01000 putative protein
Length = 415
Score = 30.4 bits (67), Expect = 1.1
Identities = 27/95 (28%), Positives = 42/95 (43%), Gaps = 8/95 (8%)
Query: 188 KKLQERVKKQQETVGKKIGTVRRIAGMNRTEWVENADRLVAG----FLEMFEEGCHKMGT 243
KK E +KKQ V + +G +N+ + E +D+ + E F+ G K
Sbjct: 138 KKALEYLKKQSNKVKQGVGNGATQKYVNK--YKEESDKCILAVDLALNESFKNGKRKAKI 195
Query: 244 AIRDRIQEQLKAQQLKSLLYDEWDDDVDDEFYEDE 278
+++LK + K + D DD DDE EDE
Sbjct: 196 GAESEKKKRLKIWKGKRAVEDSDSDDSDDE--EDE 228
>At2g47570 60S ribosomal protein L18
Length = 187
Score = 30.4 bits (67), Expect = 1.1
Identities = 22/83 (26%), Positives = 43/83 (51%), Gaps = 4/83 (4%)
Query: 130 GKFKETKRTEGISTSDIIMRIIKDYNQYVMRNLDRGYSR---KELGVSYVKEKRLRMNMG 186
GK K+TKRTE + D+ ++++ N+Y++R + ++ K L +S V + L ++
Sbjct: 8 GKSKKTKRTEP-KSDDVYLKLLVKANRYLVRRTESKFNAVILKRLFMSKVNKAPLSLSRL 66
Query: 187 LKKLQERVKKQQETVGKKIGTVR 209
++ + + K VG VR
Sbjct: 67 VRYMDGKDGKIAVIVGTVTDDVR 89
>At4g09950 AIG1-like protein
Length = 336
Score = 29.6 bits (65), Expect = 1.9
Identities = 25/119 (21%), Positives = 54/119 (45%), Gaps = 5/119 (4%)
Query: 152 KDYNQYVMRNLDRGYSRKELGVSYVKEKRLRMNMGLKKLQERVKKQQETVG----KKIGT 207
KD + + NL S+K G SY+ + + ++E+ K+ +E G ++I
Sbjct: 189 KDRQVHDLLNLVEQISKKNNGKSYMADLSHELRENEATIKEKQKQIEEMKGWSSKQEISQ 248
Query: 208 VRRIAGMNRTEWVENADRLVAGFL-EMFEEGCHKMGTAIRDRIQEQLKAQQLKSLLYDE 265
+++ + E +E ++ L E E+ ++ A +R + + K +++ L DE
Sbjct: 249 MKKELEKSHNEMLEGIKEKISNQLKESLEDVKEQLAKAQAEREETEKKMNEIQKLSSDE 307
>At3g10570 putative cytochrome P450
Length = 513
Score = 29.3 bits (64), Expect = 2.5
Identities = 18/58 (31%), Positives = 28/58 (48%)
Query: 224 DRLVAGFLEMFEEGCHKMGTAIRDRIQEQLKAQQLKSLLYDEWDDDVDDEFYEDESVE 281
+ LV+ E G GTAI I + + +++S LYDE V D E++ V+
Sbjct: 304 EELVSLCSEFLNGGTDTTGTAIEWGIAQLIVNPEIQSRLYDEIKSTVGDREVEEKDVD 361
>At4g00830 unknown protein
Length = 495
Score = 28.9 bits (63), Expect = 3.3
Identities = 16/52 (30%), Positives = 28/52 (53%), Gaps = 3/52 (5%)
Query: 234 FEEGCHKMGTAIRDRIQEQLKAQQLKSLLYDEWDDDVDDEFYEDESVEYYSD 285
FEEG + + + D ++E+ + + D+ DDDV ++ E+ VE Y D
Sbjct: 13 FEEGSY---SEMEDEVEEEQVEEYEEEEEEDDDDDDVGNQNAEEREVEDYGD 61
>At3g56300 cysteine-tRNA ligase-like protein
Length = 489
Score = 28.9 bits (63), Expect = 3.3
Identities = 40/182 (21%), Positives = 69/182 (36%), Gaps = 24/182 (13%)
Query: 12 NDKPVRVYADGI--YDLFHFGHARSLEQAKKSFPNTYLLVGCCSDEITHKYKGKTVMNEQ 69
N + +Y GI YD H GHAR A SF Y + ++T+ V N
Sbjct: 28 NPGKIGIYVCGITAYDYSHIGHAR----AAVSFDLLYRYLRHLGYQVTY------VRNFT 77
Query: 70 ERYESLRHCKWVDEVIPDVPWVINQEFIDKHKIDYVAHDALPYADTSGAGNDVYEFVKAI 129
+ + ++C + P ++ F +++ +D A L + + + +K I
Sbjct: 78 DVDDKAKNC-------GEKPLDLSNRFCEEYLLDMAALQCLLPTHQPRVSDHMEQIIKMI 130
Query: 130 GKFKETKRTEGISTSDIIMRIIK--DYNQYVMRNLDRGYSRKELGVSYVKEKRLRMNMGL 187
K E + D+ + K Y Q + LD + K + V KR + L
Sbjct: 131 EKIIENGCGYAVG-GDVFFSVDKSPSYGQLSGQRLDHTQAGKRVAVD--SRKRNPADFAL 187
Query: 188 KK 189
+K
Sbjct: 188 RK 189
>At1g54440 hypothetical protein
Length = 738
Score = 28.9 bits (63), Expect = 3.3
Identities = 25/128 (19%), Positives = 54/128 (41%), Gaps = 11/128 (8%)
Query: 163 DRGYSRKELGVSYV---KEKRLRMNMGLKKLQERVKKQQETVGKKI-------GTVRRIA 212
D G S L + V K+ G+ L+ +++ + V K I GT +
Sbjct: 543 DTGASASSLSLEKVCVDDSKKQSSGFGVLPLKRKLESDKTVVEKNIEPKIEKTGTEASAS 602
Query: 213 GMNRTE-WVENADRLVAGFLEMFEEGCHKMGTAIRDRIQEQLKAQQLKSLLYDEWDDDVD 271
++ + V+++ + +GF + + + +++ ++ ++ D+ DDD D
Sbjct: 603 SLSSKKVCVDDSKKQSSGFGVLLSKRKFESDNKVKEEVKVSKSKPDKVIIVVDDDDDDDD 662
Query: 272 DEFYEDES 279
DE YE +
Sbjct: 663 DESYEQST 670
>At4g16820 triacylglycerol lipase like protein
Length = 601
Score = 28.5 bits (62), Expect = 4.3
Identities = 10/21 (47%), Positives = 15/21 (70%)
Query: 74 SLRHCKWVDEVIPDVPWVINQ 94
S+R KW+D+V PD+ W+ Q
Sbjct: 198 SVRLPKWIDDVAPDLRWMTKQ 218
>At2g46010 hypothetical protein
Length = 942
Score = 28.5 bits (62), Expect = 4.3
Identities = 15/52 (28%), Positives = 32/52 (60%), Gaps = 1/52 (1%)
Query: 151 IKDYNQYVMRNLDRGYSRKELGVSY-VKEKRLRMNMGLKKLQERVKKQQETV 201
+K N + +NL+R R +L + ++EK+LR++ +++E V +QQ+ +
Sbjct: 690 LKKINGLLAKNLERKRIRPDLVLRLQIEEKKLRLSDLQSRVREEVDRQQQEI 741
>At4g10480 putative alpha NAC
Length = 212
Score = 28.1 bits (61), Expect = 5.6
Identities = 15/36 (41%), Positives = 22/36 (60%), Gaps = 6/36 (16%)
Query: 244 AIRDRIQEQLKAQQLKSLLY------DEWDDDVDDE 273
A+ D I+EQ+K Q+ ++ DE DDDVDD+
Sbjct: 13 ALMDAIKEQMKLQKENDVVVEDVKDGDEDDDDVDDD 48
>At3g58370 putative protein
Length = 219
Score = 28.1 bits (61), Expect = 5.6
Identities = 21/74 (28%), Positives = 34/74 (45%), Gaps = 1/74 (1%)
Query: 131 KFKETKRTEGISTSDIIMRIIKDYNQYVMRNLDRGYSRKELGVSYVKEKRLRMNMGLKKL 190
KF+ R + +++ +IK Q D + VSYV+ LR++ KKL
Sbjct: 102 KFRPKNRHLKSTYMTVLLGLIKTLCQLPEELTDDDLDEASVAVSYVENGGLRLDWLEKKL 161
Query: 191 QE-RVKKQQETVGK 203
E + KK++ GK
Sbjct: 162 AEVKAKKKKVETGK 175
>At5g15070 unknown protein
Length = 1049
Score = 27.7 bits (60), Expect = 7.4
Identities = 19/70 (27%), Positives = 32/70 (45%), Gaps = 8/70 (11%)
Query: 17 RVYADGIYDLFHFGHARSLEQAKKSFPNTYLLVGCCSDEITHKYKGKTVMNEQERYESLR 76
R++A G +++ HFG LE +S+P L+ Y + + + Y SLR
Sbjct: 35 RIHAFGEFEIIHFGDKVILEDPVESWPICDCLIAF--------YSSGYPLEKVQAYSSLR 86
Query: 77 HCKWVDEVIP 86
V+E+ P
Sbjct: 87 KPFLVNELDP 96
>At5g04660 cytochrom P450 - like protein
Length = 512
Score = 27.7 bits (60), Expect = 7.4
Identities = 18/55 (32%), Positives = 25/55 (44%)
Query: 224 DRLVAGFLEMFEEGCHKMGTAIRDRIQEQLKAQQLKSLLYDEWDDDVDDEFYEDE 278
+ LV E G GTAI I + + +++S LYDE V D+ DE
Sbjct: 302 EELVTLCSEFLNGGTDTTGTAIEWGIAQLIANPEIQSRLYDEIKSTVGDDRRVDE 356
>At4g34450 Nonclathrin coat protein gamma - like protein
Length = 831
Score = 27.7 bits (60), Expect = 7.4
Identities = 35/140 (25%), Positives = 62/140 (44%), Gaps = 10/140 (7%)
Query: 91 VINQEFIDKHKIDYVAHDALPYADTSGAGNDVYEF---VKAIGKFKET------KRTEGI 141
V+N E + ++ A ++LPY D+ G V+E V A+GKF T +
Sbjct: 632 VLNIEAEEFSEVTSKALNSLPY-DSPGQAFVVFEKPAGVPAVGKFSNTLTFVVKEVHVDP 690
Query: 142 STSDIIMRIIKDYNQYVMRNLDRGYSRKELGVSYVKEKRLRMNMGLKKLQERVKKQQETV 201
ST + ++D Q + G ++GVS + M+ +++ E Q+E++
Sbjct: 691 STGEAEDDGVEDEYQLEDLEVVAGDYMVKVGVSNFRNAWESMDEEDERVDEYGLGQRESL 750
Query: 202 GKKIGTVRRIAGMNRTEWVE 221
G+ + V + GM E E
Sbjct: 751 GEAVKAVMDLLGMQTCEGTE 770
>At1g16150
Length = 779
Score = 27.7 bits (60), Expect = 7.4
Identities = 10/31 (32%), Positives = 23/31 (73%)
Query: 251 EQLKAQQLKSLLYDEWDDDVDDEFYEDESVE 281
E++++ KS ++++ DDD DD+ +D+++E
Sbjct: 713 ERIRSSSYKSEIHNDDDDDDDDDDEDDQAME 743
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.317 0.136 0.398
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 6,518,758
Number of Sequences: 26719
Number of extensions: 280502
Number of successful extensions: 1068
Number of sequences better than 10.0: 28
Number of HSP's better than 10.0 without gapping: 9
Number of HSP's successfully gapped in prelim test: 19
Number of HSP's that attempted gapping in prelim test: 1049
Number of HSP's gapped (non-prelim): 33
length of query: 285
length of database: 11,318,596
effective HSP length: 98
effective length of query: 187
effective length of database: 8,700,134
effective search space: 1626925058
effective search space used: 1626925058
T: 11
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.6 bits)
S2: 59 (27.3 bits)
Medicago: description of AC131248.8


