

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC131240.9 - phase: 0
(430 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
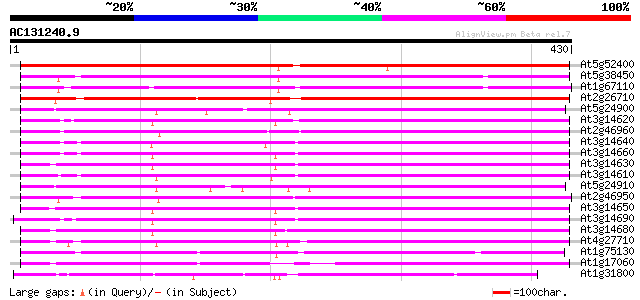
Score E
Sequences producing significant alignments: (bits) Value
At5g52400 cytochrome P-450-like protein 565 e-161
At5g38450 cytochrome P450-like protein 324 7e-89
At1g67110 unknown protein 311 3e-85
At2g26710 putative cytochrome P450 294 6e-80
At5g24900 cytochrome P450-like protein 283 1e-76
At3g14620 putative cytochrome P450 283 2e-76
At2g46960 putative cytochrome P450 280 1e-75
At3g14640 putative cytochrome P450 279 2e-75
At3g14660 putative cytochrome P450 279 2e-75
At3g14630 putative cytochrome P450 279 2e-75
At3g14610 putative cytochrome P450 278 3e-75
At5g24910 cytochrome P450-like protein 277 9e-75
At2g46950 putative cytochrome P450 276 2e-74
At3g14650 putative cytochrome P450 273 1e-73
At3g14690 putative cytochrome P450 270 1e-72
At3g14680 putative cytochrome P450 269 2e-72
At4g27710 cytochrome P450 - like protein 263 2e-70
At1g75130 cytochrome P450, putative 257 8e-69
At1g17060 putative cytochrome P450 214 1e-55
At1g31800 unknown protein 137 9e-33
>At5g52400 cytochrome P-450-like protein
Length = 530
Score = 565 bits (1456), Expect = e-161
Identities = 272/434 (62%), Positives = 333/434 (76%), Gaps = 18/434 (4%)
Query: 9 GKVFIYWLGTEPFLYIANAEFLKKMSTEVMAKRWGKPSVFRNDRDPMFGSGLVMVEGNDW 68
GKVF+YWLG EPF+Y+A+ EFL MS V+ K WGKP+VF+ DR+PMFG+GLVMVEG+DW
Sbjct: 102 GKVFVYWLGIEPFVYVADPEFLSVMSKGVLGKSWGKPNVFKKDREPMFGTGLVMVEGDDW 161
Query: 69 VRHRHIVAPAFNPLNLKVMASMMIESTNQMIDRWTSQINFGNHEIDMEKEIITIAGEIIA 128
RHRHI+ PAF PLNLKVM +MM+ES + M+DRW QIN GN E DME EII AGEIIA
Sbjct: 162 TRHRHIITPAFAPLNLKVMTNMMVESVSNMLDRWGIQINSGNPEFDMESEIIGTAGEIIA 221
Query: 129 KTSFGAEDENAKEVFDKLRALQMTLFNTNRYVGVPFGKYFNVKKNLEAKKLGKEIDKLLL 188
KTSFG EN +V LRA+Q LFN+NRYVGVPF + K+ ++AK LG EID LLL
Sbjct: 222 KTSFGVTGENGTQVLKNLRAVQFALFNSNRYVGVPFSNILSYKQTVKAKGLGHEIDGLLL 281
Query: 189 SIVEARKNSPKQNSQK--DLLGLLLKENNEDGKLGKTLTSREVVDECKTFFFGGHETTAL 246
S + RK S + + DLLG+LLK + + T++E+VDECKTFFF GHETTAL
Sbjct: 282 SFINKRKISLAEGDDQGHDLLGMLLKADQKG-----NFTAKELVDECKTFFFAGHETTAL 336
Query: 247 AITWTLLLLATHEDWQNQLREEIKEVVGNNELDITMLSGLKK-----------MKCVMNE 295
A+TWT +LLA H +WQ+ +REEI+EV+G+++++ L+GLKK M VMNE
Sbjct: 337 ALTWTFMLLAIHPEWQDTIREEIREVIGDSKIEYNKLAGLKKVHYQTQYTTIHMSWVMNE 396
Query: 296 VLRLYPPAPNVQRQAREDIQVDDVTVPNGTNMWIDVVAMHHDPELWGDDVNEFKPERFMD 355
VLRLYPPAPN QRQAR DI+V+ +PNGTN+WIDVVAMHHD ELWGDDVNEFKPERF
Sbjct: 397 VLRLYPPAPNAQRQARNDIEVNGRVIPNGTNIWIDVVAMHHDVELWGDDVNEFKPERFDG 456
Query: 356 DVNGGCKHKMGYLPFGFGGRMCVGRNLTFMEYKIVLTILLSNFTFKVSPSYQHSPAIMLS 415
+++GGCK+KMGY+PFGFGGRMC+GRNLT MEYKIVL+++LS F VSP Y+HSP MLS
Sbjct: 457 NLHGGCKNKMGYMPFGFGGRMCIGRNLTTMEYKIVLSLVLSRFEISVSPGYRHSPTYMLS 516
Query: 416 LRPAHGLPLIVQPL 429
LRP +GLPLI++PL
Sbjct: 517 LRPGYGLPLIIRPL 530
>At5g38450 cytochrome P450-like protein
Length = 518
Score = 324 bits (830), Expect = 7e-89
Identities = 166/430 (38%), Positives = 259/430 (59%), Gaps = 16/430 (3%)
Query: 9 GKVFIYWLGTEPFLYIANAEFLKKMSTE---VMAKRWGKPSVFRNDRDPMFGSGLVMVEG 65
GK FI W GT+P L + E +K++ + V + W + +N G GL+M G
Sbjct: 94 GKRFIVWNGTDPRLCLTETELIKELLMKHNGVSGRSWLQQQGTKN----FIGRGLLMANG 149
Query: 66 NDWVRHRHIVAPAFNPLNLKVMASMMIESTNQMIDRWTSQINFGNHEIDMEKEIITIAGE 125
DW RH+ APAF LK A M+E T+++++R ++ G +E+++ +E+ + +
Sbjct: 150 QDWHHQRHLAAPAFTGERLKGYARHMVECTSKLVERLRKEVGEGANEVEIGEEMHKLTAD 209
Query: 126 IIAKTSFGAEDENAKEVFDKLRALQMTLFNTNRYVGVPFGKYFNVKKNLEAKKLGKEIDK 185
II++T FG+ E KE+F+ L LQ R++ P ++ K N E K L KE+++
Sbjct: 210 IISRTKFGSSFEKGKELFNHLTVLQRRCAQATRHLCFPGSRFLPSKYNREIKSLKKEVER 269
Query: 186 LLLSIVEARKNSPKQNSQK----DLLGLLLKENNEDGKLGKTLTSRE-VVDECKTFFFGG 240
LL+ I+++R++ + DLLGLLL E + D + + ++DECKTFFF G
Sbjct: 270 LLIEIIQSRRDCAEMGRSSTHGDDLLGLLLNEMDIDKNNNNNNNNLQLIMDECKTFFFAG 329
Query: 241 HETTALAITWTLLLLATHEDWQNQLREEIKEVVGNNEL-DITMLSGLKKMKCVMNEVLRL 299
HETTAL +TWT +LLA + WQ ++REE++EV G N L + LS L + V+NE LRL
Sbjct: 330 HETTALLLTWTTMLLADNPTWQEKVREEVREVFGRNGLPSVDQLSKLTSLSKVINESLRL 389
Query: 300 YPPAPNVQRQAREDIQVDDVTVPNGTNMWIDVVAMHHDPELWGDDVNEFKPERFMDDVNG 359
YPPA + R A ED+++ D+T+P G ++WI V+A+HH ELWG D N+F PERF
Sbjct: 390 YPPATLLPRMAFEDLKLGDLTIPKGLSIWIPVLAIHHSEELWGKDANQFNPERFGGRPFA 449
Query: 360 GCKHKMGYLPFGFGGRMCVGRNLTFMEYKIVLTILLSNFTFKVSPSYQHSPAIMLSLRPA 419
+H ++PF G R C+G+ ME KI+L L+S F F +S +Y+H+P ++L+++P
Sbjct: 450 SGRH---FIPFAAGPRNCIGQQFALMEAKIILATLISKFNFTISKNYRHAPIVVLTIKPK 506
Query: 420 HGLPLIVQPL 429
+G+ +I++PL
Sbjct: 507 YGVQVILKPL 516
>At1g67110 unknown protein
Length = 512
Score = 311 bits (798), Expect = 3e-85
Identities = 163/430 (37%), Positives = 256/430 (58%), Gaps = 21/430 (4%)
Query: 9 GKVFIYWLGTEPFLYIANAEFLKKMSTE---VMAKRWGKPSVFRNDRDPMFGSGLVMVEG 65
GK FI W GTEP L + E +K++ T+ V K W + + G GL+M G
Sbjct: 95 GKRFIMWNGTEPRLCLTETEMIKELLTKHNPVTGKSW----LQQQGTKGFIGRGLLMANG 150
Query: 66 NDWVRHRHIVAPAFNPLNLKVMASMMIESTNQMIDRWTSQINFGNHEIDMEKEIITIAGE 125
W RH+ APAF LK A M+E T M +R ++ E+++ +E+ + +
Sbjct: 151 EAWHHQRHMAAPAFTRDRLKGYAKHMVECTKMMAERLRKEVG---EEVEIGEEMRRLTAD 207
Query: 126 IIAKTSFGAEDENAKEVFDKLRALQMTLFNTNRYVGVPFGKYFNVKKNLEAKKLGKEIDK 185
II++T FG+ + KE+F L LQ R++ P ++ K N E K L E+++
Sbjct: 208 IISRTEFGSSCDKGKELFSLLTVLQRLCAQATRHLCFPGSRFLPSKYNREIKSLKTEVER 267
Query: 186 LLLSIVEARKNSPKQNSQK----DLLGLLLKENNEDGKLGKTLTSREVVDECKTFFFGGH 241
LL+ I+++RK+S + DLLGLLL + + + L + ++DECKTFFF GH
Sbjct: 268 LLMEIIDSRKDSVEIGRSSSYGDDLLGLLLNQMDSNKN---NLNVQMIMDECKTFFFTGH 324
Query: 242 ETTALAITWTLLLLATHEDWQNQLREEIKEVVGNNEL-DITMLSGLKKMKCVMNEVLRLY 300
ETT+L +TWTL+LLA + WQ+ +R+E+++V G + + + LS L + V+NE LRLY
Sbjct: 325 ETTSLLLTWTLMLLAHNPTWQDNVRDEVRQVCGQDGVPSVEQLSSLTSLNKVINESLRLY 384
Query: 301 PPAPNVQRQAREDIQVDDVTVPNGTNMWIDVVAMHHDPELWGDDVNEFKPERFMDDVNGG 360
PPA + R A EDI++ D+ +P G ++WI V+A+HH ELWG+D NEF PERF
Sbjct: 385 PPATLLPRMAFEDIKLGDLIIPKGLSIWIPVLAIHHSNELWGEDANEFNPERFTTRSFAS 444
Query: 361 CKHKMGYLPFGFGGRMCVGRNLTFMEYKIVLTILLSNFTFKVSPSYQHSPAIMLSLRPAH 420
+H ++PF G R C+G+ ME KI+L +L+S F+F +S +Y+H+P ++L+++P +
Sbjct: 445 SRH---FMPFAAGPRNCIGQTFAMMEAKIILAMLVSKFSFAISENYRHAPIVVLTIKPKY 501
Query: 421 GLPLIVQPLN 430
G+ L+++PL+
Sbjct: 502 GVQLVLKPLD 511
>At2g26710 putative cytochrome P450
Length = 520
Score = 294 bits (753), Expect = 6e-80
Identities = 149/434 (34%), Positives = 262/434 (60%), Gaps = 27/434 (6%)
Query: 9 GKVFIYWLGTEPFLYIANAEFLKKM--STEVMAKRWGKPSVFRNDRDPMFGSGLVMVEGN 66
G F+ W G L +A+ + ++++ +E K P V + + G GL+ ++G
Sbjct: 96 GATFLVWFGPTFRLTVADPDLIREIFSKSEFYEKNEAHPLVKQLE-----GDGLLSLKGE 150
Query: 67 DWVRHRHIVAPAFNPLNLKVMASMMIESTNQMIDRWTSQINF-GNHEIDMEKEIITIAGE 125
W HR I++P F+ NLK++ ++++S M+D+W+ +++ G E+D+ + + +
Sbjct: 151 KWAHHRKIISPTFHMENLKLLVPVVLKSVTDMVDKWSDKLSENGEVEVDVYEWFQILTED 210
Query: 126 IIAKTSFGAEDENAKEVFDKLRALQMTLF-NTNRYVGVPFGKYFNVKKNLEAKKLGKEID 184
+I++T+FG+ E+ + VF +L+A QM L + V +P ++F + NL++ KL KEI
Sbjct: 211 VISRTAFGSSYEDGRAVF-RLQAQQMLLCAEAFQKVFIPGYRFFPTRGNLKSWKLDKEIR 269
Query: 185 KLLLSIVEARKNSP--------KQNSQKDLLGLLLKENNEDGKLGKTLTSREVVDECKTF 236
K LL ++E R+ + K+ + KDLLGL+++ N +T +++V+ECK+F
Sbjct: 270 KSLLKLIERRRQNAIDGEGEECKEPAAKDLLGLMIQAKN--------VTVQDIVEECKSF 321
Query: 237 FFGGHETTALAITWTLLLLATHEDWQNQLREEIKEVVGNNELDIT-MLSGLKKMKCVMNE 295
FF G +TT+ +TWT +LL+ H +WQ + R+E+ V G+ ++ + LK + ++NE
Sbjct: 322 FFAGKQTTSNLLTWTTILLSMHPEWQAKARDEVLRVCGSRDVPTKDHVVKLKTLSMILNE 381
Query: 296 VLRLYPPAPNVQRQAREDIQVDDVTVPNGTNMWIDVVAMHHDPELWGDDVNEFKPERFMD 355
LRLYPP R+A+ D+++ +P GT + I ++A+HHD +WG+DVNEF P RF D
Sbjct: 382 SLRLYPPIVATIRRAKSDVKLGGYKIPCGTELLIPIIAVHHDQAIWGNDVNEFNPARFAD 441
Query: 356 DVNGGCKHKMGYLPFGFGGRMCVGRNLTFMEYKIVLTILLSNFTFKVSPSYQHSPAIMLS 415
V KH +G++PFG G R C+G+NL ++ K+ L +++ FTF ++P+YQH+P +++
Sbjct: 442 GVPRAAKHPVGFIPFGLGVRTCIGQNLAILQAKLTLAVMIQRFTFHLAPTYQHAPTVLML 501
Query: 416 LRPAHGLPLIVQPL 429
L P HG P+ + L
Sbjct: 502 LYPQHGAPITFRRL 515
>At5g24900 cytochrome P450-like protein
Length = 525
Score = 283 bits (724), Expect = 1e-76
Identities = 145/430 (33%), Positives = 249/430 (57%), Gaps = 15/430 (3%)
Query: 9 GKVFIYWLGTEPFLYIANAEFLKKMSTEVMAKRWGKPSVFRNDRDPMFGSGLVMVEGNDW 68
G+++ Y G + LYI + E +K++S + G+ + +P+ G+G++ G W
Sbjct: 98 GRIYTYSTGLKQHLYINHPEMVKELS-QTNTLNLGRITHITKRLNPILGNGIITSNGPHW 156
Query: 69 VRHRHIVAPAFNPLNLKVMASMMIESTNQMIDRWTSQINFGNH---EIDMEKEIITIAGE 125
R I+A F +K M +M+ES M+++W + G +I +++++ ++ +
Sbjct: 157 AHQRRIIAYEFTHDKIKGMVGLMVESAMPMLNKWEEMVKRGGEMGCDIRVDEDLKDVSAD 216
Query: 126 IIAKTSFGAEDENAKEVFDKLRAL------QMTLFNTNRYVGVPFGKYFNVKKNLEAKKL 179
+IAK FG+ K +F +R L + LF N + + FG + +++A ++
Sbjct: 217 VIAKACFGSSFSKGKAIFSMIRDLLTAITKRSVLFRFNGFTDMVFGSKKHGDVDIDALEM 276
Query: 180 GKEIDKLLLSIVEARKNSPKQNSQKDLLGLLLKE--NNEDGKL-GKTLTSREVVDECKTF 236
E++ + V+ R+ K +KDL+ L+L+ + DG L K+ R VVD CK+
Sbjct: 277 --ELESSIWETVKEREIECKDTHKKDLMQLILEGAMRSCDGNLWDKSAYRRFVVDNCKSI 334
Query: 237 FFGGHETTALAITWTLLLLATHEDWQNQLREEIKEVVGNNELDITMLSGLKKMKCVMNEV 296
+F GH++TA++++W L+LLA + WQ ++R+EI N D + LK + V+ E
Sbjct: 335 YFAGHDSTAVSVSWCLMLLALNPSWQVKIRDEILSSCKNGIPDAESIPNLKTVTMVIQET 394
Query: 297 LRLYPPAPNVQRQAREDIQVDDVTVPNGTNMWIDVVAMHHDPELWGDDVNEFKPERFMDD 356
+RLYPPAP V R+A +DI++ D+ VP G +W + A+H DPE+WG D N+FKPERF +
Sbjct: 395 MRLYPPAPIVGREASKDIRLGDLVVPKGVCIWTLIPALHRDPEIWGPDANDFKPERFSEG 454
Query: 357 VNGGCKHKMGYLPFGFGGRMCVGRNLTFMEYKIVLTILLSNFTFKVSPSYQHSPAIMLSL 416
++ CK+ Y+PFG G R CVG+N ME K+++++++S F+F +SP+YQHSP+ L +
Sbjct: 455 ISKACKYPQSYIPFGLGPRTCVGKNFGMMEVKVLVSLIVSKFSFTLSPTYQHSPSHKLLV 514
Query: 417 RPAHGLPLIV 426
P HG+ + V
Sbjct: 515 EPQHGVVIRV 524
>At3g14620 putative cytochrome P450
Length = 515
Score = 283 bits (723), Expect = 2e-76
Identities = 149/427 (34%), Positives = 245/427 (56%), Gaps = 14/427 (3%)
Query: 9 GKVFIYWLGTEPFLYIANAEFLKKMSTEVMAKRWGKPSVFRNDRDPMFGSGLVMVEGNDW 68
GK W+G + + E +K + V + KP V + +F +G+ + EG W
Sbjct: 97 GKTSYMWMGPIASVIVTKPEHIKDVLNRVYD--FPKPPV--HPIVELFATGVALYEGEKW 152
Query: 69 VRHRHIVAPAFNPLNLKVMASMMIESTNQMIDRWTSQINF--GNHEIDMEKEIITIAGEI 126
+HR I+ P+F+ LK+M ES ++MI +W + ++EID+ + + ++
Sbjct: 153 SKHRKIINPSFHLEKLKIMIPAFYESCSEMISKWEKLVTEQGSSNEIDVWPYLGDLTSDV 212
Query: 127 IAKTSFGAEDENAKEVFDKLRALQMTLFNTNRYVGVPFGKYFNVKKNLEAKKLGKEIDKL 186
I++T+FG+ E K +F+ + +P ++ K NL +++ KE+
Sbjct: 213 ISRTAFGSSYEEGKRIFELQEEQGRRVLKALELAFIPGMRFLPTKNNLRMRQINKEVKSR 272
Query: 187 LLSIVEARKNSPKQNS--QKDLLGLLLKENNEDGKLGKTLTSREVVDECKTFFFGGHETT 244
L I+ R+ + DLLG+LL+ N+ D ++ +VV+EC+ F F G ETT
Sbjct: 273 LREIIMKRQRGMDTGEAPKNDLLGILLESNSGD----HGMSIEDVVEECRLFHFAGQETT 328
Query: 245 ALAITWTLLLLATHEDWQNQLREEIKEVVG-NNELDITMLSGLKKMKCVMNEVLRLYPPA 303
A+ + WT+++L+ H+ WQ+Q REEI +V+G NN+ + LS LK M ++NEVLRLYPP
Sbjct: 329 AVLLVWTMIMLSHHQKWQDQAREEILKVIGKNNKPNFDALSRLKTMSMILNEVLRLYPPG 388
Query: 304 PNVQRQAREDIQV-DDVTVPNGTNMWIDVVAMHHDPELWGDDVNEFKPERFMDDVNGGCK 362
+ R ++ ++ +D+T+P G + I V+ +H DPELWG+DV+EF PERF D ++ K
Sbjct: 389 ILLGRTVEKETKLGEDMTLPGGAQVVIPVLMVHRDPELWGEDVHEFNPERFADGISKATK 448
Query: 363 HKMGYLPFGFGGRMCVGRNLTFMEYKIVLTILLSNFTFKVSPSYQHSPAIMLSLRPAHGL 422
+++ +LPFG+G R C G+N ME K+ L ++L F+F++SPSY H+P +L+L P G
Sbjct: 449 NQVSFLPFGWGPRFCPGQNFALMEAKMALVLILQRFSFELSPSYTHAPHTVLTLHPQFGA 508
Query: 423 PLIVQPL 429
PLI L
Sbjct: 509 PLIFHML 515
>At2g46960 putative cytochrome P450
Length = 519
Score = 280 bits (716), Expect = 1e-75
Identities = 157/429 (36%), Positives = 244/429 (56%), Gaps = 12/429 (2%)
Query: 9 GKVFIYWLGTEPFLYIANAEFLKKMSTEVMAKRWGKPSVFRNDRDPMFGS-GLVMVEGND 67
G+ F+YW GTEP + I++ E K M + + + S R + + GS GLV +EG D
Sbjct: 93 GETFLYWNGTEPRICISDPELAKTMLSNKLG--FFVKSKARPEAVKLVGSKGLVFIEGAD 150
Query: 68 WVRHRHIVAPAFNPLNLKVMASMMIESTNQMIDRWTSQINFGNHEI-----DMEKEIITI 122
WVRHR I+ PAF+ LK+M ++M++ T +M++ W + E +M +E +
Sbjct: 151 WVRHRRILNPAFSIDRLKIMTTVMVDCTLKMLEEWRKESTKEETEHPKIKKEMNEEFQRL 210
Query: 123 AGEIIAKTSFGAEDENAKEVFDKLRALQMTLFNTNRYVGVPFGKYFNVKKNLEAKKLGKE 182
+IIA ++FG+ EVF L+ + V +P +Y N+ KL ++
Sbjct: 211 TADIIATSAFGSSYVEGIEVFRSQMELKRCYTTSLNQVSIPGTQYLPTPSNIRVWKLERK 270
Query: 183 IDKLLLSIVEARKNSPKQNSQKDLLGLLLKENNEDGKLGKTLTSREVVDECKTFFFGGHE 242
+D + I+ +R S K + DLLG+LLK N +GK K ++ E++ EC+TFFFGGHE
Sbjct: 271 MDNSIKRIISSRLQS-KSDYGDDLLGILLKAYNTEGKERK-MSIEEIIHECRTFFFGGHE 328
Query: 243 TTALAITWTLLLLATHEDWQNQLREEIKEVVGNNEL-DITMLSGLKKMKCVMNEVLRLYP 301
TT+ + WT +LL+ H+DWQ +LREEI + G + D S LK M V+ E LRLY
Sbjct: 329 TTSNLLAWTTMLLSLHQDWQEKLREEIFKECGKEKTPDSETFSKLKLMNMVIMESLRLYG 388
Query: 302 PAPNVQRQAREDIQVDDVTVPNGTNMWIDVVAMHHDPELWGDDVNEFKPERFMDDVNGGC 361
P + R+A +I++ D+ +P GT + I ++ MH D LWG D ++F P RF + V+
Sbjct: 389 PVSALAREASVNIKLGDLEIPKGTTVVIPLLKMHSDKTLWGSDADKFNPMRFANGVSRAA 448
Query: 362 KHKMGYLPFGFGGRMCVGRNLTFMEYKIVLTILLSNFTF-KVSPSYQHSPAIMLSLRPAH 420
H L F G R C+G+N +E K VLT++L F F + Y+H+P ++++P +
Sbjct: 449 NHPNALLAFSVGPRACIGQNFVMIEAKTVLTMILQRFRFISLCDEYKHTPVDNVTIQPQY 508
Query: 421 GLPLIVQPL 429
GLP+++QPL
Sbjct: 509 GLPVMLQPL 517
>At3g14640 putative cytochrome P450
Length = 514
Score = 279 bits (714), Expect = 2e-75
Identities = 139/427 (32%), Positives = 249/427 (57%), Gaps = 11/427 (2%)
Query: 9 GKVFIYWLGTEPFLYIANAEFLKKMSTEVMAKRWGKPSVFRNDRDPMFGSGLVMVEGNDW 68
G+ F WLG +P + I + E +K++ +V + K F R + +G++ +G+ W
Sbjct: 93 GRTFFTWLGPKPTITIMDPELIKEVFNKVYD--YPKAQTFLLGR--LIATGIINYDGDKW 148
Query: 69 VRHRHIVAPAFNPLNLKVMASMMIESTNQMIDRWTSQIN---FGNHEIDMEKEIITIAGE 125
+HR I+ PAF+ +K M +S + ++ W+ ++ + E+D+ ++++ G+
Sbjct: 149 AKHRRIINPAFHIEKIKNMVPAFHQSCSDVVGEWSKLVSDKGSSSCEVDVWPWLVSMTGD 208
Query: 126 IIAKTSFGAEDENAKEVFDKLRALQMTLFNTNRYVGVPFGKYFNVKKNLEAKKLGKEIDK 185
+I++T+FG+ + + +F+ L + V +P +Y K N K +EI
Sbjct: 209 VISRTAFGSSYKEGQRIFELQAELVHLILQAFWKVYIPGYRYLPTKSNRRMKAAAREIQV 268
Query: 186 LLLSIVEAR---KNSPKQNSQKDLLGLLLKENNEDGKLGKTLTSREVVDECKTFFFGGHE 242
+L IV R + + K DLLG+LL+ N K G +++ +V++ECK F+F G E
Sbjct: 269 ILKGIVNKRLRAREAGKAAPNDDLLGILLESNLGQAK-GNGMSTEDVMEECKLFYFAGQE 327
Query: 243 TTALAITWTLLLLATHEDWQNQLREEIKEVVGNNELDITMLSGLKKMKCVMNEVLRLYPP 302
TT++ + W ++LL+ H+DWQ + REE+K+V G+ E D LS LK M ++ EVLRLYPP
Sbjct: 328 TTSVLLVWAMVLLSHHQDWQARAREEVKQVFGDKEPDTECLSQLKVMTMILYEVLRLYPP 387
Query: 303 APNVQRQAREDIQVDDVTVPNGTNMWIDVVAMHHDPELWGDDVNEFKPERFMDDVNGGCK 362
++ R +++++ D+T+P G ++ + ++ + DP LWG D EFKPERF D ++ K
Sbjct: 388 VTHLTRAIDKEMKLGDLTLPAGVHISLPIMLVQRDPMLWGTDAAEFKPERFKDGLSKATK 447
Query: 363 HKMGYLPFGFGGRMCVGRNLTFMEYKIVLTILLSNFTFKVSPSYQHSPAIMLSLRPAHGL 422
++ + PF +G R+C+G+N +E K+ + ++L FTF++SPSY H+P ++++ P G
Sbjct: 448 SQVSFFPFAWGPRICIGQNFAMLEAKMAMALILQTFTFELSPSYVHAPQTVVTIHPQFGA 507
Query: 423 PLIVQPL 429
LI++ L
Sbjct: 508 HLILRKL 514
>At3g14660 putative cytochrome P450
Length = 512
Score = 279 bits (713), Expect = 2e-75
Identities = 141/425 (33%), Positives = 245/425 (57%), Gaps = 9/425 (2%)
Query: 9 GKVFIYWLGTEPFLYIANAEFLKKMSTEVMAKRWGKPSVFRNDRDPMFGSGLVMVEGNDW 68
G+ F W G P + I + E +K++ +V + K F R + +GLV +G+ W
Sbjct: 93 GRTFFTWFGPIPTITIMDPEQIKEVFNKVYD--FQKAHTFPLGR--LIAAGLVSYDGDKW 148
Query: 69 VRHRHIVAPAFNPLNLKVMASMMIESTNQMIDRWTSQINF--GNHEIDMEKEIITIAGEI 126
+HR I+ PAF+ +K M +S ++++ W + + E+D+ ++++ ++
Sbjct: 149 TKHRRIINPAFHLEKIKNMVPAFHQSCSEIVGEWDKLVTDKQSSCEVDIWPWLVSMTADV 208
Query: 127 IAKTSFGAEDENAKEVFDKLRALQMTLFNTNRYVGVPFGKYFNVKKNLEAKKLGKEIDKL 186
I++T+FG+ + + +F+ L + R +P +YF K N K +EI +
Sbjct: 209 ISRTAFGSSYKEGQRIFELQAELAQLIIQAFRKAIIPGYRYFPTKGNRRMKAAAREIKFI 268
Query: 187 LLSIVEARKNSPKQNS--QKDLLGLLLKENNEDGKLGKTLTSREVVDECKTFFFGGHETT 244
L IV R + + DLLG+LL+ N K G +++ E+++ECK F+F G ETT
Sbjct: 269 LRGIVNKRLRAREAGEAPSDDLLGILLESNLGQTK-GNGMSTEELMEECKLFYFAGQETT 327
Query: 245 ALAITWTLLLLATHEDWQNQLREEIKEVVGNNELDITMLSGLKKMKCVMNEVLRLYPPAP 304
+ + WT++LL+ H+DWQ + REE+K+V G+ E D L+ LK M ++ EVLRLYPP
Sbjct: 328 TVLLVWTMVLLSQHQDWQARAREEVKQVFGDKEPDAEGLNQLKVMTMILYEVLRLYPPVV 387
Query: 305 NVQRQAREDIQVDDVTVPNGTNMWIDVVAMHHDPELWGDDVNEFKPERFMDDVNGGCKHK 364
+ R +++Q+ D+T+P G + + ++ + D ELWG+D EFKP+RF D ++ K++
Sbjct: 388 QLTRAIHKEMQLGDLTLPGGVQISLPILLIQRDRELWGNDAGEFKPDRFKDGLSKATKNQ 447
Query: 365 MGYLPFGFGGRMCVGRNLTFMEYKIVLTILLSNFTFKVSPSYQHSPAIMLSLRPAHGLPL 424
+ + PF +G R+C+G+N +E K+ +T++L F+F++SPSY H+P +L+ P G PL
Sbjct: 448 VSFFPFAWGPRICIGQNFALLEAKMAMTLILRKFSFELSPSYVHAPYTVLTTHPQFGAPL 507
Query: 425 IVQPL 429
I+ L
Sbjct: 508 ILHKL 512
>At3g14630 putative cytochrome P450
Length = 508
Score = 279 bits (713), Expect = 2e-75
Identities = 146/425 (34%), Positives = 241/425 (56%), Gaps = 9/425 (2%)
Query: 9 GKVFIYWLGTEPFLYIANAEFLKKMSTEVMAKRWGKPSVFRNDRDPMFGSGLVMVEGNDW 68
GK F W G P + I N + +K EV K + + GL +G+ W
Sbjct: 89 GKTFFTWSGPIPAITIMNPQLIK----EVYNKFYDFEKTHTFPLTSLLTDGLANADGDKW 144
Query: 69 VRHRHIVAPAFNPLNLKVMASMMIESTNQMIDRWTSQINF--GNHEIDMEKEIITIAGEI 126
V+HR I+ PAF+ +K M +S +++ W ++ + E+D+ I+ + G++
Sbjct: 145 VKHRKIINPAFHFEKIKNMVPTFYKSCIEVMCEWEKLVSDKGSSCELDVWPWIVNMTGDV 204
Query: 127 IAKTSFGAEDENAKEVFDKLRALQMTLFNTNRYVGVPFGKYFNVKKNLEAKKLGKEIDKL 186
I++T+FG+ + + +F L + +P ++F K N K + KEI +
Sbjct: 205 ISRTAFGSSYKEGQRIFILQAELAHLIILALGKNYIPAYRHFPTKNNRRMKTIVKEIQVI 264
Query: 187 LLSIVEARKNSPKQNS--QKDLLGLLLKENNEDGKLGKTLTSREVVDECKTFFFGGHETT 244
L I+ R+ + DLLG+LLK N+E K G L E+++ECK F+F G ETT
Sbjct: 265 LRGIISHREKARDAGEAPSDDLLGILLKSNSEQSK-GNGLNMEEIMEECKLFYFAGQETT 323
Query: 245 ALAITWTLLLLATHEDWQNQLREEIKEVVGNNELDITMLSGLKKMKCVMNEVLRLYPPAP 304
++ + WT++LL+ H+DWQ + REE+ +V G+N+ D+ ++ LK M ++ EVLRLYPP
Sbjct: 324 SVLLAWTMVLLSQHQDWQARAREEVMQVFGHNKPDLQGINQLKVMTMIIYEVLRLYPPVI 383
Query: 305 NVQRQAREDIQVDDVTVPNGTNMWIDVVAMHHDPELWGDDVNEFKPERFMDDVNGGCKHK 364
+ R ++I++ D+T+P G + + V+ +H D +LWGDD EFKPERF D + K++
Sbjct: 384 QMNRATHKEIKLGDMTLPGGIQVHMPVLLIHRDTKLWGDDAAEFKPERFKDGIAKATKNQ 443
Query: 365 MGYLPFGFGGRMCVGRNLTFMEYKIVLTILLSNFTFKVSPSYQHSPAIMLSLRPAHGLPL 424
+ +LPFG+G R+C+G+N +E K+ L ++L F+F++SPSY HSP + ++ P G L
Sbjct: 444 VCFLPFGWGPRICIGQNFALLEAKMALALILQRFSFELSPSYVHSPYRVFTIHPQCGAHL 503
Query: 425 IVQPL 429
I+ L
Sbjct: 504 ILHKL 508
>At3g14610 putative cytochrome P450
Length = 512
Score = 278 bits (712), Expect = 3e-75
Identities = 138/426 (32%), Positives = 248/426 (57%), Gaps = 10/426 (2%)
Query: 9 GKVFIYWLGTEPFLYIANAEFLKKMSTEVMAKRWGKPSVFRNDRDPMFGSGLVMVEGNDW 68
GK F W+G P + I N E +K++ +V + K S F R + GL +G+ W
Sbjct: 92 GKTFFIWIGPLPTIVITNPEQIKEVFNKV--NDFEKASTFPLIR--LLAGGLASYKGDKW 147
Query: 69 VRHRHIVAPAFNPLNLKVMASMMIESTNQMIDRWTSQINFGNH--EIDMEKEIITIAGEI 126
HR I+ PAF+ +K M ++++ +W E+D+ ++ + ++
Sbjct: 148 ASHRRIINPAFHLEKIKNMIPAFYHCCSEVVCQWEKLFTDKESPLEVDVWPWLVNMTADV 207
Query: 127 IAKTSFGAEDENAKEVFDKLRALQMTLFNTNRYVGVPFGKYFNVKKNLEAKKLGKEIDKL 186
I+ T+FG+ + + +F L + + +P +++ K N K + +E+D +
Sbjct: 208 ISHTAFGSSYKEGQRIFQLQGELAELIAQAFKKSYIPGSRFYPTKSNRRMKAIDREVDVI 267
Query: 187 LLSIVEARKNSPK--QNSQKDLLGLLLKENNEDGKLGKTLTSREVVDECKTFFFGGHETT 244
L IV R+ + + + + DLLG+LL+ N+E+ + G ++ +V+ ECK F+F G ETT
Sbjct: 268 LRGIVSKREKAREAGEPANDDLLGILLESNSEESQ-GNGMSVEDVMKECKLFYFAGQETT 326
Query: 245 ALAITWTLLLLATHEDWQNQLREEIKEVVG-NNELDITMLSGLKKMKCVMNEVLRLYPPA 303
++ + WT++LL+ H+DWQ + REE+ +V+G NN+ D+ L+ LK M + NEVLRLYPP
Sbjct: 327 SVLLVWTMVLLSHHQDWQARAREEVMQVLGENNKPDMESLNNLKVMTMIFNEVLRLYPPV 386
Query: 304 PNVQRQAREDIQVDDVTVPNGTNMWIDVVAMHHDPELWGDDVNEFKPERFMDDVNGGCKH 363
++R +++++ ++T+P G +++ + + D ELWGDD +FKPERF D ++ K+
Sbjct: 387 AQLKRVVNKEMKLGELTLPAGIQIYLPTILVQRDTELWGDDAADFKPERFRDGLSKATKN 446
Query: 364 KMGYLPFGFGGRMCVGRNLTFMEYKIVLTILLSNFTFKVSPSYQHSPAIMLSLRPAHGLP 423
++ + PFG+G R+C+G+N +E K+ + ++L F+F++SPSY H+P +++ RP G
Sbjct: 447 QVSFFPFGWGPRICIGQNFAMLEAKMAMALILQKFSFELSPSYVHAPQTVMTTRPQFGAH 506
Query: 424 LIVQPL 429
LI+ L
Sbjct: 507 LILHKL 512
>At5g24910 cytochrome P450-like protein
Length = 532
Score = 277 bits (708), Expect = 9e-75
Identities = 148/433 (34%), Positives = 244/433 (56%), Gaps = 20/433 (4%)
Query: 9 GKVFIYWLGTEPFLYIANAEFLKKMSTEVMAKRWGKPSVFRNDRDPMFGSGLVMVEGNDW 68
G+V+ Y G + LY+ + E +K+++ + GK S + G G++ G W
Sbjct: 102 GRVYTYSTGVKQHLYMNHPELVKELN-QANTLNLGKVSYVTKRLKSILGRGVITSNGPHW 160
Query: 69 VRHRHIVAPAFNPLNLKVMASMMIESTNQMIDRWTSQINFGNH---EIDMEKEIITIAGE 125
R I+AP F +K M +++ES M+ +W + +I +++++ + +
Sbjct: 161 AHQRRIIAPEFFLDKVKGMVGLVVESAMPMLSKWEEMMKREGEMVCDIIVDEDLRAASAD 220
Query: 126 IIAKTSFGAEDENAKEVFDKLRALQMT------LFNTNRYVGVPFGKYFNVKKNLEAK-- 177
+I++ FG+ KE+F KLR LQ LF+ N + V FG KK+ K
Sbjct: 221 VISRACFGSSFSKGKEIFSKLRCLQKAITHNNILFSLNGFTDVVFG----TKKHGNGKID 276
Query: 178 KLGKEIDKLLLSIVEARKNSPKQNSQKDLLGLLLK--ENNEDGKLGKTLTSRE--VVDEC 233
+L + I+ L+ V+ R+ + +KDL+ L+L+ ++ DG L S + VVD C
Sbjct: 277 ELERHIESLIWETVKERERECVGDHKKDLMQLILEGARSSCDGNLEDKTQSYKSFVVDNC 336
Query: 234 KTFFFGGHETTALAITWTLLLLATHEDWQNQLREEIKEVVGNNELDITMLSGLKKMKCVM 293
K+ +F GHET+A+A++W L+LLA + WQ ++R+E+ N D +S LK + V+
Sbjct: 337 KSIYFAGHETSAVAVSWCLMLLALNPSWQTRIRDEVFLHCKNGIPDADSISNLKTVTMVI 396
Query: 294 NEVLRLYPPAPNVQRQAREDIQVDDVTVPNGTNMWIDVVAMHHDPELWGDDVNEFKPERF 353
E LRLYPPA V R+A ED ++ ++ VP G +W + +H DPE+WG D NEF PERF
Sbjct: 397 QETLRLYPPAAFVSREALEDTKLGNLVVPKGVCIWTLIPTLHRDPEIWGADANEFNPERF 456
Query: 354 MDDVNGGCKHKMGYLPFGFGGRMCVGRNLTFMEYKIVLTILLSNFTFKVSPSYQHSPAIM 413
+ V+ CKH ++PFG G R+C+G+N ME K+++++++S F+F +SP+YQHSP
Sbjct: 457 SEGVSKACKHPQSFVPFGLGTRLCLGKNFGMMELKVLVSLIVSRFSFTLSPTYQHSPVFR 516
Query: 414 LSLRPAHGLPLIV 426
+ + P HG+ + V
Sbjct: 517 MLVEPQHGVVIRV 529
>At2g46950 putative cytochrome P450
Length = 517
Score = 276 bits (705), Expect = 2e-74
Identities = 150/428 (35%), Positives = 240/428 (56%), Gaps = 12/428 (2%)
Query: 9 GKVFIYWLGTEPFLYIANAEFLKKMSTE---VMAKRWGKPSVFRNDRDPMFGSGLVMVEG 65
G+ F+YW GT+P L I++ E K++ + +K KP + + + G+GL+ V G
Sbjct: 95 GETFLYWQGTDPRLCISDHELAKQILSNKFVFFSKSKTKPEILK-----LSGNGLIFVNG 149
Query: 66 NDWVRHRHIVAPAFNPLNLKVMASMMIESTNQMIDRWTSQINFGNHE--IDMEKEIITIA 123
DWVRHR I+ PAF+ LK+M +M++ T +M W Q N E + + +E +
Sbjct: 150 LDWVRHRRILNPAFSMDKLKLMTQLMVDCTFRMFLEWKKQRNGVETEQFVLISREFKRLT 209
Query: 124 GEIIAKTSFGAEDENAKEVFDKLRALQMTLFNTNRYVGVPFGKYFNVKKNLEAKKLGKEI 183
+IIA +FG+ EVF LQ + P +Y NL+ KL ++
Sbjct: 210 ADIIATAAFGSSYAEGIEVFKSQLELQKCCAAALTDLYFPGIQYLPTPSNLQIWKLDMKV 269
Query: 184 DKLLLSIVEARKNSPKQNSQKDLLGLLLKENNEDGKLGKTLTSREVVDECKTFFFGGHET 243
+ + I++AR S ++ DLLG++L + + + K ++ E+++ECKTFFF GHET
Sbjct: 270 NSSIKRIIDARLTSESKDYGNDLLGIMLTAASSN-ESEKKMSIDEIIEECKTFFFAGHET 328
Query: 244 TALAITWTLLLLATHEDWQNQLREEIKEVVGNNEL-DITMLSGLKKMKCVMNEVLRLYPP 302
TA +TW+ +LL+ H+DWQ +LREE+ G +++ D S LK M V E LRLY P
Sbjct: 329 TANLLTWSTMLLSLHQDWQEKLREEVFNECGKDKIPDAETCSKLKLMNTVFMESLRLYGP 388
Query: 303 APNVQRQAREDIQVDDVTVPNGTNMWIDVVAMHHDPELWGDDVNEFKPERFMDDVNGGCK 362
N+ R A ED+++ ++ +P GT + + + MH D +WG D ++F P RF + ++
Sbjct: 389 VLNLLRLASEDMKLGNLEIPKGTTIILPIAKMHRDKAVWGSDADKFNPMRFANGLSRAAN 448
Query: 363 HKMGYLPFGFGGRMCVGRNLTFMEYKIVLTILLSNFTFKVSPSYQHSPAIMLSLRPAHGL 422
H L F G R C+G+N ME K VL ++L F +S Y+H+PA L+L+P + L
Sbjct: 449 HPNALLAFSMGPRACIGQNFAIMEAKTVLAMILQRFRLNLSADYKHAPADHLTLQPQYDL 508
Query: 423 PLIVQPLN 430
P+I++P++
Sbjct: 509 PVILEPID 516
>At3g14650 putative cytochrome P450
Length = 512
Score = 273 bits (698), Expect = 1e-73
Identities = 135/425 (31%), Positives = 242/425 (56%), Gaps = 9/425 (2%)
Query: 9 GKVFIYWLGTEPFLYIANAEFLKKMSTEVMAKRWGKPSVFRNDRDPMFGSGLVMVEGNDW 68
G+ F W G P + I + E + TEV+ K + + +G++ +G+ W
Sbjct: 93 GRTFFTWFGAIPTITIMDPEQI----TEVLNKVYDFQKAHTFPLGRLIATGVLSYDGDKW 148
Query: 69 VRHRHIVAPAFNPLNLKVMASMMIESTNQMIDRWTSQINF--GNHEIDMEKEIITIAGEI 126
+HR I+ PAF+ +K M +S ++++ +W ++ + E+D+ ++++ ++
Sbjct: 149 AKHRRIINPAFHLEKIKNMVPAFHQSCSEIVCKWDKLVSDKESSCEVDVWPGLVSMTADV 208
Query: 127 IAKTSFGAEDENAKEVFDKLRALQMTLFNTNRYVGVPFGKYFNVKKNLEAKKLGKEIDKL 186
I++T+FG+ + +F+ L + T R +P Y K N K +EI +
Sbjct: 209 ISRTAFGSSCVEGQRIFELQAELAQLIIQTVRKAFIPGYSYLPTKGNRRMKAKAREIQVI 268
Query: 187 LLSIVEARKNSPKQNS--QKDLLGLLLKENNEDGKLGKTLTSREVVDECKTFFFGGHETT 244
L IV R + + DLLG+LL+ N K G +++ ++++ECK F+F G ETT
Sbjct: 269 LRGIVNKRLRAREAGEAPNDDLLGILLESNLGQTK-GNGMSTEDLMEECKLFYFVGQETT 327
Query: 245 ALAITWTLLLLATHEDWQNQLREEIKEVVGNNELDITMLSGLKKMKCVMNEVLRLYPPAP 304
++ + WT++LL+ H+DWQ + REE+K+V G+ E D L+ LK M ++ EVLRLYPP P
Sbjct: 328 SVLLVWTMVLLSQHQDWQARAREEVKQVFGDKEPDAEGLNQLKVMTMILYEVLRLYPPIP 387
Query: 305 NVQRQAREDIQVDDVTVPNGTNMWIDVVAMHHDPELWGDDVNEFKPERFMDDVNGGCKHK 364
+ R +++++ D+T+P G + + ++ + D ELWG+D EFKP+RF D ++ K++
Sbjct: 388 QLSRAIHKEMELGDLTLPGGVLINLPILLVQRDTELWGNDAGEFKPDRFKDGLSKATKNQ 447
Query: 365 MGYLPFGFGGRMCVGRNLTFMEYKIVLTILLSNFTFKVSPSYQHSPAIMLSLRPAHGLPL 424
+ PF +G R+C+G+N +E K+ + ++L F+F++SPSY H+P + ++ P G PL
Sbjct: 448 ASFFPFAWGSRICIGQNFALLEAKMAMALILQRFSFELSPSYVHAPYTVFTIHPQFGAPL 507
Query: 425 IVQPL 429
I+ L
Sbjct: 508 IMHKL 512
>At3g14690 putative cytochrome P450
Length = 512
Score = 270 bits (689), Expect = 1e-72
Identities = 133/430 (30%), Positives = 242/430 (55%), Gaps = 9/430 (2%)
Query: 4 IFVCVGKVFIYWLGTEPFLYIANAEFLKKMSTEVMAKRWGKPSVFRNDRDPMFGSGLVMV 63
+F G+ + W G P + I + E +K++ +V + KP F + GL
Sbjct: 88 MFKTYGRTYFTWFGPIPTITIMDPEQIKEVFNKVYD--FQKPHTF--PLATIIAKGLANY 143
Query: 64 EGNDWVRHRHIVAPAFNPLNLKVMASMMIESTNQMIDRWTSQINF--GNHEIDMEKEIIT 121
+G+ W +HR I+ PAF+ +K M +S +++ W ++ + E+D+ +++
Sbjct: 144 DGDKWAKHRRIINPAFHIEKIKNMVPAFHQSCREVVGEWDQLVSDKGSSCEVDVWPGLVS 203
Query: 122 IAGEIIAKTSFGAEDENAKEVFDKLRALQMTLFNTNRYVGVPFGKYFNVKKNLEAKKLGK 181
+ ++I++T+FG+ + + +F+ L + R +P Y K N K +
Sbjct: 204 MTADVISRTAFGSSYKEGQRIFELQAELAQLIIQAFRKAFIPGYSYLPTKSNRRMKAAAR 263
Query: 182 EIDKLLLSIVEARKNSPKQNS--QKDLLGLLLKENNEDGKLGKTLTSREVVDECKTFFFG 239
EI +L IV R + + DLLG+LL+ N + G +++ ++++ECK F+F
Sbjct: 264 EIQVILRGIVNKRLRAREAGEAPSDDLLGILLESNLRQTE-GNGMSTEDLMEECKLFYFA 322
Query: 240 GHETTALAITWTLLLLATHEDWQNQLREEIKEVVGNNELDITMLSGLKKMKCVMNEVLRL 299
G ETT++ + WT++LL+ H+DWQ + REE+K+V G+ E D L+ LK M ++ EVLRL
Sbjct: 323 GQETTSVLLVWTMVLLSQHQDWQARAREEVKQVFGDKEPDAEGLNQLKVMTMILYEVLRL 382
Query: 300 YPPAPNVQRQAREDIQVDDVTVPNGTNMWIDVVAMHHDPELWGDDVNEFKPERFMDDVNG 359
YPP + R +++++ D+T+P G + + ++ + HD ELWG+D EF P+RF D ++
Sbjct: 383 YPPVTQLTRAIHKELKLGDLTLPGGVQISLPILLVQHDIELWGNDAAEFNPDRFKDGLSK 442
Query: 360 GCKHKMGYLPFGFGGRMCVGRNLTFMEYKIVLTILLSNFTFKVSPSYQHSPAIMLSLRPA 419
K ++ + PF +G R+C+G+N +E K+ + ++L F+F++SPSY H+P ++++ P
Sbjct: 443 ATKSQVSFFPFAWGPRICIGQNFALLEAKMAMALILRRFSFEISPSYVHAPYTVITIHPQ 502
Query: 420 HGLPLIVQPL 429
G LI+ L
Sbjct: 503 FGAQLIMHKL 512
>At3g14680 putative cytochrome P450
Length = 512
Score = 269 bits (688), Expect = 2e-72
Identities = 135/425 (31%), Positives = 243/425 (56%), Gaps = 9/425 (2%)
Query: 9 GKVFIYWLGTEPFLYIANAEFLKKMSTEVMAKRWGKPSVFRNDRDPMFGSGLVMVEGNDW 68
G+ + W G P + I + E +K EV K + + G+GLV +G+ W
Sbjct: 93 GRTNLTWFGPIPTITIMDPEQIK----EVFNKVYDFQKAHTFPLSKILGTGLVSYDGDKW 148
Query: 69 VRHRHIVAPAFNPLNLKVMASMMIESTNQMIDRWTSQINF--GNHEIDMEKEIITIAGEI 126
+HR I+ PAF+ +K M + ES ++++ W ++ + E+D+ + ++ ++
Sbjct: 149 AQHRRIINPAFHLEKIKNMVHVFHESCSELVGEWDKLVSDKGSSCEVDVWPGLTSMTADV 208
Query: 127 IAKTSFGAEDENAKEVFDKLRALQMTLFNTNRYVGVPFGKYFNVKKNLEAKKLGKEIDKL 186
I++T+FG+ +F+ L + + +P Y K N K +EI +
Sbjct: 209 ISRTAFGSSYREGHRIFELQAELAQLVMQAFQKFFIPGYIYLPTKGNRRMKTAAREIQDI 268
Query: 187 LLSIVEARKNSPKQNS--QKDLLGLLLKENNEDGKLGKTLTSREVVDECKTFFFGGHETT 244
L I+ R+ + + +DLLG+LL E+N G +++ ++++ECK F+ G ETT
Sbjct: 269 LRGIINKRERARESGEAPSEDLLGILL-ESNLGQTEGNGMSTEDMMEECKLFYLAGQETT 327
Query: 245 ALAITWTLLLLATHEDWQNQLREEIKEVVGNNELDITMLSGLKKMKCVMNEVLRLYPPAP 304
++ + WT++LL+ H+DWQ + REE+K+V G+ + D L+ LK M ++ EVLRLYPP
Sbjct: 328 SVLLVWTMVLLSQHQDWQARAREEVKQVFGDKQPDTEGLNQLKVMTMILYEVLRLYPPVV 387
Query: 305 NVQRQAREDIQVDDVTVPNGTNMWIDVVAMHHDPELWGDDVNEFKPERFMDDVNGGCKHK 364
+ R +++++ D+T+P G + + V+ +H D ELWG+D EFKPERF D ++ K++
Sbjct: 388 QLTRAIHKEMKLGDLTLPGGVQISLPVLLVHRDTELWGNDAGEFKPERFKDGLSKATKNQ 447
Query: 365 MGYLPFGFGGRMCVGRNLTFMEYKIVLTILLSNFTFKVSPSYQHSPAIMLSLRPAHGLPL 424
+ + PF +G R+C+G+N T +E K+ ++++L F+F++SPSY H+P +++L P G L
Sbjct: 448 VSFFPFAWGPRICIGQNFTLLEAKMAMSLILQRFSFELSPSYVHAPYTIITLYPQFGAHL 507
Query: 425 IVQPL 429
++ L
Sbjct: 508 MLHKL 512
>At4g27710 cytochrome P450 - like protein
Length = 518
Score = 263 bits (671), Expect = 2e-70
Identities = 145/436 (33%), Positives = 239/436 (54%), Gaps = 28/436 (6%)
Query: 9 GKVFIYWLGTEPFLYIANAEFLKKMSTEVMAKRWG-------KPSVFRNDRDPMFGSGLV 61
G F++W GT+P +YI+N E K +V++ ++G +P VF +FG GL
Sbjct: 95 GDTFLFWTGTKPTIYISNHELAK----QVLSSKFGFTIIPVKRPEVFI-----LFGKGLS 145
Query: 62 MVEGNDWVRHRHIVAPAFNPLNLKVMASMMIESTNQMIDRWTSQINFGNH--EIDMEKEI 119
++G+DW+RHR I+ PAF+ LK M M + T ++ + W Q G +I++ KE
Sbjct: 146 FIQGDDWIRHRRILNPAFSMDRLKAMTQPMGDCTLRIFEEWRKQRRNGEVLIKIEISKEF 205
Query: 120 ITIAGEIIAKTSFGAEDENAKEVFDKLRALQMTLFNTNRYVGVPFGKYFNVKKNLEAKKL 179
+ +IIA T+FG+ E+ L+ ++ V +P +Y NL+ +L
Sbjct: 206 HKLTADIIATTAFGSSYAEGIELCRSQTELEKYYISSLTNVFIPGTQYLPTPTNLKLWEL 265
Query: 180 GKEIDKLLLSIVEARKNSPKQNSQ--KDLLGLLL---KENNEDGKLGKTLTSREVVDECK 234
K++ + I+++R S + DLLG++L K N + K+ E+++ECK
Sbjct: 266 HKKVKNSIKRIIDSRLKSKCKTYGYGDDLLGVMLTAAKSNEYERKMRMD----EIIEECK 321
Query: 235 TFFFGGHETTALAITWTLLLLATHEDWQNQLREEIKEVVGNNEL-DITMLSGLKKMKCVM 293
F++ G TT++ +TWT +LL+ H+ WQ +LREE+ G +++ D S LK M V+
Sbjct: 322 NFYYAGQGTTSILLTWTTMLLSLHQGWQEKLREEVFNECGKDKIPDTDTFSKLKLMNMVL 381
Query: 294 NEVLRLYPPAPNVQRQAREDIQVDDVTVPNGTNMWIDVVAMHHDPELWGDDVNEFKPERF 353
E LRLY P + R+A +D++V + +P GT++ I ++ MH D +WG+D +F P RF
Sbjct: 382 MESLRLYGPVIKISREATQDMKVGHLEIPKGTSIIIPLLKMHRDKAIWGEDAEQFNPLRF 441
Query: 354 MDDVNGGCKHKMGYLPFGFGGRMCVGRNLTFMEYKIVLTILLSNFTFKVSPSYQHSPAIM 413
+ ++ H LPF G R C+ +N +E K VLT++L F +SP Y+H+P
Sbjct: 442 ENGISQATIHPNALLPFSIGPRACIAKNFAMVEAKTVLTMILQQFQLSLSPEYKHTPVDH 501
Query: 414 LSLRPAHGLPLIVQPL 429
L P +GLP+++ PL
Sbjct: 502 FDLFPQYGLPVMLHPL 517
>At1g75130 cytochrome P450, putative
Length = 505
Score = 257 bits (657), Expect = 8e-69
Identities = 146/423 (34%), Positives = 238/423 (55%), Gaps = 19/423 (4%)
Query: 9 GKVFIYWLGTEPFLYIANAEFLKK-MSTEVMAKRWGKPSVFRNDRDPMFGSGLVMVEGND 67
GK F+YW G++P + ++ +++ ++T R G + + ++ GL + G+
Sbjct: 90 GKTFLYWFGSKPVVATSDPRLIREALTTGGSFDRIGHNPLSKL----LYAQGLPGLRGDQ 145
Query: 68 WVRHRHIVAPAFNPLNLKVMASMMIESTNQMIDRWTSQINFGNH-EIDMEKEIITIAGEI 126
W HR I AF LK M+ ST ++++W N G E+++ KE+ ++ E+
Sbjct: 146 WAFHRRIAKQAFTMEKLKRWVPQMVTSTMMLMEKWEDMRNGGEEIELEVHKEMHNLSAEM 205
Query: 127 IAKTSFGAEDENAKEVFDKLRALQMTLFNTNRY-VGVPFGKYFNVKKNLEAKKLGKEIDK 185
+++T+FG E K +F+ L+ M LF R+ V +P ++F K N E ++ K+I
Sbjct: 206 LSRTAFGNSVEEGKGIFE-LQERMMRLFYLVRWSVYIPGFRFFPSKTNREIWRIEKQIRV 264
Query: 186 LLLSIVEARKNSPKQNSQ--KDLLGLLLKENNEDGKLGKTLTSREVVDECKTFFFGGHET 243
+L ++E K + +++ + + +N ++ KLG EV DECKTF+F ET
Sbjct: 265 SILKLIENNKTAVEKSGTLLQAFMSPYTNQNGQEEKLG----IEEVTDECKTFYFAAKET 320
Query: 244 TALAITWTLLLLATHEDWQNQLREEIKEVVGNNELD-ITMLSGLKKMKCVMNEVLRLYPP 302
TA +T+ L+LLA +++WQN REE+ V+G L + +L LK + ++NE LRLYPP
Sbjct: 321 TANLMTFVLVLLAMNQEWQNIAREEVICVLGQTGLPTLDILQDLKTLSMIINETLRLYPP 380
Query: 303 APNVQRQAREDIQVDDVTVPNGTNMWIDVVAMHHDPELWGDDVNEFKPERFMDDVNGGCK 362
A + R + ++ D+ +P GT +++ VVAMHHD E WGDD EF P RF D K
Sbjct: 381 AMTLNRDTLKRAKLGDLDIPAGTQLYLSVVAMHHDKETWGDDAEEFNPRRFEDPK----K 436
Query: 363 HKMGYLPFGFGGRMCVGRNLTFMEYKIVLTILLSNFTFKVSPSYQHSPAIMLSLRPAHGL 422
+PFG G R CVG+NL E K VL +L ++F++SPSY H+P + ++L+P +G
Sbjct: 437 QSALLVPFGLGPRTCVGQNLAVNEAKTVLATILKYYSFRLSPSYAHAPVLFVTLQPQNGA 496
Query: 423 PLI 425
L+
Sbjct: 497 HLL 499
>At1g17060 putative cytochrome P450
Length = 476
Score = 214 bits (544), Expect = 1e-55
Identities = 129/423 (30%), Positives = 218/423 (51%), Gaps = 43/423 (10%)
Query: 9 GKVFIYWLGTEPFLYIANAEFLKKMSTEVMAKRWGKPSVFRNDRDPMFGSGLVMVEGNDW 68
GK W G P + + + E L+ E+M+K P + +F SGL+ EG W
Sbjct: 95 GKKCFTWYGPYPNVIVMDPETLR----EIMSKHELFPKPKIGSHNHVFLSGLLNHEGPKW 150
Query: 69 VRHRHIVAPAFNPLNLKVMASMMIESTNQMIDRWTSQINF-GNHEIDMEKEIITIAGEII 127
+HR I+ PAF NLK + S +M++ W + G E+D + ++
Sbjct: 151 SKHRSILNPAFRIDNLKSILPAFNSSCKEMLEEWERLASAKGTMELDSWTHCHDLTRNML 210
Query: 128 AKTSFGAEDENAKEVFDKLRALQMTL-FNTNRYVGVPFGKYFNVKKNLEAKKLGKEIDKL 186
A+ SFG ++ ++F+ ++ Q+ L R V +P K+ K N ++ +++ +
Sbjct: 211 ARASFGDSYKDGIKIFE-IQQEQIDLGLLAIRAVYIPGSKFLPTKFNRRLRETERDMRAM 269
Query: 187 LLSIVEARKNSPKQNSQKDLLGLLLKENNEDGKLGKTLTSREVVDECKTFFFGGHETTAL 246
+++E ++ K+ G+ G+ +TS V
Sbjct: 270 FKAMIETKEEEIKR-----------------GRAGQNVTSSLFV---------------- 296
Query: 247 AITWTLLLLATHEDWQNQLREEIKEVVGNNELDITMLSGLKKMKCVMNEVLRLYPPAPNV 306
WTL+ L+ H+DWQN+ R+EI + GNNE D LS LK + +++EVLRLY PA
Sbjct: 297 ---WTLVALSQHQDWQNKARDEISQAFGNNEPDFEGLSHLKVVTMILHEVLRLYSPAYFT 353
Query: 307 QRQAREDIQVDDVTVPNGTNMWIDVVAMHHDPELWGDDVNEFKPERFMDDVNGGCKHKMG 366
R +++++++ ++P G + I ++ +HHD +LWGDDV EFKPERF + V G K ++
Sbjct: 354 CRITKQEVKLERFSLPEGVVVTIPMLLVHHDSDLWGDDVKEFKPERFANGVAGATKGRLS 413
Query: 367 YLPFGFGGRMCVGRNLTFMEYKIVLTILLSNFTFKVSPSYQHSPAIMLSLRPAHGLPLIV 426
+LPF G R C+G+N + ++ K+ L ++L F+ ++SPSY H+P + P HG LI+
Sbjct: 414 FLPFSSGPRTCIGQNFSMLQAKLFLAMVLQRFSVELSPSYTHAPFPAATTFPQHGAHLII 473
Query: 427 QPL 429
+ L
Sbjct: 474 RKL 476
>At1g31800 unknown protein
Length = 595
Score = 137 bits (346), Expect = 9e-33
Identities = 108/421 (25%), Positives = 194/421 (45%), Gaps = 32/421 (7%)
Query: 4 IFVCVGKVFIYWLGTEPFLYIANAEFLKKMSTEVMAKRWGKPSVFRNDRDPMFGSGLVMV 63
+F+ G +F G + FL +++ K + + AK + K + D + G GL+
Sbjct: 135 LFLTYGGIFRLTFGPKSFLIVSDPSIAKHILKD-NAKAYSK-GILAEILDFVMGKGLIPA 192
Query: 64 EGNDWVRHRHIVAPAFNPLNLKVMASMMIESTNQMIDRWTSQINFGNHEIDMEKEIITIA 123
+G W R R + PA + + M S+ E+++++ + + G E++ME +
Sbjct: 193 DGEIWRRRRRAIVPALHQKYVAAMISLFGEASDRLCQKLDAAALKGE-EVEMESLFSRLT 251
Query: 124 GEIIAKTSFGAEDENA-------KEVFDKLRALQMTLFNTNRYVGVPFGKYFNVKKNLEA 176
+II K F + ++ + V+ LR + + +P K + ++ A
Sbjct: 252 LDIIGKAVFNYDFDSLTNDTGVIEAVYTVLREAEDRSVSPIPVWDIPIWKDISPRQRKVA 311
Query: 177 KKLGKEIDKLLLSIVEARKNSPKQN---------SQKD--LLGLLLKENNEDGKLGKTLT 225
L K I+ L ++ K ++ +++D +L LL G ++
Sbjct: 312 TSL-KLINDTLDDLIATCKRMVEEEELQFHEEYMNERDPSILHFLLAS-------GDDVS 363
Query: 226 SREVVDECKTFFFGGHETTALAITWTLLLLATHEDWQNQLREEIKEVVGNNELDITMLSG 285
S+++ D+ T GHET+A +TWT LL T +L+EE+ V+G+ I +
Sbjct: 364 SKQLRDDLMTMLIAGHETSAAVLTWTFYLLTTEPSVVAKLQEEVDSVIGDRFPTIQDMKK 423
Query: 286 LKKMKCVMNEVLRLYPPAPNVQRQAREDIQVDDVTVPNGTNMWIDVVAMHHDPELWGDDV 345
LK VMNE LRLYP P + R++ ++ + + + G +++I V +H P W DD
Sbjct: 424 LKYTTRVMNESLRLYPQPPVLIRRSIDNDILGEYPIKRGEDIFISVWNLHRSPLHW-DDA 482
Query: 346 NEFKPERF-MDDVN-GGCKHKMGYLPFGFGGRMCVGRNLTFMEYKIVLTILLSNFTFKVS 403
+F PER+ +D N YLPFG G R C+G E + + +L+ F F+++
Sbjct: 483 EKFNPERWPLDGPNPNETNQNFSYLPFGGGPRKCIGDMFASFENVVAIAMLIRRFNFQIA 542
Query: 404 P 404
P
Sbjct: 543 P 543
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.321 0.138 0.416
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 10,120,537
Number of Sequences: 26719
Number of extensions: 449855
Number of successful extensions: 1978
Number of sequences better than 10.0: 262
Number of HSP's better than 10.0 without gapping: 210
Number of HSP's successfully gapped in prelim test: 52
Number of HSP's that attempted gapping in prelim test: 1130
Number of HSP's gapped (non-prelim): 288
length of query: 430
length of database: 11,318,596
effective HSP length: 102
effective length of query: 328
effective length of database: 8,593,258
effective search space: 2818588624
effective search space used: 2818588624
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.8 bits)
S2: 61 (28.1 bits)
Medicago: description of AC131240.9


