

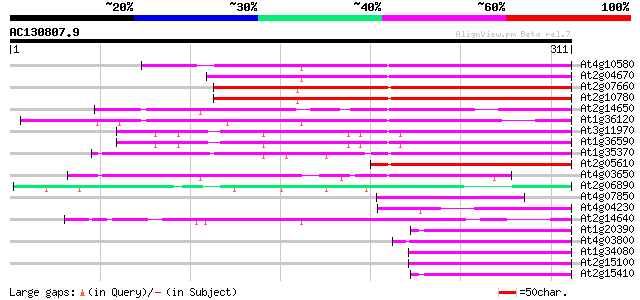
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC130807.9 + phase: 0 /pseudo
(311 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
At4g10580 putative reverse-transcriptase -like protein 169 1e-42
At2g04670 putative retroelement pol polyprotein 169 2e-42
At2g07660 putative retroelement pol polyprotein 164 6e-41
At2g10780 pseudogene 164 7e-41
At2g14650 putative retroelement pol polyprotein 144 5e-35
At1g36120 putative reverse transcriptase gb|AAD22339.1 122 3e-28
At3g11970 hypothetical protein 108 4e-24
At1g36590 hypothetical protein 108 4e-24
At1g35370 hypothetical protein 105 2e-23
At2g05610 putative retroelement pol polyprotein 97 9e-21
At4g03650 putative reverse transcriptase 93 2e-19
At2g06890 putative retroelement integrase 69 4e-12
At4g07850 putative polyprotein 65 6e-11
At4g04230 putative transposon protein 62 3e-10
At2g14640 putative retroelement pol polyprotein 57 1e-08
At1g20390 hypothetical protein 55 6e-08
At4g03800 54 8e-08
At1g34080 putative protein 50 2e-06
At2g15100 putative retroelement pol polyprotein 49 3e-06
At2g15410 putative retroelement pol polyprotein 49 5e-06
>At4g10580 putative reverse-transcriptase -like protein
Length = 1240
Score = 169 bits (429), Expect = 1e-42
Identities = 99/248 (39%), Positives = 136/248 (53%), Gaps = 20/248 (8%)
Query: 74 CVSALGLDLSDMNGEMVVETPTKGSVTTSLVCLRCPLSMFGHDFEMDLVCLHLSGMDVIL 133
C S LD D G+ + V + G DL+ + DVIL
Sbjct: 331 CFSCGSLDHKDAGGQFLAVLGRAKGVDIQIA---------GESMPADLIISPVELYDVIL 381
Query: 134 GMNWLEYNHVHINCFSKSVCFSSAEEE---------SGA*FLSTKQLKQMERDGILMFSL 184
GM+WL+Y VH++ V F E SG+ +S Q ++M G + +
Sbjct: 382 GMDWLDYYRVHLDWHRGRVFFERPEGRLVYQGVRPISGSLVISAVQAEKMIEKGCEAYLV 441
Query: 185 MASLSIE-NQAMIDKL*VVCDFPEVLPDEIPDMPPKREVEFSIDLVPRTKPVSMAPYRMS 243
S+ Q + + VV +F +V + +PP + F+I+L P T P+S APYRM+
Sbjct: 442 TISMPESVGQVAVSDIRVVQEFQDVF-QSLQGLPPSQSDPFTIELEPGTAPLSKAPYRMA 500
Query: 244 ASELAELKKRLEDLLENKFVRPSVSPWGAPVFLVMKKDGSMRLCIDYRQLNKVTIKNRYP 303
+E+AELKK+L+DLL F+RPS SPWGAPV V KKDGS RLCIDYR+LN+VT+KNRYP
Sbjct: 501 PAEMAELKKQLKDLLGKGFIRPSTSPWGAPVLFVKKKDGSFRLCIDYRELNRVTVKNRYP 560
Query: 304 LPRIDDLM 311
LPRID+L+
Sbjct: 561 LPRIDELL 568
>At2g04670 putative retroelement pol polyprotein
Length = 1411
Score = 169 bits (428), Expect = 2e-42
Identities = 92/212 (43%), Positives = 126/212 (59%), Gaps = 11/212 (5%)
Query: 110 LSMFGHDFEMDLVCLHLSGMDVILGMNWLEYNHVHINCFSKSVCFSSAEEE--------- 160
+ + G DL+ + DVILGM+WL++ VH++C V F E
Sbjct: 384 IQIAGESMPADLIISPVELYDVILGMDWLDHYRVHLDCHRGRVSFERPEGRLVYQGVRPT 443
Query: 161 SGA*FLSTKQLKQMERDGILMFSLMASLSIE-NQAMIDKL*VVCDFPEVLPDEIPDMPPK 219
SG+ +S Q K+M G + + S+ Q + + V+ +F +V + +PP
Sbjct: 444 SGSLVISAVQAKKMIEKGCEAYLVTISMPESLGQVAVSDIRVIQEFEDVF-QSLQGLPPS 502
Query: 220 REVEFSIDLVPRTKPVSMAPYRMSASELAELKKRLEDLLENKFVRPSVSPWGAPVFLVMK 279
R F+I+L P T P+S APYRM+ +E+ ELKK+LEDLL F+RPS SPWGAPV V K
Sbjct: 503 RSDPFTIELEPGTAPLSKAPYRMAPAEMTELKKQLEDLLGKGFIRPSTSPWGAPVLFVKK 562
Query: 280 KDGSMRLCIDYRQLNKVTIKNRYPLPRIDDLM 311
KDGS RLCIDYR LN VT+KN+YPLPRID+L+
Sbjct: 563 KDGSFRLCIDYRGLNWVTVKNKYPLPRIDELL 594
>At2g07660 putative retroelement pol polyprotein
Length = 949
Score = 164 bits (415), Expect = 6e-41
Identities = 92/208 (44%), Positives = 127/208 (60%), Gaps = 11/208 (5%)
Query: 114 GHDFEMDLVCLHLSGMDVILGMNWLEYNHVHINCFSKSVCFSSAE---------EESGA* 164
G + DL+ + L DVILGM+WL HI+C + + F E SG+
Sbjct: 25 GVNMPADLIIVPLKKHDVILGMDWLGKYKGHIDCHRERIQFERDEGMLKFQGIRTTSGSL 84
Query: 165 FLSTKQLKQMERDGILMF-SLMASLSIENQAMIDKL*VVCDFPEVLPDEIPDMPPKREVE 223
+S Q ++M G + + + + + A + + +V +F +V + +PP R
Sbjct: 85 VISAIQAERMLGKGCEAYLATITTKEVGASAELKDILIVNEFSDVFA-AVSGVPPDRSDP 143
Query: 224 FSIDLVPRTKPVSMAPYRMSASELAELKKRLEDLLENKFVRPSVSPWGAPVFLVMKKDGS 283
F+I+L P T P+S APYRM+ +E+AELKK+LE+LL F+RPS SPWGAPV V KKDGS
Sbjct: 144 FTIELEPGTTPISKAPYRMAPAEMAELKKQLEELLAKGFIRPSSSPWGAPVLFVKKKDGS 203
Query: 284 MRLCIDYRQLNKVTIKNRYPLPRIDDLM 311
RLCIDYR LNKVT+KN+YPLPRID+LM
Sbjct: 204 FRLCIDYRGLNKVTVKNKYPLPRIDELM 231
>At2g10780 pseudogene
Length = 1611
Score = 164 bits (414), Expect = 7e-41
Identities = 91/208 (43%), Positives = 127/208 (60%), Gaps = 11/208 (5%)
Query: 114 GHDFEMDLVCLHLSGMDVILGMNWLEYNHVHINCFSKSVCFSSAE---------EESGA* 164
G + DL+ + L DVILGM+WL HI+C + F E SG+
Sbjct: 530 GVNMPADLIIVPLKKHDVILGMDWLGKYKAHIDCHRGRIQFERDEGMLKFQGIRTTSGSL 589
Query: 165 FLSTKQLKQMERDGILMF-SLMASLSIENQAMIDKL*VVCDFPEVLPDEIPDMPPKREVE 223
+S Q ++M G + + + + + A + + +V +F +V + +PP R
Sbjct: 590 VISAIQAERMLGKGCEAYLATITTKEVGASAELKDIPIVNEFSDVFA-AVSGVPPDRSDP 648
Query: 224 FSIDLVPRTKPVSMAPYRMSASELAELKKRLEDLLENKFVRPSVSPWGAPVFLVMKKDGS 283
F+I+L P T P+S APYRM+ +E+A+LKK+LE+LL+ F+RPS SPWGAPV V KKDGS
Sbjct: 649 FTIELEPGTTPISKAPYRMAPAEMAKLKKQLEELLDKGFIRPSSSPWGAPVLFVKKKDGS 708
Query: 284 MRLCIDYRQLNKVTIKNRYPLPRIDDLM 311
RLCIDYR LNKVT+KN+YPLPRID+LM
Sbjct: 709 FRLCIDYRGLNKVTVKNKYPLPRIDELM 736
>At2g14650 putative retroelement pol polyprotein
Length = 1328
Score = 144 bits (364), Expect = 5e-35
Identities = 97/267 (36%), Positives = 138/267 (51%), Gaps = 30/267 (11%)
Query: 48 GTCYINNTPLVAIIDTGATHCFIAFDCVSALGLDLSDMNGEMVVETPTKGSVTTSLV--C 105
GT + + D+GA+HCFI + SA ++ GE + G +++
Sbjct: 304 GTLLVGGVEAHVLFDSGASHCFITPE--SASRGNIRGDPGEQLGAVKVAGGQFVAVLGRT 361
Query: 106 LRCPLSMFGHDFEMDLVCLHLSGMDVILGMNWLEYNHVHINCFSKSVCFSSAEEESGA*F 165
+ + G DL+ + DVILGM+WL++ VH++C V F E
Sbjct: 362 KGVDIQIAGESMPADLIISPVELYDVILGMDWLDHYRVHLDCHRGRVSFERPEG------ 415
Query: 166 LSTKQLKQMERDGILMFSLMASLSIENQAMIDK-L*VVCDFPEVLPDEIPDMPPKREVEF 224
S G L+ S +++ + MI+K +F +V + +PP R F
Sbjct: 416 -SLVYQGVRPTSGSLVIS-----AVQAEKMIEKGCEAYLEFEDVF-QSLQGLPPSRSDPF 468
Query: 225 SIDLVPRTKPVSMAPYRMSASELAELKKRLEDLLENKFVRPSVSPWGAPVFLVMKKDGSM 284
+I+L P T P+S APYRM+ +E+AELKK+LEDLL GAPV V KKDGS
Sbjct: 469 TIELEPGTAPLSKAPYRMAPAEMAELKKQLEDLL------------GAPVLFVKKKDGSF 516
Query: 285 RLCIDYRQLNKVTIKNRYPLPRIDDLM 311
RLCIDYR LN VT+KN+YPLPRID+L+
Sbjct: 517 RLCIDYRGLNWVTVKNKYPLPRIDELL 543
>At1g36120 putative reverse transcriptase gb|AAD22339.1
Length = 1235
Score = 122 bits (305), Expect = 3e-28
Identities = 96/328 (29%), Positives = 152/328 (46%), Gaps = 44/328 (13%)
Query: 7 FNCNEEGHIGSQCKQPKRAPTTGRVFAMAGTQTENEDRLIR----GTCYINNTPLVA--- 59
F + G GS + G + ++ E + GT ++ L++
Sbjct: 288 FRAGQGGRAGSAADDSSTSTAGGAAQSAGAARSAAECAAVSTYCSGTTWLYYADLISGRC 347
Query: 60 ----IIDTGATHCFIAFDCVSALGLDLSDMNGEMVVETPTKGSVTTSLVCLRCPLSMFGH 115
+ D+GA+HCFI + SA ++ GE G +++ +++
Sbjct: 348 RGHVLFDSGASHCFITPE--SASRDNIRGEPGEQFGAVKIAGGQFLTVLGRAKGVNIQIE 405
Query: 116 DFEM--DLVCLHLSGMDVILGMNWLEYNHVHINCFSKSVCFSSAEEE---------SGA* 164
+ M DL+ + D ILGM+WL++ VH++C V F E S +
Sbjct: 406 EESMPADLIINPVVLYDAILGMDWLDHYRVHLDCHRGRVSFERPEGRLVYQGVRPTSRSL 465
Query: 165 FLSTKQLKQMERDGILMFSLMASLSIE-NQAMIDKL*VVCDFPEVLPDEIPDMPPKREVE 223
+S Q+++M G + + S+S Q + + VV +F +V + +PP R
Sbjct: 466 VISEVQVEKMIEKGCEAYLVTISMSESVGQVAVSDIWVVQEFEDVF-QSLQGLPPSRSDP 524
Query: 224 FSIDLVPRTKPVSMAPYRMSASELAELKKRLEDLLENKFVRPSVSPWGAPVFLVMKKDGS 283
F+I+L T P+S PYRM +E+AELKK+LEDLL F+RPS S WGAP
Sbjct: 525 FTIELELGTAPLSKTPYRMVPAEIAELKKQLEDLLGKGFIRPSTSRWGAP---------- 574
Query: 284 MRLCIDYRQLNKVTIKNRYPLPRIDDLM 311
LN+VT+KN+YPLPRID+L+
Sbjct: 575 --------GLNRVTLKNKYPLPRIDELL 594
>At3g11970 hypothetical protein
Length = 1499
Score = 108 bits (270), Expect = 4e-24
Identities = 86/282 (30%), Positives = 142/282 (49%), Gaps = 37/282 (13%)
Query: 60 IIDTGATHCFIAFDCVSALG--LDLSDMNGEMVVE---TPTKGSVTTSLVCLRCPLSMFG 114
+ID+G+TH F+ + + LG +D + + V + +G VT L+
Sbjct: 404 LIDSGSTHNFLDPNTAAKLGCKVDTAGLTRVSVADGRKLRVEGKVTDFSWKLQTTT---- 459
Query: 115 HDFEMDLVCLHLSGMDVILGMNWLE--------YNHVHINC-FSKSVCFSSAEEESGA*F 165
F+ D++ + L G+D++LG+ WLE + + + F+
Sbjct: 460 --FQSDILLIPLQGIDMVLGVQWLETLGRISWEFKKLEMRFKFNNQKVLLHGLTSGSVRE 517
Query: 166 LSTKQLKQMERDGILMFSLMA---SLSIENQ-----AMIDKL*VVCDFPEVLPDEIPD-- 215
+ ++L++++ D + + L S S E + A+ +L EVL +E PD
Sbjct: 518 VKAQKLQKLQEDQVQLAMLCVQEVSESTEGELCTINALTSELGEESVVEEVL-NEYPDIF 576
Query: 216 -----MPPKREVE-FSIDLVPRTKPVSMAPYRMSASELAELKKRLEDLLENKFVRPSVSP 269
+PP RE I L+ + PV+ PYR S + E+ K +EDLL N V+ S SP
Sbjct: 577 IEPTALPPFREKHNHKIKLLEGSNPVNQRPYRYSIHQKNEIDKLVEDLLTNGTVQASSSP 636
Query: 270 WGAPVFLVMKKDGSMRLCIDYRQLNKVTIKNRYPLPRIDDLM 311
+ +PV LV KKDG+ RLC+DYR+LN +T+K+ +P+P I+DLM
Sbjct: 637 YASPVVLVKKKDGTWRLCVDYRELNGMTVKDSFPIPLIEDLM 678
>At1g36590 hypothetical protein
Length = 1499
Score = 108 bits (270), Expect = 4e-24
Identities = 86/282 (30%), Positives = 142/282 (49%), Gaps = 37/282 (13%)
Query: 60 IIDTGATHCFIAFDCVSALG--LDLSDMNGEMVVE---TPTKGSVTTSLVCLRCPLSMFG 114
+ID+G+TH F+ + + LG +D + + V + +G VT L+
Sbjct: 404 LIDSGSTHNFLDPNTAAKLGCKVDTAGLTRVSVADGRKLRVEGKVTDFSWKLQTTT---- 459
Query: 115 HDFEMDLVCLHLSGMDVILGMNWLE--------YNHVHINC-FSKSVCFSSAEEESGA*F 165
F+ D++ + L G+D++LG+ WLE + + + F+
Sbjct: 460 --FQSDILLIPLQGIDMVLGVQWLETLGRISWEFKKLEMRFKFNNQKVLLHGLTSGSVRE 517
Query: 166 LSTKQLKQMERDGILMFSLMA---SLSIENQ-----AMIDKL*VVCDFPEVLPDEIPD-- 215
+ ++L++++ D + + L S S E + A+ +L EVL +E PD
Sbjct: 518 VKAQKLQKLQEDQVQLAMLCVQEVSESTEGELCTINALTSELGEESVVEEVL-NEYPDIF 576
Query: 216 -----MPPKREVE-FSIDLVPRTKPVSMAPYRMSASELAELKKRLEDLLENKFVRPSVSP 269
+PP RE I L+ + PV+ PYR S + E+ K +EDLL N V+ S SP
Sbjct: 577 IEPTALPPFREKHNHKIKLLEGSNPVNQRPYRYSIHQKNEIDKLVEDLLTNGTVQASSSP 636
Query: 270 WGAPVFLVMKKDGSMRLCIDYRQLNKVTIKNRYPLPRIDDLM 311
+ +PV LV KKDG+ RLC+DYR+LN +T+K+ +P+P I+DLM
Sbjct: 637 YASPVVLVKKKDGTWRLCVDYRELNGMTVKDSFPIPLIEDLM 678
>At1g35370 hypothetical protein
Length = 1447
Score = 105 bits (263), Expect = 2e-23
Identities = 83/295 (28%), Positives = 144/295 (48%), Gaps = 37/295 (12%)
Query: 46 IRGTCYINNTPLVAIIDTGATHCFIAFDCVSALGLDLSDMN-GEMVVETPTKGSVTTSLV 104
++GT ++ L +ID+G+TH FI + LG + ++ V K +V +
Sbjct: 371 VKGT--VDKRDLFILIDSGSTHNFIDSTVAAKLGCHVESAGLTKVAVADGRKLNVDGQIK 428
Query: 105 CLRCPLSMFGHDFEMDLVCLHLSGMDVILGMNWLE--------YNHVHINCFSKSV---- 152
L F+ D++ + L G+D++LG+ WLE + + + F K+
Sbjct: 429 GFTWKLQ--STTFQSDILLIPLQGVDMVLGVQWLETLGRISWEFKKLEMQFFYKNQRVWL 486
Query: 153 --CFSSAEEESGA*FLSTKQLKQM-------------ERDGILMFSLMASLSIENQAMID 197
+ + + A L Q Q+ E I S + S +E + +
Sbjct: 487 HGIITGSVRDIKAHKLQKTQADQIQLAMVCVREVVSDEEQEIGSISALTSDVVEESVVQN 546
Query: 198 KL*VVCDFPEVLPDEIPDMPPKREV-EFSIDLVPRTKPVSMAPYRMSASELAELKKRLED 256
+V +FP+V E D+PP RE + I L+ PV+ PYR + E+ K ++D
Sbjct: 547 ---IVEEFPDVFA-EPTDLPPFREKHDHKIKLLEGANPVNQRPYRYVVHQKDEIDKIVQD 602
Query: 257 LLENKFVRPSVSPWGAPVFLVMKKDGSMRLCIDYRQLNKVTIKNRYPLPRIDDLM 311
++++ ++ S SP+ +PV LV KKDG+ RLC+DY +LN +T+K+R+ +P I+DLM
Sbjct: 603 MIKSGTIQVSSSPFASPVVLVKKKDGTWRLCVDYTELNGMTVKDRFLIPLIEDLM 657
>At2g05610 putative retroelement pol polyprotein
Length = 780
Score = 97.4 bits (241), Expect = 9e-21
Identities = 48/112 (42%), Positives = 76/112 (67%), Gaps = 2/112 (1%)
Query: 201 VVCDFPEVLPDEIPDMPPKREV-EFSIDLVPRTKPVSMAPYRMSASELAELKKRLEDLLE 259
VV +FP++ E D+PP R + I+L+ + PV+ PYR + E+ K ++D+L
Sbjct: 10 VVTEFPDIFV-EPTDLPPFRAPHDHKIELLEDSNPVNQRPYRYVVHQKNEIDKIVDDMLA 68
Query: 260 NKFVRPSVSPWGAPVFLVMKKDGSMRLCIDYRQLNKVTIKNRYPLPRIDDLM 311
+ ++ S SP+ +PV LV KKDG+ RLC+DYR+LN +T+K+R+P+P I+DLM
Sbjct: 69 SGTIQASSSPYASPVVLVKKKDGTWRLCVDYRELNGMTVKDRFPIPLIEDLM 120
>At4g03650 putative reverse transcriptase
Length = 839
Score = 92.8 bits (229), Expect = 2e-19
Identities = 74/253 (29%), Positives = 120/253 (47%), Gaps = 21/253 (8%)
Query: 33 AMAGTQTENEDRLIRGTCYINNTPLVAIIDTGATHCFIAFDCVSALGLDLSDMNGEMVVE 92
A+ T E E+ L R + +P V T HC + SA ++ GE +
Sbjct: 300 ALVETAAEVEEDLQRQV--VGVSPAVQPKKTQQQHCIASLPPESASRGNIRGDPGEQLGA 357
Query: 93 TPTKGSVTTSLV--CLRCPLSMFGHDFEMDLVCLHLSGMDVILGMNWLEYNHVHINCFSK 150
G +++ + + G DL+ H+ DVILGM+WL++ VH++C
Sbjct: 358 VKVAGGQFLAVLGGAKGVDIQIAGESLPADLIISHVELYDVILGMDWLDHYRVHLDCHRG 417
Query: 151 SVCFSSAEEESGA*FLSTKQLKQMERDGILMF---SLMASLSIENQAMIDKL*VVCDFPE 207
V F E +G ++ +R+G+ + A + Q + + VV +F
Sbjct: 418 RVSFERPEGSAG---------RENDREGLRGLPGDDIYAGVC--GQVAVSDIRVVQEFQY 466
Query: 208 VLPDEIPDMPPKREVEFSIDLVPRTKPVSMAPYRMSASELAELKKRLEDLLENKFVRPSV 267
V + +PP R F+I+L P+T P+S APYRM+ +++AELKK+LEDLL VR +
Sbjct: 467 VF-QSLQGLPPSRSDPFTIELEPKTAPLSKAPYRMAPAKMAELKKQLEDLLAEADVRKTA 525
Query: 268 --SPWGAPVFLVM 278
+ +G F+VM
Sbjct: 526 FRTRYGHFEFVVM 538
>At2g06890 putative retroelement integrase
Length = 1215
Score = 68.6 bits (166), Expect = 4e-12
Identities = 81/380 (21%), Positives = 141/380 (36%), Gaps = 110/380 (28%)
Query: 3 DIVCFNCNEEGHIGSQC---------------------------KQPKRAPTTGRVFAMA 35
D+ C+ C +GH ++C K+ + P G +
Sbjct: 156 DVRCYKCQGKGHYANECPNKRVMILLDNGEIEPEEEIPDSPSSLKENEELPAQGELLVAR 215
Query: 36 GT--------QTENEDRLIRGTCYINNTPLVAIIDTGATHCFIAFDCVSALGLDLSDMNG 87
T + E L C+++ IID G+ + V LGL + +G
Sbjct: 216 RTLSVQTKTDEQEQRKNLFHTRCHVHGKVCSLIIDGGSCTNVASETMVKKLGLKWLNDSG 275
Query: 88 EMVVETPTKGSVTTSLVCLRCPLSMFGHDFEMDLVC--LHLSGMDVILGMNWLEYNHVHI 145
+M V K V +V + +E +++C L + ++LG W V
Sbjct: 276 KMRV----KNQVVVPIVIGK---------YEDEILCDVLPMEAGHILLGRPWQSDRKVMH 322
Query: 146 NCFS----------KSVCFSSAEEESGA*FLSTKQLKQM-------------------ER 176
+ F+ K++ S E + KQ K+ +
Sbjct: 323 DGFTNRHSFEFKGGKTILVSMTPHEVYQDQIHLKQKKEQVVKQPNFFAKSGEVKSAYSSK 382
Query: 177 DGILMFSLMASL-SIENQAMI---DKL*VVCDFPEVLPDEIP-DMPPKREVEFSIDLVPR 231
+L+F +L S+ N A + + ++ D+ +V P++ P +PP R +E ID VP
Sbjct: 383 QPMLLFVFKEALTSLTNFAPVLPSEMTSLLQDYKDVFPEDNPKGLPPIRGIEHQIDFVPG 442
Query: 232 TKPVSMAPYRMSASELAELKKRLEDLLENKFVRPSVSPWGAPVFLVMKKDGSMRLCIDYR 291
+ YR + E EL++ +KDGS R+C D R
Sbjct: 443 ASLPNRPAYRTNPVETKELQR--------------------------QKDGSWRMCFDCR 476
Query: 292 QLNKVTIKNRYPLPRIDDLM 311
+N VT+K +P+PR+DD++
Sbjct: 477 AINNVTVKYCHPIPRLDDML 496
>At4g07850 putative polyprotein
Length = 1138
Score = 64.7 bits (156), Expect = 6e-11
Identities = 34/83 (40%), Positives = 49/83 (58%), Gaps = 1/83 (1%)
Query: 204 DFPEVLPDEIPD-MPPKREVEFSIDLVPRTKPVSMAPYRMSASELAELKKRLEDLLENKF 262
D+ +V P+E P+ +PP R +E ID VP + YR + E EL+K++ +L+E
Sbjct: 447 DYQDVFPEENPEGLPPIRGIEHQIDFVPGASLPNRPAYRTNPVETKELEKQVNELMERGH 506
Query: 263 VRPSVSPWGAPVFLVMKKDGSMR 285
+ S+SP PV LV KKDGS R
Sbjct: 507 ICESMSPCAVPVLLVPKKDGSWR 529
>At4g04230 putative transposon protein
Length = 315
Score = 62.4 bits (150), Expect = 3e-10
Identities = 37/111 (33%), Positives = 54/111 (48%), Gaps = 22/111 (19%)
Query: 205 FPEVLPDEIPDMPPKREVEFSI----DLVPRTKPVSMAPYRMSASELAELKKRLEDLLEN 260
F E L I D EV+ + DL P P + P +
Sbjct: 86 FKECLSTGIGDPELSAEVQVVLHWYKDLFPEEIPPGLPP------------------IHK 127
Query: 261 KFVRPSVSPWGAPVFLVMKKDGSMRLCIDYRQLNKVTIKNRYPLPRIDDLM 311
++R S+SP PV LV KKDG+ R+C+D R +N +TIK R+P+PR+ D++
Sbjct: 128 GYIRESLSPCAVPVLLVPKKDGTWRMCLDCRAINNITIKYRHPIPRLYDML 178
>At2g14640 putative retroelement pol polyprotein
Length = 945
Score = 57.0 bits (136), Expect = 1e-08
Identities = 72/297 (24%), Positives = 124/297 (41%), Gaps = 56/297 (18%)
Query: 31 VFAMAGTQTENEDRLIRGTCYINNTPLVAIIDTGATHCFIAFDCVSALGLDLSDMNGEMV 90
++A+ G T + R +R T + N L+A+I++G+TH FI V MN +
Sbjct: 306 LYALEGVDTTSTIR-VRATIHRNR--LIALINSGSTHNFIGEKAVRG-------MNLKAT 355
Query: 91 VETPTKGSVTTS--LVCLR----CPLSMFGHDFEMDLVCLHLSGMDVILGMNWLE-YNHV 143
P V LVC P+ M G F + L L L G+D+ +G+ WL
Sbjct: 356 TTKPFTVRVVNGMPLVCRSRYEAIPVVMGGVVFPVTLYALPLMGLDLAMGVQWLSTLGPT 415
Query: 144 HINCFSKSVCFSSAEEE--------SGA*FLSTKQLKQMERDGILMFSL-MASLSIENQA 194
N +++ F A +E +G + K + + R G +F++ MA S +
Sbjct: 416 LCNWKEQTLQFHWAGDEVRLMGIKPTGLRGVEHKTITKKARMGHTIFAITMAHNSSDPNT 475
Query: 195 MIDKL*VVCDFPEVLPDEIPDMPPKREVEFSIDLVPRTKPVSMAPYRMSASELAELKKRL 254
++ + + L + +P +R + I L T PV++ PYR + + E+ +
Sbjct: 476 PDGEIKKLIEEFAGLFETPTQLPSERPIVHRIALKKGTDPVNVRPYRYAFYQKDEMSRA- 534
Query: 255 EDLLENKFVRPSVSPWGAPVFLVMKKDGSMRLCIDYRQLNKVTIKNRYPLPRIDDLM 311
+RPS SP+ +PV L R+P+P +DD++
Sbjct: 535 ------GIIRPSSSPFLSPVLL-----------------------ERFPIPTVDDML 562
>At1g20390 hypothetical protein
Length = 1791
Score = 54.7 bits (130), Expect = 6e-08
Identities = 30/90 (33%), Positives = 46/90 (50%), Gaps = 3/90 (3%)
Query: 223 EFSIDLVPRTKPVSMAPYRMSASELAELKKRLEDLLE-NKFVRPSVSPWGAPVFLVMKKD 281
E ++D P KPV ++ + + +E LL+ + + W A +V KK+
Sbjct: 768 ELNVD--PTFKPVKQKRRKLGPERARAVNEEVEKLLKAGQIIEVKYPEWLANPVVVKKKN 825
Query: 282 GSMRLCIDYRQLNKVTIKNRYPLPRIDDLM 311
G R+C+DY LNK K+ YPLP ID L+
Sbjct: 826 GKWRVCVDYTDLNKACPKDSYPLPHIDRLV 855
>At4g03800
Length = 637
Score = 54.3 bits (129), Expect = 8e-08
Identities = 31/100 (31%), Positives = 53/100 (53%), Gaps = 2/100 (2%)
Query: 213 IPDMPPKREVEFSIDLVPRTKPVSMAPYRMSASELAELKKRLEDLLENKFVRPSVSP-WG 271
+PD+ P + +++ PR KP+ ++ + +++ LL+ +R P W
Sbjct: 321 MPDIDPSI-ICHELNVDPRFKPLKQKRRKLGVERAKAVNGKIDKLLKIGSIREVQYPDWV 379
Query: 272 APVFLVMKKDGSMRLCIDYRQLNKVTIKNRYPLPRIDDLM 311
A +V KK+G R+CID+ LNK K+ +PLP ID L+
Sbjct: 380 AITVVVKKKNGKDRVCIDFTDLNKACPKDSFPLPHIDRLV 419
>At1g34080 putative protein
Length = 811
Score = 49.7 bits (117), Expect = 2e-06
Identities = 27/91 (29%), Positives = 45/91 (48%), Gaps = 1/91 (1%)
Query: 222 VEFSIDLVPRTKPVSMAPYRMSASELAELKKRLEDLLENKFVRPSVSP-WGAPVFLVMKK 280
+ +++ P KPV ++ + + LL+ +R P W A +V KK
Sbjct: 636 ISHDLNVDPTLKPVKQKRRKLGPDRAEAVNAEVVRLLKAGLIREVKYPEWLANPVVVKKK 695
Query: 281 DGSMRLCIDYRQLNKVTIKNRYPLPRIDDLM 311
+G R+C+D+ LNK K+ +PLP ID L+
Sbjct: 696 NGKWRVCVDFTDLNKACPKDFFPLPHIDRLV 726
>At2g15100 putative retroelement pol polyprotein
Length = 1329
Score = 48.9 bits (115), Expect = 3e-06
Identities = 28/91 (30%), Positives = 45/91 (48%), Gaps = 1/91 (1%)
Query: 222 VEFSIDLVPRTKPVSMAPYRMSASELAELKKRLEDLLENKFVRPSVSP-WGAPVFLVMKK 280
+ +++ P KPV ++ + + LLE +R P W A +V KK
Sbjct: 420 ISHELNVDPTFKPVKQKRRKLGPDRAQAVNIEVVRLLEVGRIREVKYPEWLANPVVVKKK 479
Query: 281 DGSMRLCIDYRQLNKVTIKNRYPLPRIDDLM 311
+G R+C+D+ LNK K+ +PLP ID L+
Sbjct: 480 NGKWRVCVDFTDLNKACSKDFFPLPHIDRLV 510
>At2g15410 putative retroelement pol polyprotein
Length = 1787
Score = 48.5 bits (114), Expect = 5e-06
Identities = 27/90 (30%), Positives = 45/90 (50%), Gaps = 3/90 (3%)
Query: 223 EFSIDLVPRTKPVSMAPYRMSASELAELKKRLEDLLE-NKFVRPSVSPWGAPVFLVMKKD 281
E ++D P KP+ ++ ++ + ++ LL+ V W +V KK+
Sbjct: 773 ELNVD--PTYKPLKQKRRKLGPDRTKDVNEEVKKLLDAGSIVEVRYPDWLRNPVVVKKKN 830
Query: 282 GSMRLCIDYRQLNKVTIKNRYPLPRIDDLM 311
G R+CID+ LNK K+ +PLP ID L+
Sbjct: 831 GKWRVCIDFTDLNKACPKDSFPLPHIDRLV 860
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.326 0.140 0.430
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 7,132,879
Number of Sequences: 26719
Number of extensions: 301552
Number of successful extensions: 975
Number of sequences better than 10.0: 53
Number of HSP's better than 10.0 without gapping: 42
Number of HSP's successfully gapped in prelim test: 11
Number of HSP's that attempted gapping in prelim test: 880
Number of HSP's gapped (non-prelim): 81
length of query: 311
length of database: 11,318,596
effective HSP length: 99
effective length of query: 212
effective length of database: 8,673,415
effective search space: 1838763980
effective search space used: 1838763980
T: 11
A: 40
X1: 15 ( 7.1 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 40 (21.6 bits)
S2: 60 (27.7 bits)
Medicago: description of AC130807.9


