

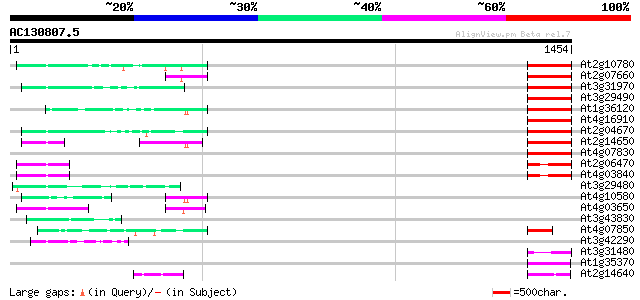
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC130807.5 + phase: 0 /pseudo
(1454 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
At2g10780 pseudogene 163 8e-40
At2g07660 putative retroelement pol polyprotein 155 1e-37
At3g31970 hypothetical protein 154 5e-37
At3g29490 hypothetical protein 152 1e-36
At1g36120 putative reverse transcriptase gb|AAD22339.1 150 5e-36
At4g16910 retrotransposon like protein 150 7e-36
At2g04670 putative retroelement pol polyprotein 148 3e-35
At2g14650 putative retroelement pol polyprotein 146 1e-34
At4g07830 putative reverse transcriptase 145 1e-34
At2g06470 putative retroelement pol polyprotein 119 1e-26
At4g03840 putative transposon protein 114 5e-25
At3g29480 hypothetical protein 84 8e-16
At4g10580 putative reverse-transcriptase -like protein 82 2e-15
At4g03650 putative reverse transcriptase 75 4e-13
At3g43830 putative protein 74 8e-13
At4g07850 putative polyprotein 68 3e-11
At3g42290 putative protein 68 3e-11
At3g31480 hypothetical protein 66 2e-10
At1g35370 hypothetical protein 61 5e-09
At2g14640 putative retroelement pol polyprotein 60 9e-09
>At2g10780 pseudogene
Length = 1611
Score = 163 bits (412), Expect = 8e-40
Identities = 156/579 (26%), Positives = 237/579 (39%), Gaps = 114/579 (19%)
Query: 17 AQAVEQQPQASNGEVR-----MLETFLRNHPPAFKGMYDPDGAQSWLKEVERIFRAMQCS 71
++ VE PQ ++G R +LE R F G DP A W ++R F++ +C
Sbjct: 149 SEVVEVNPQPNHGRKRGDYLSLLEHVSRLGTRHFMGSTDPIVADEWRSRLKRNFKSTRCP 208
Query: 72 EVKKVRFGTHMLAEEADDWWVSLLPVLEQDEAVVTWAVFRREFLNRYFLEDV*GRKEIEF 131
E + H L +A +WW+++ + + V ++A F EF +YF + R E +
Sbjct: 209 EDYQRDIAVHFLEGDAHNWWLTV--EKRRGDEVRSFADFEDEFNKKYFPPEAWDRLECAY 266
Query: 132 LELK*GDISVVEYAAKFVELAKFYPHYTAETAEFSKCIKFENGLRANIKRAIGYQQIRIF 191
L+L G+ +V EY +F L ++ E E ++ +F GLR I+ +
Sbjct: 267 LDLVQGNRTVREYDEEFNRLRRYVGRELEE--EQAQLRRFIRGLRIEIRNHCLVRTFNSV 324
Query: 192 SDLVSRCRICEEDTKAHYKVMSERRGKGRQSRP---KPYSAPADKGKQRLNDERRPNRRD 248
S+LV R + EE + E R R+ P + PAD K+R D+ + D
Sbjct: 325 SELVERAAMIEEG-------IEEERYLNREKAPIRNNQSTKPAD--KKRKFDKVDNTKSD 375
Query: 249 ASAEIVCFKCGEKGHKSNVCTKDEKKCFRCGQKGHLLADCKRGDV-----------VCYN 297
A C CG K H S C K C RCG K H + C + + C+
Sbjct: 376 AKTG-ECVTCG-KNH-SGTCWKAIGACGRCGSKDHAIQSCPKMEPGQSKVLGEETRTCFY 432
Query: 298 CNEEGHISTQCTQPKKDRTGGKVFALTGTQTTNEDRLIRGTCFFNSTPLIAIIDTGATHC 357
C + GH+ +C + ++ G+ R RG N P A+
Sbjct: 433 CGKTGHLKRECPKLTAEKQAGQ-------------RDNRGG---NGLPPPPKRQAVASRV 476
Query: 358 FIALESAYKLGLIVSDMKGEMVVETPAKGSVTTSLVCLRCPTSMF--------GRDFVVD 409
+ E A G + G G V + PT + G + D
Sbjct: 477 YELSEEANDAGNFRAITGGFRKEPNTDYGMVRAAGGQAMYPTGLVRGISVVVNGVNMPAD 536
Query: 410 LVCLPLTGMDVIFGMNWLEYNRVHINCFSKTMHF-------------------------- 443
L+ +PL DVI GM+WL + HI+C + F
Sbjct: 537 LIIVPLKKHDVILGMDWLGKYKAHIDCHRGRIQFERDEGMLKFQGIRTTSGSLVISAIQA 596
Query: 444 ---------------------SSAEEEEV-----FPD---EIPDVPPEREVEFSIDLVPG 474
+SAE +++ F D + VPP+R F+I+L PG
Sbjct: 597 ERMLGKGCEAYLATITTKEVGASAELKDIPIVNEFSDVFAAVSGVPPDRSDPFTIELEPG 656
Query: 475 TKPMSMAPYRMSSSELAELKKQLED*LDKKFVRPNVSPW 513
T P+S APYRM+ +E+A+LKKQLE+ LDK F+RP+ SPW
Sbjct: 657 TTPISKAPYRMAPAEMAKLKKQLEELLDKGFIRPSSSPW 695
Score = 152 bits (384), Expect = 1e-36
Identities = 68/113 (60%), Positives = 90/113 (79%)
Query: 1342 RTDGQSERTIQSLEDLLRVCVLEQGGTWDSHLPLIEFTYNNGYHSSIGMAPFEALYGQRC 1401
+TD QSERTIQ+LED+LR CVL+ GG W+ +L L+EF YNN + +SIGM+P+EALYG+ C
Sbjct: 1330 QTDEQSERTIQTLEDMLRACVLDWGGNWEKYLRLVEFAYNNSFQASIGMSPYEALYGRAC 1389
Query: 1402 RTPLCWFESGERVVLGPEIVQQTTEKVKMIQEKMKVSHSRQKSYHDKRRKDLE 1454
RTPLCW GER + GP IV +TTE++K ++ K+K + RQKSY +KRRK+LE
Sbjct: 1390 RTPLCWTPVGERRLFGPTIVDETTERMKFLKIKLKEAQDRQKSYANKRRKELE 1442
>At2g07660 putative retroelement pol polyprotein
Length = 949
Score = 155 bits (393), Expect = 1e-37
Identities = 70/113 (61%), Positives = 91/113 (79%)
Query: 1342 RTDGQSERTIQSLEDLLRVCVLEQGGTWDSHLPLIEFTYNNGYHSSIGMAPFEALYGQRC 1401
+TDGQSERTIQ+LED+LR CVL+ GG W+ +L LIEF YNN + +SIGM+P+EALYG+ C
Sbjct: 722 QTDGQSERTIQTLEDMLRACVLDWGGNWEKYLRLIEFAYNNSFQASIGMSPYEALYGRAC 781
Query: 1402 RTPLCWFESGERVVLGPEIVQQTTEKVKMIQEKMKVSHSRQKSYHDKRRKDLE 1454
RTPLCW GER + GP IV +TTE++K ++ K+K + RQKSY +KRRK+LE
Sbjct: 782 RTPLCWTPVGERRLFGPTIVDETTERMKFLKIKLKEAQDRQKSYANKRRKELE 834
Score = 83.2 bits (204), Expect = 1e-15
Identities = 55/166 (33%), Positives = 78/166 (46%), Gaps = 55/166 (33%)
Query: 403 GRDFVVDLVCLPLTGMDVIFGMNWLEYNRVHINCFSKTMHF------------------- 443
G + DL+ +PL DVI GM+WL + HI+C + + F
Sbjct: 25 GVNMPADLIIVPLKKHDVILGMDWLGKYKGHIDCHRERIQFERDEGMLKFQGIRTTSGSL 84
Query: 444 ----------------------------SSAEEEEV-----FPD---EIPDVPPEREVEF 467
+SAE +++ F D + VPP+R F
Sbjct: 85 VISAIQAERMLGKGCEAYLATITTKEVGASAELKDILIVNEFSDVFAAVSGVPPDRSDPF 144
Query: 468 SIDLVPGTKPMSMAPYRMSSSELAELKKQLED*LDKKFVRPNVSPW 513
+I+L PGT P+S APYRM+ +E+AELKKQLE+ L K F+RP+ SPW
Sbjct: 145 TIELEPGTTPISKAPYRMAPAEMAELKKQLEELLAKGFIRPSSSPW 190
>At3g31970 hypothetical protein
Length = 1329
Score = 154 bits (388), Expect = 5e-37
Identities = 68/113 (60%), Positives = 91/113 (80%)
Query: 1342 RTDGQSERTIQSLEDLLRVCVLEQGGTWDSHLPLIEFTYNNGYHSSIGMAPFEALYGQRC 1401
+TDGQSERTIQ+LED+LR+CVL++GG W HL L+EF YNN Y +SI MAPFEALYG+ C
Sbjct: 1071 QTDGQSERTIQTLEDMLRMCVLDRGGHWADHLSLVEFAYNNSYQASIRMAPFEALYGRPC 1130
Query: 1402 RTPLCWFESGERVVLGPEIVQQTTEKVKMIQEKMKVSHSRQKSYHDKRRKDLE 1454
RTPLCW + GER + G + V +TTE++++++ MK + RQ+SY DKRR++LE
Sbjct: 1131 RTPLCWTQVGERSIYGADYVLETTERIRVLKLNMKEAQDRQRSYADKRRRELE 1183
Score = 104 bits (260), Expect = 3e-22
Identities = 110/423 (26%), Positives = 168/423 (39%), Gaps = 49/423 (11%)
Query: 31 VRMLETFLRNHPPAFKGMYDPDGAQSWLKEVERIFRAMQCSEVKKVRFGTHMLAEEADDW 90
+RM+E R F G P+ A SW VE F + +C +V H L +A W
Sbjct: 144 LRMMEQLQRIGTRYFFGGTSPEEADSWKSRVEHNFGSSRCPAEYRVDLAVHFLEGDAHLW 203
Query: 91 WVSLLPVLEQDEAVVTWAVFRREFLNRYFLEDV*GRKEIEFLELK*GDISVVEYAAKFVE 150
W S+ + +A ++WA F EF +YF ++ R E FLEL G+ SV EY +F
Sbjct: 204 WRSV--TARRRQAHMSWADFVAEFNAKYFPQEALDRMEARFLELTQGERSVREYEREFNR 261
Query: 151 LAKFYPHYTAETAEFSKCIKFENGLRANIKRAIGYQQIRIFSDLVSRCRICEEDTKAHYK 210
L + + + ++ +F GLR +++ Q + LV EED +
Sbjct: 262 LLVYAGRGMQD--DQAQMRRFLRGLRPDLRVRCRVLQYATKAALVETAAEVEEDLQRQVV 319
Query: 211 VMSERRGKGRQSRPKPYSAPADKGKQRLNDERRPNRRDASAEIVCFKCGEKGHKSNVCTK 270
+S + + + AP+ GK +R+ E+GH C K
Sbjct: 320 GVSP---AVKPKKTQQQVAPSKGGKPAQGQKRK----------------ERGHLRPNCPK 360
Query: 271 DEKKCFRCGQKGHLLADCKRGDVVCYNCNEEGHISTQCTQPKKDRTGGKVFALTGTQTTN 330
++ Q + G V + HI+ A TT
Sbjct: 361 LQRMAVAVVQPA-----VQHGAQVQQRVQQLAHIA----------------AAPQGYTTR 399
Query: 331 EDRLIRGTCFFNSTPLIAIIDTGATHCFIALESAYKLGLIVSDMKGEMVVETPAKGSVTT 390
E I T T + D+GA+HCFI ESA + G I D ++ A G
Sbjct: 400 E---IGSTSKRAITEAHVLFDSGASHCFITPESASR-GNIRGDPGEQLGAVKVAGGQFLA 455
Query: 391 SLVCLR-CPTSMFGRDFVVDLVCLPLTGMDVIFGMNWLEYNRVHINCFSKTMHFSSAEEE 449
L + + G VDL+ P+ DVI GM+WL++ RVH++C + F E
Sbjct: 456 VLGRAKGVDIQIAGESMPVDLIISPVELYDVILGMDWLDHYRVHLDCHRGRVSFERPEGR 515
Query: 450 EVF 452
V+
Sbjct: 516 LVY 518
>At3g29490 hypothetical protein
Length = 438
Score = 152 bits (385), Expect = 1e-36
Identities = 68/113 (60%), Positives = 89/113 (78%)
Query: 1342 RTDGQSERTIQSLEDLLRVCVLEQGGTWDSHLPLIEFTYNNGYHSSIGMAPFEALYGQRC 1401
+TDGQ ERTIQ+LED+LR+CVL+ GG W HL L+EF YNN Y + IGMAPFEALYG+ C
Sbjct: 83 QTDGQFERTIQTLEDMLRMCVLDWGGHWADHLSLVEFAYNNSYQAGIGMAPFEALYGRPC 142
Query: 1402 RTPLCWFESGERVVLGPEIVQQTTEKVKMIQEKMKVSHSRQKSYHDKRRKDLE 1454
RTPLCW + GER + G + VQ+TTE++++++ MK + RQ SY DKRR++LE
Sbjct: 143 RTPLCWTQVGERSIYGADYVQETTERIRVLKLNMKEAQDRQWSYADKRRRELE 195
>At1g36120 putative reverse transcriptase gb|AAD22339.1
Length = 1235
Score = 150 bits (379), Expect = 5e-36
Identities = 66/113 (58%), Positives = 91/113 (80%)
Query: 1342 RTDGQSERTIQSLEDLLRVCVLEQGGTWDSHLPLIEFTYNNGYHSSIGMAPFEALYGQRC 1401
+TDGQSERTIQ+LED+L++CVL+ GG W HL L++F YNN Y +SIGMAPFEALYG+ C
Sbjct: 1006 QTDGQSERTIQTLEDMLQMCVLDWGGHWADHLSLVKFAYNNSYQASIGMAPFEALYGRPC 1065
Query: 1402 RTPLCWFESGERVVLGPEIVQQTTEKVKMIQEKMKVSHSRQKSYHDKRRKDLE 1454
RT LCW + GE+ + G + VQ+TTE++++++ MK + RQ+SY DKRR++LE
Sbjct: 1066 RTLLCWTQVGEKSIYGADYVQETTERIRVLKLNMKEAQDRQRSYADKRRRELE 1118
Score = 99.8 bits (247), Expect = 1e-20
Identities = 124/483 (25%), Positives = 182/483 (37%), Gaps = 115/483 (23%)
Query: 93 SLLPVLEQDEAVVTWAVFRREFLNRYFLEDV*GRKEIEFLELK*GDISVVEYAAKFVELA 152
S L ++EQ + + T F EF +YF ++ R E FLEL G++SV EY +F L
Sbjct: 142 SYLRMMEQLQRIGT--DFVAEFNAKYFPQEALDRMEARFLELTKGELSVREYGREFKRLL 199
Query: 153 KFYPHYTAETAEFSKCIKFENGLRANIKRAIGYQQIRIFSDLVSRCRICEEDTKAHYKVM 212
+ + + ++ +F GLR DL RCR+ + TKA
Sbjct: 200 VYAGRGMED--DQAQMRRFLRGLRP---------------DLRVRCRVSQYATKAALTAA 242
Query: 213 SERRGKGRQSRPKPYSAPADKGKQRLNDER--RPNRRDASAEIVCFKCGEKGHKSNVCTK 270
RQ S K +Q++ + +P + F+ G+ G +
Sbjct: 243 EVEEDLQRQVVAVSPSVQPKKIQQQVAPSKGGKPAQVQKRKWDHPFRAGQGGRAGSAA-- 300
Query: 271 DEKKCFRCGQKGHLLADCKRGDVVCYNCNEEGHISTQCTQPKKDRTGGKVFALTGTQTTN 330
D+ G A R C +ST C+ GT
Sbjct: 301 DDSSTSTAGGAAQS-AGAARSAAEC------AAVSTYCS---------------GTTWLY 338
Query: 331 EDRLIRGTCFFNSTPLIAIIDTGATHCFIALESAYKLGLIVSDMKGEMVVETPAK----G 386
LI G C + + D+GA+HCFI ESA + +++GE + A G
Sbjct: 339 YADLISGRCRGH-----VLFDSGASHCFITPESASR-----DNIRGEPGEQFGAVKIAGG 388
Query: 387 SVTTSLVCLRCPTSMFGRDFV-VDLVCLPLTGMDVIFGMNWLEYNRVHINCFSKTMHFSS 445
T L + + + DL+ P+ D I GM+WL++ RVH++C + F
Sbjct: 389 QFLTVLGRAKGVNIQIEEESMPADLIINPVVLYDAILGMDWLDHYRVHLDCHRGRVSFER 448
Query: 446 AEEEEVFP------------------------------------------------DEIP 457
E V+ E
Sbjct: 449 PEGRLVYQGVRPTSRSLVISEVQVEKMIEKGCEAYLVTISMSESVGQVAVSDIWVVQEFE 508
Query: 458 DV-------PPEREVEFSIDLVPGTKPMSMAPYRMSSSELAELKKQLED*LDKKFVRPNV 510
DV PP R F+I+L GT P+S PYRM +E+AELKKQLED L K F+RP+
Sbjct: 509 DVFQSLQGLPPSRSDPFTIELELGTAPLSKTPYRMVPAEIAELKKQLEDLLGKGFIRPST 568
Query: 511 SPW 513
S W
Sbjct: 569 SRW 571
>At4g16910 retrotransposon like protein
Length = 687
Score = 150 bits (378), Expect = 7e-36
Identities = 67/113 (59%), Positives = 90/113 (79%)
Query: 1342 RTDGQSERTIQSLEDLLRVCVLEQGGTWDSHLPLIEFTYNNGYHSSIGMAPFEALYGQRC 1401
+TDGQSERTIQ+LED+LR CVL+ GG W+ +L L+EF YNN + +SIGM+P+EALYG+
Sbjct: 414 QTDGQSERTIQTLEDMLRACVLDWGGNWEKYLRLVEFAYNNSFQASIGMSPYEALYGRAG 473
Query: 1402 RTPLCWFESGERVVLGPEIVQQTTEKVKMIQEKMKVSHSRQKSYHDKRRKDLE 1454
RTPLCW GER + GP +V +TT+K+K ++ K+K + RQKSY +KRRK+LE
Sbjct: 474 RTPLCWTPVGERRLFGPAVVDETTKKMKFLKIKLKEAQDRQKSYANKRRKELE 526
>At2g04670 putative retroelement pol polyprotein
Length = 1411
Score = 148 bits (373), Expect = 3e-35
Identities = 67/113 (59%), Positives = 90/113 (79%)
Query: 1342 RTDGQSERTIQSLEDLLRVCVLEQGGTWDSHLPLIEFTYNNGYHSSIGMAPFEALYGQRC 1401
+TDGQSERTIQ+LED+LR+CVL+ GG W HL L+EF YNN Y +SIGMAPFEALYG+ C
Sbjct: 1153 QTDGQSERTIQTLEDMLRMCVLDWGGHWADHLSLVEFAYNNSYQASIGMAPFEALYGRPC 1212
Query: 1402 RTPLCWFESGERVVLGPEIVQQTTEKVKMIQEKMKVSHSRQKSYHDKRRKDLE 1454
TPL W + ER + G + VQ+TTE++++++ MK + +RQ+SY DKRR++LE
Sbjct: 1213 WTPLRWTQVEERSIYGADYVQETTERIRVLKLNMKEAQARQRSYADKRRRELE 1265
Score = 124 bits (312), Expect = 3e-28
Identities = 129/499 (25%), Positives = 201/499 (39%), Gaps = 85/499 (17%)
Query: 31 VRMLETFLRNHPPAFKGMYDPDGAQSWLKEVERIFRAMQCSEVKKVRFGTHMLAEEADDW 90
+RM+E R F P+ A SW V+R F + +C +V H L +A W
Sbjct: 124 LRMMEQLQRIGTWYFSSGTSPEEADSWRSRVQRNFGSSRCPAEYRVDLAVHFLEGDAHLW 183
Query: 91 WVSLLPVLEQDEAVVTWAVFRREFLNRYFLEDV*GRKEIEFLELK*GDISVVEYAAKFVE 150
W S+ + +A ++WA F EF +YF ++ R E FLEL G+ SV EY +F
Sbjct: 184 WRSV--TARRRQADMSWADFVAEFNAKYFPQEALDRMEARFLELTQGEWSVREYDREFNR 241
Query: 151 LAKFYPHYTAETAEFSKCIKFENGLRANIKRAIGYQQIRIFSDLVSRCRICEEDTKAHYK 210
L + + + ++ +F GLR +++ Q + LV EED +
Sbjct: 242 LLAYAGRGMED--DQAQMRRFLRGLRPDLRVQCRVSQYATKAALVETAAEVEEDLQRQVV 299
Query: 211 VMS---ERRGKGRQSRPKPYSAPADKGKQRLNDERRPNRRDASAEIVCFKCGEKGHKSNV 267
+S + + +Q P PA K++ + R + G G+
Sbjct: 300 GVSPAVQTKNTQQQVTPSKGGKPAQGQKRKWDHPSRAGQG-----------GRAGY---- 344
Query: 268 CTKDEKKCFRCGQKGHLLADCKRGDVVCYNCNEEGHISTQCTQPKKDRTGGKVFALTGTQ 327
F CG H +ADC E GH+ + GG+ A+ G +
Sbjct: 345 --------FSCGSLDHTVADC----------TERGHLHVKVA-------GGQFLAVLG-R 378
Query: 328 TTNEDRLIRGTCFFNSTPLIAIIDT-----------GATHCFIALESAYKLGLIVSDMKG 376
D I G S P II H + L+ ++ + +G
Sbjct: 379 AKGVDIQIAG----ESMPADLIISPVELYDVILGMDWLDHYRVHLD-CHRGRVSFERPEG 433
Query: 377 EMVVE--TPAKGSVTTSLVCLRCPTSMFGRDFVVDLVCLPLTGMDVIFGMNWLEYNRVHI 434
+V + P GS+ S V + ++V + G + + ++
Sbjct: 434 RLVYQGVRPTSGSLVISAVQAKKMIEKGCEAYLVTISMPESLGQVAVSDIRVIQ------ 487
Query: 435 NCFSKTMHFSSAEEEEVFPDEIPDVPPEREVEFSIDLVPGTKPMSMAPYRMSSSELAELK 494
E E+VF + +PP R F+I+L PGT P+S APYRM+ +E+ ELK
Sbjct: 488 ------------EFEDVF-QSLQGLPPSRSDPFTIELEPGTAPLSKAPYRMAPAEMTELK 534
Query: 495 KQLED*LDKKFVRPNVSPW 513
KQLED L K F+RP+ SPW
Sbjct: 535 KQLEDLLGKGFIRPSTSPW 553
>At2g14650 putative retroelement pol polyprotein
Length = 1328
Score = 146 bits (368), Expect = 1e-34
Identities = 67/113 (59%), Positives = 89/113 (78%)
Query: 1342 RTDGQSERTIQSLEDLLRVCVLEQGGTWDSHLPLIEFTYNNGYHSSIGMAPFEALYGQRC 1401
+T GQSERTIQ+LED+LR+CVL+ GG W HL L+EF YNN Y +SIGMAPFEALY + C
Sbjct: 1073 QTYGQSERTIQTLEDMLRMCVLDWGGHWADHLSLVEFAYNNSYPASIGMAPFEALYERPC 1132
Query: 1402 RTPLCWFESGERVVLGPEIVQQTTEKVKMIQEKMKVSHSRQKSYHDKRRKDLE 1454
RTPLC + GER + G + VQ+TTE++++++ MK + RQ+SY DKRR++LE
Sbjct: 1133 RTPLCLTQVGERSIYGADYVQETTERIRVLKLNMKEAQDRQRSYADKRRRELE 1185
Score = 91.3 bits (225), Expect = 4e-18
Identities = 65/198 (32%), Positives = 88/198 (43%), Gaps = 36/198 (18%)
Query: 337 GTCFFNSTPLIAIIDTGATHCFIALESAYKLGLIVSDMKGEMVVETPAKGSVTTSLVCLR 396
GT + D+GA+HCFI ESA + G I D ++ A G L +
Sbjct: 304 GTLLVGGVEAHVLFDSGASHCFITPESASR-GNIRGDPGEQLGAVKVAGGQFVAVLGRTK 362
Query: 397 -CPTSMFGRDFVVDLVCLPLTGMDVIFGMNWLEYNRVHINCFSKTMHFSSAEEEEVFPD- 454
+ G DL+ P+ DVI GM+WL++ RVH++C + F E V+
Sbjct: 363 GVDIQIAGESMPADLIISPVELYDVILGMDWLDHYRVHLDCHRGRVSFERPEGSLVYQGV 422
Query: 455 --------------------------EIPDV-------PPEREVEFSIDLVPGTKPMSMA 481
E DV PP R F+I+L PGT P+S A
Sbjct: 423 RPTSGSLVISAVQAEKMIEKGCEAYLEFEDVFQSLQGLPPSRSDPFTIELEPGTAPLSKA 482
Query: 482 PYRMSSSELAELKKQLED 499
PYRM+ +E+AELKKQLED
Sbjct: 483 PYRMAPAEMAELKKQLED 500
Score = 61.6 bits (148), Expect = 3e-09
Identities = 39/112 (34%), Positives = 55/112 (48%), Gaps = 4/112 (3%)
Query: 31 VRMLETFLRNHPPAFKGMYDPDGAQSWLKEVERIFRAMQCSEVKKVRFGTHMLAEEADDW 90
+RM+E R F G P+ A SW VER F + +C ++ H L +A W
Sbjct: 142 LRMMEQLQRIGTGYFSGGTSPEVADSWRSRVERNFGSSRCPAEYRIDLAVHFLEGDAHLW 201
Query: 91 WVSLLPVLEQDEAVVTWAVFRREFLNRYFLEDV*GRKEIEFLELK--*GDIS 140
W S+ + +A ++WA F EF +YF ++ R E FLEL GDIS
Sbjct: 202 WRSV--TARRRQADISWADFVAEFNAKYFPQEALDRMEAHFLELTQGRGDIS 251
>At4g07830 putative reverse transcriptase
Length = 611
Score = 145 bits (367), Expect = 1e-34
Identities = 67/113 (59%), Positives = 89/113 (78%), Gaps = 1/113 (0%)
Query: 1342 RTDGQSERTIQSLEDLLRVCVLEQGGTWDSHLPLIEFTYNNGYHSSIGMAPFEALYGQRC 1401
+TDGQSERTIQ+LED+LR+CVL+ G W HL L+EF YNN Y +SIGMAPFE LYG+ C
Sbjct: 338 QTDGQSERTIQTLEDMLRMCVLDWRGHWADHLSLVEFAYNNSYQASIGMAPFEVLYGRPC 397
Query: 1402 RTPLCWFESGERVVLGPEIVQQTTEKVKMIQEKMKVSHSRQKSYHDKRRKDLE 1454
RT LCW + GER + G + VQ+ TE++++++ MK + +RQ+SY DKRRK+LE
Sbjct: 398 RT-LCWTQVGERSIYGADYVQEITERIRVLKLNMKEAQNRQRSYADKRRKELE 449
>At2g06470 putative retroelement pol polyprotein
Length = 899
Score = 119 bits (299), Expect = 1e-26
Identities = 59/113 (52%), Positives = 76/113 (67%), Gaps = 19/113 (16%)
Query: 1342 RTDGQSERTIQSLEDLLRVCVLEQGGTWDSHLPLIEFTYNNGYHSSIGMAPFEALYGQRC 1401
+TDGQSERTIQ+LED+LR CVL+ GG W+ +L L ALYG+ C
Sbjct: 708 QTDGQSERTIQTLEDMLRACVLDWGGNWEKYLTL-------------------ALYGRAC 748
Query: 1402 RTPLCWFESGERVVLGPEIVQQTTEKVKMIQEKMKVSHSRQKSYHDKRRKDLE 1454
RTPLCW GER + GP IV +TTE++K ++ K+K +H RQKSY +KRRK+LE
Sbjct: 749 RTPLCWTPVGERRLFGPTIVDETTERMKFLKIKLKEAHDRQKSYANKRRKELE 801
Score = 62.4 bits (150), Expect = 2e-09
Identities = 41/143 (28%), Positives = 70/143 (48%), Gaps = 7/143 (4%)
Query: 17 AQAVEQQPQASNGEVR-----MLETFLRNHPPAFKGMYDPDGAQSWLKEVERIFRAMQCS 71
++ VE PQ ++G R +LE R F G DP A W ++R F++ +C
Sbjct: 114 SEVVEVNPQPNHGRKRGDYLSLLEHVSRLGTRHFMGSTDPIVADEWRSRLKRNFKSTRCP 173
Query: 72 EVKKVRFGTHMLAEEADDWWVSLLPVLEQDEAVVTWAVFRREFLNRYFLEDV*GRKEIEF 131
E + H L +A +WW+++ + + V ++A F EF +YF + R E +
Sbjct: 174 EDYQRDIAVHFLEGDAHNWWLTV--EKRRGDEVRSFADFEDEFNKKYFPPEAWDRLECAY 231
Query: 132 LELK*GDISVVEYAAKFVELAKF 154
L+L G+ +V EY +F L ++
Sbjct: 232 LDLVQGNRTVREYDEEFNRLRRY 254
>At4g03840 putative transposon protein
Length = 973
Score = 114 bits (284), Expect = 5e-25
Identities = 57/113 (50%), Positives = 74/113 (65%), Gaps = 19/113 (16%)
Query: 1342 RTDGQSERTIQSLEDLLRVCVLEQGGTWDSHLPLIEFTYNNGYHSSIGMAPFEALYGQRC 1401
+TDGQSERTIQ+LED+LR C L+ GG W+ +L L ALYG+ C
Sbjct: 711 QTDGQSERTIQTLEDMLRACALDWGGNWEKYLRL-------------------ALYGRAC 751
Query: 1402 RTPLCWFESGERVVLGPEIVQQTTEKVKMIQEKMKVSHSRQKSYHDKRRKDLE 1454
RTPLCW GER + GP IV +TTE++K ++ K+K + RQKSY +KRRK+LE
Sbjct: 752 RTPLCWTPVGERRLFGPIIVDETTERMKFLKIKLKEAQDRQKSYANKRRKELE 804
Score = 62.8 bits (151), Expect = 1e-09
Identities = 41/142 (28%), Positives = 69/142 (47%), Gaps = 7/142 (4%)
Query: 18 QAVEQQPQASNGEVR-----MLETFLRNHPPAFKGMYDPDGAQSWLKEVERIFRAMQCSE 72
+ VE PQ ++G R +LE R F G DP A W ++R F++ +C E
Sbjct: 115 EVVEVNPQPNHGRKRGDYLSLLEHVSRLGTRHFMGSTDPIVADEWRSRLKRNFKSTRCPE 174
Query: 73 VKKVRFGTHMLAEEADDWWVSLLPVLEQDEAVVTWAVFRREFLNRYFLEDV*GRKEIEFL 132
+ H L +A +WW+++ + + V ++A F EF +YF + R E +L
Sbjct: 175 DYQRDIAVHFLERDAHNWWLTV--EKRRGDEVRSFADFEDEFNKKYFPPEAWDRLECAYL 232
Query: 133 ELK*GDISVVEYAAKFVELAKF 154
+L G+ +V EY +F L ++
Sbjct: 233 DLVQGNRTVREYDEEFNRLRRY 254
>At3g29480 hypothetical protein
Length = 718
Score = 83.6 bits (205), Expect = 8e-16
Identities = 103/449 (22%), Positives = 160/449 (34%), Gaps = 104/449 (23%)
Query: 8 AIAGALEAIAQA------VEQQPQASNGE-----VRMLETFLRNHPPAFKGMYDPDGAQS 56
A AG + QA E Q +A+ E +RM+E R F G P+ A S
Sbjct: 3 AAAGGAQVPVQAPVAPRVAEVQQRAAVAEEVPSYLRMMEQLQRIGIGYFSGGTSPEEADS 62
Query: 57 WLKEVERIFRAMQCSEVKKVRFGTHMLAEEADDWWVSLLPVLEQDEAVVTWAVFRREFLN 116
W VER F + +C +V H L +A WW S+ + +A ++WA F EF
Sbjct: 63 WRSRVERNFGSSRCPVEYRVDLAVHFLEGDAHLWWKSV--TTRRRQANMSWADFVAEFNA 120
Query: 117 RYFLEDV*GRKEIEFLELK*GDISVVEYAAKFVELAKFYPHYTAETAEFSKCIKFENGLR 176
+YF ++ R E FLEL G+ SV EY +F
Sbjct: 121 KYFPQEALDRMEAHFLELTQGERSVREYDREF---------------------------- 152
Query: 177 ANIKRAIGYQQIRIFSDLVSRCRICEEDTKAHYKVMSERRGKGRQSRPKPYSAPADKGKQ 236
++ ++++ S + K H +V + GK Q + + + P+ G+
Sbjct: 153 ---------NRLLVYAECRSTRIPVVQPKKTHQQVAPSKGGKPAQGQKRSWDHPSRAGQG 203
Query: 237 RLNDERRPNRRDASAEIVCFKCGEKGHKSNVCTKDEKKCFRCGQKGHLLADC-KRGDV-V 294
G G CF CG H +ADC +R +
Sbjct: 204 ----------------------GRAG------------CFFCGSLDHKVADCTQRAETRE 229
Query: 295 CYNCNEEGHISTQCTQPKKDRTGGKVFALTGTQTTNEDRLIRGTCFFNSTPLIAIIDTGA 354
Y+C + H+ C PK R V Q + ++ P G
Sbjct: 230 YYHCRKRRHLRPNC--PKLQRMAVAVVHAAVQQGAQVQQNVQQLAHIAPAP------QGY 281
Query: 355 THCFIALESAYKLGLIVSDMKGEMVVETPAKGSVTTSLVCLRCPTSMFGRDFVVDLVCLP 414
T I + + + V + G+ + V + P ++F P
Sbjct: 282 TTREIGVTNNRAI-TAVKVVGGQFLAVLGRAKGVDIQIARESMPANLF---------LSP 331
Query: 415 LTGMDVIFGMNWLEYNRVHINCFSKTMHF 443
DVI GM+WL+Y RVH++C + F
Sbjct: 332 EELYDVILGMDWLDYYRVHLDCHRGRVSF 360
>At4g10580 putative reverse-transcriptase -like protein
Length = 1240
Score = 82.0 bits (201), Expect = 2e-15
Identities = 52/166 (31%), Positives = 72/166 (43%), Gaps = 55/166 (33%)
Query: 403 GRDFVVDLVCLPLTGMDVIFGMNWLEYNRVHINCFSKTMHFSSAEEEEVF---------- 452
G DL+ P+ DVI GM+WL+Y RVH++ + F E V+
Sbjct: 362 GESMPADLIISPVELYDVILGMDWLDYYRVHLDWHRGRVFFERPEGRLVYQGVRPISGSL 421
Query: 453 ------------------------PDEIPDV---------------------PPEREVEF 467
P+ + V PP + F
Sbjct: 422 VISAVQAEKMIEKGCEAYLVTISMPESVGQVAVSDIRVVQEFQDVFQSLQGLPPSQSDPF 481
Query: 468 SIDLVPGTKPMSMAPYRMSSSELAELKKQLED*LDKKFVRPNVSPW 513
+I+L PGT P+S APYRM+ +E+AELKKQL+D L K F+RP+ SPW
Sbjct: 482 TIELEPGTAPLSKAPYRMAPAEMAELKKQLKDLLGKGFIRPSTSPW 527
Score = 57.4 bits (137), Expect = 6e-08
Identities = 56/237 (23%), Positives = 91/237 (37%), Gaps = 37/237 (15%)
Query: 31 VRMLETFLRNHPPAFKGMYDPDGAQSWLKEVERIFRAMQCSEVKKVRFGTHMLAEEADDW 90
+RM+E R F G P+ A SW V R F + +C +V H L +A W
Sbjct: 138 LRMMEQLQRIDTGYFSGGTSPEEADSWRSRVGRNFGSSRCPAEYRVDLAVHFLEGDAHLW 197
Query: 91 WVSLLPVLEQDEAVVTWAVFRREFLNRYFLEDV*GRKEIEFLELK*GDISVVEYAAKFVE 150
W S+ Q + ++WA F EF +YF ++ ++ YA + +E
Sbjct: 198 WRSVTARRRQTD--MSWADFVAEFKAKYFPQE-----------------ALDPYAGQGME 238
Query: 151 LAKFYPHYTAETAEFSKCIKFENGLRANIKRAIGYQQIRIFSDLVSRCRICEEDTKAHYK 210
+ ++ +F GLR +++ Q + LV EED +
Sbjct: 239 ------------DDQAQMRRFLRGLRPDLRVRCRVSQYATKAALVETAAEVEEDFQRQVV 286
Query: 211 VMS---ERRGKGRQSRPKPYSAPADKGKQRLNDERRPNRRDASAEIVCFKCGEKGHK 264
+S + + +Q P PA K++ + P+R CF CG HK
Sbjct: 287 GVSPVVQPKKTQQQVTPSKGGKPAQGQKRKWD---HPSRAGQGGRARCFSCGSLDHK 340
>At4g03650 putative reverse transcriptase
Length = 839
Score = 74.7 bits (182), Expect = 4e-13
Identities = 59/192 (30%), Positives = 87/192 (44%), Gaps = 9/192 (4%)
Query: 18 QAVEQQPQASNGE-----VRMLETFLRNHPPAFKGMYDPDGAQSWLKEVERIFRAMQCSE 72
+ VE Q +A+ E +RM++ R F G P+ A SW +VER F + +C
Sbjct: 124 RVVEVQQRAAVAEEVPSYLRMMKQLQRIGTEYFSGGTSPEEADSWRSQVERNFGSSRCPA 183
Query: 73 VKKVRFGTHMLAEEADDWWVSLLPVLEQDEAVVTWAVFRREFLNRYFLEDV*GRKEIEFL 132
+V H L +A WW S+ + +A ++WA F EF +YF + R E FL
Sbjct: 184 EYRVDLTVHFLEGDAHLWWRSV--TARRRQADMSWADFMAEFNAKYFPREALDRMEARFL 241
Query: 133 ELK*GDISVVEYAAKFVELAKFYPHYTAETAEFSKCIKFENGLRANIKRAIGYQQIRIFS 192
EL G SV EY KF L + + + ++ +F GLR N++ Q +
Sbjct: 242 ELTQGVRSVREYDRKFNRLLVYAGRGMED--DQAQMRRFLRGLRPNLRVRCRVSQYATKA 299
Query: 193 DLVSRCRICEED 204
LV EED
Sbjct: 300 ALVETAAEVEED 311
Score = 67.8 bits (164), Expect = 4e-11
Identities = 44/142 (30%), Positives = 63/142 (43%), Gaps = 37/142 (26%)
Query: 403 GRDFVVDLVCLPLTGMDVIFGMNWLEYNRVHINCFSKTMHFSSAE--------------- 447
G DL+ + DVI GM+WL++ RVH++C + F E
Sbjct: 381 GESLPADLIISHVELYDVILGMDWLDHYRVHLDCHRGRVSFERPEGSAGRENDREGLRGL 440
Query: 448 ----------------------EEEVFPDEIPDVPPEREVEFSIDLVPGTKPMSMAPYRM 485
E + + +PP R F+I+L P T P+S APYRM
Sbjct: 441 PGDDIYAGVCGQVAVSDIRVVQEFQYVFQSLQGLPPSRSDPFTIELEPKTAPLSKAPYRM 500
Query: 486 SSSELAELKKQLED*LDKKFVR 507
+ +++AELKKQLED L + VR
Sbjct: 501 APAKMAELKKQLEDLLAEADVR 522
>At3g43830 putative protein
Length = 329
Score = 73.6 bits (179), Expect = 8e-13
Identities = 62/248 (25%), Positives = 95/248 (38%), Gaps = 46/248 (18%)
Query: 45 FKGMYDPDGAQSWLKEVERIFRAMQCSEVKKVRFGTHMLAEEADDWWVSLLPVLEQDEAV 104
F G DP A +W +E F++M+C + L EA +WW + EQ V
Sbjct: 116 FNGGTDPVKAYNWRNMLECKFKSMRCPVEFWRDLASCYLRGEAQEWW-ERVKQREQVGCV 174
Query: 105 VTWAVFRREFLNRYFLEDV*GRKEIEFLELK*GDISVVEYAAKFVELAKFYPHYTAETAE 164
W+ F+ EF RY E+ E++FL L+ G +V +Y +F L +F + E
Sbjct: 175 DQWSFFKEEFTRRYLPEETIDDLEMKFLRLQQGTKTVRKYEKEFHSLERF---ERRKRGE 231
Query: 165 FSKCIKFENGLRANIKRAIGYQQIRIFSDLVSRCRICE---EDTKAHYKVMSERRGKGRQ 221
KF +GLR +I+ + DLV + E E+ + ++ E+ G
Sbjct: 232 HELIHKFISGLRVDIRLCCHVRDFDNMIDLVEKAASLEIGLEEEARYKRIAQEKEAMG-- 289
Query: 222 SRPKPYSAPADKGKQRLNDERRPNRRDASAEIVCFKCGEKGHKSNVCTKDEKKCFRCGQK 281
G+ GH C E C+RCG
Sbjct: 290 -----------------------------------SYGQTGHSKRRC--QEVTCYRCGVA 312
Query: 282 GHLLADCK 289
GH+ DC+
Sbjct: 313 GHIARDCR 320
Score = 33.5 bits (75), Expect = 0.92
Identities = 14/33 (42%), Positives = 17/33 (51%), Gaps = 2/33 (6%)
Query: 279 GQKGHLLADCKRGDVVCYNCNEEGHISTQCTQP 311
GQ GH C+ +V CY C GHI+ C P
Sbjct: 292 GQTGHSKRRCQ--EVTCYRCGVAGHIARDCRMP 322
>At4g07850 putative polyprotein
Length = 1138
Score = 68.2 bits (165), Expect = 3e-11
Identities = 97/478 (20%), Positives = 178/478 (36%), Gaps = 91/478 (19%)
Query: 71 SEVKKVRFGTHMLAEEADDWWVSLLPVLEQ--DEAVVTWA----VFRREFLNRYFLEDV* 124
+E KV+ + A WW ++ + D+++ TW + +R F+ ++ ++
Sbjct: 91 TEANKVKVAATEFYDYALSWWDQVVTTKRRLGDDSIETWNQLKNIMKRRFVPSHYHRELH 150
Query: 125 GRKEIEFLELK*GDISVVEYAAKFVELAKFYPHYTAETAEFSKCIKFENGLRANIK---R 181
R L G+ +V EY F E+ + + +F GL +I
Sbjct: 151 QRLR----NLVQGNRTVEEY---FKEMETLMLRADVQEECEATMSRFMGGLNRDILDRFE 203
Query: 182 AIGYQQIRIFSDLVSRCRICEEDTKAHYKVMSERRGKGRQSRPKPYSAPAD---KGKQRL 238
I Y+ + + H VM E++ K R ++P S+ + K
Sbjct: 204 VIHYENL---------------EELFHKAVMFEKQIKRRSAKPSYNSSKPSYQREEKSGF 248
Query: 239 NDERRPNRRDASAEIVCFKCGEKGHKSNVCTKDEKKCFRCGQKGHLLADCKRGDVVCYNC 298
E +P + EI KG + V + KCF+C GH ++C ++
Sbjct: 249 QKEYKPFVKPKVEEI-----SSKGKEKEVTRTRDLKCFKCHGLGHYASECSNKRIMIIR- 302
Query: 299 NEEGHISTQCTQPKK----DRTGGKVFALT--------GTQTTNEDRLIRGTCFFNSTPL 346
+ G + ++ +P++ + G++ + + L C
Sbjct: 303 -DSGEVESEDEKPEESDVEEAPKGELLVTMRVLSVLNKAEEQAQRENLFHTRCLIKGKVC 361
Query: 347 IAIIDTGATHCFIALESAYKLGLIVS-----------DMKGEMVVETPAKGSVTTSLVCL 395
IID G+ + KLGL + GEM V T V +
Sbjct: 362 SLIIDGGSCTNVASETMVQKLGLEEFPHPKPYKLQWLNESGEMAV---------TRQVQV 412
Query: 396 RCPTSMFGRDFVVDLVCLPLTGMDVIFGMNWLEYNRVHINCFSKTMHFSSAEEEEVFPDE 455
+ + + D+ LPL V+ G W + + ++VFP+E
Sbjct: 413 PLAIGKYEDEILCDI--LPLEASHVLLGRPWQSDRK---------------DYQDVFPEE 455
Query: 456 IPD-VPPEREVEFSIDLVPGTKPMSMAPYRMSSSELAELKKQLED*LDKKFVRPNVSP 512
P+ +PP R +E ID VPG + YR + E EL+KQ+ + +++ + ++SP
Sbjct: 456 NPEGLPPIRGIEHQIDFVPGASLPNRPAYRTNPVETKELEKQVNELMERGHICESMSP 513
Score = 55.5 bits (132), Expect = 2e-07
Identities = 26/64 (40%), Positives = 39/64 (60%)
Query: 1342 RTDGQSERTIQSLEDLLRVCVLEQGGTWDSHLPLIEFTYNNGYHSSIGMAPFEALYGQRC 1401
+TDGQ+E ++L LLR + + TW+ LP +EF YN+ HS+ +PF+ +YG
Sbjct: 927 QTDGQTEVVNRTLSTLLRALIKKNLKTWEDCLPHVEFAYNHSVHSATKFSPFQIVYGFNP 986
Query: 1402 RTPL 1405
TPL
Sbjct: 987 ITPL 990
>At3g42290 putative protein
Length = 462
Score = 68.2 bits (165), Expect = 3e-11
Identities = 69/253 (27%), Positives = 102/253 (40%), Gaps = 45/253 (17%)
Query: 54 AQSWLKEVERIFRAMQCSEVKKVRFGTHMLAEEADDWWVSLLPVLEQDEAVVTWAVFRRE 113
A SW V+R F + +C +V H L E+A WW S+ + +A ++WA F E
Sbjct: 163 ADSWRSRVKRNFSSSRCLMEYRVDLAVHFLEEDAHLWWKSV--TARRRQADMSWADFVVE 220
Query: 114 FLNRYFLEDV*GRKEIEFLELK*GDISVVEYAAKFVELAKFYPHYTAETAEFSKCIKFEN 173
F +YFL+D R E+ FLEL + SV EY +F + Y + + + ++ +F
Sbjct: 221 FNAKYFLQDELDRMEVRFLELTLVERSVWEYDREFNRVL-VYAGWGMKDGQ-AELRRFMR 278
Query: 174 GLRANIKRAIGYQQIRIFSDLVSRCRICEEDTKAHYKVMSERRGKGRQSRPKPYSAPADK 233
GLR DL RCR+ + KA + + +P +
Sbjct: 279 GLR---------------PDLRVRCRMSQYALKAALVETAAEVAPRKGGKPM-------Q 316
Query: 234 GKQRLNDERRPNRRDASAEIVCFKCGEKGHKSNVCTKDEKKCFRCGQKGHLLADCKRGDV 293
GK+R +R CF CG HK+ Q H+ A +
Sbjct: 317 GKKR--SWNHLSRAGQGGRAGCFSCGNLAHKN------------VQQLAHIPAAPQ---- 358
Query: 294 VCYNCNEEGHIST 306
CYN E G ST
Sbjct: 359 -CYNTFEAGGTST 370
>At3g31480 hypothetical protein
Length = 338
Score = 65.9 bits (159), Expect = 2e-10
Identities = 36/113 (31%), Positives = 56/113 (48%), Gaps = 42/113 (37%)
Query: 1342 RTDGQSERTIQSLEDLLRVCVLEQGGTWDSHLPLIEFTYNNGYHSSIGMAPFEALYGQRC 1401
+TDG+ E+TIQ+LED+LR+
Sbjct: 92 KTDGKFEKTIQTLEDMLRM----------------------------------------- 110
Query: 1402 RTPLCWFESGERVVLGPEIVQQTTEKVKMIQEKMKVSHSRQKSYHDKRRKDLE 1454
TPLCW + GER + G V +TTE++++++ MK +H RQ+SY D RR+++E
Sbjct: 111 -TPLCWTQVGERSIYGANYVHETTERIQVLKLNMKEAHDRQRSYADMRRREVE 162
>At1g35370 hypothetical protein
Length = 1447
Score = 60.8 bits (146), Expect = 5e-09
Identities = 33/111 (29%), Positives = 55/111 (48%)
Query: 1342 RTDGQSERTIQSLEDLLRVCVLEQGGTWDSHLPLIEFTYNNGYHSSIGMAPFEALYGQRC 1401
++DGQ+E + LE+ LR + W+ LPL E+ YN YHSS M PFE +YGQ
Sbjct: 1220 QSDGQTEVVNRCLENYLRCMCHARPHLWNKWLPLAEYWYNTNYHSSSQMTPFELVYGQAP 1279
Query: 1402 RTPLCWFESGERVVLGPEIVQQTTEKVKMIQEKMKVSHSRQKSYHDKRRKD 1452
L + +V + +Q+ + ++ + + R K + D+ R +
Sbjct: 1280 PIHLPYLPGKSKVAVVARSLQERENMLLFLKFHLMRAQHRMKQFADQHRTE 1330
Score = 39.7 bits (91), Expect = 0.013
Identities = 55/254 (21%), Positives = 91/254 (35%), Gaps = 83/254 (32%)
Query: 335 IRGTCFFNSTPLIAIIDTGATHCFIALESAYKLGLIVSDMKGEMVVETPAK-----GSVT 389
++GT + L +ID+G+TH FI A KLG V V + G +
Sbjct: 371 VKGTV--DKRDLFILIDSGSTHNFIDSTVAAKLGCHVESAGLTKVAVADGRKLNVDGQIK 428
Query: 390 TSLVCLRCPTSMFGRDFVVDLVCLPLTGMDVIFGMNWL--------EYNRVHINCFSKTM 441
L+ T F D++ +PL G+D++ G+ WL E+ ++ + F K
Sbjct: 429 GFTWKLQSTT------FQSDILLIPLQGVDMVLGVQWLETLGRISWEFKKLEMQFFYKNQ 482
Query: 442 ------------------------------------HFSSAEEEEV-------------- 451
S EE+E+
Sbjct: 483 RVWLHGIITGSVRDIKAHKLQKTQADQIQLAMVCVREVVSDEEQEIGSISALTSDVVEES 542
Query: 452 ----FPDEIPDV-------PPEREV-EFSIDLVPGTKPMSMAPYRMSSSELAELKKQLED 499
+E PDV PP RE + I L+ G P++ PYR + E+ K ++D
Sbjct: 543 VVQNIVEEFPDVFAEPTDLPPFREKHDHKIKLLEGANPVNQRPYRYVVHQKDEIDKIVQD 602
Query: 500 *LDKKFVRPNVSPW 513
+ ++ + SP+
Sbjct: 603 MIKSGTIQVSSSPF 616
>At2g14640 putative retroelement pol polyprotein
Length = 945
Score = 60.1 bits (144), Expect = 9e-09
Identities = 38/114 (33%), Positives = 61/114 (53%), Gaps = 4/114 (3%)
Query: 1342 RTDGQSERTIQSLEDLLRVCVLEQGGTWDSHLPLIEFTYNNGYHSSIGMAPFEALYGQRC 1401
+TDGQ+E + +E LR V W S +P E+ YN +H+S GM PF+ALYG R
Sbjct: 685 QTDGQTEVVNRCIEQFLRCFVHYHPKQWSSFIPWAEYWYNTTFHASTGMTPFQALYG-RP 743
Query: 1402 RTPLCWFESGERVVLGP--EIVQQTTEKVKMIQEKMKVSHSRQKSYHDKRRKDL 1453
+P+ +E G VV G E + E + +++ + +++ K D R +D+
Sbjct: 744 PSPIPAYELGS-VVCGELNEQMAARDELLAELKQHLVTANNCMKQQADSRLRDV 796
Score = 45.4 bits (106), Expect = 2e-04
Identities = 42/131 (32%), Positives = 60/131 (45%), Gaps = 5/131 (3%)
Query: 320 VFALTGTQTTNEDRLIRGTCFFNSTPLIAIIDTGATHCFIALESAYKLGLIVSDMKGEMV 379
++AL G TT+ R +R T N LIA+I++G+TH FI ++ + L + K V
Sbjct: 306 LYALEGVDTTSTIR-VRATIHRNR--LIALINSGSTHNFIGEKAVRGMNLKATTTKPFTV 362
Query: 380 VETPAKGSVTTSLVCLRCPTSMFGRDFVVDLVCLPLTGMDVIFGMNWLE-YNRVHINCFS 438
V S P M G F V L LPL G+D+ G+ WL N
Sbjct: 363 RVVNGMPLVCRSRY-EAIPVVMGGVVFPVTLYALPLMGLDLAMGVQWLSTLGPTLCNWKE 421
Query: 439 KTMHFSSAEEE 449
+T+ F A +E
Sbjct: 422 QTLQFHWAGDE 432
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.351 0.154 0.543
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 28,763,807
Number of Sequences: 26719
Number of extensions: 1130178
Number of successful extensions: 5083
Number of sequences better than 10.0: 84
Number of HSP's better than 10.0 without gapping: 52
Number of HSP's successfully gapped in prelim test: 33
Number of HSP's that attempted gapping in prelim test: 4686
Number of HSP's gapped (non-prelim): 288
length of query: 1454
length of database: 11,318,596
effective HSP length: 112
effective length of query: 1342
effective length of database: 8,326,068
effective search space: 11173583256
effective search space used: 11173583256
T: 11
A: 40
X1: 14 ( 7.1 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 38 (21.9 bits)
S2: 67 (30.4 bits)
Medicago: description of AC130807.5


