

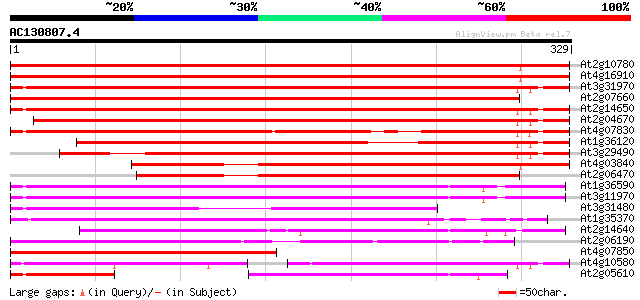
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC130807.4 - phase: 0
(329 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
At2g10780 pseudogene 360 e-100
At4g16910 retrotransposon like protein 352 2e-97
At3g31970 hypothetical protein 345 1e-95
At2g07660 putative retroelement pol polyprotein 345 1e-95
At2g14650 putative retroelement pol polyprotein 335 2e-92
At2g04670 putative retroelement pol polyprotein 322 1e-88
At4g07830 putative reverse transcriptase 291 3e-79
At1g36120 putative reverse transcriptase gb|AAD22339.1 258 2e-69
At3g29490 hypothetical protein 258 4e-69
At4g03840 putative transposon protein 236 1e-62
At2g06470 putative retroelement pol polyprotein 219 2e-57
At1g36590 hypothetical protein 206 2e-53
At3g11970 hypothetical protein 205 3e-53
At3g31480 hypothetical protein 191 6e-49
At1g35370 hypothetical protein 180 1e-45
At2g14640 putative retroelement pol polyprotein 162 2e-40
At2g06190 putative Ty3-gypsy-like retroelement pol polyprotein 151 5e-37
At4g07850 putative polyprotein 149 3e-36
At4g10580 putative reverse-transcriptase -like protein 115 3e-26
At2g05610 putative retroelement pol polyprotein 82 5e-16
>At2g10780 pseudogene
Length = 1611
Score = 360 bits (924), Expect = e-100
Identities = 177/346 (51%), Positives = 241/346 (69%), Gaps = 18/346 (5%)
Query: 1 MDFVMSLPNTTRG-HDSIWVVVDRLRKSAHFIPINIKYPVAQLAEIYIHNIVKLHGVPSS 59
MDFV LP + H+++WVVVDRL KSAHF+ I+ K +AE YI IV+LHG+P S
Sbjct: 1237 MDFVTGLPTGIKSKHNAVWVVVDRLTKSAHFMAISDKDGAEIIAEKYIDEIVRLHGIPVS 1296
Query: 60 IVSDRDPRFTSRFWKSLQDALGSKLRLSSAYHPQTDGQSKRTIQSLEYLLRVCVLEEGGA 119
IVSDRD RFTS+FWK+ Q ALG+++ LS+AYHPQTD QS+RTIQ+LE +LR CVL+ GG
Sbjct: 1297 IVSDRDTRFTSKFWKAFQKALGTRVNLSTAYHPQTDEQSERTIQTLEDMLRACVLDWGGN 1356
Query: 120 WDSHLPLIEFTYNNSYHSSIGMAPFKALYGRRCRTPLCWFESGESVVLGPDLVHETTEKV 179
W+ +L L+EF YNNS+ +SIGM+P++ALYGR CRTPLCW GE + GP +V ETTE++
Sbjct: 1357 WEKYLRLVEFAYNNSFQASIGMSPYEALYGRACRTPLCWTPVGERRLFGPTIVDETTERM 1416
Query: 180 KMIRKKMKASQSRQKSYHDKRRKALEFQEGDHVFLRVTPMTGVGHALKSRKLTPKFIGPY 239
K ++ K+K +Q RQKSY +KRRK LEFQ GD V+L+ G G +KL+P+++GPY
Sbjct: 1417 KFLKIKLKEAQDRQKSYANKRRKELEFQVGDLVYLKAMTYKGAGRFTSRKKLSPRYVGPY 1476
Query: 240 QISERVGTVAYRVGLPPHLSNLHDVFHVSQLRKYVADPSHVIPRDDVQVRDNLTVETLP- 298
++ ERVG VAY++ LPP L+ H+VFHVSQLRK ++D + +++N+TVE P
Sbjct: 1477 KVIERVGAVAYKLDLPPKLNAFHNVFHVSQLRKCLSDQEESVEDIPPGLKENMTVEAWPV 1536
Query: 299 ----------------LVKVVWDGATGESLTWELESKMRDSYPKLF 328
L+KV+W+ E TWE E+KM+ ++P+ F
Sbjct: 1537 RIMDRMTKGTRGKARDLLKVLWNCRGREEYTWETENKMKANFPEWF 1582
>At4g16910 retrotransposon like protein
Length = 687
Score = 352 bits (903), Expect = 2e-97
Identities = 174/346 (50%), Positives = 237/346 (68%), Gaps = 18/346 (5%)
Query: 1 MDFVMSLPNTTRG-HDSIWVVVDRLRKSAHFIPINIKYPVAQLAEIYIHNIVKLHGVPSS 59
MDFV LP + H+++WVVVDRL KSAHF+ I+ K +AE YI IV+LHG+P S
Sbjct: 321 MDFVTGLPTGIKSKHNAVWVVVDRLTKSAHFMAISDKDAAEIIAEKYIDEIVRLHGIPVS 380
Query: 60 IVSDRDPRFTSRFWKSLQDALGSKLRLSSAYHPQTDGQSKRTIQSLEYLLRVCVLEEGGA 119
IVSDRD RFTS+FWK Q LG+++ LS+AYHPQTDGQS+RTIQ+LE +LR CVL+ GG
Sbjct: 381 IVSDRDTRFTSKFWKPFQKVLGTRVNLSTAYHPQTDGQSERTIQTLEDMLRACVLDWGGN 440
Query: 120 WDSHLPLIEFTYNNSYHSSIGMAPFKALYGRRCRTPLCWFESGESVVLGPDLVHETTEKV 179
W+ +L L+EF YNNS+ +SIGM+P++ALYGR RTPLCW GE + GP +V ETT+K+
Sbjct: 441 WEKYLRLVEFAYNNSFQASIGMSPYEALYGRAGRTPLCWTPVGERRLFGPAVVDETTKKM 500
Query: 180 KMIRKKMKASQSRQKSYHDKRRKALEFQEGDHVFLRVTPMTGVGHALKSRKLTPKFIGPY 239
K ++ K+K +Q RQKSY +KRRK LEFQ GD V+L+ G G +KL P+++GPY
Sbjct: 501 KFLKIKLKEAQDRQKSYANKRRKELEFQVGDLVYLKAMTYKGAGRFTSRKKLRPRYVGPY 560
Query: 240 QISERVGTVAYRVGLPPHLSNLHDVFHVSQLRKYVADPSHVIPRDDVQVRDNLTVETLP- 298
++ ERVG VAY++ LPP L H+VFHVSQLRK +++ + +++N+TVE P
Sbjct: 561 KVIERVGAVAYKLDLPPKLDAFHNVFHVSQLRKCLSEQEESMEDVPPGLKENMTVEAWPV 620
Query: 299 ----------------LVKVVWDGATGESLTWELESKMRDSYPKLF 328
L+K++W+ E TWE E+KM+ ++P+ F
Sbjct: 621 RIMDQMKKGTRGKSMDLLKILWNCGGREEYTWETETKMKANFPEWF 666
>At3g31970 hypothetical protein
Length = 1329
Score = 345 bits (886), Expect = 1e-95
Identities = 174/347 (50%), Positives = 238/347 (68%), Gaps = 22/347 (6%)
Query: 1 MDFVMSLPNTTRGHDSIWVVVDRLRKSAHFIPINIKYPVAQLAEIYIHNIVKLHGVPSSI 60
MDFV+ LP +R D+IWV+VDRL KSAHF+ I LA+ Y+ IV+LHGVP SI
Sbjct: 980 MDFVVGLP-VSRTKDAIWVIVDRLTKSAHFLAIRKTDGAVLLAKKYVSEIVELHGVPVSI 1038
Query: 61 VSDRDPRFTSRFWKSLQDALGSKLRLSSAYHPQTDGQSKRTIQSLEYLLRVCVLEEGGAW 120
VSDRD +FTS FW++ Q +G+K+++S+AYHPQTDGQS+RTIQ+LE +LR+CVL+ GG W
Sbjct: 1039 VSDRDSKFTSAFWRAFQGEMGTKVQMSTAYHPQTDGQSERTIQTLEDMLRMCVLDRGGHW 1098
Query: 121 DSHLPLIEFTYNNSYHSSIGMAPFKALYGRRCRTPLCWFESGESVVLGPDLVHETTEKVK 180
HL L+EF YNNSY +SI MAPF+ALYGR CRTPLCW + GE + G D V ETTE+++
Sbjct: 1099 ADHLSLVEFAYNNSYQASIRMAPFEALYGRPCRTPLCWTQVGERSIYGADYVLETTERIR 1158
Query: 181 MIRKKMKASQSRQKSYHDKRRKALEFQEGDHVFLRVTPMTGVGHALKSRKLTPKFIGPYQ 240
+++ MK +Q RQ+SY DKRR+ LEF+ GD V+L++ + G ++ KLTP+++GP++
Sbjct: 1159 VLKLNMKEAQDRQRSYADKRRRELEFEVGDRVYLKMAMLRGPNRSISETKLTPRYMGPFR 1218
Query: 241 ISERVGTVAYRVGLPPHLSNLHDVFHVSQLRKYVADPSHVIPRDDVQVRDNLTVET---- 296
I ERVG VAYR+ LP + H VFHVS LRK + V+ + ++ N+T+E
Sbjct: 1219 IVERVGPVAYRLELPDVMRAFHKVFHVSMLRKCLHKDDEVLAKILEDLQPNMTLEARPVR 1278
Query: 297 -------------LPLVKVVW--DGATGESLTWELESKMRDSYPKLF 328
+PL+KV+W DG T E TWE E++M+ S+ K F
Sbjct: 1279 ILERRIKELRRKKIPLIKVLWNCDGVTEE--TWEPEARMKASFKKWF 1323
>At2g07660 putative retroelement pol polyprotein
Length = 949
Score = 345 bits (886), Expect = 1e-95
Identities = 165/300 (55%), Positives = 222/300 (74%), Gaps = 1/300 (0%)
Query: 1 MDFVMSLPNTTRG-HDSIWVVVDRLRKSAHFIPINIKYPVAQLAEIYIHNIVKLHGVPSS 59
MDFV LP + H+++WVVVDRL KSAHF+ I+ K +AE YI I++LHG+P S
Sbjct: 629 MDFVTRLPTGIKSKHNAVWVVVDRLTKSAHFMAISDKDGAEIIAEKYIDEIMRLHGIPVS 688
Query: 60 IVSDRDPRFTSRFWKSLQDALGSKLRLSSAYHPQTDGQSKRTIQSLEYLLRVCVLEEGGA 119
IVSDRD RFTS+FW + Q ALG+++ LS+AYHPQTDGQS+RTIQ+LE +LR CVL+ GG
Sbjct: 689 IVSDRDTRFTSKFWNAFQKALGTRVNLSTAYHPQTDGQSERTIQTLEDMLRACVLDWGGN 748
Query: 120 WDSHLPLIEFTYNNSYHSSIGMAPFKALYGRRCRTPLCWFESGESVVLGPDLVHETTEKV 179
W+ +L LIEF YNNS+ +SIGM+P++ALYGR CRTPLCW GE + GP +V ETTE++
Sbjct: 749 WEKYLRLIEFAYNNSFQASIGMSPYEALYGRACRTPLCWTPVGERRLFGPTIVDETTERM 808
Query: 180 KMIRKKMKASQSRQKSYHDKRRKALEFQEGDHVFLRVTPMTGVGHALKSRKLTPKFIGPY 239
K ++ K+K +Q RQKSY +KRRK LEFQ GD V+L+ G G +KL+P+++GPY
Sbjct: 809 KFLKIKLKEAQDRQKSYANKRRKELEFQVGDLVYLKAMTYKGAGRFTSRKKLSPRYVGPY 868
Query: 240 QISERVGTVAYRVGLPPHLSNLHDVFHVSQLRKYVADPSHVIPRDDVQVRDNLTVETLPL 299
++ ERVG VAY++ LPP L+ H+VFHVSQLRKY++D + +++N+TVE P+
Sbjct: 869 KVIERVGAVAYKLDLPPKLNVFHNVFHVSQLRKYLSDQEESVEDIPPGLKENMTVEAWPV 928
>At2g14650 putative retroelement pol polyprotein
Length = 1328
Score = 335 bits (859), Expect = 2e-92
Identities = 171/347 (49%), Positives = 236/347 (67%), Gaps = 22/347 (6%)
Query: 1 MDFVMSLPNTTRGHDSIWVVVDRLRKSAHFIPINIKYPVAQLAEIYIHNIVKLHGVPSSI 60
+DFV+ LP +R D+IWV+VDRL KSAHF+ I A LA+ Y+ IVKLHGVP SI
Sbjct: 982 IDFVVGLP-VSRTKDAIWVIVDRLTKSAHFLAIRKTDGAAVLAKKYVSEIVKLHGVPVSI 1040
Query: 61 VSDRDPRFTSRFWKSLQDALGSKLRLSSAYHPQTDGQSKRTIQSLEYLLRVCVLEEGGAW 120
VSDRD +FTS FW++ Q +G+K+++S+AYHPQT GQS+RTIQ+LE +LR+CVL+ GG W
Sbjct: 1041 VSDRDSKFTSAFWRAFQAEMGTKVQMSTAYHPQTYGQSERTIQTLEDMLRMCVLDWGGHW 1100
Query: 121 DSHLPLIEFTYNNSYHSSIGMAPFKALYGRRCRTPLCWFESGESVVLGPDLVHETTEKVK 180
HL L+EF YNNSY +SIGMAPF+ALY R CRTPLC + GE + G D V ETTE+++
Sbjct: 1101 ADHLSLVEFAYNNSYPASIGMAPFEALYERPCRTPLCLTQVGERSIYGADYVQETTERIR 1160
Query: 181 MIRKKMKASQSRQKSYHDKRRKALEFQEGDHVFLRVTPMTGVGHALKSRKLTPKFIGPYQ 240
+++ MK +Q RQ+SY DKRR+ LEF+ GD V+L++ + G ++ KL+P+++GP++
Sbjct: 1161 VLKLNMKEAQDRQRSYADKRRRELEFEVGDRVYLKMAMLRGPNRSISETKLSPRYMGPFR 1220
Query: 241 ISERVGTVAYRVGLPPHLSNLHDVFHVSQLRKYVADPSHVIPRDDVQVRDNLTVET---- 296
I ERVG VAYR+ LP + H VFHVS LRK + V+ + ++ N+T+E
Sbjct: 1221 IVERVGPVAYRLELPDVMRAFHKVFHVSMLRKCLHKDDEVLAKIPEDLQPNMTLEARPVR 1280
Query: 297 -------------LPLVKVVW--DGATGESLTWELESKMRDSYPKLF 328
+PL+KV+W DG T E TWE E++M+ + K F
Sbjct: 1281 VLERRIKELRRKKIPLIKVLWDCDGVTKE--TWEPEARMKARFKKWF 1325
>At2g04670 putative retroelement pol polyprotein
Length = 1411
Score = 322 bits (826), Expect = 1e-88
Identities = 162/333 (48%), Positives = 226/333 (67%), Gaps = 21/333 (6%)
Query: 15 DSIWVVVDRLRKSAHFIPINIKYPVAQLAEIYIHNIVKLHGVPSSIVSDRDPRFTSRFWK 74
D+IWV++DRL KSAHF+ I A LA+ Y+ IVKLHGVP SIVSDRD +FT FW+
Sbjct: 1075 DAIWVIMDRLTKSAHFLAIRKTDGAAVLAKKYVSEIVKLHGVPVSIVSDRDSKFTFAFWR 1134
Query: 75 SLQDALGSKLRLSSAYHPQTDGQSKRTIQSLEYLLRVCVLEEGGAWDSHLPLIEFTYNNS 134
+ Q +G+K+++S+AYHPQTDGQS+RTIQ+LE +LR+CVL+ GG W HL L+EF YNNS
Sbjct: 1135 AFQAKMGTKVQMSTAYHPQTDGQSERTIQTLEDMLRMCVLDWGGHWADHLSLVEFAYNNS 1194
Query: 135 YHSSIGMAPFKALYGRRCRTPLCWFESGESVVLGPDLVHETTEKVKMIRKKMKASQSRQK 194
Y +SIGMAPF+ALYGR C TPL W + E + G D V ETTE++++++ MK +Q+RQ+
Sbjct: 1195 YQASIGMAPFEALYGRPCWTPLRWTQVEERSIYGADYVQETTERIRVLKLNMKEAQARQR 1254
Query: 195 SYHDKRRKALEFQEGDHVFLRVTPMTGVGHALKSRKLTPKFIGPYQISERVGTVAYRVGL 254
SY DKRR+ LEF+ GD V+L++ + G ++ KL+P+++GP++I ERVG VAYR+ L
Sbjct: 1255 SYADKRRRELEFEVGDRVYLKMAMLRGPNRSILETKLSPRYMGPFRIVERVGPVAYRLEL 1314
Query: 255 PPHLSNLHDVFHVSQLRKYVADPSHVIPRDDVQVRDNLTVET-----------------L 297
P + H VFHV LRK + V+ + ++ N+T+E +
Sbjct: 1315 PDVMRAFHKVFHVLMLRKCLHKDDEVLVKIPEDLQPNMTLEARPVRVLERRIKELRRKKI 1374
Query: 298 PLVKVVW--DGATGESLTWELESKMRDSYPKLF 328
PL+KV+W DG T E TWE E++M+ + K F
Sbjct: 1375 PLIKVLWDCDGVTEE--TWEPEARMKARFKKWF 1405
>At4g07830 putative reverse transcriptase
Length = 611
Score = 291 bits (746), Expect = 3e-79
Identities = 162/347 (46%), Positives = 219/347 (62%), Gaps = 43/347 (12%)
Query: 1 MDFVMSLPNTTRGHDSIWVVVDRLRKSAHFIPINIKYPVAQLAEIYIHNIVKLHGVPSSI 60
MDFV+ LP +R D+IWV+VDRL KSAHF+ I A LA+ Y+ IVKLHGVP SI
Sbjct: 247 MDFVVGLP-VSRTKDAIWVIVDRLTKSAHFLAIRKTDGAAVLAKKYVSEIVKLHGVPVSI 305
Query: 61 VSDRDPRFTSRFWKSLQDALGSKLRLSSAYHPQTDGQSKRTIQSLEYLLRVCVLEEGGAW 120
VSDRD +FTS FW++ Q +G+K+++S+AYHPQTDGQS+RTIQ+LE +LR+CVL+ G W
Sbjct: 306 VSDRDSKFTSAFWRAFQAEMGTKVQMSTAYHPQTDGQSERTIQTLEDMLRMCVLDWRGHW 365
Query: 121 DSHLPLIEFTYNNSYHSSIGMAPFKALYGRRCRTPLCWFESGESVVLGPDLVHETTEKVK 180
HL L+EF YNNSY +SIGMAPF+ LYGR CRT LCW + GE + G D V E TE+++
Sbjct: 366 ADHLSLVEFAYNNSYQASIGMAPFEVLYGRPCRT-LCWTQVGERSIYGADYVQEITERIR 424
Query: 181 MIRKKMKASQSRQKSYHDKRRKALEFQEGDHVFLRVTPMTGVGHALKSRKLTPKFIGPYQ 240
+++ MK +Q+RQ+SY DKRRK LEF+ GD V + GH +S
Sbjct: 425 VLKLNMKEAQNRQRSYADKRRKELEFEVGDSV-------SQDGHVARS------------ 465
Query: 241 ISERVGTVAYRVGLPPHLSNLHDVFHVSQLRKYVADPSHVIPRDDVQVRDNLTVET---- 296
+RVG VA+R+ L + H VFHVS LRK + V+ + ++ N+T+E
Sbjct: 466 -EQRVGPVAFRLELSDVMRAFHKVFHVSMLRKCLHKDDEVLAKIPEDLQPNMTLEARPVR 524
Query: 297 -------------LPLVKVV--WDGATGESLTWELESKMRDSYPKLF 328
+PL+KV+ DG T E TWE E++++ + K F
Sbjct: 525 VLERRIKELRRKKIPLIKVLRNCDGVTEE--TWEPEARLKARFKKWF 569
>At1g36120 putative reverse transcriptase gb|AAD22339.1
Length = 1235
Score = 258 bits (660), Expect = 2e-69
Identities = 137/308 (44%), Positives = 191/308 (61%), Gaps = 50/308 (16%)
Query: 40 AQLAEIYIHNIVKLHGVPSSIVSDRDPRFTSRFWKSLQDALGSKLRLSSAYHPQTDGQSK 99
A L + Y+ IVKLHGVP SI+S RD +FTS FW++ Q +G+K+++S+AYHPQTDGQS+
Sbjct: 953 AVLPKKYVSEIVKLHGVPVSILSHRDSKFTSAFWRAFQVEMGTKVQMSTAYHPQTDGQSE 1012
Query: 100 RTIQSLEYLLRVCVLEEGGAWDSHLPLIEFTYNNSYHSSIGMAPFKALYGRRCRTPLCWF 159
RTIQ+LE +L++CVL+ GG W HL L++F YNNSY +SIGMAPF+ALYGR CRT LCW
Sbjct: 1013 RTIQTLEDMLQMCVLDWGGHWADHLSLVKFAYNNSYQASIGMAPFEALYGRPCRTLLCWT 1072
Query: 160 ESGESVVLGPDLVHETTEKVKMIRKKMKASQSRQKSYHDKRRKALEFQEGDHVFLRVTPM 219
+ GE + G D V ETTE++++++ MK +Q RQ+SY DKRR+ LEF+ G
Sbjct: 1073 QVGEKSIYGADYVQETTERIRVLKLNMKEAQDRQRSYADKRRRELEFEVGT--------- 1123
Query: 220 TGVGHALKSRKLTPKFIGPYQISERVGTVAYRVGLPPHLSNLHDVFHVSQLRKYVADPSH 279
+I ERVG VAYR+ LP + H+VFHVS LRK +
Sbjct: 1124 --------------------EIVERVGPVAYRLELPDVMRAFHNVFHVSMLRKCLHKDDE 1163
Query: 280 VIPRDDVQVRDNLTVET-----------------LPLVKVVW--DGATGESLTWELESKM 320
V+ + ++ N+T+E +P++KV+W DG T E TWE E+++
Sbjct: 1164 VLAKIPEDLQPNMTLEARPVRVLERRIKEVRRKKIPMIKVLWDCDGVTEE--TWEPEARI 1221
Query: 321 RDSYPKLF 328
+ + K F
Sbjct: 1222 KARFKKWF 1229
>At3g29490 hypothetical protein
Length = 438
Score = 258 bits (658), Expect = 4e-69
Identities = 136/318 (42%), Positives = 194/318 (60%), Gaps = 41/318 (12%)
Query: 30 FIPINIKYPVAQLAEIYIHNIVKLHGVPSSIVSDRDPRFTSRFWKSLQDALGSKLRLSSA 89
F+ I A LA+ Y+ IVKLHGVP+ +G+K+++S+
Sbjct: 40 FLAIRKTDGAAVLAKKYVSEIVKLHGVPAE--------------------MGTKVQMSTP 79
Query: 90 YHPQTDGQSKRTIQSLEYLLRVCVLEEGGAWDSHLPLIEFTYNNSYHSSIGMAPFKALYG 149
YHPQTDGQ +RTIQ+LE +LR+CVL+ GG W HL L+EF YNNSY + IGMAPF+ALYG
Sbjct: 80 YHPQTDGQFERTIQTLEDMLRMCVLDWGGHWADHLSLVEFAYNNSYQAGIGMAPFEALYG 139
Query: 150 RRCRTPLCWFESGESVVLGPDLVHETTEKVKMIRKKMKASQSRQKSYHDKRRKALEFQEG 209
R CRTPLCW + GE + G D V ETTE++++++ MK +Q RQ SY DKRR+ LEF+ G
Sbjct: 140 RPCRTPLCWTQVGERSIYGADYVQETTERIRVLKLNMKEAQDRQWSYADKRRRELEFEVG 199
Query: 210 DHVFLRVTPMTGVGHALKSRKLTPKFIGPYQISERVGTVAYRVGLPPHLSNLHDVFHVSQ 269
D V+L++ + G ++ KL+ +++GP++I ERVG VAY + LP + H VFHVS
Sbjct: 200 DRVYLKMAMLRGPNRSISETKLSLRYMGPFRIVERVGPVAYMLELPDVMRAFHKVFHVSM 259
Query: 270 LRKYVADPSHVIPRDDVQVRDNLTVET-----------------LPLVKVVW--DGATGE 310
LRK + V+ + ++ N+T+E + L+KV+W DG T E
Sbjct: 260 LRKCLHKDDEVLAKIPEDLQPNMTLEARQVRVLERRIKELQRKKISLIKVLWDCDGVTEE 319
Query: 311 SLTWELESKMRDSYPKLF 328
TW+ E++M+ + K F
Sbjct: 320 --TWQPEARMKARFKKWF 335
>At4g03840 putative transposon protein
Length = 973
Score = 236 bits (603), Expect = 1e-62
Identities = 121/274 (44%), Positives = 170/274 (61%), Gaps = 36/274 (13%)
Query: 72 FWKSLQDALGSKLRLSSAYHPQTDGQSKRTIQSLEYLLRVCVLEEGGAWDSHLPLIEFTY 131
FWK+ Q ALG+++ LS+AYHPQTDGQS+RTIQ+LE +LR C L+ GG W+ +L L
Sbjct: 690 FWKAFQKALGTRVNLSTAYHPQTDGQSERTIQTLEDMLRACALDWGGNWEKYLRL----- 744
Query: 132 NNSYHSSIGMAPFKALYGRRCRTPLCWFESGESVVLGPDLVHETTEKVKMIRKKMKASQS 191
ALYGR CRTPLCW GE + GP +V ETTE++K ++ K+K +Q
Sbjct: 745 --------------ALYGRACRTPLCWTPVGERRLFGPIIVDETTERMKFLKIKLKEAQD 790
Query: 192 RQKSYHDKRRKALEFQEGDHVFLRVTPMTGVGHALKSRKLTPKFIGPYQISERVGTVAYR 251
RQKSY +KRRK LEFQ D V+L+ G G +KL+P+++GPY++ ERVG VAY+
Sbjct: 791 RQKSYANKRRKELEFQVEDLVYLKAMTYKGAGRFTSRKKLSPRYVGPYKVIERVGAVAYK 850
Query: 252 VGLPPHLSNLHDVFHVSQLRKYVADPSHVIPRDDVQVRDNLTVETLP------------- 298
+ LPP L+ H+VFHVSQLRK +++ + +++N+TVE P
Sbjct: 851 LDLPPKLNAFHNVFHVSQLRKCLSNQEESVEDVPPGLKENMTVEAWPVQIMDRMTKGTRG 910
Query: 299 ----LVKVVWDGATGESLTWELESKMRDSYPKLF 328
L+KV+W+ E TWE E+KM+ ++ + F
Sbjct: 911 KSRDLLKVLWNCGGREQYTWETENKMKANFSEWF 944
>At2g06470 putative retroelement pol polyprotein
Length = 899
Score = 219 bits (557), Expect = 2e-57
Identities = 108/225 (48%), Positives = 149/225 (66%), Gaps = 19/225 (8%)
Query: 75 SLQDALGSKLRLSSAYHPQTDGQSKRTIQSLEYLLRVCVLEEGGAWDSHLPLIEFTYNNS 134
+ Q ALG+++ LS+AYHPQTDGQS+RTIQ+LE +LR CVL+ GG W+ +L L
Sbjct: 690 AFQKALGTRVNLSTAYHPQTDGQSERTIQTLEDMLRACVLDWGGNWEKYLTL-------- 741
Query: 135 YHSSIGMAPFKALYGRRCRTPLCWFESGESVVLGPDLVHETTEKVKMIRKKMKASQSRQK 194
ALYGR CRTPLCW GE + GP +V ETTE++K ++ K+K + RQK
Sbjct: 742 -----------ALYGRACRTPLCWTPVGERRLFGPTIVDETTERMKFLKIKLKEAHDRQK 790
Query: 195 SYHDKRRKALEFQEGDHVFLRVTPMTGVGHALKSRKLTPKFIGPYQISERVGTVAYRVGL 254
SY +KRRK LEFQ GD V+L+ G G +KL+P+++GPY++ ERVG VAY++ L
Sbjct: 791 SYANKRRKELEFQVGDLVYLKAMTYKGAGRFTSRKKLSPRYVGPYKVIERVGAVAYKLDL 850
Query: 255 PPHLSNLHDVFHVSQLRKYVADPSHVIPRDDVQVRDNLTVETLPL 299
PP L+ H+VFHVSQLRK ++D + +++N+TVE P+
Sbjct: 851 PPKLNAFHNVFHVSQLRKCLSDQEESVEDVPPGLKENMTVEAWPV 895
>At1g36590 hypothetical protein
Length = 1499
Score = 206 bits (523), Expect = 2e-53
Identities = 122/345 (35%), Positives = 184/345 (52%), Gaps = 25/345 (7%)
Query: 1 MDFVMSLPNTTRGHDSIWVVVDRLRKSAHFIPINIKYPVAQLAEIYIHNIVKLHGVPSSI 60
MDF+ LP + G I VVVDRL K+AHFI ++ Y +A+ Y+ N+ KLHG P+SI
Sbjct: 1158 MDFIEGLP-VSGGKTVIMVVVDRLSKAAHFIALSHPYSALTVAQAYLDNVFKLHGCPTSI 1216
Query: 61 VSDRDPRFTSRFWKSLQDALGSKLRLSSAYHPQTDGQSKRTIQSLEYLLRVCVLEEGGAW 120
VSDRD FTS FW+ G L+L+SAYHPQ+DGQ++ + LE LR + W
Sbjct: 1217 VSDRDVVFTSEFWREFFTLQGVALKLTSAYHPQSDGQTEVVNRCLETYLRCMCHDRPQLW 1276
Query: 121 DSHLPLIEFTYNNSYHSSIGMAPFKALYGRRCRTPLCWFESGESVVLGPDLVHETTEKVK 180
L L E+ YN +YHSS M PF+ +YG+ L + V + + E + +
Sbjct: 1277 SKWLALAEYWYNTNYHSSSRMTPFEIVYGQVPPVHLPYLPGESKVAVVARSLQEREDMLL 1336
Query: 181 MIRKKMKASQSRQKSYHDKRRKALEFQEGDHVFLRVTPMTGVGHALK-SRKLTPKFIGPY 239
++ + +Q R K + D+ R EF+ GD+V++++ P ++ ++KL+PK+ GPY
Sbjct: 1337 FLKFHLMRAQHRMKQFADQHRTEREFEIGDYVYVKLQPYRQQSVVMRANQKLSPKYFGPY 1396
Query: 240 QISERVGTVAYRVGLPPHLSNLHDVFHVSQLRKYVAD------------------PSHVI 281
+I +R G VAY++ LP + S +H VFHVSQL+ V + P V+
Sbjct: 1397 KIIDRCGEVAYKLALPSY-SQVHPVFHVSQLKVLVGNVSTTVHLPSVMQDVFEKVPEKVV 1455
Query: 282 PRDDVQVRDNLTVETLPLVKVVWDGATGESLTWELESKMRDSYPK 326
R V N + + V V W E TWE ++ ++P+
Sbjct: 1456 ERKMV----NRQGKAVTKVLVKWSNEPLEEATWEFLFDLQKTFPE 1496
>At3g11970 hypothetical protein
Length = 1499
Score = 205 bits (521), Expect = 3e-53
Identities = 122/345 (35%), Positives = 183/345 (52%), Gaps = 25/345 (7%)
Query: 1 MDFVMSLPNTTRGHDSIWVVVDRLRKSAHFIPINIKYPVAQLAEIYIHNIVKLHGVPSSI 60
MDF+ LP + G I VVVDRL K+AHFI ++ Y +A Y+ N+ KLHG P+SI
Sbjct: 1158 MDFIEGLP-VSGGKTVIMVVVDRLSKAAHFIALSHPYSALTVAHAYLDNVFKLHGCPTSI 1216
Query: 61 VSDRDPRFTSRFWKSLQDALGSKLRLSSAYHPQTDGQSKRTIQSLEYLLRVCVLEEGGAW 120
VSDRD FTS FW+ G L+L+SAYHPQ+DGQ++ + LE LR + W
Sbjct: 1217 VSDRDVVFTSEFWREFFTLQGVALKLTSAYHPQSDGQTEVVNRCLETYLRCMCHDRPQLW 1276
Query: 121 DSHLPLIEFTYNNSYHSSIGMAPFKALYGRRCRTPLCWFESGESVVLGPDLVHETTEKVK 180
L L E+ YN +YHSS M PF+ +YG+ L + V + + E + +
Sbjct: 1277 SKWLALAEYWYNTNYHSSSRMTPFEIVYGQVPPVHLPYLPGESKVAVVARSLQEREDMLL 1336
Query: 181 MIRKKMKASQSRQKSYHDKRRKALEFQEGDHVFLRVTPMTGVGHALK-SRKLTPKFIGPY 239
++ + +Q R K + D+ R EF+ GD+V++++ P ++ ++KL+PK+ GPY
Sbjct: 1337 FLKFHLMRAQHRMKQFADQHRTEREFEIGDYVYVKLQPYRQQSVVMRANQKLSPKYFGPY 1396
Query: 240 QISERVGTVAYRVGLPPHLSNLHDVFHVSQLRKYVAD------------------PSHVI 281
+I +R G VAY++ LP + S +H VFHVSQL+ V + P V+
Sbjct: 1397 KIIDRCGEVAYKLALPSY-SQVHPVFHVSQLKVLVGNVSTTVHLPSVMQDVFEKVPEKVV 1455
Query: 282 PRDDVQVRDNLTVETLPLVKVVWDGATGESLTWELESKMRDSYPK 326
R V N + + V V W E TWE ++ ++P+
Sbjct: 1456 ERKMV----NRQGKAVTKVLVKWSNEPLEEATWEFLFDLQKTFPE 1496
>At3g31480 hypothetical protein
Length = 338
Score = 191 bits (484), Expect = 6e-49
Identities = 103/251 (41%), Positives = 149/251 (59%), Gaps = 43/251 (17%)
Query: 1 MDFVMSLPNTTRGHDSIWVVVDRLRKSAHFIPINIKYPVAQLAEIYIHNIVKLHGVPSSI 60
MDFV+ LP +R D+IWV+ DRL SA F+ I VA LA Y+ IVKLHGV SI
Sbjct: 1 MDFVVGLP-MSRTKDAIWVIGDRLTNSADFLAIRKTDGVAVLANKYVSEIVKLHGVHVSI 59
Query: 61 VSDRDPRFTSRFWKSLQDALGSKLRLSSAYHPQTDGQSKRTIQSLEYLLRVCVLEEGGAW 120
VSD+D +FTS FW + Q +G+K+++S+AYHP+TDG+ ++TIQ+LE +LR+
Sbjct: 60 VSDKDSKFTSAFWIAFQAEMGTKVQISTAYHPKTDGKFEKTIQTLEDMLRM--------- 110
Query: 121 DSHLPLIEFTYNNSYHSSIGMAPFKALYGRRCRTPLCWFESGESVVLGPDLVHETTEKVK 180
TPLCW + GE + G + VHETTE+++
Sbjct: 111 ---------------------------------TPLCWTQVGERSIYGANYVHETTERIQ 137
Query: 181 MIRKKMKASQSRQKSYHDKRRKALEFQEGDHVFLRVTPMTGVGHALKSRKLTPKFIGPYQ 240
+++ MK + RQ+SY D RR+ +EF+ GD V+L++ + + KL+PK++GP++
Sbjct: 138 VLKLNMKEAHDRQRSYADMRRREVEFEVGDRVYLKMDMLQSPKRFILETKLSPKYMGPFR 197
Query: 241 ISERVGTVAYR 251
I ERVG VAYR
Sbjct: 198 IVERVGPVAYR 208
>At1g35370 hypothetical protein
Length = 1447
Score = 180 bits (456), Expect = 1e-45
Identities = 115/321 (35%), Positives = 168/321 (51%), Gaps = 22/321 (6%)
Query: 1 MDFVMSLPNTTRGHDSIWVVVDRLRKSAHFIPINIKYPVAQLAEIYIHNIVKLHGVPSSI 60
MDF+ LPN+ G I VVVDRL K+AHF+ + Y +A+ ++ N+ K HG P+SI
Sbjct: 1129 MDFIEGLPNSG-GKSVIMVVVDRLSKAAHFVALAHPYSALTVAQAFLDNVYKHHGCPTSI 1187
Query: 61 VSDRDPRFTSRFWKSLQDALGSKLRLSSAYHPQTDGQSKRTIQSLEYLLRVCVLEEGGAW 120
VSDRD FTS FWK G +LR+SSAYHPQ+DGQ++ + LE LR W
Sbjct: 1188 VSDRDVLFTSDFWKEFFKLQGVELRMSSAYHPQSDGQTEVVNRCLENYLRCMCHARPHLW 1247
Query: 121 DSHLPLIEFTYNNSYHSSIGMAPFKALYGRRCRTPLCWFESGESVVLGPDLVHETTEKVK 180
+ LPL E+ YN +YHSS M PF+ +YG+ L + V + + E +
Sbjct: 1248 NKWLPLAEYWYNTNYHSSSQMTPFELVYGQAPPIHLPYLPGKSKVAVVARSLQERENMLL 1307
Query: 181 MIRKKMKASQSRQKSYHDKRRKALEFQEGDHVFLRVTPMTGVGHALK-SRKLTPKFIGPY 239
++ + +Q R K + D+ R F GD V++++ P L+ ++KL+PK+ GPY
Sbjct: 1308 FLKFHLMRAQHRMKQFADQHRTERTFDIGDFVYVKLQPYRQQSVVLRVNQKLSPKYFGPY 1367
Query: 240 QISER-----VGTVAYRVGLPPHLSNLHDVFHVSQLRKYVADPSHVIPRDDVQVRDNLTV 294
+I E+ VG V LP S L D+F + P +++ R V+ +
Sbjct: 1368 KIIEKCGEVMVGNVTTSTQLP---SVLPDIFEKA--------PEYILERKLVKRQGR--A 1414
Query: 295 ETLPLVKVVWDGATGESLTWE 315
T+ LVK W G E TW+
Sbjct: 1415 ATMVLVK--WIGEPVEEATWK 1433
>At2g14640 putative retroelement pol polyprotein
Length = 945
Score = 162 bits (411), Expect = 2e-40
Identities = 108/307 (35%), Positives = 163/307 (52%), Gaps = 28/307 (9%)
Query: 42 LAEIYIHNIVKLHGVPSSIVSDRDPRFTSRFWKSLQDALGSKLRLSSAYHPQTDGQSKRT 101
+A ++ ++VKLHG+P SIVSD DP F S FW+ +KL +S+AYHPQTDGQ++
Sbjct: 634 VAAKFVSHVVKLHGIPRSIVSDCDPIFMSLFWQEFWKLSRTKLWMSTAYHPQTDGQTEVV 693
Query: 102 IQSLEYLLRVCVLEEGGAWDSHLPLIEFTYNNSYHSSIGMAPFKALYGRRCRTPLCWFES 161
+ +E LR V W S +P E+ YN ++H+S GM PF+ALYGR +P+ +E
Sbjct: 694 NRCIEQFLRCFVHYHPKQWSSFIPWAEYWYNTTFHASTGMTPFQALYGRP-PSPIPAYEL 752
Query: 162 GESVVLGP--DLVHETTEKVKMIRKKMKASQSRQKSYHDKRRKALEFQEGDHVFLRVTP- 218
G SVV G + + E + +++ + + + K D R + + FQ GD V LR+ P
Sbjct: 753 G-SVVCGELNEQMAARDELLAELKQHLVTANNCMKQQADSRLRDVSFQVGDWVLLRIQPY 811
Query: 219 MTGVGHALKSRKLTPKFIGPYQISERVGTVAYRVGLPPHLSNLHDVFHVSQLRKYVADPS 278
S+KL+ +F GP+Q++ + G VAYR+ LP + +H VFHVS L+ +V D
Sbjct: 812 RQKTLFRRSSQKLSHRFYGPFQVASKHGEVAYRLTLPEG-TRIHPVFHVSLLKPWVGDGE 870
Query: 279 ---------------HVIPRDDVQVR----DNLTVETLPLVKVVWDGATGESLTWELESK 319
+ P ++VR D V L V W+G E TWE +
Sbjct: 871 PDMGQLPPLRNNGELKLQPTAVLEVRWRSQDKKRVADL---LVQWEGLHIEDATWEEYDQ 927
Query: 320 MRDSYPK 326
+ S+P+
Sbjct: 928 LAASFPE 934
>At2g06190 putative Ty3-gypsy-like retroelement pol polyprotein
Length = 280
Score = 151 bits (381), Expect = 5e-37
Identities = 95/296 (32%), Positives = 149/296 (50%), Gaps = 21/296 (7%)
Query: 1 MDFVMSLPNTTRGHDSIWVVVDRLRKSAHFIPINIKYPVAQLAEIYIHNIVKLHGVPSSI 60
MDFV+ LP T RG DS++VVVDR K HFI + +A+++ +V+LHGVP SI
Sbjct: 1 MDFVLGLPRTQRGVDSVFVVVDRFSKMTHFIACKKTADASNIAKLFFKEVVRLHGVPKSI 60
Query: 61 VSDRDPRFTSRFWKSLQDALGSKLRLSSAYHPQTDGQSKRTIQSLEYLLRVCVLEEGGAW 120
SDRD +F S FW +L G+ L SS H Q DGQ++ T ++L ++R + W
Sbjct: 61 TSDRDTKFLSHFWSTLWRMFGTALNRSSTPHTQIDGQTEVTNRTLGNMVRSICGDNPKQW 120
Query: 121 DSHLPLIEFTYNNSYHSSIGMAPFKALYGRRCRTPLCWFESGESVVLGPDLVHETTEKVK 180
D LP IEF YN+ H + + G T + E +
Sbjct: 121 DLALPQIEFAYNSVVH-VVDLVKLPKALGASAET----------------MAEEILVVKE 163
Query: 181 MIRKKMKASQSRQKSYHDKRRKALEFQEGDHVFLRVTPMTGVGHALKSRKLTPKFIGPYQ 240
+++ K++A+ + K DKRR+ F+EGD V V G K+ P+ GP++
Sbjct: 164 VVKAKLEATGKKNKVAADKRRRFKVFKEGDDVM--VLLRKGRFAVGTYNKVKPRKYGPFK 221
Query: 241 ISERVGTVAYRVGLPPHLSNLHDVFHVSQLRKYVADPSHVIPRDDVQVRDNLTVET 296
+ ++ AY V LP + N+ + F+V+ + +Y AD + P ++++ + ET
Sbjct: 222 VLRKINDNAYVVALPKSM-NISNTFNVADIHEYHAD-GVLYPEENLRTSSSEVEET 275
>At4g07850 putative polyprotein
Length = 1138
Score = 149 bits (375), Expect = 3e-36
Identities = 73/156 (46%), Positives = 100/156 (63%)
Query: 1 MDFVMSLPNTTRGHDSIWVVVDRLRKSAHFIPINIKYPVAQLAEIYIHNIVKLHGVPSSI 60
MDFV+ LP T G DSI+VVVDR K AHFIP + +A ++ +V+LHG+P +I
Sbjct: 835 MDFVVGLPRTRTGKDSIFVVVDRFSKMAHFIPCHKTDDAMHIANLFFREVVRLHGMPKTI 894
Query: 61 VSDRDPRFTSRFWKSLQDALGSKLRLSSAYHPQTDGQSKRTIQSLEYLLRVCVLEEGGAW 120
VSDRD +F S FWK+L LG+KL S+ HPQTDGQ++ ++L LLR + + W
Sbjct: 895 VSDRDTKFLSYFWKTLWSKLGTKLLFSTTCHPQTDGQTEVVNRTLSTLLRALIKKNLKTW 954
Query: 121 DSHLPLIEFTYNNSYHSSIGMAPFKALYGRRCRTPL 156
+ LP +EF YN+S HS+ +PF+ +YG TPL
Sbjct: 955 EDCLPHVEFAYNHSVHSATKFSPFQIVYGFNPITPL 990
>At4g10580 putative reverse-transcriptase -like protein
Length = 1240
Score = 115 bits (288), Expect = 3e-26
Identities = 67/184 (36%), Positives = 102/184 (55%), Gaps = 22/184 (11%)
Query: 164 SVVLGPDLVHETTEKVKMIRKKMKASQSRQKSYHDKRRKALEFQEGDHVFLRVTPMTGVG 223
+ VL V E K+ + MK +Q RQ+SY DKRR+ LEF+ GD V+L++ + G
Sbjct: 1054 AAVLAKKFVSEIV-KLHGVPLNMKEAQDRQRSYADKRRRELEFEVGDRVYLKMAMLRGPN 1112
Query: 224 HALKSRKLTPKFIGPYQISERVGTVAYRVGLPPHLSNLHDVFHVSQLRKYVADPSHVIPR 283
++ KL+P+++GP++I ERV VAYR+ LP + H VFHVS LRK + + +
Sbjct: 1113 RSISETKLSPRYMGPFKIVERVEPVAYRLELPDVMRAFHKVFHVSMLRKCLHKDDEALAK 1172
Query: 284 DDVQVRDNLTVET-----------------LPLVKVVW--DGATGESLTWELESKMRDSY 324
++ N+T+E +PL+KV+W DG T E TWE E++M+ +
Sbjct: 1173 IPEDLQPNMTLEARPVRVLERRIKELRQKKIPLIKVLWDCDGVTEE--TWEPEARMKARF 1230
Query: 325 PKLF 328
K F
Sbjct: 1231 KKWF 1234
Score = 64.7 bits (156), Expect = 7e-11
Identities = 46/143 (32%), Positives = 73/143 (50%), Gaps = 5/143 (3%)
Query: 1 MDFVMSLPNTTRGHDSIWVVVDRLRKSAHFIPINIKYPVAQLAEIYIHNIVKLHGVPSSI 60
MD V+ L +R D+IWV+VDRL KSAHF+ I A LA+ ++ IVKLHGVP ++
Sbjct: 1017 MDLVVGL-RVSRTKDAIWVIVDRLTKSAHFLAIRKTDGAAVLAKKFVSEIVKLHGVPLNM 1075
Query: 61 --VSDRDPRFTSRFWKSLQDALGSKLRLSSAYHPQTDGQSKRTIQSLEYLLRVCVLE--E 116
DR + + + L+ +G ++ L A + T S Y+ ++E E
Sbjct: 1076 KEAQDRQRSYADKRRRELEFEVGDRVYLKMAMLRGPNRSISETKLSPRYMGPFKIVERVE 1135
Query: 117 GGAWDSHLPLIEFTYNNSYHSSI 139
A+ LP + ++ +H S+
Sbjct: 1136 PVAYRLELPDVMRAFHKVFHVSM 1158
>At2g05610 putative retroelement pol polyprotein
Length = 780
Score = 81.6 bits (200), Expect = 5e-16
Identities = 49/156 (31%), Positives = 85/156 (54%), Gaps = 5/156 (3%)
Query: 141 MAPFKALYGRRCRTPLCWFESGESVVLGPDLVHETTEKVKMIRKKMKASQSRQKSYHDKR 200
M P++A+YG+ L + V + + E + ++ + +Q R K D+
Sbjct: 625 MTPYEAVYGQPPPLHLPYLPGESKVAVVARSMQERESMILFLKFHLMRAQHRMKQLADQH 684
Query: 201 RKALEFQEGDHVFLRVTPMTGVGHALKS-RKLTPKFIGPYQISERVGTVAYRVGLPPHLS 259
EF+ GD+VF+++ P ++S +KL+PK+ GPY++ +R G VAY++ LP + S
Sbjct: 685 ITEREFEVGDYVFVKLQPYRQQSVVMRSTQKLSPKYFGPYKVIDRCGEVAYKLQLPAN-S 743
Query: 260 NLHDVFHVSQLRKY---VADPSHVIPRDDVQVRDNL 292
+H VFHVSQLR V +H++ + +R+NL
Sbjct: 744 QVHPVFHVSQLRVLVGTVTTSTHLLRCYLMSLRENL 779
Score = 58.9 bits (141), Expect = 4e-09
Identities = 31/61 (50%), Positives = 39/61 (63%), Gaps = 1/61 (1%)
Query: 1 MDFVMSLPNTTRGHDSIWVVVDRLRKSAHFIPINIKYPVAQLAEIYIHNIVKLHGVPSSI 60
MDF+ LP + G I VVVDRL K+AHFI + Y +A+ Y+ N+ KLHG PSSI
Sbjct: 549 MDFIDGLP-LSNGKTVIMVVVDRLSKAAHFIALAHPYSAMTVAQAYLDNVFKLHGCPSSI 607
Query: 61 V 61
V
Sbjct: 608 V 608
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.321 0.136 0.414
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 7,530,153
Number of Sequences: 26719
Number of extensions: 310570
Number of successful extensions: 864
Number of sequences better than 10.0: 49
Number of HSP's better than 10.0 without gapping: 42
Number of HSP's successfully gapped in prelim test: 7
Number of HSP's that attempted gapping in prelim test: 776
Number of HSP's gapped (non-prelim): 54
length of query: 329
length of database: 11,318,596
effective HSP length: 100
effective length of query: 229
effective length of database: 8,646,696
effective search space: 1980093384
effective search space used: 1980093384
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.8 bits)
S2: 60 (27.7 bits)
Medicago: description of AC130807.4


