

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC130807.11 - phase: 0
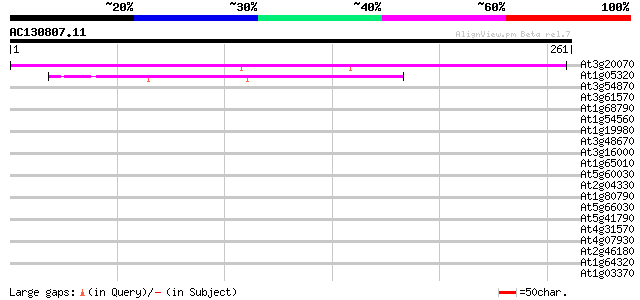
(261 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
At3g20070 unknown protein 199 2e-51
At1g05320 unknown protein 47 8e-06
At3g54870 kinesin-like protein 36 0.018
At3g61570 unknown protein 36 0.024
At1g68790 putative nuclear matrix constituent protein 1 (NMCP1) 36 0.024
At1g54560 35 0.053
At1g19980 unknown protein 35 0.053
At3g48670 unknown protein 34 0.069
At3g16000 myosin heavy chain like protein 34 0.090
At1g65010 hypothetical protein 34 0.090
At5g60030 KED - like protein 33 0.12
At2g04330 hypothetical protein 33 0.12
At1g80790 hypothetical protein 33 0.12
At5g66030 Golgi-localized protein GRIP 33 0.15
At5g41790 myosin heavy chain-like protein 33 0.20
At4g31570 putative protein 33 0.20
At4g07930 putative protein 33 0.20
At2g46180 unknown protein 33 0.20
At1g64320 bZIP transcription factor, putative 33 0.20
At1g03370 unknown protein 33 0.20
>At3g20070 unknown protein
Length = 282
Score = 199 bits (505), Expect = 2e-51
Identities = 114/280 (40%), Positives = 169/280 (59%), Gaps = 21/280 (7%)
Query: 1 MEALYKKLYSKYTTLKTNKLSELEDVHKEQELKFLKFVSAAEDVIDHLRTENDKLLGQIN 60
MEALY KLY KYT L+ K SE ++++KEQE KFL FVSA+E++++HLR EN L +
Sbjct: 1 MEALYAKLYDKYTKLQKKKYSEYDEINKEQEEKFLTFVSASEELMEHLRGENQSSLEMVE 60
Query: 61 DLGNELTSVRVAKDNELVEHKRLLLEEKKKNEALFEEVEKLQKLLK-------EGTSGDL 113
L NE+ S+R +D++ +E ++LL+EE+ KN++L EEV KL++L++ E SG
Sbjct: 61 KLRNEIISIRSGRDDKFLECQKLLMEEELKNKSLSEEVVKLKELVQEEHPRNYEDQSGKK 120
Query: 114 SNRKVVNNTSNNSSIRMTRKRMRQEQDALDIEARCIPSENEGVD--------------TQ 159
RK + + + R R+ ++ D+ + I ++ +
Sbjct: 121 QKRKTPESARVTTRSMIKRSRLSEDLVETDMVSPDISKHHKAKEPLLVSQPQCCRTTYDG 180
Query: 160 ESDHQNWLVHALFEYTLDMKLSTDCQTGRLCLSAMHQSSGYSFSISWISRAPGEEAELLY 219
S + AL ++ L MKLST+ + R C+ A H ++G SFS+++I+ GEE+ELLY
Sbjct: 181 SSSSASCTFQALGKHLLGMKLSTNNKGKRACIVASHPTTGLSFSLTFINNPNGEESELLY 240
Query: 220 HVLSLGTLERLAPEWMREDIMFSPTMCPIFFERVTHVINL 259
SLGT +R+APEWMRE I FS +MCPIFFERV+ VI L
Sbjct: 241 KPASLGTFQRVAPEWMREVIKFSTSMCPIFFERVSRVIKL 280
>At1g05320 unknown protein
Length = 841
Score = 47.4 bits (111), Expect = 8e-06
Identities = 44/175 (25%), Positives = 81/175 (46%), Gaps = 13/175 (7%)
Query: 19 KLSELEDVHKEQELKFLKFVSAAEDVIDHLRTENDKLLGQINDLG---NELTSVRVAKDN 75
KLS LE K Q+ K L+ ED+ L +E ++L QI+ L N++ + + N
Sbjct: 524 KLSVLE-AEKYQQAKELQIT--IEDLTKQLTSERERLRSQISSLEEEKNQVNEIYQSTKN 580
Query: 76 ELVEHKRLLLEEKKKNEALFEEVEKLQKLLKEGT-------SGDLSNRKVVNNTSNNSSI 128
ELV+ + L +K K++ + ++EKL L+ E + ++ ++ V + +S
Sbjct: 581 ELVKLQAQLQVDKSKSDDMVSQIEKLSALVAEKSVLESKFEQVEIHLKEEVEKVAELTSK 640
Query: 129 RMTRKRMRQEQDALDIEARCIPSENEGVDTQESDHQNWLVHALFEYTLDMKLSTD 183
K ++D L+ +A + E + T S+ + L H E +K S +
Sbjct: 641 LQEHKHKASDRDVLEEKAIQLHKELQASHTAISEQKEALSHKHSELEATLKKSQE 695
Score = 28.1 bits (61), Expect = 5.0
Identities = 30/133 (22%), Positives = 56/133 (41%), Gaps = 7/133 (5%)
Query: 23 LEDVHKEQELKFLKFVSAAEDVIDHLRTENDKLLGQINDLGNELTSVRVAKDNELVEHKR 82
L D + + ++K + E +D + + L Q + +L + + +N + EHKR
Sbjct: 405 LADTNNQLKIK----IQELEGYLDSEKETAIEKLNQKDTEAKDLITKLKSHENVIEEHKR 460
Query: 83 LLLEEKKKNEALFEEVEKLQKLLKEGT-SGDLSNRKVVNNTSNNSSIRMTRKRMRQEQDA 141
+LE + EVE + LLK T + + N +I++ +K Q +
Sbjct: 461 QVLEASGVADTRKVEVE--EALLKLNTLESTIEELEKENGDLAEVNIKLNQKLANQGSET 518
Query: 142 LDIEARCIPSENE 154
D +A+ E E
Sbjct: 519 DDFQAKLSVLEAE 531
>At3g54870 kinesin-like protein
Length = 1070
Score = 36.2 bits (82), Expect = 0.018
Identities = 30/111 (27%), Positives = 54/111 (48%), Gaps = 14/111 (12%)
Query: 20 LSELEDVHKEQELKFLKFVSAAEDVIDHLRT---ENDKLLGQINDLGNELTSVRVAKDNE 76
+S+ + E ++K + E +D L+T + D LL Q LG E+ ++
Sbjct: 690 ISQENEEANELKIKLEELSQMYESTVDELQTVKLDYDDLLQQKEKLGEEVRDMK------ 743
Query: 77 LVEHKRLLLEEKKKNEALFEEVEKLQKLLKEGTSGDLSNRKVVNNTSNNSS 127
+RLLLEEK++ + + E+ KL+K L+E + R + + S S+
Sbjct: 744 ----ERLLLEEKQRKQ-MESELSKLKKNLRESENVVEEKRYMKEDLSKGSA 789
>At3g61570 unknown protein
Length = 712
Score = 35.8 bits (81), Expect = 0.024
Identities = 27/103 (26%), Positives = 44/103 (42%), Gaps = 5/103 (4%)
Query: 51 ENDKLLGQINDLGNELTSVRVAKDNELVEHKRLLLEEKKKNEALFEEVEKLQKLLKEGTS 110
E DK ++ L L + ++ E RL+ E ++ NE ++ L+K LK+ S
Sbjct: 361 ERDKARQELKRLKQHLLEKETEESEKMDEDSRLIEELRQTNEYQRSQISHLEKSLKQAIS 420
Query: 111 GDLSNRKVVNNTSNNSSIRMTRKRMRQEQDALDIEARCIPSEN 153
NR SN++ IR + + L R I S+N
Sbjct: 421 NQEDNR-----LSNDNQIRKLKDTVDDLNQKLTNCLRTIESKN 458
Score = 27.3 bits (59), Expect = 8.5
Identities = 19/95 (20%), Positives = 43/95 (45%)
Query: 68 SVRVAKDNELVEHKRLLLEEKKKNEALFEEVEKLQKLLKEGTSGDLSNRKVVNNTSNNSS 127
+ +AK+ E + +L L+E++K F+E + +L K TS ++S + +
Sbjct: 198 ATELAKEREKLRDFQLSLQEERKRSESFKEELESMRLDKNKTSMEISKMRSELDAKLLEI 257
Query: 128 IRMTRKRMRQEQDALDIEARCIPSENEGVDTQESD 162
+ K QE A+ + N+ ++ + ++
Sbjct: 258 KHLQMKLTGQESHAIGPGMEHLKEVNKALEKENNE 292
>At1g68790 putative nuclear matrix constituent protein 1 (NMCP1)
Length = 1085
Score = 35.8 bits (81), Expect = 0.024
Identities = 35/145 (24%), Positives = 67/145 (46%), Gaps = 16/145 (11%)
Query: 13 TTLKTNKLSELED---VHKEQELKFLKFVSAAEDVIDHLRTENDKLLGQINDLGNELTSV 69
TT + +++ E + + KE+ ++FL+ S + ID ++ E + LL + +L +
Sbjct: 473 TTKQESRIREEHESLRITKEERVEFLRLQSELKQQIDKVKQEEELLLKEREELKQD--KE 530
Query: 70 RVAKDNELVEHKRLLLEEKKKNEALFEEVEKLQKL---------LKEGTSGDLSNRKVVN 120
R K+ E ++ KR + ++ A EE EKL+ L +E TS D R++
Sbjct: 531 RFEKEWEALDKKRANITREQNEVA--EENEKLRNLQISEKHRLKREEMTSRDNLKRELDG 588
Query: 121 NTSNNSSIRMTRKRMRQEQDALDIE 145
S + + ++ LD+E
Sbjct: 589 VKMQKESFEADMEDLEMQKRNLDME 613
>At1g54560
Length = 1529
Score = 34.7 bits (78), Expect = 0.053
Identities = 33/136 (24%), Positives = 65/136 (47%), Gaps = 11/136 (8%)
Query: 25 DVHKEQELKFLKFVSAAEDVIDHLRTENDKLLGQINDLGNELTSVRVAKDNELVEHKRLL 84
D+ +E+ + K S+ E++ + N L+ + E + + +V ++L
Sbjct: 917 DLEEEKNQEIKKLQSSLEEMRKKVDETNGLLVKE-----REAAKKAIEEAPPVVTETQVL 971
Query: 85 LEEKKKNEALFEEVEKLQ-KLLKEGTSGDLSNRKV----VNNTSNNSSIRMTRKRMRQEQ 139
+E+ +K EAL EEVE L+ L +E D + RK ++ + T K+ +Q Q
Sbjct: 972 VEDTQKIEALTEEVEGLKANLEQEKQRADDATRKFDEAQESSEDRKKKLEDTEKKAQQLQ 1031
Query: 140 DALD-IEARCIPSENE 154
+++ +E +C E+E
Sbjct: 1032 ESVTRLEEKCNNLESE 1047
>At1g19980 unknown protein
Length = 342
Score = 34.7 bits (78), Expect = 0.053
Identities = 25/112 (22%), Positives = 50/112 (44%), Gaps = 6/112 (5%)
Query: 1 MEALYKKLYSKYTTLKTNKLSELEDVHKEQELKFLKFVSAAEDVIDHLRTENDKL----- 55
+EA +KL +Y L + K E+ D+ +E + +F + D L+ ++D++
Sbjct: 143 LEAKIRKLKLEYEKLASEKKCEVSDLLRENGFAWNQFKCIESEFTDKLKRKDDEIVQANT 202
Query: 56 -LGQINDLGNELTSVRVAKDNELVEHKRLLLEEKKKNEALFEEVEKLQKLLK 106
+ + +L S KD + K + E + + EE+ KL + L+
Sbjct: 203 KISSLISYQEQLQSSNQEKDETISRLKAKMAEMETNSTKKDEEISKLTRDLE 254
>At3g48670 unknown protein
Length = 647
Score = 34.3 bits (77), Expect = 0.069
Identities = 41/156 (26%), Positives = 69/156 (43%), Gaps = 19/156 (12%)
Query: 51 ENDKLLGQINDLGNELTSVRVAKDNELVEHKRLLLEEKKKNEALFEEV-----EKLQKLL 105
EN + G + + ELT K LV++ R L+EEKKK+ EE+ E+L +L+
Sbjct: 242 ENLRKTGDLKTIA-ELTEEEARKQELLVQNLRQLVEEKKKDMKEIEELCSVKSEELNQLM 300
Query: 106 KEGTSGDLSNRKVVNNTSNNSSIRMTRKRMRQEQDALDIEARCIPSENEGVDTQESDHQN 165
+E + + +N T +++ D + R + SE + ++ + ++
Sbjct: 301 EEKEKNQQKHYRELNAIQER-----TMSHIQKIVDDHEKLKRLLESERKKLEIKCNELAK 355
Query: 166 WLVHALFEYTLDMKLSTDCQ-----TGRLCLSAMHQ 196
VH T MKLS D + L L+AM Q
Sbjct: 356 REVH---NGTERMKLSEDLEQNASKNSSLELAAMEQ 388
>At3g16000 myosin heavy chain like protein
Length = 727
Score = 33.9 bits (76), Expect = 0.090
Identities = 39/169 (23%), Positives = 68/169 (40%), Gaps = 14/169 (8%)
Query: 36 KFVSAAEDVIDHLRTENDKLLGQINDLGNELTSVRVAKDNELVEHKRLLLEEKKKNE--- 92
K + A+ D + L T D+ + L +EL V ++R L EK+KNE
Sbjct: 449 KTLQASRDRVSDLETMLDESRALCSKLESELAIVHEEWKEAKERYERNLDAEKQKNEISA 508
Query: 93 ---ALFEEVEKLQKLLKEGTSGDLSNRKVVNNTSNNSSIRMTRKRMRQEQDALDIEARCI 149
AL +++ + K EG + +L V N + + + K++ L+ E + +
Sbjct: 509 SELALEKDLRRRVKDELEGVTHELKESSVKNQSLQKELVEI-YKKVETSNKELEEEKKTV 567
Query: 150 PSENEGVDTQESDHQNWLVHALFEYTLDMKLSTDCQTGRLCLSAMHQSS 198
S N+ V E L E L TD + L M++++
Sbjct: 568 LSLNKEVKGMEK-------QILMEREARKSLETDLEEAVKSLDEMNKNT 609
>At1g65010 hypothetical protein
Length = 1318
Score = 33.9 bits (76), Expect = 0.090
Identities = 20/97 (20%), Positives = 44/97 (44%)
Query: 42 EDVIDHLRTENDKLLGQINDLGNELTSVRVAKDNELVEHKRLLLEEKKKNEALFEEVEKL 101
E +++ R E D L ++ + NE + + + + + + + +++N + EEV +L
Sbjct: 479 EKMLEDARNEIDSLKSTVDSIQNEFENSKAGWEQKELHLMGCVKKSEEENSSSQEEVSRL 538
Query: 102 QKLLKEGTSGDLSNRKVVNNTSNNSSIRMTRKRMRQE 138
LLKE + ++ + NN + + QE
Sbjct: 539 VNLLKESEEDACARKEEEASLKNNLKVAEGEVKYLQE 575
>At5g60030 KED - like protein
Length = 292
Score = 33.5 bits (75), Expect = 0.12
Identities = 30/127 (23%), Positives = 57/127 (44%), Gaps = 4/127 (3%)
Query: 40 AAEDVIDHLRTENDKLLGQINDLGNELTSVRVAKDNELVEHKRLLLEEKKKNEALFEEV- 98
+ E V R E + D+ +E + ++ + E + E+KKK E+V
Sbjct: 106 SVESVYGRERDEKKMKKSKDADVVDEKVNEKLEAEQRSEERRERKKEKKKKKNNKDEDVV 165
Query: 99 -EKL-QKLLKEGTSGDLSNRKVVNNTSNN-SSIRMTRKRMRQEQDALDIEARCIPSENEG 155
EK+ +KL E S D RK + NN + ++++ EQ + +I+ + + +
Sbjct: 166 DEKVKEKLEDEQKSADRKERKKKKSKKNNDEDVVDEKEKLEDEQKSAEIKEKKKNKDEDV 225
Query: 156 VDTQESD 162
VD +E +
Sbjct: 226 VDEKEKE 232
Score = 28.1 bits (61), Expect = 5.0
Identities = 23/73 (31%), Positives = 35/73 (47%), Gaps = 5/73 (6%)
Query: 79 EHKRLLLEEKKKN--EALFEEVEKLQKLLKEGTSGDLSNRKVVNNTSNNSSIRMTR--KR 134
E K ++EKKKN E + +E EK +KL E SG+ K S+ + R K+
Sbjct: 208 EQKSAEIKEKKKNKDEDVVDEKEK-EKLEDEQRSGERKKEKKKKRKSDEEIVSEERKSKK 266
Query: 135 MRQEQDALDIEAR 147
R+ + + E R
Sbjct: 267 KRKSDEEMGSEER 279
>At2g04330 hypothetical protein
Length = 564
Score = 33.5 bits (75), Expect = 0.12
Identities = 36/175 (20%), Positives = 68/175 (38%), Gaps = 23/175 (13%)
Query: 13 TTLKTNKLSELEDVHKEQELKFLKFVSAAEDV-IDHLRTENDKLLGQINDLGNELTSVRV 71
TT T + L+D LK + E++ D + EN+K G+ + EL
Sbjct: 97 TTAPTTESPMLDDSTFYDALKHIPAEETQENMQTDEVEDENEKEEGKETETNKELACANP 156
Query: 72 AKDNELVEHKRLLLEEKKKNEALFEEVEKLQKLLKEGTSGDLSNRKVVNNTSNNSSIRMT 131
++ E + ++EE+++ + +E ++K +++ D + V+
Sbjct: 157 VEEAERQDDGLAVIEEEEERSSASDEDVNVEKSVEDEGDEDERDEDVIVEK--------- 207
Query: 132 RKRMRQEQDALDIEARCIPSENEGVDTQESDHQN--WLVHALFEYTLDMKLSTDC 184
+E R I + VD +E+ + EYT MKL+T C
Sbjct: 208 -----------PVEERTIDEDIANVDMEEAMAMQPLGMYFPASEYTKKMKLATRC 251
>At1g80790 hypothetical protein
Length = 634
Score = 33.5 bits (75), Expect = 0.12
Identities = 22/134 (16%), Positives = 63/134 (46%), Gaps = 2/134 (1%)
Query: 33 KFLKFVSAAEDVIDHLRTENDKLLGQINDLGNELTSVRVAKDNELVEHKRLLLEEKKKNE 92
K F ++ I + D L +I +L ++ + + + +R+L+E+ + +
Sbjct: 237 KLRSFSDITKEEIQNKSIVVDDLANKIAMTNEDLNKLQYMNNEKTLSLRRVLIEKDELDR 296
Query: 93 ALFEEVEKLQKLLKEGTSGDLSNRKVVNN--TSNNSSIRMTRKRMRQEQDALDIEARCIP 150
+E +K+Q+L +E + ++ + N + +++++ K++ ++Q ++E + +
Sbjct: 297 VYKQETKKMQELSREKINRIFREKERLTNELEAKMNNLKIWSKQLDKKQALTELERQKLD 356
Query: 151 SENEGVDTQESDHQ 164
+ + D S Q
Sbjct: 357 EDKKKSDVMNSSLQ 370
>At5g66030 Golgi-localized protein GRIP
Length = 788
Score = 33.1 bits (74), Expect = 0.15
Identities = 34/143 (23%), Positives = 63/143 (43%), Gaps = 16/143 (11%)
Query: 20 LSELEDVHKEQELKFLKFVSAAEDVIDHLRTENDKLLGQINDLGNELTSVRVAKDNELVE 79
LS DV K+ + AAE ++HLR + + + ++ + V D E+ E
Sbjct: 94 LSREIDVEKQTRV-------AAEQALEHLREAYSEADAKSQEYSSKFSQVEQKLDQEIKE 146
Query: 80 HKRLLLEEKKKNEALFEEV-EKLQKLLKEGTSGDLSNRKVVNN----TSNNSSIRMTRKR 134
+ K L + +++Q++ KE D R+V +S +SS++ +R
Sbjct: 147 RDEKYADLDAKFTRLHKRAKQRIQEIQKEKDDLDARFREVNETAERASSQHSSMQQELER 206
Query: 135 MRQEQD----ALDIEARCIPSEN 153
RQ+ + A+D E + + S N
Sbjct: 207 TRQQANEALKAMDAERQQLRSAN 229
>At5g41790 myosin heavy chain-like protein
Length = 1305
Score = 32.7 bits (73), Expect = 0.20
Identities = 23/86 (26%), Positives = 40/86 (45%), Gaps = 9/86 (10%)
Query: 7 KLYSKYTTLKT--NKLSELEDVHKEQELKFLKFVSAAEDVI-------DHLRTENDKLLG 57
KL T+K ++L EL+D HKE+E + V +A+ + D+ E L
Sbjct: 373 KLEQAQNTIKELMDELGELKDRHKEKESELSSLVKSADQQVADMKQSLDNAEEEKKMLSQ 432
Query: 58 QINDLGNELTSVRVAKDNELVEHKRL 83
+I D+ NE+ + + E ++L
Sbjct: 433 RILDISNEIQEAQKTIQEHMSESEQL 458
Score = 30.0 bits (66), Expect = 1.3
Identities = 32/112 (28%), Positives = 55/112 (48%), Gaps = 11/112 (9%)
Query: 2 EALYKKLYSKYTTLKTNKLSELEDVHKE-----QELKFLKFVSAA-EDVIDHLRTENDKL 55
E++ +++ +K +L ELE + K+ +EL+ K + D I+ +E L
Sbjct: 924 ESILEEINGLSEKIKGREL-ELETLGKQRSELDEELRTKKEENVQMHDKINVASSEIMAL 982
Query: 56 LGQINDLGNELTSVRVAKDNELVEHKRLLLEEKKKNEALFEEVEKLQKLLKE 107
IN+L NEL S++V K E +R EK++ L ++ +QK L E
Sbjct: 983 TELINNLKNELDSLQVQKSETEAELER----EKQEKSELSNQITDVQKALVE 1030
>At4g31570 putative protein
Length = 2712
Score = 32.7 bits (73), Expect = 0.20
Identities = 27/108 (25%), Positives = 48/108 (44%), Gaps = 3/108 (2%)
Query: 8 LYSKYTTLKTNKLSELEDV---HKEQELKFLKFVSAAEDVIDHLRTENDKLLGQINDLGN 64
L K K N+L E+E+ HK + + ++ AE+ + +R+E ++
Sbjct: 1296 LLRKQLEAKGNELMEIEESLLHHKTKIAGLRESLTQAEESLVAVRSELQDKSNELEQSEQ 1355
Query: 65 ELTSVRVAKDNELVEHKRLLLEEKKKNEALFEEVEKLQKLLKEGTSGD 112
L S R + + K L+++ ++L E KLQK +E S D
Sbjct: 1356 RLLSTREKLSIAVTKGKGLIVQRDNVKQSLAEASAKLQKCSEELNSKD 1403
Score = 30.0 bits (66), Expect = 1.3
Identities = 35/122 (28%), Positives = 52/122 (41%), Gaps = 31/122 (25%)
Query: 14 TLKTNKLSELEDVHKEQELKFLKFVSAAEDVIDHLRTENDKLLGQINDLGNELTSVRVAK 73
T ++ S L++ + K LKF E HL ENDKL ++ L +++
Sbjct: 694 TYLLSEYSNLKEGYTLLNNKLLKFQGEKE----HLVEENDKLTQELLTLQEHMST----- 744
Query: 74 DNELVEHKRLLLEEKKKNEALFEEVEKLQKLLKEGTSGDLSNRKVVNNTSNNSSIRMTRK 133
VE +R LE + L E + +L KL +E NTS SSI + +
Sbjct: 745 ----VEEERTHLEVE-----LREAIARLDKLAEE-------------NTSLTSSIMVEKA 782
Query: 134 RM 135
RM
Sbjct: 783 RM 784
>At4g07930 putative protein
Length = 277
Score = 32.7 bits (73), Expect = 0.20
Identities = 29/116 (25%), Positives = 52/116 (44%), Gaps = 10/116 (8%)
Query: 41 AEDVIDHLRTENDKLLGQINDLGNELTSVRVAKDNELVE----HKRLLLEEKKKNEALFE 96
A + I L + DKL ++ DL + + A + VE + +LL K K
Sbjct: 103 ARNQIHRLNEKKDKLSKRVLDLTSTAQGAKKAVHDAKVELAAAYSKLLAGIKDK------ 156
Query: 97 EVEKLQKLLKEGTSGDLSNRKVVNNTSNNSSIRMTRKRMRQEQDALDIEARCIPSE 152
V K + + EG + ++ + + + ++I T +R R + + D+EARC E
Sbjct: 157 WVAKKEFTVLEGQAAEVESNLALIDQIMKAAIDQTMERPRLQAELDDLEARCASKE 212
>At2g46180 unknown protein
Length = 725
Score = 32.7 bits (73), Expect = 0.20
Identities = 33/153 (21%), Positives = 71/153 (45%), Gaps = 12/153 (7%)
Query: 6 KKLYSKYTTLKTNKLSELEDVHK-----EQELKFLKFVSAAEDVIDHLRTENDKLLGQIN 60
+KL+ K T + LS L++ +++++ K + E ++ R E DK ++
Sbjct: 318 RKLFPKSTEDLSRHLSSLDEEKAGTFPGKEDME--KSLQRLEKELEEARREKDKARQELK 375
Query: 61 DLGNELTSVRVAKDNELVEHKRLLLEEKKKNEALFEEVEKLQKLLKEGTSGDLSNRKVVN 120
L L + ++ E RL+ E ++ NE ++ L+K L++ ++N++ +
Sbjct: 376 RLKQHLLEKETEESEKMDEDSRLIDELRQTNEYQRSQILGLEKALRQ----TMANQEEI- 430
Query: 121 NTSNNSSIRMTRKRMRQEQDALDIEARCIPSEN 153
+S++ IR ++ + L R I S+N
Sbjct: 431 KSSSDLEIRKSKGIIEDLNQKLANCLRTIDSKN 463
Score = 27.3 bits (59), Expect = 8.5
Identities = 23/81 (28%), Positives = 42/81 (51%), Gaps = 3/81 (3%)
Query: 83 LLLEEKKKNEALFEEVEKLQKLLKEGTSGDLSNRKVVNNTSNNSSIRMTRKRMR-QEQDA 141
LL EE+K+NE EE++ L +L KE T + SN+ + + IR + ++ EQ A
Sbjct: 219 LLQEERKQNETFKEELQSL-RLDKEKTLME-SNKVRRELDAKLAEIRQLQMKLNGGEQHA 276
Query: 142 LDIEARCIPSENEGVDTQESD 162
I + N+ ++ + ++
Sbjct: 277 FGISRENLKEVNKALEKENNE 297
>At1g64320 bZIP transcription factor, putative
Length = 476
Score = 32.7 bits (73), Expect = 0.20
Identities = 24/95 (25%), Positives = 47/95 (49%), Gaps = 8/95 (8%)
Query: 19 KLSELEDVHKEQELKFLKFVSAAEDVIDHLRTENDKLLGQINDLGNELTSVRVAKDNELV 78
++SEL+ +H E + K + A +D ++ KL+ + +D+ L++ +
Sbjct: 164 RISELDSLHMEMKTKSAHEMEDASKKLDTEVSDQKKLVKEQDDIIRRLSA--------KI 215
Query: 79 EHKRLLLEEKKKNEALFEEVEKLQKLLKEGTSGDL 113
+ ++ LL+E+K F E +K K G+S DL
Sbjct: 216 KDQQRLLKEQKDTIDKFAEDQKQSKRWSFGSSRDL 250
Score = 30.4 bits (67), Expect = 1.00
Identities = 30/113 (26%), Positives = 56/113 (49%), Gaps = 13/113 (11%)
Query: 8 LYSKYTTLKTNKLSELEDV-----HKEQELKFLKFVSAA-EDVIDHLRTENDK----LLG 57
L +K+ + S +ED+ + EQE+ FL+ +A ++ + E + L+
Sbjct: 37 LVNKFGNSELGLTSRIEDLKCQLKNLEQEIGFLRARNAGLAGNLEVTKVEEKERVKGLMD 96
Query: 58 QINDLGNELTSVRVAKDN---ELVEHKRLLLEEKKKNEALFEEVEKLQKLLKE 107
Q+N + +EL S+R KD +L + + E K + ++L EE E+ + L E
Sbjct: 97 QVNGMKHELESLRSQKDESEAKLEKKVEEVTETKMQLKSLKEETEEERNRLSE 149
>At1g03370 unknown protein
Length = 1859
Score = 32.7 bits (73), Expect = 0.20
Identities = 35/118 (29%), Positives = 54/118 (45%), Gaps = 14/118 (11%)
Query: 54 KLLGQINDLGNEL--------TSVRVAKDNELVEHKRLLLEEKKKNEALFEEVEKLQKLL 105
KL+ ++NDL NEL V+ A L + L K++ EA E+ +K ++LL
Sbjct: 500 KLVPRVNDLQNELQVWTDWANQKVKEATGRLLKDQPELKALRKEREEA--EQYKKEKQLL 557
Query: 106 KEGTSGDLSNR--KVVNNTSNNSSIRMTRKRMRQEQDALDIEARCIPSENEGVDTQES 161
+E T LS + N TS T R+ EQ L E ++ + V++ ES
Sbjct: 558 EENTRKRLSEMDFALKNATSQLEKAFNTAHRLELEQSILKKEMEA--AKIKAVESAES 613
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.315 0.131 0.364
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 5,745,364
Number of Sequences: 26719
Number of extensions: 243308
Number of successful extensions: 1088
Number of sequences better than 10.0: 131
Number of HSP's better than 10.0 without gapping: 23
Number of HSP's successfully gapped in prelim test: 108
Number of HSP's that attempted gapping in prelim test: 1008
Number of HSP's gapped (non-prelim): 184
length of query: 261
length of database: 11,318,596
effective HSP length: 97
effective length of query: 164
effective length of database: 8,726,853
effective search space: 1431203892
effective search space used: 1431203892
T: 11
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 42 (22.0 bits)
S2: 59 (27.3 bits)
Medicago: description of AC130807.11


