

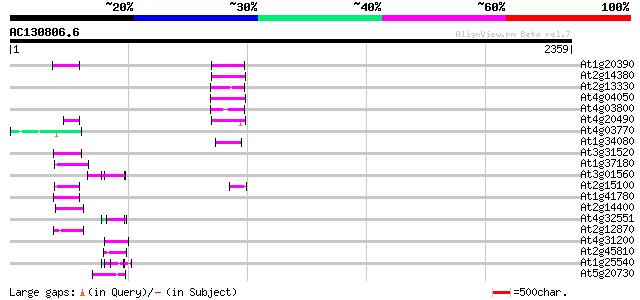
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC130806.6 + phase: 0 /pseudo
(2359 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
At1g20390 hypothetical protein 81 8e-15
At2g14380 putative retroelement pol polyprotein 80 1e-14
At2g13330 F14O4.9 77 1e-13
At4g04050 putative transposon protein 75 5e-13
At4g03800 75 5e-13
At4g20490 putative protein 69 4e-11
At4g03770 hypothetical protein 68 6e-11
At1g34080 putative protein 67 9e-11
At3g31520 hypothetical protein 64 1e-09
At1g37180 hypothetical protein 62 4e-09
At3g01560 unknown protein 55 6e-07
At2g15100 putative retroelement pol polyprotein 55 6e-07
At1g41780 hypothetical protein 55 6e-07
At2g14400 putative retroelement pol polyprotein 53 2e-06
At4g32551 Leunig protein 52 4e-06
At2g12870 putative retroelement pol polyprotein 51 7e-06
At4g31200 predicted protein 50 1e-05
At2g45810 putative ATP-dependent RNA helicase 50 2e-05
At1g25540 hypothetical protein 50 2e-05
At5g20730 auxin response factor 7 (ARF7) 48 6e-05
>At1g20390 hypothetical protein
Length = 1791
Score = 80.9 bits (198), Expect = 8e-15
Identities = 44/138 (31%), Positives = 72/138 (51%)
Query: 847 LSFSDEELPAEGRNHNRALHISVNCKTDALSNVLVDTGSSLNVLSKTTYAQLAYQDAPLR 906
+SF+D +L HN L + + ++ VL+DTGSS++++ K + D ++
Sbjct: 553 ISFTDVDLEGLDTPHNDPLVVELIISDSRVTRVLIDTGSSVDLIFKDVLTAMNITDRQIK 612
Query: 907 RSGVMVKAFDGSRKDVLGEVVLPITVGPQVFQVNFQVMDIQASYSCLLGRPWIHEAGAVT 966
+ FDG +G + LPI VG + V F V+ A Y+ +LG PWIH+ A+
Sbjct: 613 PVSKPLAGFDGDFVMTIGTIKLPIFVGGLIAWVKFVVIGKPAVYNVILGTPWIHQMQAIP 672
Query: 967 STLHQKLKFVKNGKLVTV 984
ST HQ +KF + + T+
Sbjct: 673 STYHQCVKFPTHNGIFTL 690
Score = 52.8 bits (125), Expect = 2e-06
Identities = 31/117 (26%), Positives = 58/117 (49%), Gaps = 7/117 (5%)
Query: 181 SVQVPHKFKIPDFEKYKGSSCPEEHLKMY---VRRMPAYAQD-DQILIYYFQESLTGPAS 236
S++ K K+ E Y G + P+E L + + R + D F E LTGPA
Sbjct: 165 SIRGAQKIKL---ESYNGRNDPKEFLTSFNVAINRAELTIDNFDAGRCQIFIEHLTGPAH 221
Query: 237 KWYTNLDKTRVQTFRDLCEAFVEQYSYNVDMTPDRSDLQAMTQGDKETFKEYAQRWR 293
W++ L + +F L +F++ Y+ ++ +DL +++QG KE+ + + R++
Sbjct: 222 NWFSRLKPNSIDSFHQLTSSFLKHYAPLIENQTSNADLWSISQGAKESLRSFVDRFK 278
>At2g14380 putative retroelement pol polyprotein
Length = 764
Score = 80.1 bits (196), Expect = 1e-14
Identities = 45/144 (31%), Positives = 75/144 (51%)
Query: 847 LSFSDEELPAEGRNHNRALHISVNCKTDALSNVLVDTGSSLNVLSKTTYAQLAYQDAPLR 906
L+F+ E+L HN L I ++ ++ +L+DTGSS+NV+ K ++ D ++
Sbjct: 330 LTFTSEDLFGVDLPHNDPLVIELHIGESEVTRILIDTGSSVNVVFKDVLQKMKVHDRHIK 389
Query: 907 RSGVMVKAFDGSRKDVLGEVVLPITVGPQVFQVNFQVMDIQASYSCLLGRPWIHEAGAVT 966
S + FDG+ G + LPI +G F V+D Y+ +LG PWIH+ A+
Sbjct: 390 PSVRPLTGFDGNTMMTNGTIKLPIYLGGAATWHKFVVVDKPTIYNIILGTPWIHDMQAIP 449
Query: 967 STLHQKLKFVKNGKLVTVNGEEAL 990
S+ HQ +K + + T+ G + L
Sbjct: 450 SSYHQCIKIPTSIGIETIRGNQNL 473
>At2g13330 F14O4.9
Length = 889
Score = 77.0 bits (188), Expect = 1e-13
Identities = 47/145 (32%), Positives = 79/145 (54%), Gaps = 2/145 (1%)
Query: 845 NNLSFSDEELPAEGRNHNRALHISVNCKTDALSNVLVDTGSSLNVLSKTTYAQLAYQDAP 904
+ ++F + E+ + H+ AL I ++ LS ++VDTGSS++VL + + + D+
Sbjct: 45 DGITFWESEITDLDKPHDDALVIRIDVGNYELSCIMVDTGSSVDVLFYDAFKRTGHLDSK 104
Query: 905 LRRSGVMVKAFDGSRKDVLGEVVLPITVGPQVFQV-NFQVMDIQASYSCLLGRPWIHEAG 963
L+ + F G +G + LP T+ V Q+ NF V+D +A ++ +LGRPW+H
Sbjct: 105 LQGRKTPLTGFAGDTTFSIGTIQLP-TIARGVRQLTNFLVVDKKAPFNAILGRPWLHVMK 163
Query: 964 AVTSTLHQKLKFVKNGKLVTVNGEE 988
AV ST HQ +KF + V G +
Sbjct: 164 AVPSTYHQCIKFPSYKGIAVVYGSQ 188
>At4g04050 putative transposon protein
Length = 681
Score = 75.1 bits (183), Expect = 5e-13
Identities = 42/145 (28%), Positives = 76/145 (51%)
Query: 845 NNLSFSDEELPAEGRNHNRALHISVNCKTDALSNVLVDTGSSLNVLSKTTYAQLAYQDAP 904
+ ++F + E + H+ AL I ++ LS++++DTGSS++VL + ++ + D+
Sbjct: 519 HQITFWESETTDLNKPHDDALVIRIDVGNYELSHIMIDTGSSVDVLFYDAFKRMGHLDSE 578
Query: 905 LRRSGVMVKAFDGSRKDVLGEVVLPITVGPQVFQVNFQVMDIQASYSCLLGRPWIHEAGA 964
L+ + F G LG + LP +F V++ +A ++ +LGRPW+H A
Sbjct: 579 LQGRKTPLTGFAGDTTFSLGTIQLPTIARGVRRLTSFLVVNKKAPFNAILGRPWLHAMKA 638
Query: 965 VTSTLHQKLKFVKNGKLVTVNGEEA 989
V ST HQ +KF + + V+ A
Sbjct: 639 VPSTYHQCIKFPSDKGIAVVSAANA 663
>At4g03800
Length = 637
Score = 75.1 bits (183), Expect = 5e-13
Identities = 48/144 (33%), Positives = 74/144 (51%), Gaps = 14/144 (9%)
Query: 844 CNNLSFSDEELPAEGRNHNRALHISVNCKTDALSNVLVDTGSSLNVLSKTTYAQLAYQDA 903
C+++SF +EE R H+ AL I+++ +S +LVDTGSS++++ + L
Sbjct: 182 CSSVSFDEEETRHIERPHDDALIITLDVANFKISRILVDTGSSVDLIFLGPPSPLV---- 237
Query: 904 PLRRSGVMVKAFDGSRKDVLGEVVLPITVGPQVFQVNFQVMDIQASYSCLLGRPWIHEAG 963
AF LG + LP+ G V+F V D A+Y+ +LG PWI +
Sbjct: 238 ----------AFTSESAMSLGTIKLPVLAGKMSKIVDFVVFDKPATYNIILGTPWIFQMK 287
Query: 964 AVTSTLHQKLKFVKNGKLVTVNGE 987
AV ST HQ LKF + + T+ G+
Sbjct: 288 AVPSTYHQWLKFPTSNGVETIWGD 311
>At4g20490 putative protein
Length = 853
Score = 68.6 bits (166), Expect = 4e-11
Identities = 41/155 (26%), Positives = 72/155 (46%), Gaps = 14/155 (9%)
Query: 849 FSDEELPAEGRNHNRALHISVNCKTDALSNVLVDTGSSLNVLSKTTYAQLAYQDAPLRRS 908
F+ E+ HN AL I + + + +L+DTGSS+N++ T ++ ++ +
Sbjct: 465 FTSEDASGIQHPHNDALVIKLVMEDFDVERILIDTGSSVNLMFLKTLFKMGISESKIMPK 524
Query: 909 GVMVKAFDGSRKDVLGEVVLPITVGPQVFQVNFQVMDIQASYSCLLGRPWIHEAGAVTST 968
+ +DG K +GE+ L + VG + F V+D + Y+ +LG PWI+ A+ ST
Sbjct: 525 IRPMTGYDGEAKMSIGEIKLQVQVGGITQKTKFVVIDSEPIYNAILGSPWIYSMKAIPST 584
Query: 969 --------------LHQKLKFVKNGKLVTVNGEEA 989
L + + K G L+ + G A
Sbjct: 585 NLHTIRQPEGLTDMLRDREEAAKEGNLIAIKGRRA 619
Score = 45.4 bits (106), Expect = 4e-04
Identities = 23/68 (33%), Positives = 36/68 (52%)
Query: 227 FQESLTGPASKWYTNLDKTRVQTFRDLCEAFVEQYSYNVDMTPDRSDLQAMTQGDKETFK 286
F E LTG A ++ L+ + F L AF++QY + SDL +MTQ + ET
Sbjct: 225 FVEGLTGNALTCFSRLEANSIDNFTQLSTAFLKQYRVFIQPGASSSDLWSMTQENGETLN 284
Query: 287 EYAQRWRD 294
+Y R+++
Sbjct: 285 DYLGRFKE 292
>At4g03770 hypothetical protein
Length = 464
Score = 68.2 bits (165), Expect = 6e-11
Identities = 84/325 (25%), Positives = 131/325 (39%), Gaps = 41/325 (12%)
Query: 4 MAELIKTMAETQTQAQAQIQAQ---AQTQAQAVTEAQARSQAPPPPPPVRTQAEASSSWT 60
+AEL MA+ Q + Q QA AQ A ++ Q R+ R A
Sbjct: 34 LAELRSMMAQLQQKVNDQEQANRSLAQQLEAATSQGQIRT--------TRFGARHLQDRR 85
Query: 61 LCAD-TPTRSAPQRSTPWFPPF--TAGEIFRPITCEAQMP------THQYTAQVPLPAMR 111
AD PTR P TA EI R T A + T + Q+P P
Sbjct: 86 AAADLNPTRFVFHTPGNTTRPVRRTAPEIGRDRTEPAILGNRETKRTERNKPQLPPPRAE 145
Query: 112 VTPATMTYSAPVIHTIPQTEEPIFHSGNAEAYEEVSDLREKYDELRRDMKALHEKGKFGK 171
VT A + EE I + +E+S ++ + +MK + +
Sbjct: 146 VTEADHIG----VSDDKDLEENIRWAEEYATEQEISAIKLSLAKAENEMKLVRSQMHNAV 201
Query: 172 TA---YDLCLVPSVQVPHKFKIPD----------FEKYKGSSCPEEHLKMYV----RRMP 214
++ D L S P I + E + GSS P+ HLK+++ R
Sbjct: 202 SSAPNIDRILEESHNTPFTHMISNAIISDPGKLRIEYFNGSSDPKGHLKLFIISVARAKF 261
Query: 215 AYAQDDQILIYYFQESLTGPASKWYTNLDKTRVQTFRDLCEAFVEQYSYNVDMTPDRSDL 274
+ D L + F E L GPA W+T L V +F++L F++QYS +D + +DL
Sbjct: 262 RPEERDAGLCHLFVEHLKGPALNWFTRLKGNSVDSFQELSTLFLKQYSVLIDPSTSDADL 321
Query: 275 QAMTQGDKETFKEYAQRWRDTAAQV 299
+++Q E +++ ++R T A+V
Sbjct: 322 WSLSQQPNEPLRDFLAKFRSTLAKV 346
>At1g34080 putative protein
Length = 811
Score = 67.4 bits (163), Expect = 9e-11
Identities = 36/112 (32%), Positives = 61/112 (54%)
Query: 864 ALHISVNCKTDALSNVLVDTGSSLNVLSKTTYAQLAYQDAPLRRSGVMVKAFDGSRKDVL 923
AL IS++ + +L+DTGSS++++ T ++ ++ + + +F L
Sbjct: 476 ALVISLDVGNCEVQRILIDTGSSVDLIFLDTLVRMGISKKDIKGAPSPLVSFTIETSMSL 535
Query: 924 GEVVLPITVGPQVFQVNFQVMDIQASYSCLLGRPWIHEAGAVTSTLHQKLKF 975
G + LP+T V + F V D A+Y+ +LG PW++E AV ST HQ +KF
Sbjct: 536 GTITLPVTAQGVVKMIEFTVFDRPAAYNIILGTPWLYEMKAVPSTYHQCVKF 587
Score = 31.2 bits (69), Expect = 7.5
Identities = 27/137 (19%), Positives = 55/137 (39%), Gaps = 12/137 (8%)
Query: 175 DLCLVPSVQVPHKFKIPDFEKYKGSSCPE-EHLKMYVRRMPAYAQDDQILIYYFQE---- 229
D+ + +PH + +S P+ E + RR P + ++ I FQ+
Sbjct: 153 DIRRITPRMIPHLIRYEKRPHQATTSAPKVEKIIEATRRTPFTLRISKLRIREFQDFKLP 212
Query: 230 --SLTGPASKWYTNLD-----KTRVQTFRDLCEAFVEQYSYNVDMTPDRSDLQAMTQGDK 282
+ G ++ T+ + + F+ L AF++QY Y + + L ++Q
Sbjct: 213 GYNGKGDPKEYLTSFQVIAGREGSIDNFKQLSTAFIKQYEYFIKSDVTEAQLWNLSQAAD 272
Query: 283 ETFKEYAQRWRDTAAQV 299
E + Y +++ QV
Sbjct: 273 EPLRTYNTAFKEIMVQV 289
>At3g31520 hypothetical protein
Length = 528
Score = 63.9 bits (154), Expect = 1e-09
Identities = 37/119 (31%), Positives = 64/119 (53%), Gaps = 7/119 (5%)
Query: 185 PHKFKIPDFEKYKGSSCPEEHLKMYV----RRMPAYAQDDQILIYYFQESLTGPASKWYT 240
P K +I E + GSS P+ HLK ++ R + D L + F E L GPA W++
Sbjct: 239 PGKLRI---EYFNGSSDPKRHLKSFIISVARTKFRPEERDAGLCHLFVEHLKGPALCWFS 295
Query: 241 NLDKTRVQTFRDLCEAFVEQYSYNVDMTPDRSDLQAMTQGDKETFKEYAQRWRDTAAQV 299
L+ + +F++L F++QYS +D+ +DL +++Q E +++ + R T A+V
Sbjct: 296 RLEGNSMDSFQELSTLFLKQYSVLIDLGTSDADLWSLSQQPNEPLRDFLAKLRSTLAKV 354
>At1g37180 hypothetical protein
Length = 661
Score = 62.0 bits (149), Expect = 4e-09
Identities = 42/148 (28%), Positives = 72/148 (48%), Gaps = 9/148 (6%)
Query: 188 FKIPDFEKYKGSSCPEEHLKMY---VRRMPAYA-QDDQILIYYFQESLTGPASKWYTNLD 243
FK+P Y G P EHL + VRR+P ++D L F E+L+GPA W+T L+
Sbjct: 314 FKLP---VYNGKGDPNEHLTSFQVIVRRVPLEPYEEDAGLCKLFSENLSGPALTWFTQLE 370
Query: 244 KTRVQTFRDLCEAFVEQYSYNVDMTPDRSDLQAMTQGDKETFKEYAQRWRDTAAQVSPRI 303
+ + F+ L AF++QY Y + + L ++Q + + Y +++ Q+ P +
Sbjct: 371 EGSIDNFKQLSTAFIKQYGYFIKSDITEAHLWNLSQSADDPLRTYIIVFKEIMVQI-PSL 429
Query: 304 EEKEMTKLFLKTLNHFYYKKMVGSTPKS 331
+ + LK K+ S+PK+
Sbjct: 430 SD-STAQFALKNGTFSVEKESSKSSPKA 456
>At3g01560 unknown protein
Length = 511
Score = 54.7 bits (130), Expect = 6e-07
Identities = 33/88 (37%), Positives = 39/88 (43%), Gaps = 5/88 (5%)
Query: 399 PTSHPFQQTNNHP--QIPQY--PQIPQYPQIPQYPQFPQNPSPQNTQQQNFQQQPYQQQP 454
P SHP +N P Q PQ P P Y PQ PQ+PQ P P + +Q PYQ Q
Sbjct: 289 PPSHPQPPPSNPPPYQAPQTQTPHQPSYQSPPQQPQYPQQPPPSSGYNPE-EQPPYQMQS 347
Query: 455 YQQFPYQQYPQQNFQQQPYQQRPQQPRP 482
Y P +Q P P QP+P
Sbjct: 348 YPPNPPRQQPPAGSTPSQQFYNPPQPQP 375
Score = 50.4 bits (119), Expect = 1e-05
Identities = 49/176 (27%), Positives = 72/176 (40%), Gaps = 20/176 (11%)
Query: 326 GSTPKSFAEMVGMGVQLEEGVREGRLVKDATPASGTK-KTGNHFPRKKEQEVGMVTHGR- 383
GST ++ + V+++ GV+ + ++ A +K + N + V
Sbjct: 182 GSTDGKMRQLKNILVEVQSGVQLLKDKQEILEAQLSKHQVSNQHAKTHSLHVDPTAQSPA 241
Query: 384 --PQQTYPAYQHIAAITPTSHPFQQTNNH--PQIP------QYPQIPQYPQIPQYPQFPQ 433
P Q +P + T+ P Q ++ PQ+P Q P P P PQ P P
Sbjct: 242 PVPMQQFPLTSFPQPPSSTAAPSQPPSSQLPPQLPTQFSSQQEPYCPP-PSHPQPP--PS 298
Query: 434 NPSPQNTQQQNFQQQP-YQQQPYQ-QFPYQQYPQQNF---QQQPYQQRPQQPRPPR 484
NP P Q QP YQ P Q Q+P Q P + +Q PYQ + P PPR
Sbjct: 299 NPPPYQAPQTQTPHQPSYQSPPQQPQYPQQPPPSSGYNPEEQPPYQMQSYPPNPPR 354
Score = 47.8 bits (112), Expect = 8e-05
Identities = 35/105 (33%), Positives = 48/105 (45%), Gaps = 6/105 (5%)
Query: 386 QTYPAYQHIAAITPTSHPFQQ--TNNHPQIPQYPQIPQYPQIPQYPQFPQNPSPQNTQQQ 443
+T+ + A +P P QQ + PQ P P P Q P PQ P+ ++QQ+
Sbjct: 227 KTHSLHVDPTAQSPAPVPMQQFPLTSFPQPPSSTAAPSQPPSSQLP--PQLPTQFSSQQE 284
Query: 444 NFQQQP-YQQQPYQQFPYQQYPQQNFQQQP-YQQRPQQPRPPRMP 486
+ P + Q P P Q PQ QP YQ PQQP+ P+ P
Sbjct: 285 PYCPPPSHPQPPPSNPPPYQAPQTQTPHQPSYQSPPQQPQYPQQP 329
Score = 44.7 bits (104), Expect = 7e-04
Identities = 31/86 (36%), Positives = 37/86 (42%), Gaps = 4/86 (4%)
Query: 399 PTSHPFQQTNNHPQIPQYPQIPQYPQIPQYPQFPQNPSPQNTQQQNFQQQP-YQQQPYQQ 457
PT QQ P P +PQ P P P Q PQ +P Q+ QQP Y QQP
Sbjct: 276 PTQFSSQQEPYCPP-PSHPQPP--PSNPPPYQAPQTQTPHQPSYQSPPQQPQYPQQPPPS 332
Query: 458 FPYQQYPQQNFQQQPYQQRPQQPRPP 483
Y Q +Q Q Y P + +PP
Sbjct: 333 SGYNPEEQPPYQMQSYPPNPPRQQPP 358
>At2g15100 putative retroelement pol polyprotein
Length = 1329
Score = 54.7 bits (130), Expect = 6e-07
Identities = 31/73 (42%), Positives = 43/73 (58%), Gaps = 4/73 (5%)
Query: 923 LGEVVLPITVGPQVFQVNFQVMDIQASYSCLLGRPWIHEAGAVTSTLHQKLKF-VKNGKL 981
LG VVLP+T V V F V D A+Y+ +LG PW++E V ST HQ +KF GK+
Sbjct: 353 LGTVVLPVTAQGVVKMVEFTVFDRPAAYNVILGTPWLYEMKVVPSTYHQCVKFPTPVGKM 412
Query: 982 VTVNGEEALLVSH 994
++ E ++SH
Sbjct: 413 TGISTE---VISH 422
Score = 49.3 bits (116), Expect = 3e-05
Identities = 32/108 (29%), Positives = 51/108 (46%), Gaps = 7/108 (6%)
Query: 188 FKIPDFEKYKGSSCPEEHLKMY---VRRMPAYA-QDDQILIYYFQESLTGPASKWYTNLD 243
FK+P Y G +EHL + R+P ++D L F E+L G A W+T L+
Sbjct: 173 FKLP---VYNGKGDLKEHLTSFQVIAGRVPLEPHEEDAGLCKLFSENLFGLALTWFTQLE 229
Query: 244 KTRVQTFRDLCEAFVEQYSYNVDMTPDRSDLQAMTQGDKETFKEYAQR 291
+ + F+ L AF++QY Y ++ + L +Q E + Y R
Sbjct: 230 EGSIDNFKQLSTAFIKQYEYFINSDITEAHLWNFSQSADEPLRTYIYR 277
>At1g41780 hypothetical protein
Length = 246
Score = 54.7 bits (130), Expect = 6e-07
Identities = 33/112 (29%), Positives = 55/112 (48%), Gaps = 7/112 (6%)
Query: 185 PHKFKIPDFEKYKGSSCPEEHLKMY----VRRMPAYAQDDQILIYYFQESLTGPASKWYT 240
P K +I E + GSS P+ HLK + R + D L + F E L GP W++
Sbjct: 138 PEKLRI---EYFSGSSDPKGHLKSFKISVARAKFKPEESDAGLCHLFVEHLKGPVLDWFS 194
Query: 241 NLDKTRVQTFRDLCEAFVEQYSYNVDMTPDRSDLQAMTQGDKETFKEYAQRW 292
L+ V +F++L F++QYS +D DL +++Q E ++ ++
Sbjct: 195 RLEGNSVDSFQELSTLFLKQYSVLIDPGTSDVDLWSLSQQPNEPLSDFLTKF 246
>At2g14400 putative retroelement pol polyprotein
Length = 1466
Score = 52.8 bits (125), Expect = 2e-06
Identities = 31/124 (25%), Positives = 61/124 (49%), Gaps = 4/124 (3%)
Query: 192 DFEKYKGSSCPEEHLKMYV----RRMPAYAQDDQILIYYFQESLTGPASKWYTNLDKTRV 247
+ E Y G P+ +L ++ R A +D F E+L G A W+T L+ +
Sbjct: 161 NLESYNGLEDPKGYLAAFLIAAGRVDLNEADEDVRYCKLFSENLCGQALMWFTQLEPGSI 220
Query: 248 QTFRDLCEAFVEQYSYNVDMTPDRSDLQAMTQGDKETFKEYAQRWRDTAAQVSPRIEEKE 307
F +L F++QYS +D + +DL ++QG ET + + +++ +++S ++
Sbjct: 221 SNFNELSVVFLKQYSILMDKSISDTDLWNLSQGPNETLRAFITKFKYVLSKLSRISQQSA 280
Query: 308 MTKL 311
++ L
Sbjct: 281 LSAL 284
Score = 34.7 bits (78), Expect = 0.68
Identities = 16/62 (25%), Positives = 37/62 (58%)
Query: 846 NLSFSDEELPAEGRNHNRALHISVNCKTDALSNVLVDTGSSLNVLSKTTYAQLAYQDAPL 905
++ F ++E R+H+ AL ++++ +S +L+DTGSS++++ +T ++ A +
Sbjct: 408 SILFDEKETQHLERSHDDALVVTLDVANFEVSRILIDTGSSVDLIFLSTLERMGISRADV 467
Query: 906 RR 907
R
Sbjct: 468 NR 469
>At4g32551 Leunig protein
Length = 931
Score = 52.0 bits (123), Expect = 4e-06
Identities = 33/88 (37%), Positives = 38/88 (42%), Gaps = 2/88 (2%)
Query: 406 QTNNHPQIPQYPQIPQYPQIPQYPQFPQNPSPQNTQQQ--NFQQQPYQQQPYQQFPYQQY 463
Q + HPQ+ Q Q Q QI Q Q QQQ + Q QQQ QQ QQ
Sbjct: 92 QQSQHPQVSQQQQQQQQQQIQMQQLLLQRAQQQQQQQQQQHHHHQQQQQQQQQQQQQQQQ 151
Query: 464 PQQNFQQQPYQQRPQQPRPPRMPINPIP 491
QQ Q QP Q+ QQ P+ P P
Sbjct: 152 QQQQHQNQPPSQQQQQQSTPQHQQQPTP 179
Score = 44.7 bits (104), Expect = 7e-04
Identities = 30/95 (31%), Positives = 38/95 (39%)
Query: 386 QTYPAYQHIAAITPTSHPFQQTNNHPQIPQYPQIPQYPQIPQYPQFPQNPSPQNTQQQNF 445
+T + +AA + + Q Q+PQ+ Q Q Q Q QQ
Sbjct: 66 RTNEKHSEVAASYIETQMIKAREQQLQQSQHPQVSQQQQQQQQQQIQMQQLLLQRAQQQQ 125
Query: 446 QQQPYQQQPYQQFPYQQYPQQNFQQQPYQQRPQQP 480
QQQ Q +QQ QQ QQ QQQ QQ QP
Sbjct: 126 QQQQQQHHHHQQQQQQQQQQQQQQQQQQQQHQNQP 160
Score = 42.4 bits (98), Expect = 0.003
Identities = 28/72 (38%), Positives = 32/72 (43%), Gaps = 6/72 (8%)
Query: 421 QYPQIPQYPQFPQNPSPQNTQQQNFQQ------QPYQQQPYQQFPYQQYPQQNFQQQPYQ 474
Q Q Q+PQ Q Q QQ QQ Q QQQ QQ + Q QQ QQQ Q
Sbjct: 89 QQLQQSQHPQVSQQQQQQQQQQIQMQQLLLQRAQQQQQQQQQQHHHHQQQQQQQQQQQQQ 148
Query: 475 QRPQQPRPPRMP 486
Q+ QQ + P
Sbjct: 149 QQQQQQQHQNQP 160
Score = 34.3 bits (77), Expect = 0.89
Identities = 22/58 (37%), Positives = 26/58 (43%)
Query: 405 QQTNNHPQIPQYPQIPQYPQIPQYPQFPQNPSPQNTQQQNFQQQPYQQQPYQQFPYQQ 462
QQ ++H Q Q Q Q Q Q Q QN P QQQ Q QQ QQ P ++
Sbjct: 129 QQQHHHHQQQQQQQQQQQQQQQQQQQQHQNQPPSQQQQQQSTPQHQQQPTPQQQPQRR 186
>At2g12870 putative retroelement pol polyprotein
Length = 411
Score = 51.2 bits (121), Expect = 7e-06
Identities = 38/135 (28%), Positives = 63/135 (46%), Gaps = 15/135 (11%)
Query: 185 PHKFKIPDFEKYKGSSCPEEHLKMYVRRMPAYAQDDQI--------LIYYFQESLTGPAS 236
P K +I + + GSS P+ HLK ++ YA D+ L + F E L GP
Sbjct: 74 PGKLRI---DYFNGSSYPKGHLKSFI----IYAARDKFKPEERDAGLCHLFVEHLKGPVL 126
Query: 237 KWYTNLDKTRVQTFRDLCEAFVEQYSYNVDMTPDRSDLQAMTQGDKETFKEYAQRWRDTA 296
+ L+ V +F +L F++Q+S +D SDL +++Q E + ++R T
Sbjct: 127 VLFLRLEGNFVDSFEELSTLFLKQFSVLIDPGTLDSDLWSLSQQPNEPLMDLLTKFRSTL 186
Query: 297 AQVSPRIEEKEMTKL 311
A V I+ ++ L
Sbjct: 187 ASVEGIIDVAALSAL 201
>At4g31200 predicted protein
Length = 650
Score = 50.4 bits (119), Expect = 1e-05
Identities = 38/106 (35%), Positives = 47/106 (43%), Gaps = 8/106 (7%)
Query: 399 PTSHPFQQTNNHPQIPQYPQIPQYPQIPQYPQFPQN-----PSPQNTQQQNFQQQPYQQQ 453
P + QQ + Q Q PQ +PQ PQYP P N P + QQ +Q +QQQ
Sbjct: 15 PYAQQQQQQGPNFQQQQQPQFGFHPQHPQYPS-PMNASGFIPPHPSMQQFPYQHPMHQQQ 73
Query: 454 PYQQFPYQQYPQQNFQQQPYQQRPQQPRPPRMPINPIPVTYAELLP 499
Q P+ +PQ QQQP P P P +P P P Y P
Sbjct: 74 QPQHLPHPPHPQMFGQQQPQAFLPHLP-PHHLP-PPFPGPYDSAPP 117
Score = 41.2 bits (95), Expect = 0.007
Identities = 40/144 (27%), Positives = 50/144 (33%), Gaps = 9/144 (6%)
Query: 403 PFQQTNNHPQIPQYPQIPQYPQ-------IPQYPQFPQNPSPQNTQQQNFQQQPYQQQPY 455
P Q PQ +PQ PQYP IP +P Q P Q+ Q Q Q P+
Sbjct: 25 PNFQQQQQPQFGFHPQHPQYPSPMNASGFIPPHPSMQQFPY-QHPMHQQQQPQHLPHPPH 83
Query: 456 QQFPYQQYPQQNFQQ-QPYQQRPQQPRPPRMPINPIPVTYAELLPGLLKKNLVQTRTAPP 514
Q QQ PQ P+ P P P P P EL + K + P
Sbjct: 84 PQMFGQQQPQAFLPHLPPHHLPPPFPGPYDSAPPPPPPADPELQKRIDKLVEYSVKNGPE 143
Query: 515 IPEKLPSWYRLDQTCDFHEGGRGH 538
+ + + F GG GH
Sbjct: 144 FEAMMRDRQKDNPDYAFLFGGEGH 167
Score = 40.0 bits (92), Expect = 0.016
Identities = 35/114 (30%), Positives = 45/114 (38%), Gaps = 19/114 (16%)
Query: 403 PFQQTNNHPQIPQYPQIPQYPQIPQYPQFPQNPSPQNTQQQNFQQQPYQQQPYQQFPYQQ 462
P+ Q Q P + Q Q PQ +PQ PQ PSP N P+ QQFPYQ
Sbjct: 15 PYAQQQQQ-QGPNFQQ-QQQPQFGFHPQHPQYPSPMNASGFI---PPHPSM--QQFPYQ- 66
Query: 463 YPQQNFQQQPYQQRPQQPRPPRMPINPIPVTYAELLPGLLKKNLVQTRTAPPIP 516
+P QQ +P +P P P + + P +L PP P
Sbjct: 67 HPMH-----------QQQQPQHLPHPPHPQMFGQQQPQAFLPHLPPHHLPPPFP 109
>At2g45810 putative ATP-dependent RNA helicase
Length = 528
Score = 49.7 bits (117), Expect = 2e-05
Identities = 38/97 (39%), Positives = 45/97 (46%), Gaps = 5/97 (5%)
Query: 394 IAAITPTSHPFQQTNNHPQIPQYPQIPQYPQI-PQYPQFPQNPSPQNTQQQNFQQQPYQQ 452
I A P P Q+ N P PQ QY Q +PQ PQ PQ Q Q+ QQ Q+
Sbjct: 15 IGAAGPGPDPNFQSRN----PNPPQPQQYLQSRTPFPQQPQPQPPQYLQSQSDAQQYVQR 70
Query: 453 QPYQQFPYQQYPQQNFQQQPYQQRPQQPRPPRMPINP 489
QQ QQ QQ QQQ QQ Q R ++P +P
Sbjct: 71 GYPQQIQQQQQLQQQQQQQQQQQEQQWSRRAQLPGDP 107
>At1g25540 hypothetical protein
Length = 809
Score = 49.7 bits (117), Expect = 2e-05
Identities = 41/120 (34%), Positives = 51/120 (42%), Gaps = 12/120 (10%)
Query: 399 PTSHPFQQTNNHPQIPQYPQIPQYPQIPQYPQFPQNPSPQNTQQQNFQQQPYQQQPYQQF 458
P QQ H Q Q QI Q Q Q+ Q Q P Q QQQ+ QQQ Q Q Q
Sbjct: 652 PNQQQQQQQQLHQQQQQQQQIQQQQQQQQHLQQQQMPQLQQQQQQHQQQQQQQHQLSQLQ 711
Query: 459 PYQQYPQQNFQQQPYQQRPQ-----QPRPPRMPINPIPVTYAELLPGLLKKNLVQTRTAP 513
+QQ QQ QQQ Q Q Q + P+N + + L N +Q +T+P
Sbjct: 712 HHQQQQQQQQQQQQQHQLTQLQHHHQQQQQASPLNQMQQQTSPL-------NQMQQQTSP 764
Score = 44.7 bits (104), Expect = 7e-04
Identities = 34/103 (33%), Positives = 39/103 (37%), Gaps = 4/103 (3%)
Query: 384 PQQTYPAYQHIAAITPTSHPFQQTNNHPQIPQYPQIPQYPQIPQYPQFPQNPSPQNTQQQ 443
P Q Q + QQ Q Q Q+PQ Q Q Q Q Q +Q Q
Sbjct: 652 PNQQQQQQQQLHQQQQQQQQIQQQQQQQQHLQQQQMPQLQQQQQQHQQQQQQQHQLSQLQ 711
Query: 444 NFQQQPYQQQPYQQ----FPYQQYPQQNFQQQPYQQRPQQPRP 482
+ QQQ QQQ QQ Q + QQ Q P Q QQ P
Sbjct: 712 HHQQQQQQQQQQQQQHQLTQLQHHHQQQQQASPLNQMQQQTSP 754
Score = 44.7 bits (104), Expect = 7e-04
Identities = 29/58 (50%), Positives = 31/58 (53%), Gaps = 4/58 (6%)
Query: 423 PQIPQYPQFPQNP-SPQNTQQQNFQQQPYQQQPYQQFPYQQYPQQNFQQQPYQQRPQQ 479
PQIP Q Q Q QQQ QQQ QQQ QQ QQ PQ QQQ +QQ+ QQ
Sbjct: 649 PQIPNQQQQQQQQLHQQQQQQQQIQQQQQQQQHLQQ---QQMPQLQQQQQQHQQQQQQ 703
Score = 43.5 bits (101), Expect = 0.001
Identities = 33/77 (42%), Positives = 37/77 (47%), Gaps = 9/77 (11%)
Query: 411 PQIPQYPQIPQYPQIPQYPQFPQNPSPQNTQQQNFQQQ--PYQQQPYQQFPYQQYPQ--- 465
PQIP Q Q Q+ Q Q Q Q QQQ+ QQQ P QQ QQ QQ Q
Sbjct: 649 PQIPNQQQ-QQQQQLHQQQQQQQQIQQQQQQQQHLQQQQMPQLQQQQQQHQQQQQQQHQL 707
Query: 466 ---QNFQQQPYQQRPQQ 479
Q+ QQQ QQ+ QQ
Sbjct: 708 SQLQHHQQQQQQQQQQQ 724
Score = 38.1 bits (87), Expect = 0.062
Identities = 33/104 (31%), Positives = 39/104 (36%), Gaps = 9/104 (8%)
Query: 385 QQTYPAYQHIAAITPTSHPFQQTNNHPQIPQYPQIPQ--YPQIPQYPQFPQNPSPQNTQQ 442
QQ QH+ QQ + Q Q Q+ Q + Q Q Q Q Q TQ
Sbjct: 673 QQQQQQQQHLQQQQMPQLQQQQQQHQQQQQQQHQLSQLQHHQQQQQQQQQQQQQHQLTQL 732
Query: 443 QNFQQQPYQQQPYQQFPYQQYPQQNFQQQP-------YQQRPQQ 479
Q+ QQ Q P Q Q P QQQ QQ+PQQ
Sbjct: 733 QHHHQQQQQASPLNQMQQQTSPLNQMQQQTSPLNQMQQQQQPQQ 776
Score = 32.7 bits (73), Expect = 2.6
Identities = 22/50 (44%), Positives = 25/50 (50%), Gaps = 9/50 (18%)
Query: 432 PQNPSPQNTQQQNFQQQPYQQQPYQQFPYQQYPQQNFQQQPYQQRPQQPR 481
PQ P+ Q QQQ QQ QQQ QQ Q QQQ QQ+ Q P+
Sbjct: 649 PQIPNQQQQQQQQLHQQQQQQQQIQQ--------QQQQQQHLQQQ-QMPQ 689
>At5g20730 auxin response factor 7 (ARF7)
Length = 1164
Score = 48.1 bits (113), Expect = 6e-05
Identities = 43/142 (30%), Positives = 62/142 (43%), Gaps = 10/142 (7%)
Query: 350 RLVKDATPASGTKKTGNHFPRKKEQEVGMVTHGRPQQTYPAYQHIAAITPTS--HPFQQT 407
+L+ TP G + F K+ Q+ M +P T Q + + +S H QQ+
Sbjct: 462 KLLSFQTPHGGISSSNLQF-NKQNQQAPMSQLPQPPTTLSQQQQLQQLLHSSLNHQQQQS 520
Query: 408 NNHPQIPQYPQIPQYPQIPQYPQFPQNPSPQNTQQQNFQQQPYQQQPYQQFPYQQYPQQN 467
+ Q Q + Q Q+ N S QQQ QQQ QQQ QQ +QQ QQ
Sbjct: 521 QSQQQQQQQQLLQQQQQLQSQQHSNNNQSQSQQQQQLLQQQ--QQQQLQQ-QHQQPLQQQ 577
Query: 468 FQQQPYQQRPQQ----PRPPRM 485
QQQ + +P Q P+P ++
Sbjct: 578 TQQQQLRTQPLQSHSHPQPQQL 599
Score = 38.9 bits (89), Expect = 0.036
Identities = 25/63 (39%), Positives = 28/63 (43%), Gaps = 2/63 (3%)
Query: 408 NNHPQIPQYPQIPQYPQIPQYPQFPQNPSPQNTQQQNFQQQPYQQQPYQQFPYQQYPQQN 467
NN Q Q Q+ Q Q Q Q Q P Q TQQQ + QP Q + Q QQ Q
Sbjct: 546 NNQSQSQQQQQLLQQQQQQQLQQQHQQPLQQQTQQQQLRTQPLQSHSHPQ--PQQLQQHK 603
Query: 468 FQQ 470
QQ
Sbjct: 604 LQQ 606
Score = 35.8 bits (81), Expect = 0.31
Identities = 42/160 (26%), Positives = 61/160 (37%), Gaps = 31/160 (19%)
Query: 385 QQTYPAYQHIAAITPTSHPFQQTNNHPQIPQYPQIPQYPQIPQYPQFPQNP--SPQNTQ- 441
+Q A+ H+ A T S + + + + P PQ Q + + Q P + QN Q
Sbjct: 667 KQLQQAHHHLGASTSQSSVIETSKSSSNLMSAP--PQETQFSRQVEQQQPPGLNGQNQQT 724
Query: 442 -----------QQNFQQ----QPY---------QQQPYQQF--PYQQYPQQNFQQQPYQQ 475
QQ FQQ QP+ QQQ QQF P Q P Q Q QQ
Sbjct: 725 LLQQKAHQAQAQQIFQQSLLEQPHIQFQLLQRLQQQQQQQFLSPQSQLPHHQLQSQQLQQ 784
Query: 476 RPQQPRPPRMPINPIPVTYAELLPGLLKKNLVQTRTAPPI 515
P + + P + + L P + + Q + PP+
Sbjct: 785 LPTLSQGHQFPSSCTNNGLSTLQPPQMLVSRPQEKQNPPV 824
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.343 0.149 0.504
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 49,323,797
Number of Sequences: 26719
Number of extensions: 2095740
Number of successful extensions: 15368
Number of sequences better than 10.0: 204
Number of HSP's better than 10.0 without gapping: 91
Number of HSP's successfully gapped in prelim test: 123
Number of HSP's that attempted gapping in prelim test: 12565
Number of HSP's gapped (non-prelim): 1311
length of query: 2359
length of database: 11,318,596
effective HSP length: 116
effective length of query: 2243
effective length of database: 8,219,192
effective search space: 18435647656
effective search space used: 18435647656
T: 11
A: 40
X1: 15 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 38 (21.5 bits)
S2: 68 (30.8 bits)
Medicago: description of AC130806.6


