

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC130806.14 - phase: 0
(394 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
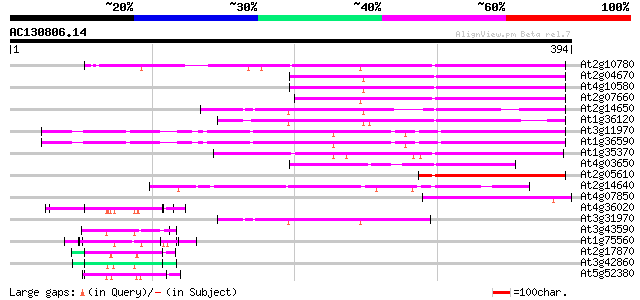
Score E
Sequences producing significant alignments: (bits) Value
At2g10780 pseudogene 172 4e-43
At2g04670 putative retroelement pol polyprotein 155 4e-38
At4g10580 putative reverse-transcriptase -like protein 154 1e-37
At2g07660 putative retroelement pol polyprotein 153 1e-37
At2g14650 putative retroelement pol polyprotein 135 5e-32
At1g36120 putative reverse transcriptase gb|AAD22339.1 113 2e-25
At3g11970 hypothetical protein 102 3e-22
At1g36590 hypothetical protein 102 3e-22
At1g35370 hypothetical protein 97 1e-20
At4g03650 putative reverse transcriptase 95 8e-20
At2g05610 putative retroelement pol polyprotein 79 3e-15
At2g14640 putative retroelement pol polyprotein 66 4e-11
At4g07850 putative polyprotein 60 2e-09
At4g36020 glycine-rich protein 57 2e-08
At3g31970 hypothetical protein 56 3e-08
At3g43590 putative protein 54 1e-07
At1g75560 DNA-binding protein 54 1e-07
At2g17870 putative glycine-rich, zinc-finger DNA-binding protein 50 2e-06
At3g42860 unknown protein (At3g42860) 50 3e-06
At5g52380 unknown protein 49 6e-06
>At2g10780 pseudogene
Length = 1611
Score = 172 bits (435), Expect = 4e-43
Identities = 122/368 (33%), Positives = 172/368 (46%), Gaps = 50/368 (13%)
Query: 53 CFKCGEKGHKSNVCTKDEKKCFRCGQKGHVLADCKRGDI-----------VCYNCNGEGH 101
C CG K H S C K C RCG K H + C + + C+ C GH
Sbjct: 381 CVTCG-KNH-SGTCWKAIGACGRCGSKDHAIQSCPKMEPGQSKVLGEETRTCFYCGKTGH 438
Query: 102 ISSQCTQPKKVRTGGKVFALAGMQTTNEDRLIRGTCFFNSIPLIAIIDTGATHCFIALEC 161
+ +C + + G+ G N +P A+ + E
Sbjct: 439 LKRECPKLTAEKQAGQRDNRGG----------------NGLPPPPKRQAVASRVYELSEE 482
Query: 162 AYKLG---LIVSDMKGE------MVVETPAKGSVTTSLVCLICPISMFGRDFEVDLVCLP 212
A G I + E MV + T LV I + + G + DL+ +P
Sbjct: 483 ANDAGNFRAITGGFRKEPNTDYGMVRAAGGQAMYPTGLVRGISVV-VNGVNMPADLIIVP 541
Query: 213 LTGMDVIFGMNWLEYNRVHINCFNKTVHFSSAE---------EESGVEILSTKEMKQLER 263
L DVI GM+WL + HI+C + F E SG ++S + +++
Sbjct: 542 LKKHDVILGMDWLGKYKAHIDCHRGRIQFERDEGMLKFQGIRTTSGSLVISAIQAERMLG 601
Query: 264 DGILMY-SLMACLSLENQAVIDKLPVVNEFPEVFSDEIPDVPPEREVEFSIDLVPRTKPV 322
G Y + + + A + +P+VNEF +VF+ + VPP+R F+I+L P T P+
Sbjct: 602 KGCEAYLATITTKEVGASAELKDIPIVNEFSDVFA-AVSGVPPDRSDPFTIELEPGTTPI 660
Query: 323 SMAPYRMSASELAELKNQLEDLLDKKFVRASVFPWGAPVLLAKKKDGSMRLCIAYPPLNK 382
S APYRM+ +E+A+LK QLE+LLDK F+R S PWGAPVL KKKDGS RLCI Y LNK
Sbjct: 661 SKAPYRMAPAEMAKLKKQLEELLDKGFIRPSSSPWGAPVLFVKKKDGSFRLCIDYRGLNK 720
Query: 383 VPIKNRYP 390
V +KN+YP
Sbjct: 721 VTVKNKYP 728
>At2g04670 putative retroelement pol polyprotein
Length = 1411
Score = 155 bits (392), Expect = 4e-38
Identities = 85/204 (41%), Positives = 115/204 (55%), Gaps = 11/204 (5%)
Query: 197 ISMFGRDFEVDLVCLPLTGMDVIFGMNWLEYNRVHINCFNKTVHFSSAEEE--------- 247
I + G DL+ P+ DVI GM+WL++ RVH++C V F E
Sbjct: 384 IQIAGESMPADLIISPVELYDVILGMDWLDHYRVHLDCHRGRVSFERPEGRLVYQGVRPT 443
Query: 248 SGVEILSTKEMKQLERDGILMYSLMACLSLE-NQAVIDKLPVVNEFPEVFSDEIPDVPPE 306
SG ++S + K++ G Y + + Q + + V+ EF +VF + +PP
Sbjct: 444 SGSLVISAVQAKKMIEKGCEAYLVTISMPESLGQVAVSDIRVIQEFEDVFQS-LQGLPPS 502
Query: 307 REVEFSIDLVPRTKPVSMAPYRMSASELAELKNQLEDLLDKKFVRASVFPWGAPVLLAKK 366
R F+I+L P T P+S APYRM+ +E+ ELK QLEDLL K F+R S PWGAPVL KK
Sbjct: 503 RSDPFTIELEPGTAPLSKAPYRMAPAEMTELKKQLEDLLGKGFIRPSTSPWGAPVLFVKK 562
Query: 367 KDGSMRLCIAYPPLNKVPIKNRYP 390
KDGS RLCI Y LN V +KN+YP
Sbjct: 563 KDGSFRLCIDYRGLNWVTVKNKYP 586
>At4g10580 putative reverse-transcriptase -like protein
Length = 1240
Score = 154 bits (388), Expect = 1e-37
Identities = 85/204 (41%), Positives = 116/204 (56%), Gaps = 11/204 (5%)
Query: 197 ISMFGRDFEVDLVCLPLTGMDVIFGMNWLEYNRVHINCFNKTVHFSSAEEE--------- 247
I + G DL+ P+ DVI GM+WL+Y RVH++ V F E
Sbjct: 358 IQIAGESMPADLIISPVELYDVILGMDWLDYYRVHLDWHRGRVFFERPEGRLVYQGVRPI 417
Query: 248 SGVEILSTKEMKQLERDGILMYSLMACLSLE-NQAVIDKLPVVNEFPEVFSDEIPDVPPE 306
SG ++S + +++ G Y + + Q + + VV EF +VF + +PP
Sbjct: 418 SGSLVISAVQAEKMIEKGCEAYLVTISMPESVGQVAVSDIRVVQEFQDVFQS-LQGLPPS 476
Query: 307 REVEFSIDLVPRTKPVSMAPYRMSASELAELKNQLEDLLDKKFVRASVFPWGAPVLLAKK 366
+ F+I+L P T P+S APYRM+ +E+AELK QL+DLL K F+R S PWGAPVL KK
Sbjct: 477 QSDPFTIELEPGTAPLSKAPYRMAPAEMAELKKQLKDLLGKGFIRPSTSPWGAPVLFVKK 536
Query: 367 KDGSMRLCIAYPPLNKVPIKNRYP 390
KDGS RLCI Y LN+V +KNRYP
Sbjct: 537 KDGSFRLCIDYRELNRVTVKNRYP 560
>At2g07660 putative retroelement pol polyprotein
Length = 949
Score = 153 bits (387), Expect = 1e-37
Identities = 85/200 (42%), Positives = 119/200 (59%), Gaps = 11/200 (5%)
Query: 201 GRDFEVDLVCLPLTGMDVIFGMNWLEYNRVHINCFNKTVHFSSAE---------EESGVE 251
G + DL+ +PL DVI GM+WL + HI+C + + F E SG
Sbjct: 25 GVNMPADLIIVPLKKHDVILGMDWLGKYKGHIDCHRERIQFERDEGMLKFQGIRTTSGSL 84
Query: 252 ILSTKEMKQLERDGILMY-SLMACLSLENQAVIDKLPVVNEFPEVFSDEIPDVPPEREVE 310
++S + +++ G Y + + + A + + +VNEF +VF+ + VPP+R
Sbjct: 85 VISAIQAERMLGKGCEAYLATITTKEVGASAELKDILIVNEFSDVFA-AVSGVPPDRSDP 143
Query: 311 FSIDLVPRTKPVSMAPYRMSASELAELKNQLEDLLDKKFVRASVFPWGAPVLLAKKKDGS 370
F+I+L P T P+S APYRM+ +E+AELK QLE+LL K F+R S PWGAPVL KKKDGS
Sbjct: 144 FTIELEPGTTPISKAPYRMAPAEMAELKKQLEELLAKGFIRPSSSPWGAPVLFVKKKDGS 203
Query: 371 MRLCIAYPPLNKVPIKNRYP 390
RLCI Y LNKV +KN+YP
Sbjct: 204 FRLCIDYRGLNKVTVKNKYP 223
>At2g14650 putative retroelement pol polyprotein
Length = 1328
Score = 135 bits (339), Expect = 5e-32
Identities = 94/267 (35%), Positives = 128/267 (47%), Gaps = 46/267 (17%)
Query: 135 GTCFFNSIPLIAIIDTGATHCFIALECAYKLGLIVSDMKGEMVVETPAKGSVTTSLVCLI 194
GT + + D+GA+HCFI E A + G I D GE + G +++
Sbjct: 304 GTLLVGGVEAHVLFDSGASHCFITPESASR-GNIRGD-PGEQLGAVKVAGGQFVAVLGRT 361
Query: 195 --CPISMFGRDFEVDLVCLPLTGMDVIFGMNWLEYNRVHINCFNKTVHFSSAEEE----- 247
I + G DL+ P+ DVI GM+WL++ RVH++C V F E
Sbjct: 362 KGVDIQIAGESMPADLIISPVELYDVILGMDWLDHYRVHLDCHRGRVSFERPEGSLVYQG 421
Query: 248 ----SGVEILSTKEMKQLERDGILMYSLMACLSLENQAVIDKLPVVNEFPEVFSDEIPDV 303
SG ++S + +++ G Y EF +VF + +
Sbjct: 422 VRPTSGSLVISAVQAEKMIEKGCEAYL--------------------EFEDVFQS-LQGL 460
Query: 304 PPEREVEFSIDLVPRTKPVSMAPYRMSASELAELKNQLEDLLDKKFVRASVFPWGAPVLL 363
PP R F+I+L P T P+S APYRM+ +E+AELK QLEDLL GAPVL
Sbjct: 461 PPSRSDPFTIELEPGTAPLSKAPYRMAPAEMAELKKQLEDLL------------GAPVLF 508
Query: 364 AKKKDGSMRLCIAYPPLNKVPIKNRYP 390
KKKDGS RLCI Y LN V +KN+YP
Sbjct: 509 VKKKDGSFRLCIDYRGLNWVTVKNKYP 535
>At1g36120 putative reverse transcriptase gb|AAD22339.1
Length = 1235
Score = 113 bits (283), Expect = 2e-25
Identities = 81/259 (31%), Positives = 123/259 (47%), Gaps = 39/259 (15%)
Query: 147 IIDTGATHCFIALECAYKLGLIVSDMKGEMVVETPAKGSVTTSLVCLI-----CPISMFG 201
+ D+GA+HCFI E A + +++GE + A + ++ I +
Sbjct: 352 LFDSGASHCFITPESASR-----DNIRGEPGEQFGAVKIAGGQFLTVLGRAKGVNIQIEE 406
Query: 202 RDFEVDLVCLPLTGMDVIFGMNWLEYNRVHINCFNKTVHFSSAEEE---SGVE------I 252
DL+ P+ D I GM+WL++ RVH++C V F E GV +
Sbjct: 407 ESMPADLIINPVVLYDAILGMDWLDHYRVHLDCHRGRVSFERPEGRLVYQGVRPTSRSLV 466
Query: 253 LSTKEMKQLERDGILMYSLMACLSLE-NQAVIDKLPVVNEFPEVFSDEIPDVPPEREVEF 311
+S +++++ G Y + +S Q + + VV EF +VF + +PP R F
Sbjct: 467 ISEVQVEKMIEKGCEAYLVTISMSESVGQVAVSDIWVVQEFEDVFQS-LQGLPPSRSDPF 525
Query: 312 SIDLVPRTKPVSMAPYRMSASELAELKNQLEDLLDKKFVRASVFPWGAPVLLAKKKDGSM 371
+I+L T P+S PYRM +E+AELK QLEDLL K F+R S WGA
Sbjct: 526 TIELELGTAPLSKTPYRMVPAEIAELKKQLEDLLGKGFIRPSTSRWGA------------ 573
Query: 372 RLCIAYPPLNKVPIKNRYP 390
P LN+V +KN+YP
Sbjct: 574 ------PGLNRVTLKNKYP 586
>At3g11970 hypothetical protein
Length = 1499
Score = 102 bits (255), Expect = 3e-22
Identities = 100/397 (25%), Positives = 173/397 (43%), Gaps = 61/397 (15%)
Query: 23 KPYSAPAGKGKQRLNDERRPKGRDTPAEIVCFKCGEKGHKSNVCTKDEKKCFRCGQKGHV 82
KP S K Q+ +RR KG +C+ C EK + + + FR
Sbjct: 306 KPVSQQPKKMSQQEMSDRRSKG-------LCYFCDEKYTPEHYLVHKKTQLFRMDVDEEF 358
Query: 83 LADCKRGDIVCYNCNGEGHISSQCTQPKKVRTGGKVFALAGMQTTNEDRLIRGTCFFNSI 142
+ R ++V + IS V +AG +T ++GT ++
Sbjct: 359 --EDAREELVNDDDEHMPQISVNA-----------VSGIAGYKTMR----VKGT--YDKK 399
Query: 143 PLIAIIDTGATHCFIALECAYKLGLIVSDMKGEMVVETPAKGSVTTSLVCLICPISMFGR 202
+ +ID+G+TH F+ A KLG V D G V + +
Sbjct: 400 IIFILIDSGSTHNFLDPNTAAKLGCKV-DTAGLTRVSVADGRKLRVEGKVTDFSWKLQTT 458
Query: 203 DFEVDLVCLPLTGMDVIFGMNWLE--------YNRVHINC-FNKTVHFSSAEEESGVEIL 253
F+ D++ +PL G+D++ G+ WLE + ++ + FN V +
Sbjct: 459 TFQSDILLIPLQGIDMVLGVQWLETLGRISWEFKKLEMRFKFNNQKVLLHGLTSGSVREV 518
Query: 254 STKEMKQLERDGILMYSLMACLS-------------------LENQAVIDKLPVVNEFPE 294
+++++L+ D + + M C+ L ++V+++ V+NE+P+
Sbjct: 519 KAQKLQKLQEDQVQL--AMLCVQEVSESTEGELCTINALTSELGEESVVEE--VLNEYPD 574
Query: 295 VFSDEIPDVPPEREVE-FSIDLVPRTKPVSMAPYRMSASELAELKNQLEDLLDKKFVRAS 353
+F + +PP RE I L+ + PV+ PYR S + E+ +EDLL V+AS
Sbjct: 575 IFIEPTA-LPPFREKHNHKIKLLEGSNPVNQRPYRYSIHQKNEIDKLVEDLLTNGTVQAS 633
Query: 354 VFPWGAPVLLAKKKDGSMRLCIAYPPLNKVPIKNRYP 390
P+ +PV+L KKKDG+ RLC+ Y LN + +K+ +P
Sbjct: 634 SSPYASPVVLVKKKDGTWRLCVDYRELNGMTVKDSFP 670
>At1g36590 hypothetical protein
Length = 1499
Score = 102 bits (255), Expect = 3e-22
Identities = 100/397 (25%), Positives = 173/397 (43%), Gaps = 61/397 (15%)
Query: 23 KPYSAPAGKGKQRLNDERRPKGRDTPAEIVCFKCGEKGHKSNVCTKDEKKCFRCGQKGHV 82
KP S K Q+ +RR KG +C+ C EK + + + FR
Sbjct: 306 KPVSQQPKKMSQQEMSDRRSKG-------LCYFCDEKYTPEHYLVHKKTQLFRMDVDEEF 358
Query: 83 LADCKRGDIVCYNCNGEGHISSQCTQPKKVRTGGKVFALAGMQTTNEDRLIRGTCFFNSI 142
+ R ++V + IS V +AG +T ++GT ++
Sbjct: 359 --EDAREELVNDDDEHMPQISVNA-----------VSGIAGYKTMR----VKGT--YDKK 399
Query: 143 PLIAIIDTGATHCFIALECAYKLGLIVSDMKGEMVVETPAKGSVTTSLVCLICPISMFGR 202
+ +ID+G+TH F+ A KLG V D G V + +
Sbjct: 400 IIFILIDSGSTHNFLDPNTAAKLGCKV-DTAGLTRVSVADGRKLRVEGKVTDFSWKLQTT 458
Query: 203 DFEVDLVCLPLTGMDVIFGMNWLE--------YNRVHINC-FNKTVHFSSAEEESGVEIL 253
F+ D++ +PL G+D++ G+ WLE + ++ + FN V +
Sbjct: 459 TFQSDILLIPLQGIDMVLGVQWLETLGRISWEFKKLEMRFKFNNQKVLLHGLTSGSVREV 518
Query: 254 STKEMKQLERDGILMYSLMACLS-------------------LENQAVIDKLPVVNEFPE 294
+++++L+ D + + M C+ L ++V+++ V+NE+P+
Sbjct: 519 KAQKLQKLQEDQVQL--AMLCVQEVSESTEGELCTINALTSELGEESVVEE--VLNEYPD 574
Query: 295 VFSDEIPDVPPEREVE-FSIDLVPRTKPVSMAPYRMSASELAELKNQLEDLLDKKFVRAS 353
+F + +PP RE I L+ + PV+ PYR S + E+ +EDLL V+AS
Sbjct: 575 IFIEPTA-LPPFREKHNHKIKLLEGSNPVNQRPYRYSIHQKNEIDKLVEDLLTNGTVQAS 633
Query: 354 VFPWGAPVLLAKKKDGSMRLCIAYPPLNKVPIKNRYP 390
P+ +PV+L KKKDG+ RLC+ Y LN + +K+ +P
Sbjct: 634 SSPYASPVVLVKKKDGTWRLCVDYRELNGMTVKDSFP 670
>At1g35370 hypothetical protein
Length = 1447
Score = 97.4 bits (241), Expect = 1e-20
Identities = 79/272 (29%), Positives = 132/272 (48%), Gaps = 29/272 (10%)
Query: 144 LIAIIDTGATHCFIALECAYKLGLIVSDMK-GEMVVETPAKGSVTTSLVCLICPISMFGR 202
L +ID+G+TH FI A KLG V ++ V K +V + +
Sbjct: 380 LFILIDSGSTHNFIDSTVAAKLGCHVESAGLTKVAVADGRKLNVDGQIKGFTWKLQ--ST 437
Query: 203 DFEVDLVCLPLTGMDVIFGMNWLE--------YNRVHINCF--NKTVHFSSAEEESGVEI 252
F+ D++ +PL G+D++ G+ WLE + ++ + F N+ V S +I
Sbjct: 438 TFQSDILLIPLQGVDMVLGVQWLETLGRISWEFKKLEMQFFYKNQRVWLHGIITGSVRDI 497
Query: 253 LSTKEMK-QLERDGILMYSLMACLSLENQAV--IDKLP-----------VVNEFPEVFSD 298
+ K K Q ++ + M + +S E Q + I L +V EFP+VF+
Sbjct: 498 KAHKLQKTQADQIQLAMVCVREVVSDEEQEIGSISALTSDVVEESVVQNIVEEFPDVFA- 556
Query: 299 EIPDVPPEREV-EFSIDLVPRTKPVSMAPYRMSASELAELKNQLEDLLDKKFVRASVFPW 357
E D+PP RE + I L+ PV+ PYR + E+ ++D++ ++ S P+
Sbjct: 557 EPTDLPPFREKHDHKIKLLEGANPVNQRPYRYVVHQKDEIDKIVQDMIKSGTIQVSSSPF 616
Query: 358 GAPVLLAKKKDGSMRLCIAYPPLNKVPIKNRY 389
+PV+L KKKDG+ RLC+ Y LN + +K+R+
Sbjct: 617 ASPVVLVKKKDGTWRLCVDYTELNGMTVKDRF 648
>At4g03650 putative reverse transcriptase
Length = 839
Score = 94.7 bits (234), Expect = 8e-20
Identities = 58/159 (36%), Positives = 85/159 (52%), Gaps = 9/159 (5%)
Query: 197 ISMFGRDFEVDLVCLPLTGMDVIFGMNWLEYNRVHINCFNKTVHFSSAEEESGVEILSTK 256
I + G DL+ + DVI GM+WL++ RVH++C V F E +G E +
Sbjct: 377 IQIAGESLPADLIISHVELYDVILGMDWLDHYRVHLDCHRGRVSFERPEGSAGREN-DRE 435
Query: 257 EMKQLERDGILMYSLMACLSLENQAVIDKLPVVNEFPEVFSDEIPDVPPEREVEFSIDLV 316
++ L D I + Q + + VV EF VF + +PP R F+I+L
Sbjct: 436 GLRGLPGDDIYA-------GVCGQVAVSDIRVVQEFQYVFQS-LQGLPPSRSDPFTIELE 487
Query: 317 PRTKPVSMAPYRMSASELAELKNQLEDLLDKKFVRASVF 355
P+T P+S APYRM+ +++AELK QLEDLL + VR + F
Sbjct: 488 PKTAPLSKAPYRMAPAKMAELKKQLEDLLAEADVRKTAF 526
>At2g05610 putative retroelement pol polyprotein
Length = 780
Score = 79.3 bits (194), Expect = 3e-15
Identities = 41/104 (39%), Positives = 65/104 (62%), Gaps = 2/104 (1%)
Query: 288 VVNEFPEVFSDEIPDVPPEREV-EFSIDLVPRTKPVSMAPYRMSASELAELKNQLEDLLD 346
VV EFP++F E D+PP R + I+L+ + PV+ PYR + E+ ++D+L
Sbjct: 10 VVTEFPDIFV-EPTDLPPFRAPHDHKIELLEDSNPVNQRPYRYVVHQKNEIDKIVDDMLA 68
Query: 347 KKFVRASVFPWGAPVLLAKKKDGSMRLCIAYPPLNKVPIKNRYP 390
++AS P+ +PV+L KKKDG+ RLC+ Y LN + +K+R+P
Sbjct: 69 SGTIQASSSPYASPVVLVKKKDGTWRLCVDYRELNGMTVKDRFP 112
>At2g14640 putative retroelement pol polyprotein
Length = 945
Score = 65.9 bits (159), Expect = 4e-11
Identities = 79/287 (27%), Positives = 125/287 (43%), Gaps = 37/287 (12%)
Query: 99 EGHISSQCTQPKKVRTGGK----VFALAGMQTTNEDRLIRGTCFFNSIPLIAIIDTGATH 154
+G + + + GK ++AL G+ TT+ R +R T N LIA+I++G+TH
Sbjct: 283 KGENDDEAVEEESTEAEGKPEITLYALEGVDTTSTIR-VRATIHRNR--LIALINSGSTH 339
Query: 155 CFIALECAYKLGLIVSDMKGEMVVETPAKGSVTTSLVCLICPISMFGRDFEVDLVCLPLT 214
FI + + L + K V V S I P+ M G F V L LPL
Sbjct: 340 NFIGEKAVRGMNLKATTTKPFTVRVVNGMPLVCRSRYEAI-PVVMGGVVFPVTLYALPLM 398
Query: 215 GMDVIFGMNWLE-YNRVHINCFNKTVHFSSAEEESGVEILSTK-------EMKQLERDGI 266
G+D+ G+ WL N +T+ F A +E V ++ K E K + +
Sbjct: 399 GLDLAMGVQWLSTLGPTLCNWKEQTLQFHWAGDE--VRLMGIKPTGLRGVEHKTITKKAR 456
Query: 267 LMYSLMACLSLENQA-------VIDKLPVVNEFPEVFSDEIP-DVPPEREVEFSIDLVPR 318
+ +++ A N + I KL + EF +F E P +P ER + I L
Sbjct: 457 MGHTIFAITMAHNSSDPNTPDGEIKKL--IEEFAGLF--ETPTQLPSERPIVHRIALKKG 512
Query: 319 TKPVSMAPYRMSASELAELKNQLEDLLDKKFVRASVFPWGAPVLLAK 365
T PV++ PYR + Q +++ +R S P+ +PVLL +
Sbjct: 513 TDPVNVRPYRYAF-------YQKDEMSRAGIIRPSSSPFLSPVLLER 552
>At4g07850 putative polyprotein
Length = 1138
Score = 60.1 bits (144), Expect = 2e-09
Identities = 36/109 (33%), Positives = 57/109 (52%), Gaps = 5/109 (4%)
Query: 291 EFPEVFSDEIPD-VPPEREVEFSIDLVPRTKPVSMAPYRMSASELAELKNQLEDLLDKKF 349
++ +VF +E P+ +PP R +E ID VP + YR + E EL+ Q+ +L+++
Sbjct: 447 DYQDVFPEENPEGLPPIRGIEHQIDFVPGASLPNRPAYRTNPVETKELEKQVNELMERGH 506
Query: 350 VRASVFPWGAPVLLAKKKDGSMRLCIAYPPL----NKVPIKNRYPSSKN 394
+ S+ P PVLL KKDGS R + + KV +PS K+
Sbjct: 507 ICESMSPCAVPVLLVPKKDGSWRFVASTDGVKVDEEKVKAIREWPSPKS 555
>At4g36020 glycine-rich protein
Length = 299
Score = 56.6 bits (135), Expect = 2e-08
Identities = 36/122 (29%), Positives = 50/122 (40%), Gaps = 25/122 (20%)
Query: 26 SAPAGKGKQRLNDERRPKGRDTPAEIVCFKCGEKGHKSNVC------------TKDEKKC 73
+AP G ++ N+ R R C+ CGE GH S C ++ + C
Sbjct: 75 TAPGGGSLKKENNSRGNGARRGGGGSGCYNCGELGHISKDCGIGGGGGGGERRSRGGEGC 134
Query: 74 FRCGQKGHVLADC------------KRGDIVCYNCNGEGHISSQCTQPKKVRTGGKVFAL 121
+ CG GH DC K G+ CY C GH++ CTQ K V G + A+
Sbjct: 135 YNCGDTGHFARDCTSAGNGDQRGATKGGNDGCYTCGDVGHVARDCTQ-KSVGNGDQRGAV 193
Query: 122 AG 123
G
Sbjct: 194 KG 195
Score = 51.2 bits (121), Expect = 1e-06
Identities = 23/65 (35%), Positives = 34/65 (51%), Gaps = 10/65 (15%)
Query: 53 CFKCGEKGHKSNVCT---KDEKKCFRCGQKGHVLADC-KRG------DIVCYNCNGEGHI 102
C+ CG GH + C + + C++CG GH+ DC +RG D CY C EGH
Sbjct: 232 CYSCGGVGHIARDCATKRQPSRGCYQCGGSGHLARDCDQRGSGGGGNDNACYKCGKEGHF 291
Query: 103 SSQCT 107
+ +C+
Sbjct: 292 ARECS 296
Score = 49.3 bits (116), Expect = 4e-06
Identities = 22/71 (30%), Positives = 32/71 (44%), Gaps = 15/71 (21%)
Query: 53 CFKCGEKGHKSNVCTKD------------EKKCFRCGQKGHVLADC---KRGDIVCYNCN 97
C+ CG+ GH + CT+ C+ CG GH+ DC ++ CY C
Sbjct: 200 CYTCGDVGHFARDCTQKVAAGNVRSGGGGSGTCYSCGGVGHIARDCATKRQPSRGCYQCG 259
Query: 98 GEGHISSQCTQ 108
G GH++ C Q
Sbjct: 260 GSGHLARDCDQ 270
Score = 48.1 bits (113), Expect = 8e-06
Identities = 31/113 (27%), Positives = 43/113 (37%), Gaps = 34/113 (30%)
Query: 29 AGKGKQRLNDERRPKGRDTPAEIVCFKCGEKGHKSNVCTK------DEKK--------CF 74
AG G QR G C+ CG+ GH + CT+ D++ C+
Sbjct: 150 AGNGDQR--------GATKGGNDGCYTCGDVGHVARDCTQKSVGNGDQRGAVKGGNDGCY 201
Query: 75 RCGQKGHVLADCKR------------GDIVCYNCNGEGHISSQCTQPKKVRTG 115
CG GH DC + G CY+C G GHI+ C ++ G
Sbjct: 202 TCGDVGHFARDCTQKVAAGNVRSGGGGSGTCYSCGGVGHIARDCATKRQPSRG 254
>At3g31970 hypothetical protein
Length = 1329
Score = 56.2 bits (134), Expect = 3e-08
Identities = 44/161 (27%), Positives = 73/161 (45%), Gaps = 14/161 (8%)
Query: 147 IIDTGATHCFIALECAYKLGLIVSDMKGEMVVETPAKGSVTTSLVCLI--CPISMFGRDF 204
+ D+GA+HCFI E A + G I D GE + G +++ I + G
Sbjct: 415 LFDSGASHCFITPESASR-GNIRGD-PGEQLGAVKVAGGQFLAVLGRAKGVDIQIAGESM 472
Query: 205 EVDLVCLPLTGMDVIFGMNWLEYNRVHINCFNKTVHFSSAE---------EESGVEILST 255
VDL+ P+ DVI GM+WL++ RVH++C V F E S ++S
Sbjct: 473 PVDLIISPVELYDVILGMDWLDHYRVHLDCHRGRVSFERPEGRLVYQRVRPTSRSLVISA 532
Query: 256 KEMKQLERDGILMYSLMACL-SLENQAVIDKLPVVNEFPEV 295
+ +++ G Y + + Q + + VV+EF ++
Sbjct: 533 VQAEKMIEKGCEAYLVTISMPESVGQVAVSDIRVVHEFEDI 573
>At3g43590 putative protein
Length = 551
Score = 54.3 bits (129), Expect = 1e-07
Identities = 28/70 (40%), Positives = 34/70 (48%), Gaps = 6/70 (8%)
Query: 51 IVCFKCGEKGHKSNVC---TKDEKKCFRCGQKGHVLADCKRGDIVCYNCNGEGHISSQCT 107
+ C+ CGE+GH S C TK K CF CG H C +G CY C GH + C
Sbjct: 166 VSCYSCGEQGHTSFNCPTPTKRRKPCFICGSLEHGAKQCSKGHD-CYICKKTGHRAKDC- 223
Query: 108 QPKKVRTGGK 117
P K + G K
Sbjct: 224 -PDKYKNGSK 232
Score = 53.1 bits (126), Expect = 3e-07
Identities = 22/93 (23%), Positives = 39/93 (41%), Gaps = 31/93 (33%)
Query: 51 IVCFKCGEKGHKSNVC--------------------TKDEKKCFRCGQKGHVLADC---- 86
+ C++CG+ GH C +++ +C+RCG++GH +C
Sbjct: 285 VSCYRCGQLGHSGLACGRHYEESNENDSATPERLFNSREASECYRCGEEGHFARECPNSS 344
Query: 87 -------KRGDIVCYNCNGEGHISSQCTQPKKV 112
+ +CY CNG GH + +C +V
Sbjct: 345 SISTSHGRESQTLCYRCNGSGHFARECPNSSQV 377
Score = 35.8 bits (81), Expect = 0.042
Identities = 25/114 (21%), Positives = 33/114 (28%), Gaps = 51/114 (44%)
Query: 52 VCFKCGEKGHKSNVCTKDEKK-------------------------------CFRCGQKG 80
VC +CG+ GH +C + K C+RCGQ G
Sbjct: 235 VCLRCGDFGHDMILCKYEYSKEDLKDVQCYICKSFGHLCCVEPGNSLSWAVSCYRCGQLG 294
Query: 81 HVLADC--------------------KRGDIVCYNCNGEGHISSQCTQPKKVRT 114
H C R CY C EGH + +C + T
Sbjct: 295 HSGLACGRHYEESNENDSATPERLFNSREASECYRCGEEGHFARECPNSSSIST 348
Score = 29.3 bits (64), Expect = 3.9
Identities = 11/25 (44%), Positives = 14/25 (56%)
Query: 89 GDIVCYNCNGEGHISSQCTQPKKVR 113
G + CY+C +GH S C P K R
Sbjct: 164 GWVSCYSCGEQGHTSFNCPTPTKRR 188
>At1g75560 DNA-binding protein
Length = 257
Score = 53.9 bits (128), Expect = 1e-07
Identities = 28/79 (35%), Positives = 40/79 (50%), Gaps = 10/79 (12%)
Query: 49 AEIVCFKCGEKGHKSNVCTKDEKKCFRCGQKGHVLADCKRGDI------VCYNCNGEGHI 102
AE C+ C E GH ++ C+ +E C CG+ GH DC D +C NC +GH+
Sbjct: 91 AESRCWNCREPGHVASNCS-NEGICHSCGKSGHRARDCSNSDSRAGDLRLCNNCFKQGHL 149
Query: 103 SSQCTQP---KKVRTGGKV 118
++ CT K RT G +
Sbjct: 150 AADCTNDKACKNCRTSGHI 168
Score = 52.0 bits (123), Expect = 6e-07
Identities = 22/63 (34%), Positives = 33/63 (51%), Gaps = 7/63 (11%)
Query: 50 EIVCFKCGEKGHKSNVCTKDEKK------CFRCGQKGHVLADCKRGDIVCYNCNGEGHIS 103
E +C CG+ GH++ C+ + + C C ++GH+ ADC D C NC GHI+
Sbjct: 111 EGICHSCGKSGHRARDCSNSDSRAGDLRLCNNCFKQGHLAADC-TNDKACKNCRTSGHIA 169
Query: 104 SQC 106
C
Sbjct: 170 RDC 172
Score = 49.3 bits (116), Expect = 4e-06
Identities = 29/88 (32%), Positives = 41/88 (45%), Gaps = 12/88 (13%)
Query: 52 VCFKCGEKGHKSNVCTKDEKKCFRCGQKGHVLADCKRGDIVCYNCNGEGHISSQC----- 106
+C C ++GH + CT D K C C GH+ DC R D VC C+ GH++ C
Sbjct: 139 LCNNCFKQGHLAADCTND-KACKNCRTSGHIARDC-RNDPVCNICSISGHVARHCPKGDS 196
Query: 107 ---TQPKKVRTGGKVFALAGMQTTNEDR 131
+ +VR GG G+ + DR
Sbjct: 197 NYSDRGSRVRDGG--MQRGGLSRMSRDR 222
Score = 46.6 bits (109), Expect = 2e-05
Identities = 24/79 (30%), Positives = 38/79 (47%), Gaps = 3/79 (3%)
Query: 39 ERRPKGRDTPAEIVCFKCGEKGHKSNVCTKDEKKCFRCGQKGHVLADCKRGDIVCYNCNG 98
ER P+ + + C C GH + C+ + C CG GH+ A+C + C+NC
Sbjct: 44 EREPRRAFSQGNL-CNNCKRPGHFARDCS-NVSVCNNCGLPGHIAAECT-AESRCWNCRE 100
Query: 99 EGHISSQCTQPKKVRTGGK 117
GH++S C+ + GK
Sbjct: 101 PGHVASNCSNEGICHSCGK 119
Score = 37.0 bits (84), Expect = 0.019
Identities = 24/103 (23%), Positives = 32/103 (30%), Gaps = 34/103 (33%)
Query: 46 DTPAEIVCFKCGEKGHKSNVCTKDEKKCFRCGQKGHVLADCKRGD--------------- 90
D + C C GH + C D C C GHV C +GD
Sbjct: 152 DCTNDKACKNCRTSGHIARDCRNDPV-CNICSISGHVARHCPKGDSNYSDRGSRVRDGGM 210
Query: 91 ------------------IVCYNCNGEGHISSQCTQPKKVRTG 115
I+C+NC G GH + +C + G
Sbjct: 211 QRGGLSRMSRDREGVSAMIICHNCGGRGHRAYECPSARVADRG 253
>At2g17870 putative glycine-rich, zinc-finger DNA-binding protein
Length = 301
Score = 50.1 bits (118), Expect = 2e-06
Identities = 24/73 (32%), Positives = 32/73 (42%), Gaps = 18/73 (24%)
Query: 53 CFKCGEKGHKSNVCTKD--------EKKCFRCGQKGHVLADCKR----------GDIVCY 94
C+ CG GH + VCT + C+ CG GH+ DC R G C+
Sbjct: 226 CYTCGGVGHIAKVCTSKIPSGGGGGGRACYECGGTGHLARDCDRRGSGSSGGGGGSNKCF 285
Query: 95 NCNGEGHISSQCT 107
C EGH + +CT
Sbjct: 286 ICGKEGHFARECT 298
Score = 49.7 bits (117), Expect = 3e-06
Identities = 27/87 (31%), Positives = 35/87 (40%), Gaps = 25/87 (28%)
Query: 53 CFKCGEKGHKSNVCTKDE---------------KKCFRCGQKGHVLADCKR--------G 89
C+ CGE GH + C C+ CG GH DC++ G
Sbjct: 163 CYSCGEVGHLAKDCRGGSGGNRYGGGGGRGSGGDGCYMCGGVGHFARDCRQNGGGNVGGG 222
Query: 90 DIVCYNCNGEGHISSQCTQPKKVRTGG 116
CY C G GHI+ CT K+ +GG
Sbjct: 223 GSTCYTCGGVGHIAKVCT--SKIPSGG 247
Score = 48.1 bits (113), Expect = 8e-06
Identities = 27/99 (27%), Positives = 38/99 (38%), Gaps = 26/99 (26%)
Query: 44 GRDTPAEIVCFKCGEKGHKSNVCTKDE-----------KKCFRCGQKGHVLADCK----- 87
GR + E C+ CG+ GH + C + + C+ CG+ GH+ DC+
Sbjct: 123 GRRSGGEGECYMCGDVGHFARDCRQSGGGNSGGGGGGGRPCYSCGEVGHLAKDCRGGSGG 182
Query: 88 ----------RGDIVCYNCNGEGHISSQCTQPKKVRTGG 116
G CY C G GH + C Q GG
Sbjct: 183 NRYGGGGGRGSGGDGCYMCGGVGHFARDCRQNGGGNVGG 221
Score = 44.3 bits (103), Expect = 1e-04
Identities = 22/80 (27%), Positives = 32/80 (39%), Gaps = 16/80 (20%)
Query: 53 CFKCGEKGHKSNVCTKDE--------KKCFRCGQKGHVLADCKR--------GDIVCYNC 96
C+ CG GH + C ++ C+ CG GH+ C G CY C
Sbjct: 198 CYMCGGVGHFARDCRQNGGGNVGGGGSTCYTCGGVGHIAKVCTSKIPSGGGGGGRACYEC 257
Query: 97 NGEGHISSQCTQPKKVRTGG 116
G GH++ C + +GG
Sbjct: 258 GGTGHLARDCDRRGSGSSGG 277
Score = 39.3 bits (90), Expect = 0.004
Identities = 23/89 (25%), Positives = 30/89 (32%), Gaps = 16/89 (17%)
Query: 44 GRDTPAEIVCFKCGEKGHKSNVCTKDEKKCFRCGQKGHVLADC----------------K 87
G+ E+ G K N CF CG+ GH+ DC
Sbjct: 67 GKTKAIEVTAPGGGSLNKKENSSRGSGGNCFNCGEVGHMAKDCDGGSGGKSFGGGGGRRS 126
Query: 88 RGDIVCYNCNGEGHISSQCTQPKKVRTGG 116
G+ CY C GH + C Q +GG
Sbjct: 127 GGEGECYMCGDVGHFARDCRQSGGGNSGG 155
>At3g42860 unknown protein (At3g42860)
Length = 372
Score = 49.7 bits (117), Expect = 3e-06
Identities = 33/108 (30%), Positives = 41/108 (37%), Gaps = 37/108 (34%)
Query: 45 RDTPAEIVCFKCGEKGHKSNVCT----------------KDEKKCFRCGQKGHVLADC-- 86
+ T A CFKCG+ GH S CT +C++CG++GH DC
Sbjct: 262 KSTSAAGDCFKCGKPGHWSRDCTAQSGNPKYEPGQMKSSSSSGECYKCGKQGHWSRDCTG 321
Query: 87 ----------------KRGDIVCYNCNGEGHISSQCTQP-KKVRTGGK 117
GD CY C GH S CT P + T GK
Sbjct: 322 QSSNQQFQSGQAKSTSSTGD--CYKCGKAGHWSRDCTSPAQTTNTPGK 367
Score = 45.1 bits (105), Expect = 7e-05
Identities = 24/82 (29%), Positives = 31/82 (37%), Gaps = 27/82 (32%)
Query: 53 CFKCGEKGHKSNVCTKDEK-----------KCFRCGQKGHVLADC--------------- 86
C+KCG++GH + CT CF+CG+ GH DC
Sbjct: 239 CYKCGKEGHWARDCTVQSDTGPVKSTSAAGDCFKCGKPGHWSRDCTAQSGNPKYEPGQMK 298
Query: 87 -KRGDIVCYNCNGEGHISSQCT 107
CY C +GH S CT
Sbjct: 299 SSSSSGECYKCGKQGHWSRDCT 320
>At5g52380 unknown protein
Length = 268
Score = 48.5 bits (114), Expect = 6e-06
Identities = 22/72 (30%), Positives = 33/72 (45%), Gaps = 13/72 (18%)
Query: 52 VCFKCGEKGHKSNVC------TKDEKKCFRCGQKGHVLADCK-------RGDIVCYNCNG 98
+C +C +GH C + ++K C+ CG GH L+ C C+ C G
Sbjct: 100 ICLQCRRRGHSLKNCPEKNNESSEKKLCYNCGDTGHSLSHCPYPMEDGGTKFASCFICKG 159
Query: 99 EGHISSQCTQPK 110
+GHIS C + K
Sbjct: 160 QGHISKNCPENK 171
Score = 47.0 bits (110), Expect = 2e-05
Identities = 26/79 (32%), Positives = 36/79 (44%), Gaps = 13/79 (16%)
Query: 53 CFKCGEKGHKSNVCTKDE-----KKCFRCGQKGHVLADCKRGD------IVCYNCNGEGH 101
CF C K H + +C + K C +C ++GH L +C + +CYNC GH
Sbjct: 76 CFICHSKTHIAKLCPEKSEWERNKICLQCRRRGHSLKNCPEKNNESSEKKLCYNCGDTGH 135
Query: 102 ISSQCTQPKKVRTGGKVFA 120
S C P + GG FA
Sbjct: 136 SLSHCPYP--MEDGGTKFA 152
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.320 0.137 0.414
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 9,264,054
Number of Sequences: 26719
Number of extensions: 414299
Number of successful extensions: 1512
Number of sequences better than 10.0: 97
Number of HSP's better than 10.0 without gapping: 67
Number of HSP's successfully gapped in prelim test: 30
Number of HSP's that attempted gapping in prelim test: 1168
Number of HSP's gapped (non-prelim): 233
length of query: 394
length of database: 11,318,596
effective HSP length: 101
effective length of query: 293
effective length of database: 8,619,977
effective search space: 2525653261
effective search space used: 2525653261
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.8 bits)
S2: 61 (28.1 bits)
Medicago: description of AC130806.14


