

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC130800.8 + phase: 0 /pseudo
(324 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
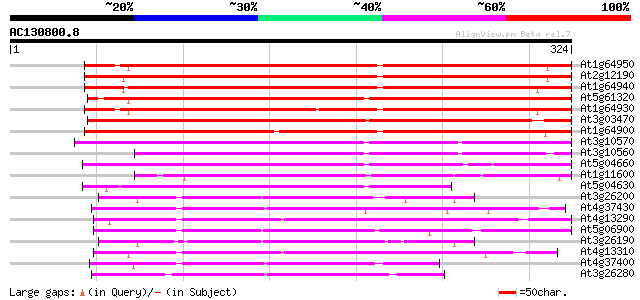
Score E
Sequences producing significant alignments: (bits) Value
At1g64950 unknown protein 266 1e-71
At2g12190 pseudogene 263 1e-70
At1g64940 hypothetical protein 254 4e-68
At5g61320 cytochrome P450 - like protein 244 5e-65
At1g64930 hypothetical protein 240 9e-64
At3g03470 cytochrome P450 nlike protein 232 2e-61
At1g64900 unknown protein 231 4e-61
At3g10570 putative cytochrome P450 144 8e-35
At3g10560 putative cytochrome P450 133 1e-31
At5g04660 cytochrom P450 - like protein 133 1e-31
At1g11600 cytochrome P450 like protein 132 3e-31
At5g04630 cytochrome P450 - like protein 123 2e-28
At3g26200 cytochrome P450 like protein 92 4e-19
At4g37430 cytochrome P450 monooxygenase (CYP91A2) 91 8e-19
At4g13290 cytochrome p450 - like protein 85 5e-17
At5g06900 cytochrome P450 84 8e-17
At3g26190 cytochrome P450, putative 83 2e-16
At4g13310 cytochrome p450 like protein 82 4e-16
At4g37400 cytochrome P450 monooxygenase -like protein 82 5e-16
At3g26280 cytochrome P450 monooxygenase (CYP71B4) 81 9e-16
>At1g64950 unknown protein
Length = 510
Score = 266 bits (679), Expect = 1e-71
Identities = 134/293 (45%), Positives = 195/293 (65%), Gaps = 17/293 (5%)
Query: 44 IETWFIVLVTLCIIFLIRAMLSFF----TTTVPLPPGPLHIPIITNFQLLQKSISQLEPF 99
+E W ++L +L + L+ L FF ++++PLPP P + P I Q L++ + L +
Sbjct: 1 MEIWLLILGSLFLSLLLN--LLFFRLRDSSSLPLPPDPNYFPFIGTIQWLRQGLGGLNNY 58
Query: 100 LKTLHAKHGPIIIVHIGSQPSIFINDHALAHHVLVQNSAIISDRPTALPTSKMLSSNQHN 159
L+++H + GPII + I S+PSIF+ D +LAH LV N A+ +DRP A P SK++SSNQHN
Sbjct: 59 LRSVHHRLGPIITLRITSRPSIFVADRSLAHQALVLNGAVFADRPPAAPISKIISSNQHN 118
Query: 160 INTAFYGPTWRTLRRNLAFEMLHPSKLKSFSEIQKWVLRTLINRLKTASESEPTDSIKVM 219
I++ YG TWR LRRNL E+LHPS+++S+S ++WVL L +R EP I V+
Sbjct: 119 ISSCLYGATWRLLRRNLTSEILHPSRVRSYSHARRWVLEILFDRFGKNRGEEP---IVVV 175
Query: 220 PHFKYAMFSLLVFMCFGERVNDEKISDIERVQRKIMLNFGRFNKLNFWPKVTRILSRNQW 279
H YAMF+LLV MCFG+++++++I +E VQR+ +L F RFN LN WPK T+++ R +W
Sbjct: 176 DHLHYAMFALLVLMCFGDKLDEKQIKQVEYVQRRQLLGFSRFNILNLWPKFTKLILRKRW 235
Query: 280 EEFLKLLKDQEDVLLPLIRARKQVKESKLN--------NVNTVVSYADTLLEL 324
EEF ++ ++Q DVLLPLIRAR+++ E + N N V SY DTLLEL
Sbjct: 236 EEFFQMRREQHDVLLPLIRARRKIVEERKNRSSEEEEDNKEYVQSYVDTLLEL 288
>At2g12190 pseudogene
Length = 512
Score = 263 bits (671), Expect = 1e-70
Identities = 130/291 (44%), Positives = 195/291 (66%), Gaps = 13/291 (4%)
Query: 44 IETWFIVLVTLCIIFLIRAML--SFFTTTVPLPPGPLHIPIITNFQLLQKSISQLEPFLK 101
+E W ++L +L + L+ +L ++++PLPP P P + Q L++ + L +L+
Sbjct: 1 MEIWLLILGSLFLSLLLNLLLFRRRDSSSLPLPPDPNFFPFLGTLQWLRQGLGGLNNYLR 60
Query: 102 TLHAKHGPIIIVHIGSQPSIFINDHALAHHVLVQNSAIISDRPTALPTSKMLSSNQHNIN 161
++H + GPII + I S+P+IF+ D +LAH LV N A+ +DRP A P SK++SSNQHNI+
Sbjct: 61 SVHHRLGPIITLRITSRPAIFVADRSLAHQALVLNGAVFADRPPAAPISKIISSNQHNIS 120
Query: 162 TAFYGPTWRTLRRNLAFEMLHPSKLKSFSEIQKWVLRTLINRLKTASESEPTDSIKVMPH 221
++ YG TWR LRRNL E+LHPS+++S+S ++WVL L +R + EP I V+ H
Sbjct: 121 SSLYGATWRLLRRNLTSEILHPSRVRSYSHARRWVLEILFDRFGKSRGEEP---IVVVDH 177
Query: 222 FKYAMFSLLVFMCFGERVNDEKISDIERVQRKIMLNFGRFNKLNFWPKVTRILSRNQWEE 281
YAMF+LLV MCFG+++++++I +E VQR+ +L F RFN LN WPK T+++ R +WEE
Sbjct: 178 LHYAMFALLVLMCFGDKLDEKQIKQVEYVQRRQLLGFSRFNILNLWPKFTKLILRKRWEE 237
Query: 282 FLKLLKDQEDVLLPLIRARKQVKESKLN--------NVNTVVSYADTLLEL 324
F ++ ++Q DVLLPLIRAR+++ E + N N V SY DTLLEL
Sbjct: 238 FFQMRREQHDVLLPLIRARRKIVEERKNRSSEEEEDNKVYVQSYVDTLLEL 288
>At1g64940 hypothetical protein
Length = 511
Score = 254 bits (649), Expect = 4e-68
Identities = 129/294 (43%), Positives = 191/294 (64%), Gaps = 18/294 (6%)
Query: 44 IETWFIVLVTLCIIFLIRAML-----SFFTTTVPLPPGPLHIPIITNFQLLQKSISQLEP 98
+E W ++L +L + L+ +L SF +++PLPP P P I + L+K + L+
Sbjct: 1 MEIWLLILGSLFLSLLVNHLLFRRRDSF--SSLPLPPDPNFFPFIGTLKWLRKGLGGLDN 58
Query: 99 FLKTLHAKHGPIIIVHIGSQPSIFINDHALAHHVLVQNSAIISDRPTALPTSKMLSSNQH 158
+L+++H GPII + I S+P+IF+ D +LAH LV N A+ +DRP A SK++SSNQH
Sbjct: 59 YLRSVHHHLGPIITLRITSRPAIFVTDRSLAHQALVLNGAVFADRPPAESISKIISSNQH 118
Query: 159 NINTAFYGPTWRTLRRNLAFEMLHPSKLKSFSEIQKWVLRTLINRLKTASESEPTDSIKV 218
NI++ YG TWR LRRNL E+LHPS+L+S+S ++WVL L R EP I V
Sbjct: 119 NISSCLYGATWRLLRRNLTSEILHPSRLRSYSHARRWVLEILFGRFGKNRGEEP---IVV 175
Query: 219 MPHFKYAMFSLLVFMCFGERVNDEKISDIERVQRKIMLNFGRFNKLNFWPKVTRILSRNQ 278
+ H YAMF+LLV MCFG+++++++I +E VQR+ +L F RFN L WPK T+++ R +
Sbjct: 176 VDHLHYAMFALLVLMCFGDKLDEKQIKQVEYVQRRQLLGFSRFNILTLWPKFTKLIYRKR 235
Query: 279 WEEFLKLLKDQEDVLLPLIRARKQV--------KESKLNNVNTVVSYADTLLEL 324
WEEF ++ +Q+DVLLPLIRAR+++ E + +N V SY DTLL++
Sbjct: 236 WEEFFQMQSEQQDVLLPLIRARRKIVDERKKRSSEEEKDNKEYVQSYVDTLLDV 289
>At5g61320 cytochrome P450 - like protein
Length = 497
Score = 244 bits (623), Expect = 5e-65
Identities = 124/284 (43%), Positives = 185/284 (64%), Gaps = 10/284 (3%)
Query: 46 TWFIVLVTLCIIFLIRAMLSFF----TTTVPLPPGPLHIPIITNFQLLQKSISQLEPFLK 101
TWF + C+ F + +L F +PLPP P P+ FQ L++ +L+
Sbjct: 3 TWFHIF---CVSFTVNFLLYLFFRRTNNNLPLPPNPNFFPMPGPFQWLRQGFDDFYSYLR 59
Query: 102 TLHAKHGPIIIVHIGSQPSIFINDHALAHHVLVQNSAIISDRPTALPTSKMLSSNQHNIN 161
++H + GPII + I S P+IF++D +LAH LV N A+ SDRP ALPT K+++SNQH I+
Sbjct: 60 SIHHRLGPIISLRIFSVPAIFVSDRSLAHKALVLNGAVFSDRPPALPTGKIITSNQHTIS 119
Query: 162 TAFYGPTWRTLRRNLAFEMLHPSKLKSFSEIQKWVLRTLINRLKTASESEPTDSIKVMPH 221
+ YG TWR LRRNL E+LHPS++KS+S ++ VL L +R++ + E I V+ H
Sbjct: 120 SGSYGATWRLLRRNLTSEILHPSRVKSYSNARRSVLENLCSRIR--NHGEEAKPIVVVDH 177
Query: 222 FKYAMFSLLVFMCFGERVNDEKISDIERVQRKIMLNFGRFNKLNFWPKVTRILSRNQWEE 281
+YAMFSLLV MCFG+++++E+I +E VQR+ ++ RFN LN +P T++ R +WEE
Sbjct: 178 LRYAMFSLLVLMCFGDKLDEEQIKQVEFVQRRELITLPRFNILNVFPSFTKLFLRKRWEE 237
Query: 282 FLKLLKDQEDVLLPLIRARKQVK-ESKLNNVNTVVSYADTLLEL 324
FL ++ ++VLLPLIR+R+++ ESK + + SY DTLL+L
Sbjct: 238 FLTFRREHKNVLLPLIRSRRKIMIESKDSGKEYIQSYVDTLLDL 281
>At1g64930 hypothetical protein
Length = 511
Score = 240 bits (612), Expect = 9e-64
Identities = 121/293 (41%), Positives = 191/293 (64%), Gaps = 18/293 (6%)
Query: 44 IETWFIVLVTLCIIFLIRAMLSFF----TTTVPLPPGPLHIPIITNFQLLQKSISQLEPF 99
+E W ++L +L + L+ L FF ++++PLPP P P + Q L++ + +
Sbjct: 1 MEIWLLILGSLSLSLLLN--LLFFRLRDSSSLPLPPAPNFFPFLGTLQWLRQGLGGFNNY 58
Query: 100 LKTLHAKHGPIIIVHIGSQPSIFINDHALAHHVLVQNSAIISDRPTALPTSKMLSSNQHN 159
++++H + GPII + I S+P+IF+ D +LAH LV N A+ +DRP A P SK+LS+NQH
Sbjct: 59 VRSVHHRLGPIITLRITSRPAIFVADGSLAHQALVLNGAVFADRPPAAPISKILSNNQHT 118
Query: 160 INTAFYGPTWRTLRRNLAFEMLHPSKLKSFSEIQKWVLRTLINRLKTASESEPTDSIKVM 219
I + YG TWR LRRN+ E+LHPS++KS+S ++ WVL L +RL+ + EP I V
Sbjct: 119 ITSCLYGVTWRLLRRNIT-EILHPSRMKSYSHVRHWVLEILFDRLRKSGGEEP---IVVF 174
Query: 220 PHFKYAMFSLLVFMCFGERVNDEKISDIERVQRKIMLNFGRFNKLNFWPKVTRILSRNQW 279
H YAMF++LV MCFG+++++++I +E VQR+++L F R++ LN PK T+++ R +W
Sbjct: 175 DHLHYAMFAVLVLMCFGDKLDEKQIKQVEYVQRQMLLGFARYSILNLCPKFTKLILRKRW 234
Query: 280 EEFLKLLKDQEDVLLPLIRARKQV--------KESKLNNVNTVVSYADTLLEL 324
EEF ++ ++Q+DVLL LI AR+++ E + N V SY DTLL++
Sbjct: 235 EEFFQMRREQQDVLLRLIYARRKIVEERKKRSSEEEEENKEYVQSYVDTLLDV 287
>At3g03470 cytochrome P450 nlike protein
Length = 511
Score = 232 bits (591), Expect = 2e-61
Identities = 119/280 (42%), Positives = 178/280 (63%), Gaps = 8/280 (2%)
Query: 46 TWFIVLVTLCIIFLIRAMLSFFTTTVPLPPGPLHIPIITNFQLLQKS-ISQLEPFLKTLH 104
T I L+ + F I L FF +T LPPGP P+I N L+K+ S + L+ L
Sbjct: 4 TTIIFLIISSLTFSIFLKLIFFFSTHKLPPGPPRFPVIGNIIWLKKNNFSDFQGVLRDLA 63
Query: 105 AKHGPIIIVHIGSQPSIFINDHALAHHVLVQNSAIISDRPTALPTSKMLSSNQHNINTAF 164
++HGPII +H+GS+PSI++ D +LAH LVQN A+ SDR ALPT+K+++SNQH+I+++
Sbjct: 64 SRHGPIITLHVGSKPSIWVTDRSLAHQALVQNGAVFSDRSLALPTTKVITSNQHDIHSSV 123
Query: 165 YGPTWRTLRRNLAFEMLHPSKLKSFSEIQKWVLRTLINRLKTASESEPTDSIKVMPHFKY 224
YG WRTLRRNL E+L PS++K+ + +KW L L++ +T + E + H ++
Sbjct: 124 YGSLWRTLRRNLTSEILQPSRVKAHAPSRKWSLEILVDLFET-EQREKGHISDALDHLRH 182
Query: 225 AMFSLLVFMCFGERVNDEKISDIERVQRKIMLNFGRFNKLNFWPKVTRILSRNQWEEFLK 284
AMF LL MCFGE++ E+I +IE Q ++++++ +F+ LN +P VT+ L R +W+EFL+
Sbjct: 183 AMFYLLALMCFGEKLRKEEIREIEEAQYQMLISYTKFSVLNIFPSVTKFLLRRKWKEFLE 242
Query: 285 LLKDQEDVLLPLIRARKQVKESKLNNVNTVVSYADTLLEL 324
L K QE V+L + AR + V+ Y DTLL L
Sbjct: 243 LRKSQESVILRYVNARSK------ETTGDVLCYVDTLLNL 276
>At1g64900 unknown protein
Length = 506
Score = 231 bits (589), Expect = 4e-61
Identities = 114/287 (39%), Positives = 186/287 (64%), Gaps = 11/287 (3%)
Query: 44 IETWFIVLVTLCIIFLIRAMLSFF-TTTVPLPPGPLHIPIITNFQLLQKSISQLEPFLKT 102
+E W ++L +L L+ +L +++ PLPP P +P + Q L++ + LE +L++
Sbjct: 1 MEIWLLILASLSGSLLLHLLLRRRNSSSPPLPPDPNFLPFLGTLQWLREGLGGLESYLRS 60
Query: 103 LHAKHGPIIIVHIGSQPSIFINDHALAHHVLVQNSAIISDRPTALPTSKMLSSNQHNINT 162
+H + GPI+ + I S+P+IF+ D +L H LV N A+ +DRP SK++ ++HNI++
Sbjct: 61 VHHRLGPIVTLRITSRPAIFVADRSLTHEALVLNGAVYADRPPPAVISKIV--DEHNISS 118
Query: 163 AFYGPTWRTLRRNLAFEMLHPSKLKSFSEIQKWVLRTLINRLKTASESEPTDSIKVMPHF 222
YG TWR LRRN+ E+LHPS+++S+S + WVL L R + EP I ++ H
Sbjct: 119 GSYGATWRLLRRNITSEILHPSRVRSYSHARHWVLEILFERFRNHGGEEP---IVLIHHL 175
Query: 223 KYAMFSLLVFMCFGERVNDEKISDIERVQRKIMLNFGRFNKLNFWPKVTRILSRNQWEEF 282
YAMF+LLV MCFG+++++++I ++E +QR +L+ +FN N WPK T+++ R +W+EF
Sbjct: 176 HYAMFALLVLMCFGDKLDEKQIKEVEFIQRLQLLSLTKFNIFNIWPKFTKLILRKRWQEF 235
Query: 283 LKLLKDQEDVLLPLIRARKQVKESKL-----NNVNTVVSYADTLLEL 324
L++ + Q DVLLPLIRAR+++ E + + + V SY DTLL+L
Sbjct: 236 LQIRRQQRDVLLPLIRARRKIVEERKRSEQEDKKDYVQSYVDTLLDL 282
>At3g10570 putative cytochrome P450
Length = 513
Score = 144 bits (362), Expect = 8e-35
Identities = 84/290 (28%), Positives = 157/290 (53%), Gaps = 7/290 (2%)
Query: 38 PNIYVNIE-TWFIVLVTLCIIFLIRAMLSFFTTTVPLPPGPLHIPIITNFQLLQKSISQL 96
P+ + NI + F ++ ++ LI S + V LPPGP P++ N +S Q
Sbjct: 7 PHTFFNISPSLFYTILISSLVLLILTRRSAKSKIVKLPPGPPGWPVVGNLFQFARSGKQF 66
Query: 97 EPFLKTLHAKHGPIIIVHIGSQPSIFINDHALAHHVLVQNSAIISDRPTALPTSKMLSSN 156
++ + K+GPI + +GS+ I I+D AL H VL+Q + + RPT PT + SSN
Sbjct: 67 YEYVDDVRKKYGPIYTLRMGSRTMIIISDSALVHDVLIQRGPMFATRPTENPTRTIFSSN 126
Query: 157 QHNINTAFYGPTWRTLRRNLAFEMLHPSKLKSFSEIQKWVLRTLINRLKTASESEPTDS- 215
+N + YGP WR+LR+N+ ML + + F +++ + L+ R+K SE++ D
Sbjct: 127 TFTVNASAYGPVWRSLRKNMVQNMLSSIRFREFGSLRQSAMDKLVERIK--SEAKDNDGL 184
Query: 216 IKVMPHFKYAMFSLLVFMCFGERVNDEKISDIERVQRKIMLNFGRFNKLNFWPKVTRILS 275
+ V+ + ++A F +L+ MCFG +++E I ++++V +K+++ +L+ + +
Sbjct: 185 VWVLRNARFAAFCILLEMCFGIEMDEESILNMDQVMKKVLITLN--PRLDDYLPILAPFY 242
Query: 276 RNQWEEFLKLLKDQEDVLLPLI-RARKQVKESKLNNVNTVVSYADTLLEL 324
+ L++ +Q D ++ LI R R+ +++ + + SY DTL +L
Sbjct: 243 SKERARALEVRCEQVDFIVKLIERRRRAIQKPGTDKTASSFSYLDTLFDL 292
>At3g10560 putative cytochrome P450
Length = 514
Score = 133 bits (335), Expect = 1e-31
Identities = 80/254 (31%), Positives = 142/254 (55%), Gaps = 10/254 (3%)
Query: 73 LPPGPLHIPIITNFQLLQKSISQLEPFLKTLHAKHGPIIIVHIGSQPSIFINDHALAHHV 132
LPPGP P+I N +S Q +++ L +GPI+ + +G++ I I+D +LAH
Sbjct: 46 LPPGPPGWPVIGNLFQFTRSGKQFFEYVEDLVKIYGPILTLRLGTRTMIIISDASLAHEA 105
Query: 133 LVQNSAIISDRPTALPTSKMLSSNQHNINTAFYGPTWRTLRRNLAFEMLHPSKLKSFSEI 192
L++ A + RP PT K+ SS++ +++A YGP WR+LRRN+ ML ++LK F +
Sbjct: 106 LIERGAQFATRPVETPTRKIFSSSEITVHSAMYGPVWRSLRRNMVQNMLSSNRLKEFGSV 165
Query: 193 QKWVLRTLINRLKTASESEPTDS-IKVMPHFKYAMFSLLVFMCFGERVNDEKISDIERVQ 251
+K + LI R+K SE+ D + V+ + +YA F +L+ +CFG + +E I ++++
Sbjct: 166 RKSAMDKLIERIK--SEARDNDGLVWVLQNSRYAAFCVLLDICFGVEMEEESIEKMDQLM 223
Query: 252 RKIMLNFGRFNKLNFWPKVTRILSRNQWEEFLKLLKDQEDVLLPLI-RARKQVKESKLNN 310
I+ KL+ + + + N+ LKL ++ D ++ I + RK ++ + +++
Sbjct: 224 TAILNAVD--PKLHDYLPILTPFNYNERNRALKLRRELVDFVVGFIEKRRKAIRTATVSS 281
Query: 311 VNTVVSYADTLLEL 324
SY DTL +L
Sbjct: 282 ----FSYLDTLFDL 291
>At5g04660 cytochrom P450 - like protein
Length = 512
Score = 133 bits (334), Expect = 1e-31
Identities = 79/285 (27%), Positives = 152/285 (52%), Gaps = 8/285 (2%)
Query: 43 NIETWFIVLVTLCIIFLIRAMLSFFTTTVPLPPGPLHIPIITNFQLLQKSISQLEPFLKT 102
+++ F ++ +F+I S + LPPGP P++ N +S + +
Sbjct: 11 SLDFTFFAIIISGFVFIITRWNSNSKKRLNLPPGPPGWPVVGNLFQFARSGKPFFEYAED 70
Query: 103 LHAKHGPIIIVHIGSQPSIFINDHALAHHVLVQNSAIISDRPTALPTSKMLSSNQHNINT 162
L +GPI + +G++ I ++D L H L+Q A+ + RP PT + S N+ +N
Sbjct: 71 LKKTYGPIFTLRMGTRTMIILSDATLVHEALIQRGALFASRPAENPTRTIFSCNKFTVNA 130
Query: 163 AFYGPTWRTLRRNLAFEMLHPSKLKSFSEIQKWVLRTLINRLKTASESEPTDS-IKVMPH 221
A YGP WR+LRRN+ ML ++LK F ++++ + LI R+K SE+ D I V+ +
Sbjct: 131 AKYGPVWRSLRRNMVQNMLSSTRLKEFGKLRQSAMDKLIERIK--SEARDNDGLIWVLKN 188
Query: 222 FKYAMFSLLVFMCFGERVNDEKISDIERVQRKIMLNFG-RFNKLNFWPKVTRILSRNQWE 280
++A F +L+ MCFG +++E I ++ + + +++ R + ++ P + S+ + +
Sbjct: 189 ARFAAFCILLEMCFGIEMDEETIEKMDEILKTVLMTVDPRID--DYLPILAPFFSKER-K 245
Query: 281 EFLKLLKDQEDVLLPLI-RARKQVKESKLNNVNTVVSYADTLLEL 324
L++ ++Q D ++ +I R R+ ++ + + SY DTL +L
Sbjct: 246 RALEVRREQVDYVVGVIERRRRAIQNPGSDKTASSFSYLDTLFDL 290
>At1g11600 cytochrome P450 like protein
Length = 510
Score = 132 bits (332), Expect = 3e-31
Identities = 89/260 (34%), Positives = 136/260 (52%), Gaps = 15/260 (5%)
Query: 73 LPPGPLHIPIITNFQLLQKSISQLEPF---LKTLHAKHGPIIIVHIGSQPSIFINDHALA 129
+PPGP P++ N L + I Q F ++ L K+GPI + +G + I I D L
Sbjct: 35 IPPGPKGWPLVGN---LLQVIFQRRHFVFLMRDLRKKYGPIFTMQMGQRTMIIITDEKLI 91
Query: 130 HHVLVQNSAIISDRPTALPTSKMLSSNQHNINTAFYGPTWRTLRRNLAFEMLHPSKLKSF 189
H LVQ + RP P M S + IN+A YG WRTLRRN E++ ++K
Sbjct: 92 HEALVQRGPTFASRPPDSPIRLMFSVGKCAINSAEYGSLWRTLRRNFVTELVTAPRVKQC 151
Query: 190 SEIQKWVLRTLINRLKTASESEPTDSIKVMPHFKYAMFSLLVFMCFGERVNDEKISDIER 249
S I+ W ++ + R+KT E+ ++VM + + S+L+ +CFG ++++EKI +IE
Sbjct: 152 SWIRSWAMQNHMKRIKT--ENVEKGFVEVMSQCRLTICSILICLCFGAKISEEKIKNIEN 209
Query: 250 VQRKIMLNFGRFNKLNFWPKVTRILSRNQWEEFLKLLKDQEDVLLPLIRARKQVKESKLN 309
V + +ML +F P T L R Q E +L K Q + L+PLIR R++ ++K N
Sbjct: 210 VLKDVML-ITSPTLPDFLPVFTP-LFRRQVREARELRKTQLECLVPLIRNRRKFVDAKEN 267
Query: 310 NVNTVVS-----YADTLLEL 324
+VS Y D+L L
Sbjct: 268 PNEEMVSPIGAAYVDSLFRL 287
>At5g04630 cytochrome P450 - like protein
Length = 509
Score = 123 bits (308), Expect = 2e-28
Identities = 68/217 (31%), Positives = 121/217 (55%), Gaps = 7/217 (3%)
Query: 43 NIETWFIVLVTL---CIIFLIRAMLSFFTTTVPLPPGPLHIPIITNFQLLQKSISQLEPF 99
N +F LVT+ C+++++ S LPPGP P++ N +S Q +
Sbjct: 6 NYVFFFFTLVTILLSCLVYILTRH-SHNPKCANLPPGPKGWPVVGNLLQFARSGKQFFEY 64
Query: 100 LKTLHAKHGPIIIVHIGSQPSIFINDHALAHHVLVQNSAIISDRPTALPTSKMLSSNQHN 159
+ + +GPI + +G + I I+D LAH L++ A + RP PT K+ SS+
Sbjct: 65 VDEMRNIYGPIFTLKMGIRTMIIISDANLAHQALIERGAQFATRPAETPTRKIFSSSDIT 124
Query: 160 INTAFYGPTWRTLRRNLAFEMLHPSKLKSFSEIQKWVLRTLINRLKTASESEPTDS-IKV 218
+++A YGP WR+LRRN+ ML ++LK F I+K + L+ ++K SE++ D + V
Sbjct: 125 VHSAMYGPVWRSLRRNMVQNMLCSNRLKEFGSIRKSAIDKLVEKIK--SEAKENDGLVWV 182
Query: 219 MPHFKYAMFSLLVFMCFGERVNDEKISDIERVQRKIM 255
+ + ++A F +L+ MCFG ++ +E I ++++ +I+
Sbjct: 183 LRNARFAAFCILLDMCFGVKMEEESIEKMDQMMTEIL 219
>At3g26200 cytochrome P450 like protein
Length = 500
Score = 92.0 bits (227), Expect = 4e-19
Identities = 64/228 (28%), Positives = 110/228 (48%), Gaps = 21/228 (9%)
Query: 52 VTLCIIFLIRAMLSFFTTTVP----LPPGPLHIPIITNFQLLQKSISQLEPFLKTLHAKH 107
++L + L+ L FF P LPPGPL +PII N L KS+ + L +
Sbjct: 3 ISLYFLLLLPLFLIFFKKLSPSKGKLPPGPLGLPIIGNLHQLGKSLHRS---FHKLSQNY 59
Query: 108 GPIIIVHIGSQPSIFINDHALAHHVLVQNSAIISDRPTALPTSKMLSSNQHNINTAFYGP 167
GP++ +H G P + ++ A VL + RP L +K+ S N +I A YG
Sbjct: 60 GPVMFLHFGVVPVVVVSTREAAEEVLKTHDLETCTRPK-LTATKLFSYNYKDIGFAQYGD 118
Query: 168 TWRTLRRNLAFEMLHPSKLKSFSEIQKWVLRTLINRLKTASESEPTDSIKVMPHFKYAMF 227
WR +R+ E+ KLK+F I++ L+N+L ++E+ + M + A+F
Sbjct: 119 DWREMRKLAMLELFSSKKLKAFRYIREEESEVLVNKLSKSAET------RTMVDLRKALF 172
Query: 228 ----SLLVFMCFGERVNDEKISDIERVQRKIM---LNFGRFNKLNFWP 268
S++ + FG+ ++ D+++V+ ++ N G F +F+P
Sbjct: 173 SYTASIVCRLAFGQNFHECDFVDMDKVEDLVLESETNLGSFAFTDFFP 220
>At4g37430 cytochrome P450 monooxygenase (CYP91A2)
Length = 500
Score = 90.9 bits (224), Expect = 8e-19
Identities = 76/284 (26%), Positives = 140/284 (48%), Gaps = 19/284 (6%)
Query: 48 FIVLVTLCIIFLIRAMLSFFTTTVPLPPGPLHIPIITNFQLLQKSISQLEPFLKTLHAKH 107
+ +L+ L + + L T LPPGP P + + L++ I +L L+ ++
Sbjct: 3 YFILLPLLFLVISYKFLYSKTQRFNLPPGPPSRPFVGHLHLMKPPIHRL---LQRYSNQY 59
Query: 108 GPIIIVHIGSQPSIFINDHALAHHVLV-QNSAIISDRPTALPTSKMLSSNQHNINTAFYG 166
GPI + GS+ + I +LA QN ++S RP L T+K ++ N + TA YG
Sbjct: 60 GPIFSLRFGSRRVVVITSPSLAQESFTGQNDIVLSSRPLQL-TAKYVAYNHTTVGTAPYG 118
Query: 167 PTWRTLRRNLAFEMLHPSKLKSFSEIQKWVLRTLINRL----KTASESEPTDSIKVMPHF 222
WR LRR + E+L +L +F I+K + ++ RL +T++ES I++ P
Sbjct: 119 DHWRNLRRICSQEILSSHRLINFQHIRKDEILRMLTRLSRYTQTSNESNDFTHIELEPLL 178
Query: 223 KYAMFSLLVFMCFGERVNDEKISDIERVQ--RKIMLNFGRFNKLNFWPKVTRILS--RNQ 278
F+ +V M G+R + +++ E + +K++ + ++ N IL N+
Sbjct: 179 SDLTFNNIVRMVTGKRYYGDDVNNKEEAELFKKLVYDIAMYSGANHSADYLPILKLFGNK 238
Query: 279 WEEFLKLL-KDQEDVLLPLIRARKQVKESKLNNVNTVVSYADTL 321
+E+ +K + K +D+L L+ ++ KE NT+V++ +L
Sbjct: 239 FEKEVKAIGKSMDDILQRLLDECRRDKEG-----NTMVNHLISL 277
>At4g13290 cytochrome p450 - like protein
Length = 490
Score = 85.1 bits (209), Expect = 5e-17
Identities = 64/283 (22%), Positives = 133/283 (46%), Gaps = 16/283 (5%)
Query: 49 IVLVTLCI-----IFLIRAMLSFFTTT-VPLPPGPLHIPIITNFQLLQKSISQLEPFLKT 102
I+LVTLC+ + L++++L TT + LPP P +P+I N L + + L++
Sbjct: 3 IILVTLCLTTLLALLLLKSILKRTTTNNLNLPPSPWRLPVIGNLHQLSLNTHRS---LRS 59
Query: 103 LHAKHGPIIIVHIGSQPSIFINDHALAHHVLVQNSAIISDRPTALPTSKMLSSNQHNINT 162
L ++GP++++H G P + ++ +AH +L I ++RP K+L + ++
Sbjct: 60 LSLRYGPLMLLHFGRTPVLIVSSADVAHDILKTYDVICANRPKTKVIDKILRGGR-DVAF 118
Query: 163 AFYGPTWRTLRRNLAFEMLHPSKLKSFSEIQKWVLRTLINRLKTASESEPTDSIKVMPHF 222
A YG W+ ++ +L ++S+ +I++ ++ +I +++ AS P + + F
Sbjct: 119 APYGEYWKQMKSICIQNLLSNKMVRSYKKIREDEIKLMIEKVENASSCSPPSPVNLSQLF 178
Query: 223 KYAMFSLLVFMCFGERVNDEKIS-DIERVQRKIMLNFGRFNKLNFWPKVTRILSRNQWEE 281
++ G + + ++ D+E + R G F + P ++ I +
Sbjct: 179 MTLTNDIICRAALGRKYSSKEDGIDVENIVRAFSALVGEFPIGEYIPSLSWIDKIRGQDH 238
Query: 282 FLKLLKDQEDVLLPLIRARKQVKESKLNNVNTVVSYADTLLEL 324
++ + + D L + VKE + N +T DTLL +
Sbjct: 239 KMEEVDKRFDEFL-----ERVVKEHEDANKDTRSDLVDTLLTI 276
>At5g06900 cytochrome P450
Length = 507
Score = 84.3 bits (207), Expect = 8e-17
Identities = 73/281 (25%), Positives = 129/281 (44%), Gaps = 16/281 (5%)
Query: 49 IVLVTLCIIFLIRAMLSFFTTTVPLPPGPLHIPIITNFQLLQKSISQLEPFLKTLHAKHG 108
I+LV L I LI+A+ + +PLPP P +PII + LL Q L L ++G
Sbjct: 10 IILVCLGITVLIQAITNRLRDRLPLPPSPTALPIIGHIHLLGPIAHQA---LHKLSIRYG 66
Query: 109 PIIIVHIGSQPSIFINDHALAHHVLVQNSAIISDRPTALPTSKMLSSNQHNINTAFYGPT 168
P++ + IGS P++ ++ +A+ +L N +RPT + L+ + +A YG
Sbjct: 67 PLMYLFIGSIPNLIVSSAEMANEILKSNELNFLNRPT-MQNVDYLTYGSADFFSAPYGLH 125
Query: 169 WRTLRRNLAFEMLHPSKLKSFSEIQKWVLRTLINRLKTASESEPTDSIKVMPHFKYAMFS 228
W+ ++R E+ L SF ++ L+ L+ R+ +E+E +S+ + K +
Sbjct: 126 WKFMKRICMVELFSSRALDSFVSVRSEELKKLLIRVLKKAEAE--ESVNLGEQLKELTSN 183
Query: 229 LLVFMCFGERVND----EKISDIERVQRKIMLNFGRFN-KLNFWPKVTRILSRNQWEEFL 283
++ M F + +D EK ++ ++ ++ G FN FW L R +
Sbjct: 184 IITRMMFRKMQSDSDGGEKSEEVIKMVVELNELAGFFNVSETFW-----FLKRLDLQGLK 238
Query: 284 KLLKDQEDVLLPLIRARKQVKESKLNNVNTVVSYADTLLEL 324
K LK+ D +I + ES N + D LL++
Sbjct: 239 KRLKNARDKYDVIIERIMEEHESSKKNATGERNMLDVLLDI 279
>At3g26190 cytochrome P450, putative
Length = 499
Score = 83.2 bits (204), Expect = 2e-16
Identities = 58/224 (25%), Positives = 110/224 (48%), Gaps = 13/224 (5%)
Query: 52 VTLCIIFLIRAMLSFFTTTVP----LPPGPLHIPIITNFQLLQKSISQLEPFLKTLHAKH 107
+ LC + L+ L F+ +P LPPGP+ +PII N L KS+ + F K L ++
Sbjct: 3 IFLCFLLLLPLFLVFYKRLLPSKGKLPPGPISLPIIGNLHQLGKSLHR--SFYK-LSQEY 59
Query: 108 GPIIIVHIGSQPSIFINDHALAHHVLVQNSAIISDRPTALPTSKMLSSNQHNINTAFYGP 167
GP++ + G P + + A VL + RP L + + + N +I A YG
Sbjct: 60 GPVMFLRFGVVPVVVFSTKEAAEEVLKTHDLETCTRPK-LSATGLFTYNFKDIGFAQYGE 118
Query: 168 TWRTLRRNLAFEMLHPSKLKSFSEIQKWVLRTLINRLKTASESEPTDSIKVMPHFKYAMF 227
WR +R+ E+ KLK+F I++ L+ ++ +++++ ++ F Y
Sbjct: 119 DWREMRKLAMLELFSSKKLKAFRYIREEESELLVKKVTESAQTQTLVDLR-KALFSYTA- 176
Query: 228 SLLVFMCFGERVNDEKISDIERVQRKIM---LNFGRFNKLNFWP 268
S++ + FG+ ++ D+++V+ ++ N G F ++F+P
Sbjct: 177 SIVCRLAFGQNFHECDFVDMDKVEELVLESETNLGSFAFIDFFP 220
>At4g13310 cytochrome p450 like protein
Length = 497
Score = 82.0 bits (201), Expect = 4e-16
Identities = 61/286 (21%), Positives = 134/286 (46%), Gaps = 29/286 (10%)
Query: 49 IVLVTLCIIFLIRAMLSFF-----TTTVPLPPGPLHIPIITNFQLLQKSISQLEPFLKTL 103
++L+TLC+ L+ +L T LPP P +P+I N L + L++L
Sbjct: 3 MILITLCLTTLLALLLKSILKRTATKNFNLPPSPWRLPVIGNLHQLSLHTHRS---LRSL 59
Query: 104 HAKHGPIIIVHIGSQPSIFINDHALAHHVLVQNSAIISDRPTALPTSKMLSSNQHNINTA 163
++GP++++H G P + ++ +AH V+ + + ++RP K+LS + ++ A
Sbjct: 60 SLRYGPLMLLHFGRTPVLIVSSADVAHDVMKTHDLVCANRPKTKVVDKILSGGR-DVAFA 118
Query: 164 FYGPTWRTLRRNLAFEMLHPSKLKSFSEIQKWVLRTLINRLKTASESEPTDSIKVMPHFK 223
YG WR ++ +L+ ++S+ +I++ ++ +I +L+ AS S + +
Sbjct: 119 PYGEYWRQMKSICIQNLLNNKMVRSYEKIREEEIKRMIEKLEKASCSSSPSPVNLSQILM 178
Query: 224 YAMFSLLVFMCFGERVNDEKIS-DIERVQRKIMLNFGRFNKLNFWPKVTRI--------- 273
++ + G + + +K D+E + R G F + P ++ I
Sbjct: 179 TLTNDIICRVALGRKYSGKKDGIDVENIVRTFAALLGEFPVGEYIPSLSWIDRIRGLDHK 238
Query: 274 --LSRNQWEEFL-KLLKDQEDVLLPLIRARKQVKESKLNNVNTVVS 316
+ +++EFL +++K+ E+ A K+ + ++ + T+ S
Sbjct: 239 MEVVDKRFDEFLERVVKEHEE-------ADKETRSDLVDKLLTIQS 277
>At4g37400 cytochrome P450 monooxygenase -like protein
Length = 501
Score = 81.6 bits (200), Expect = 5e-16
Identities = 62/206 (30%), Positives = 105/206 (50%), Gaps = 12/206 (5%)
Query: 47 WFIVLVTLCIIFLIRAMLSFFTTT--VPLPPGPLH-IPIITNFQLLQKSISQLEPFLKTL 103
++ V+V +FL+ L F + T LPP P + +PI+ + LL+ + +L L
Sbjct: 2 FYYVIVLPLALFLLAYKLFFTSKTKRFNLPPSPPYSLPILGHHNLLKPPVHRL---FHRL 58
Query: 104 HAKHGPIIIVHIGSQPSIFINDHALAHHVLV-QNSAIISDRPTALPTSKMLSSNQHNINT 162
HGPI + GS+ ++ I+ +LA QN I+S+RP L T+K ++ N + T
Sbjct: 59 SKTHGPIFSLQFGSRRAVVISSSSLATQCFTGQNDIILSNRPCFL-TAKYVAYNYTTVGT 117
Query: 163 AFYGPTWRTLRRNLAFEMLHPSKLKSFSEIQKWVLRTLINRLKTASESEPTDSIKVMPHF 222
A YG WR LRR + E+L ++L +F I+K + ++ RL E I++ P
Sbjct: 118 APYGDHWRNLRRICSLEILSSNRLTNFLHIRKDEIHRMLTRLSRDVNKE----IELEPLL 173
Query: 223 KYAMFSLLVFMCFGERVNDEKISDIE 248
F+ +V M G+R +++ + E
Sbjct: 174 SDLTFNNIVRMVTGKRYYGDEVHNEE 199
>At3g26280 cytochrome P450 monooxygenase (CYP71B4)
Length = 504
Score = 80.9 bits (198), Expect = 9e-16
Identities = 60/206 (29%), Positives = 95/206 (45%), Gaps = 10/206 (4%)
Query: 48 FIVLVTLCIIFLIRAMLSFFTTTVPLPPGPLHIPIITNFQLLQKSISQLEPFLKTLHAKH 107
F +L+ + I FL+ + LPPGP +PII N LQ L L L KH
Sbjct: 7 FFLLLLVPIFFLLIFTKKIKESKQNLPPGPAKLPIIGNLHQLQ---GLLHKCLHDLSKKH 63
Query: 108 GPIIIVHIGSQPSIFINDHALAHHVLVQNSAIISDRPTALPTSKMLSSNQHNINTAFYGP 167
GP++ + +G P + I+ A L + RP + S++ S N +I YG
Sbjct: 64 GPVMHLRLGFAPMVVISSSEAAEEALKTHDLECCSRPITM-ASRVFSRNGKDIGFGVYGD 122
Query: 168 TWRTLRRNLAFEMLHPSKLKSFSEIQKWVLRTLINRLK-TASESEPTDSIKVMPHFKYAM 226
WR LR+ E K++SF I++ +I +LK AS+ P D K++ + +
Sbjct: 123 EWRELRKLSVREFFSVKKVQSFKYIREEENDLMIKKLKELASKQSPVDLSKIL----FGL 178
Query: 227 FSLLVF-MCFGERVNDEKISDIERVQ 251
+ ++F FG+ D K D E ++
Sbjct: 179 TASIIFRTAFGQSFFDNKHVDQESIK 204
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.323 0.136 0.405
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 7,219,191
Number of Sequences: 26719
Number of extensions: 296637
Number of successful extensions: 1296
Number of sequences better than 10.0: 161
Number of HSP's better than 10.0 without gapping: 98
Number of HSP's successfully gapped in prelim test: 63
Number of HSP's that attempted gapping in prelim test: 1031
Number of HSP's gapped (non-prelim): 168
length of query: 324
length of database: 11,318,596
effective HSP length: 99
effective length of query: 225
effective length of database: 8,673,415
effective search space: 1951518375
effective search space used: 1951518375
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (22.0 bits)
S2: 60 (27.7 bits)
Medicago: description of AC130800.8


