

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC130800.11 + phase: 0
(508 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
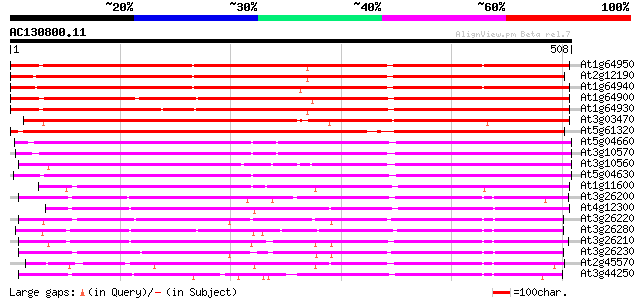
Score E
Sequences producing significant alignments: (bits) Value
At1g64950 unknown protein 613 e-176
At2g12190 pseudogene 602 e-172
At1g64940 hypothetical protein 586 e-167
At1g64900 unknown protein 564 e-161
At1g64930 hypothetical protein 552 e-157
At3g03470 cytochrome P450 nlike protein 548 e-156
At5g61320 cytochrome P450 - like protein 536 e-152
At5g04660 cytochrom P450 - like protein 350 1e-96
At3g10570 putative cytochrome P450 345 3e-95
At3g10560 putative cytochrome P450 336 2e-92
At5g04630 cytochrome P450 - like protein 334 6e-92
At1g11600 cytochrome P450 like protein 326 2e-89
At3g26200 cytochrome P450 like protein 219 3e-57
At4g12300 flavonoid 3',5'-hydroxylase -like protein 216 2e-56
At3g26220 cytochrome P450 monooxygenase (CYP71B3) 211 8e-55
At3g26280 cytochrome P450 monooxygenase (CYP71B4) 210 2e-54
At3g26210 cytochrome P450 like protein 209 2e-54
At3g26230 cytochrome P450, putative 209 3e-54
At2g45570 cytochrome P450 (CYP76C2) 206 2e-53
At3g44250 cytochrome P450-like protein 206 3e-53
>At1g64950 unknown protein
Length = 510
Score = 613 bits (1581), Expect = e-176
Identities = 305/515 (59%), Positives = 386/515 (74%), Gaps = 16/515 (3%)
Query: 1 METWFIALVSLCIIFLIRATLSSISTTKKTTATLPPGPPHIPIITSIQWLRKSFAQLEPF 60
ME W + L SL + L+ + + ++ LPP P + P I +IQWLR+ L +
Sbjct: 1 MEIWLLILGSLFLSLLLNLLFFRLRDS--SSLPLPPDPNYFPFIGTIQWLRQGLGGLNNY 58
Query: 61 LRTLHAKHGPIINLNIGSRPSIFIADRALAHHALVQNSSVFSDRPKALPTNKLMSSNQHN 120
LR++H + GPII L I SRPSIF+ADR+LAH ALV N +VF+DRP A P +K++SSNQHN
Sbjct: 59 LRSVHHRLGPIITLRITSRPSIFVADRSLAHQALVLNGAVFADRPPAAPISKIISSNQHN 118
Query: 121 ISSAFYGPTWRTLRRNLASEMLHPSKIKSFSEIRKWVLQTLINRLKTASESEDSVTVVSH 180
ISS YG TWR LRRNL SE+LHPS+++S+S R+WVL+ L +R + E+ + VV H
Sbjct: 119 ISSCLYGATWRLLRRNLTSEILHPSRVRSYSHARRWVLEILFDRF-GKNRGEEPIVVVDH 177
Query: 181 FRYAVFCLLVFMCFGERVNDEKINDIERVQRTILLNFSRFNILNFWPKVTRILLRKRWDE 240
YA+F LLV MCFG+++++++I +E VQR LL FSRFNILN WPK T+++LRKRW+E
Sbjct: 178 LHYAMFALLVLMCFGDKLDEKQIKQVEYVQRRQLLGFSRFNILNLWPKFTKLILRKRWEE 237
Query: 241 LAKLLKDQEDVLLPLIRARKHVKESRLN--------NINTVVSYVDTLMELELPQEKRKL 292
++ ++Q DVLLPLIRAR+ + E R N N V SYVDTL+ELELP EKRKL
Sbjct: 238 FFQMRREQHDVLLPLIRARRKIVEERKNRSSEEEEDNKEYVQSYVDTLLELELPDEKRKL 297
Query: 293 SEDEMISLCSEFLNGGTDTTSTALQWIMANLVKYPDVQGKLVEEITEVMGGDDAGEKEEV 352
+EDE++SLCSEFLNGGTDTT+TALQWIMANLVK PD+Q +L EEI V+G E EV
Sbjct: 298 NEDEIVSLCSEFLNGGTDTTATALQWIMANLVKNPDIQKRLYEEIKSVVG----EEANEV 353
Query: 353 KEEDLHKLRYLKCVVLEGLRRHPPGHFVLPHAVSEDVVLNGYLVPKDGTVNFMVAEMGLD 412
+EED K+ YL+ VV+EGLRRHPPGHFVLPH+V+ED VL GY VPK+GT+NFMVAE+G D
Sbjct: 354 EEEDAQKMPYLEAVVMEGLRRHPPGHFVLPHSVTEDTVLGGYKVPKNGTINFMVAEIGRD 413
Query: 413 PRVWEDPMEFKPERFLKDETFDITGSKEIKMMPFGAGRRICPGYNLALLHLEYFVANLVW 472
P+VWE+PM FKPERF+ +E DITGS+ IKMMPFGAGRRICPG LA+LHLEY+VAN+V
Sbjct: 414 PKVWEEPMAFKPERFM-EEAVDITGSRGIKMMPFGAGRRICPGIGLAMLHLEYYVANMVR 472
Query: 473 NFDWKVPEGGRVDLSEKQEFTMVMKNPLQAHISPR 507
FDWK +G VDL+EK EFT+VMK+PL+A PR
Sbjct: 473 EFDWKEVQGHEVDLTEKLEFTVVMKHPLKALAVPR 507
>At2g12190 pseudogene
Length = 512
Score = 602 bits (1552), Expect = e-172
Identities = 300/511 (58%), Positives = 382/511 (74%), Gaps = 16/511 (3%)
Query: 1 METWFIALVSLCIIFLIRATLSSISTTKKTTATLPPGPPHIPIITSIQWLRKSFAQLEPF 60
ME W + L SL + L+ L ++ LPP P P + ++QWLR+ L +
Sbjct: 1 MEIWLLILGSLFLSLLLNLLL--FRRRDSSSLPLPPDPNFFPFLGTLQWLRQGLGGLNNY 58
Query: 61 LRTLHAKHGPIINLNIGSRPSIFIADRALAHHALVQNSSVFSDRPKALPTNKLMSSNQHN 120
LR++H + GPII L I SRP+IF+ADR+LAH ALV N +VF+DRP A P +K++SSNQHN
Sbjct: 59 LRSVHHRLGPIITLRITSRPAIFVADRSLAHQALVLNGAVFADRPPAAPISKIISSNQHN 118
Query: 121 ISSAFYGPTWRTLRRNLASEMLHPSKIKSFSEIRKWVLQTLINRLKTASESEDSVTVVSH 180
ISS+ YG TWR LRRNL SE+LHPS+++S+S R+WVL+ L +R S E+ + VV H
Sbjct: 119 ISSSLYGATWRLLRRNLTSEILHPSRVRSYSHARRWVLEILFDRF-GKSRGEEPIVVVDH 177
Query: 181 FRYAVFCLLVFMCFGERVNDEKINDIERVQRTILLNFSRFNILNFWPKVTRILLRKRWDE 240
YA+F LLV MCFG+++++++I +E VQR LL FSRFNILN WPK T+++LRKRW+E
Sbjct: 178 LHYAMFALLVLMCFGDKLDEKQIKQVEYVQRRQLLGFSRFNILNLWPKFTKLILRKRWEE 237
Query: 241 LAKLLKDQEDVLLPLIRARKHVKESRLN--------NINTVVSYVDTLMELELPQEKRKL 292
++ ++Q DVLLPLIRAR+ + E R N N V SYVDTL+ELELP EKRKL
Sbjct: 238 FFQMRREQHDVLLPLIRARRKIVEERKNRSSEEEEDNKVYVQSYVDTLLELELPDEKRKL 297
Query: 293 SEDEMISLCSEFLNGGTDTTSTALQWIMANLVKYPDVQGKLVEEITEVMGGDDAGEKEEV 352
+EDE++SLCSEFLNGGTDTT+TALQWIMANLVK P++Q +L EEI V+G E +EV
Sbjct: 298 NEDEIVSLCSEFLNGGTDTTATALQWIMANLVKNPEIQKRLYEEIKSVVG----EEAKEV 353
Query: 353 KEEDLHKLRYLKCVVLEGLRRHPPGHFVLPHAVSEDVVLNGYLVPKDGTVNFMVAEMGLD 412
+EED K+ YLK VV+EGLRRHPPGHFVLPH+V+ED VL GY VPK GT+NFMVAE+G D
Sbjct: 354 EEEDAQKMPYLKAVVMEGLRRHPPGHFVLPHSVTEDTVLGGYKVPKKGTINFMVAEIGRD 413
Query: 413 PRVWEDPMEFKPERFL-KDETFDITGSKEIKMMPFGAGRRICPGYNLALLHLEYFVANLV 471
P VWE+PM FKPERF+ ++E DITGS+ IKMMPFGAGRRICPG LA+LHLEY+VAN+V
Sbjct: 414 PMVWEEPMAFKPERFMGEEEAVDITGSRGIKMMPFGAGRRICPGIGLAMLHLEYYVANMV 473
Query: 472 WNFDWKVPEGGRVDLSEKQEFTMVMKNPLQA 502
F+WK +G VDL+EK EFT+VMK+ L+A
Sbjct: 474 REFEWKEVQGHEVDLTEKFEFTVVMKHSLKA 504
>At1g64940 hypothetical protein
Length = 511
Score = 586 bits (1510), Expect = e-167
Identities = 291/515 (56%), Positives = 377/515 (72%), Gaps = 15/515 (2%)
Query: 1 METWFIALVSLCIIFLIRATLSSISTTKKTTATLPPGPPHIPIITSIQWLRKSFAQLEPF 60
ME W + L SL + L+ L ++ LPP P P I +++WLRK L+ +
Sbjct: 1 MEIWLLILGSLFLSLLVNHLLFR-RRDSFSSLPLPPDPNFFPFIGTLKWLRKGLGGLDNY 59
Query: 61 LRTLHAKHGPIINLNIGSRPSIFIADRALAHHALVQNSSVFSDRPKALPTNKLMSSNQHN 120
LR++H GPII L I SRP+IF+ DR+LAH ALV N +VF+DRP A +K++SSNQHN
Sbjct: 60 LRSVHHHLGPIITLRITSRPAIFVTDRSLAHQALVLNGAVFADRPPAESISKIISSNQHN 119
Query: 121 ISSAFYGPTWRTLRRNLASEMLHPSKIKSFSEIRKWVLQTLINRLKTASESEDSVTVVSH 180
ISS YG TWR LRRNL SE+LHPS+++S+S R+WVL+ L R + E+ + VV H
Sbjct: 120 ISSCLYGATWRLLRRNLTSEILHPSRLRSYSHARRWVLEILFGRF-GKNRGEEPIVVVDH 178
Query: 181 FRYAVFCLLVFMCFGERVNDEKINDIERVQRTILLNFSRFNILNFWPKVTRILLRKRWDE 240
YA+F LLV MCFG+++++++I +E VQR LL FSRFNIL WPK T+++ RKRW+E
Sbjct: 179 LHYAMFALLVLMCFGDKLDEKQIKQVEYVQRRQLLGFSRFNILTLWPKFTKLIYRKRWEE 238
Query: 241 LAKLLKDQEDVLLPLIRARKHV--------KESRLNNINTVVSYVDTLMELELPQEKRKL 292
++ +Q+DVLLPLIRAR+ + E +N V SYVDTL+++ELP EKRKL
Sbjct: 239 FFQMQSEQQDVLLPLIRARRKIVDERKKRSSEEEKDNKEYVQSYVDTLLDVELPDEKRKL 298
Query: 293 SEDEMISLCSEFLNGGTDTTSTALQWIMANLVKYPDVQGKLVEEITEVMGGDDAGEKEEV 352
+EDE++SLCSEFLN GTDTT+TALQWIMANLVK P++Q +L EEI ++G E +EV
Sbjct: 299 NEDEIVSLCSEFLNAGTDTTATALQWIMANLVKNPEIQRRLYEEIKSIVG----EEAKEV 354
Query: 353 KEEDLHKLRYLKCVVLEGLRRHPPGHFVLPHAVSEDVVLNGYLVPKDGTVNFMVAEMGLD 412
+E+D K+ YLK VV+EGLRRHPPGHFVLPH+V+ED VL GY VPK GT+NFMVAE+G D
Sbjct: 355 EEQDAQKMPYLKAVVMEGLRRHPPGHFVLPHSVTEDTVLGGYKVPKKGTINFMVAEIGRD 414
Query: 413 PRVWEDPMEFKPERFLKDETFDITGSKEIKMMPFGAGRRICPGYNLALLHLEYFVANLVW 472
P+VWE+PM FKPERF+ +E DITGS+ IKMMPFGAGRRICPG LA+LHLEY+VAN+V
Sbjct: 415 PKVWEEPMAFKPERFM-EEAVDITGSRGIKMMPFGAGRRICPGIGLAMLHLEYYVANMVR 473
Query: 473 NFDWKVPEGGRVDLSEKQEFTMVMKNPLQAHISPR 507
F+W+ +G VDL+EK EFT+VMK+PL+A PR
Sbjct: 474 EFEWQEVQGHEVDLTEKLEFTVVMKHPLKALAVPR 508
>At1g64900 unknown protein
Length = 506
Score = 564 bits (1453), Expect = e-161
Identities = 276/513 (53%), Positives = 376/513 (72%), Gaps = 16/513 (3%)
Query: 1 METWFIALVSLCIIFLIRATLSSISTTKKTTATLPPGPPHIPIITSIQWLRKSFAQLEPF 60
ME W + L SL L+ L +++ + LPP P +P + ++QWLR+ LE +
Sbjct: 1 MEIWLLILASLSGSLLLHLLLRRRNSS---SPPLPPDPNFLPFLGTLQWLREGLGGLESY 57
Query: 61 LRTLHAKHGPIINLNIGSRPSIFIADRALAHHALVQNSSVFSDRPKALPTNKLMSSNQHN 120
LR++H + GPI+ L I SRP+IF+ADR+L H ALV N +V++DRP +K++ ++HN
Sbjct: 58 LRSVHHRLGPIVTLRITSRPAIFVADRSLTHEALVLNGAVYADRPPPAVISKIV--DEHN 115
Query: 121 ISSAFYGPTWRTLRRNLASEMLHPSKIKSFSEIRKWVLQTLINRLKTASESEDSVTVVSH 180
ISS YG TWR LRRN+ SE+LHPS+++S+S R WVL+ L R + E+ + ++ H
Sbjct: 116 ISSGSYGATWRLLRRNITSEILHPSRVRSYSHARHWVLEILFERFRNHG-GEEPIVLIHH 174
Query: 181 FRYAVFCLLVFMCFGERVNDEKINDIERVQRTILLNFSRFNILNFWPKVTRILLRKRWDE 240
YA+F LLV MCFG+++++++I ++E +QR LL+ ++FNI N WPK T+++LRKRW E
Sbjct: 175 LHYAMFALLVLMCFGDKLDEKQIKEVEFIQRLQLLSLTKFNIFNIWPKFTKLILRKRWQE 234
Query: 241 LAKLLKDQEDVLLPLIRARKHVKESRLNNINT-----VVSYVDTLMELELPQEKRKLSED 295
++ + Q DVLLPLIRAR+ + E R + V SYVDTL++LELP+E RKL+E+
Sbjct: 235 FLQIRRQQRDVLLPLIRARRKIVEERKRSEQEDKKDYVQSYVDTLLDLELPEENRKLNEE 294
Query: 296 EMISLCSEFLNGGTDTTSTALQWIMANLVKYPDVQGKLVEEITEVMGGDDAGEKEEVKEE 355
++++LCSEFL GTDTT+TALQWIMANLVKYP++Q +L EEI V+G E +EV+EE
Sbjct: 295 DIMNLCSEFLTAGTDTTATALQWIMANLVKYPEIQERLHEEIKSVVGE----EAKEVEEE 350
Query: 356 DLHKLRYLKCVVLEGLRRHPPGHFVLPHAVSEDVVLNGYLVPKDGTVNFMVAEMGLDPRV 415
D+ K+ YLK VVLEGLRRHPPGHF+LPH+V+ED VL GY VPK+GT+NFMVAE+G DP
Sbjct: 351 DVEKMPYLKAVVLEGLRRHPPGHFLLPHSVTEDTVLGGYKVPKNGTINFMVAEIGRDPVE 410
Query: 416 WEDPMEFKPERFL-KDETFDITGSKEIKMMPFGAGRRICPGYNLALLHLEYFVANLVWNF 474
WE+PM FKPERF+ ++E D+TGS+ IKMMPFGAGRRICPG LA+LHLEY+VAN+V F
Sbjct: 411 WEEPMAFKPERFMGEEEAVDLTGSRGIKMMPFGAGRRICPGIGLAMLHLEYYVANMVREF 470
Query: 475 DWKVPEGGRVDLSEKQEFTMVMKNPLQAHISPR 507
WK +G VDL+EK EFT+VMK+PL+A PR
Sbjct: 471 QWKEVQGHEVDLTEKLEFTVVMKHPLKALAVPR 503
>At1g64930 hypothetical protein
Length = 511
Score = 552 bits (1423), Expect = e-157
Identities = 275/516 (53%), Positives = 375/516 (72%), Gaps = 17/516 (3%)
Query: 1 METWFIALVSLCIIFLIRATLSSISTTKKTTATLPPGPPHIPIITSIQWLRKSFAQLEPF 60
ME W + L SL + L+ + + ++ LPP P P + ++QWLR+ +
Sbjct: 1 MEIWLLILGSLSLSLLLNLLFFRLRDS--SSLPLPPAPNFFPFLGTLQWLRQGLGGFNNY 58
Query: 61 LRTLHAKHGPIINLNIGSRPSIFIADRALAHHALVQNSSVFSDRPKALPTNKLMSSNQHN 120
+R++H + GPII L I SRP+IF+AD +LAH ALV N +VF+DRP A P +K++S+NQH
Sbjct: 59 VRSVHHRLGPIITLRITSRPAIFVADGSLAHQALVLNGAVFADRPPAAPISKILSNNQHT 118
Query: 121 ISSAFYGPTWRTLRRNLASEMLHPSKIKSFSEIRKWVLQTLINRLKTASESEDSVTVVSH 180
I+S YG TWR LRRN+ +E+LHPS++KS+S +R WVL+ L +RL+ S E+ + V H
Sbjct: 119 ITSCLYGVTWRLLRRNI-TEILHPSRMKSYSHVRHWVLEILFDRLRK-SGGEEPIVVFDH 176
Query: 181 FRYAVFCLLVFMCFGERVNDEKINDIERVQRTILLNFSRFNILNFWPKVTRILLRKRWDE 240
YA+F +LV MCFG+++++++I +E VQR +LL F+R++ILN PK T+++LRKRW+E
Sbjct: 177 LHYAMFAVLVLMCFGDKLDEKQIKQVEYVQRQMLLGFARYSILNLCPKFTKLILRKRWEE 236
Query: 241 LAKLLKDQEDVLLPLIRARKHVKESRLN--------NINTVVSYVDTLMELELPQEKRKL 292
++ ++Q+DVLL LI AR+ + E R N V SYVDTL+++ELP EKRKL
Sbjct: 237 FFQMRREQQDVLLRLIYARRKIVEERKKRSSEEEEENKEYVQSYVDTLLDVELPDEKRKL 296
Query: 293 SEDEMISLCSEFLNGGTDTTSTALQWIMANLVKYPDVQGKLVEEITEVMGGDDAGEKEEV 352
+EDE++SLCSEFL G+DTT+T LQWIMANLVK ++Q +L EEIT V+G E + V
Sbjct: 297 NEDEIVSLCSEFLIAGSDTTATVLQWIMANLVKNQEIQERLYEEITNVVGE----EAKVV 352
Query: 353 KEEDLHKLRYLKCVVLEGLRRHPPGHFVLPHAVSEDVVLNGYLVPKDGTVNFMVAEMGLD 412
+E+D K+ YLK VV+E LRRHPPG+ VLPH+V+ED VL GY VPK GT+NF+VAE+G D
Sbjct: 353 EEKDTQKMPYLKAVVMEALRRHPPGNTVLPHSVTEDTVLGGYKVPKKGTINFLVAEIGRD 412
Query: 413 PRVWEDPMEFKPERFL-KDETFDITGSKEIKMMPFGAGRRICPGYNLALLHLEYFVANLV 471
P+VWE+PM FKPERF+ ++E DITGS+ IKMMPFGAGRRICPG LA+LHLEY+VAN+V
Sbjct: 413 PKVWEEPMAFKPERFMGEEEAVDITGSRGIKMMPFGAGRRICPGIGLAMLHLEYYVANMV 472
Query: 472 WNFDWKVPEGGRVDLSEKQEFTMVMKNPLQAHISPR 507
F WK EG VDL+EK EFT++MK+PL+A PR
Sbjct: 473 REFQWKEVEGHEVDLTEKVEFTVIMKHPLKAIAVPR 508
>At3g03470 cytochrome P450 nlike protein
Length = 511
Score = 548 bits (1412), Expect = e-156
Identities = 274/511 (53%), Positives = 375/511 (72%), Gaps = 23/511 (4%)
Query: 13 IIFLIRATLSSISTTKK----TTATLPPGPPHIPIITSIQWLRKS-FAQLEPFLRTLHAK 67
IIFLI ++L+ K +T LPPGPP P+I +I WL+K+ F+ + LR L ++
Sbjct: 6 IIFLIISSLTFSIFLKLIFFFSTHKLPPGPPRFPVIGNIIWLKKNNFSDFQGVLRDLASR 65
Query: 68 HGPIINLNIGSRPSIFIADRALAHHALVQNSSVFSDRPKALPTNKLMSSNQHNISSAFYG 127
HGPII L++GS+PSI++ DR+LAH ALVQN +VFSDR ALPT K+++SNQH+I S+ YG
Sbjct: 66 HGPIITLHVGSKPSIWVTDRSLAHQALVQNGAVFSDRSLALPTTKVITSNQHDIHSSVYG 125
Query: 128 PTWRTLRRNLASEMLHPSKIKSFSEIRKWVLQTLINRLKTASESEDSVT-VVSHFRYAVF 186
WRTLRRNL SE+L PS++K+ + RKW L+ L++ +T + ++ + H R+A+F
Sbjct: 126 SLWRTLRRNLTSEILQPSRVKAHAPSRKWSLEILVDLFETEQREKGHISDALDHLRHAMF 185
Query: 187 CLLVFMCFGERVNDEKINDIERVQRTILLNFSRFNILNFWPKVTRILLRKRWDELAKLLK 246
LL MCFGE++ E+I +IE Q +L+++++F++LN +P VT+ LLR++W E +L K
Sbjct: 186 YLLALMCFGEKLRKEEIREIEEAQYQMLISYTKFSVLNIFPSVTKFLLRRKWKEFLELRK 245
Query: 247 DQEDVLLPLIRARKHVKESRLNNINTVVSYVDTLMELELPQE------KRKLSEDEMISL 300
QE V+L + AR KE+ V+ YVDTL+ LE+P E KRKLS+ E++SL
Sbjct: 246 SQESVILRYVNARS--KET----TGDVLCYVDTLLNLEIPTEEKEGGKKRKLSDSEIVSL 299
Query: 301 CSEFLNGGTDTTSTALQWIMANLVKYPDVQGKLVEEITEVMGGDDAGEKEEVKEEDLHKL 360
CSEFLN TD T+T++QWIMA +VKYP++Q K+ EE+ V G++ E+EE++EEDL KL
Sbjct: 300 CSEFLNAATDPTATSMQWIMAIMVKYPEIQRKVYEEMKTVFAGEEE-EREEIREEDLGKL 358
Query: 361 RYLKCVVLEGLRRHPPGHFVLPHAVSEDVVLNGYLVPKDGTVNFMVAEMGLDPRVWEDPM 420
YLK V+LE LRRHPPGH++ H V+ D VL G+L+P+ GT+NFMV EMG DP++WEDP+
Sbjct: 359 SYLKAVILECLRRHPPGHYLSYHKVTHDTVLGGFLIPRQGTINFMVGEMGRDPKIWEDPL 418
Query: 421 EFKPERFLKDE---TFDITGSKEIKMMPFGAGRRICPGYNLALLHLEYFVANLVWNFDWK 477
FKPERFL++ FD+TG++EIKMMPFGAGRR+CPGY L+LLHLEY+VANLVW F+WK
Sbjct: 419 TFKPERFLENGEACDFDMTGTREIKMMPFGAGRRMCPGYALSLLHLEYYVANLVWKFEWK 478
Query: 478 VPEGGRVDLSEKQEF-TMVMKNPLQAHISPR 507
EG VDLSEKQ+F TMVMKNP +A+I PR
Sbjct: 479 CVEGEEVDLSEKQQFITMVMKNPFKANIYPR 509
>At5g61320 cytochrome P450 - like protein
Length = 497
Score = 536 bits (1381), Expect = e-152
Identities = 274/504 (54%), Positives = 350/504 (69%), Gaps = 24/504 (4%)
Query: 1 METWFIALVSLCIIFLIRATLSSISTTKKTTATLPPGPPHIPIITSIQWLRKSFAQLEPF 60
M TWF C+ F + L LPP P P+ QWLR+ F +
Sbjct: 1 MATWFHIF---CVSFTVNFLLYLFFRRTNNNLPLPPNPNFFPMPGPFQWLRQGFDDFYSY 57
Query: 61 LRTLHAKHGPIINLNIGSRPSIFIADRALAHHALVQNSSVFSDRPKALPTNKLMSSNQHN 120
LR++H + GPII+L I S P+IF++DR+LAH ALV N +VFSDRP ALPT K+++SNQH
Sbjct: 58 LRSIHHRLGPIISLRIFSVPAIFVSDRSLAHKALVLNGAVFSDRPPALPTGKIITSNQHT 117
Query: 121 ISSAFYGPTWRTLRRNLASEMLHPSKIKSFSEIRKWVLQTLINRLKTASESEDSVTVVSH 180
ISS YG TWR LRRNL SE+LHPS++KS+S R+ VL+ L +R++ E + VV H
Sbjct: 118 ISSGSYGATWRLLRRNLTSEILHPSRVKSYSNARRSVLENLCSRIRNHGEEAKPIVVVDH 177
Query: 181 FRYAVFCLLVFMCFGERVNDEKINDIERVQRTILLNFSRFNILNFWPKVTRILLRKRWDE 240
RYA+F LLV MCFG+++++E+I +E VQR L+ RFNILN +P T++ LRKRW+E
Sbjct: 178 LRYAMFSLLVLMCFGDKLDEEQIKQVEFVQRRELITLPRFNILNVFPSFTKLFLRKRWEE 237
Query: 241 LAKLLKDQEDVLLPLIRARKHVK-ESRLNNINTVVSYVDTLMELELPQEKRKLSEDEMIS 299
++ ++VLLPLIR+R+ + ES+ + + SYVDTL++LELP EKRKL+EDE++S
Sbjct: 238 FLTFRREHKNVLLPLIRSRRKIMIESKDSGKEYIQSYVDTLLDLELPDEKRKLNEDEIVS 297
Query: 300 LCSEFLNGGTDTTSTALQWIMANLVKYPDVQGKLVEEITEVMGGDDAGEKEEVKEEDLHK 359
LCSEFLN GTDTT+T LQWIMANLV + EE E++E++EE++ K
Sbjct: 298 LCSEFLNAGTDTTATTLQWIMANLV--------IGEE-----------EEKEIEEEEMKK 338
Query: 360 LRYLKCVVLEGLRRHPPGHFVLPHAVSEDVVLNGYLVPKDGTVNFMVAEMGLDPRVWEDP 419
+ YLK VVLEGLR HPPGH +LPH VSED L GY VPK GT N VA +G DP VWE+P
Sbjct: 339 MPYLKAVVLEGLRLHPPGHLLLPHRVSEDTELGGYRVPKKGTFNINVAMIGRDPTVWEEP 398
Query: 420 MEFKPERFL-KDETFDITGSKEIKMMPFGAGRRICPGYNLALLHLEYFVANLVWNFDWKV 478
MEFKPERF+ +D+ D+TGS+ IKMMPFGAGRRICPG A+LHLEYFV NLV F+WK
Sbjct: 399 MEFKPERFIGEDKEVDVTGSRGIKMMPFGAGRRICPGIGSAMLHLEYFVVNLVKEFEWKE 458
Query: 479 PEGGRVDLSEKQEFTMVMKNPLQA 502
EG VDLSEK EFT+VMK PL+A
Sbjct: 459 VEGYEVDLSEKWEFTVVMKYPLKA 482
>At5g04660 cytochrom P450 - like protein
Length = 512
Score = 350 bits (898), Expect = 1e-96
Identities = 184/508 (36%), Positives = 299/508 (58%), Gaps = 15/508 (2%)
Query: 5 FIALVSLCIIFLIRATLSSISTTKKTTATLPPGPPHIPIITSIQWLRKSFAQLEPFLRTL 64
F A++ +F+I + ++ K LPPGPP P++ ++ +S + L
Sbjct: 16 FFAIIISGFVFII----TRWNSNSKKRLNLPPGPPGWPVVGNLFQFARSGKPFFEYAEDL 71
Query: 65 HAKHGPIINLNIGSRPSIFIADRALAHHALVQNSSVFSDRPKALPTNKLMSSNQHNISSA 124
+GPI L +G+R I ++D L H AL+Q ++F+ RP PT + S N+ +++A
Sbjct: 72 KKTYGPIFTLRMGTRTMIILSDATLVHEALIQRGALFASRPAENPTRTIFSCNKFTVNAA 131
Query: 125 FYGPTWRTLRRNLASEMLHPSKIKSFSEIRKWVLQTLINRLKTASESEDS-VTVVSHFRY 183
YGP WR+LRRN+ ML +++K F ++R+ + LI R+K+ + D + V+ + R+
Sbjct: 132 KYGPVWRSLRRNMVQNMLSSTRLKEFGKLRQSAMDKLIERIKSEARDNDGLIWVLKNARF 191
Query: 184 AVFCLLVFMCFGERVNDEKINDIERVQRTILLNFSRFNILNFWPKVTRILLRKRWDELAK 243
A FC+L+ MCFG +++E I ++ + +T+L+ I ++ P + ++R L +
Sbjct: 192 AAFCILLEMCFGIEMDEETIEKMDEILKTVLMTVDP-RIDDYLPILAPFFSKERKRAL-E 249
Query: 244 LLKDQEDVLLPLI-RARKHVKESRLNNINTVVSYVDTLMELELPQEKRKLSEDEMISLCS 302
+ ++Q D ++ +I R R+ ++ + + SY+DTL +L++ K S +E+++LCS
Sbjct: 250 VRREQVDYVVGVIERRRRAIQNPGSDKTASSFSYLDTLFDLKIEGRKTTPSNEELVTLCS 309
Query: 303 EFLNGGTDTTSTALQWIMANLVKYPDVQGKLVEEITEVMGGDDAGEKEEVKEEDLHKLRY 362
EFLNGGTDTT TA++W +A L+ P++Q +L +EI +G D V E+D+ K+ +
Sbjct: 310 EFLNGGTDTTGTAIEWGIAQLIANPEIQSRLYDEIKSTVGDD-----RRVDEKDVDKMVF 364
Query: 363 LKCVVLEGLRRHPPGHFVLPHAVSEDVVLNGYLVPKDGTVNFMVAEMGLDPRVWEDPMEF 422
L+ V E LR+HPP +F L HAV E L GY +P V + + DPR+W +P +F
Sbjct: 365 LQAFVKELLRKHPPTYFSLTHAVMETTTLAGYDIPAGVNVEVYLPGISEDPRIWNNPKKF 424
Query: 423 KPERF-LKDETFDITGSKEIKMMPFGAGRRICPGYNLALLHLEYFVANLVWNFDWKV-PE 480
P+RF L E DITG +KM+PFG GRRICPG +A +H+ +A +V F+W P
Sbjct: 425 DPDRFMLGKEDADITGISGVKMIPFGVGRRICPGLAMATIHVHLMLARMVQEFEWCAHPP 484
Query: 481 GGRVDLSEKQEFTMVMKNPLQAHISPRI 508
G +D + K EFT+VMKNPL+A + PRI
Sbjct: 485 GSEIDFAGKLEFTVVMKNPLRAMVKPRI 512
>At3g10570 putative cytochrome P450
Length = 513
Score = 345 bits (885), Expect = 3e-95
Identities = 184/507 (36%), Positives = 299/507 (58%), Gaps = 18/507 (3%)
Query: 6 IALVSLCIIFLIRATLSSISTTKKTTATLPPGPPHIPIITSIQWLRKSFAQLEPFLRTLH 65
I + SL ++ L R + K LPPGPP P++ ++ +S Q ++ +
Sbjct: 21 ILISSLVLLILTRRS------AKSKIVKLPPGPPGWPVVGNLFQFARSGKQFYEYVDDVR 74
Query: 66 AKHGPIINLNIGSRPSIFIADRALAHHALVQNSSVFSDRPKALPTNKLMSSNQHNISSAF 125
K+GPI L +GSR I I+D AL H L+Q +F+ RP PT + SSN ++++
Sbjct: 75 KKYGPIYTLRMGSRTMIIISDSALVHDVLIQRGPMFATRPTENPTRTIFSSNTFTVNASA 134
Query: 126 YGPTWRTLRRNLASEMLHPSKIKSFSEIRKWVLQTLINRLKTASESEDS-VTVVSHFRYA 184
YGP WR+LR+N+ ML + + F +R+ + L+ R+K+ ++ D V V+ + R+A
Sbjct: 135 YGPVWRSLRKNMVQNMLSSIRFREFGSLRQSAMDKLVERIKSEAKDNDGLVWVLRNARFA 194
Query: 185 VFCLLVFMCFGERVNDEKINDIERVQRTILLNFSRFNILNFWPKVTRILLRKRWDELAKL 244
FC+L+ MCFG +++E I ++++V + +L+ + + ++ P + ++R L ++
Sbjct: 195 AFCILLEMCFGIEMDEESILNMDQVMKKVLITLNP-RLDDYLPILAPFYSKERARAL-EV 252
Query: 245 LKDQEDVLLPLI-RARKHVKESRLNNINTVVSYVDTLMELELPQEKRKLSEDEMISLCSE 303
+Q D ++ LI R R+ +++ + + SY+DTL +L+ S +E++SLCSE
Sbjct: 253 RCEQVDFIVKLIERRRRAIQKPGTDKTASSFSYLDTLFDLKTEGRITTPSNEELVSLCSE 312
Query: 304 FLNGGTDTTSTALQWIMANLVKYPDVQGKLVEEITEVMGGDDAGEKEEVKEEDLHKLRYL 363
FLNGGTDTT TA++W +A L+ P++Q +L +EI +G EV+E+D+ K+ +L
Sbjct: 313 FLNGGTDTTGTAIEWGIAQLIVNPEIQSRLYDEIKSTVG------DREVEEKDVDKMVFL 366
Query: 364 KCVVLEGLRRHPPGHFVLPHAVSEDVVLNGYLVPKDGTVNFMVAEMGLDPRVWEDPMEFK 423
+ VV E LR+HPP +F L H+V+E + GY VP V F + + DP++W DP +F
Sbjct: 367 QAVVKEILRKHPPTYFTLTHSVTEPTTVAGYDVPVGINVEFYLPGINEDPKLWSDPKKFN 426
Query: 424 PERFLK-DETFDITGSKEIKMMPFGAGRRICPGYNLALLHLEYFVANLVWNFDWKV-PEG 481
P+RF+ E DITG +KMMPFG GRRICPG +A +H+ +A +V F+W P
Sbjct: 427 PDRFISGKEEADITGVTGVKMMPFGIGRRICPGLAMATVHVHLMLAKMVQEFEWSAYPPE 486
Query: 482 GRVDLSEKQEFTMVMKNPLQAHISPRI 508
+D + K EFT+VMK PL+A + PR+
Sbjct: 487 SEIDFAGKLEFTVVMKKPLRAMVKPRV 513
>At3g10560 putative cytochrome P450
Length = 514
Score = 336 bits (862), Expect = 2e-92
Identities = 186/507 (36%), Positives = 295/507 (57%), Gaps = 18/507 (3%)
Query: 9 VSLCIIFLIRATLSSISTTKKTTAT---LPPGPPHIPIITSIQWLRKSFAQLEPFLRTLH 65
V L I+ +I + L T +K + LPPGPP P+I ++ +S Q ++ L
Sbjct: 18 VFLTILAIIISGLLKTITYRKHNSNHLNLPPGPPGWPVIGNLFQFTRSGKQFFEYVEDLV 77
Query: 66 AKHGPIINLNIGSRPSIFIADRALAHHALVQNSSVFSDRPKALPTNKLMSSNQHNISSAF 125
+GPI+ L +G+R I I+D +LAH AL++ + F+ RP PT K+ SS++ + SA
Sbjct: 78 KIYGPILTLRLGTRTMIIISDASLAHEALIERGAQFATRPVETPTRKIFSSSEITVHSAM 137
Query: 126 YGPTWRTLRRNLASEMLHPSKIKSFSEIRKWVLQTLINRLKTASESEDS-VTVVSHFRYA 184
YGP WR+LRRN+ ML +++K F +RK + LI R+K+ + D V V+ + RYA
Sbjct: 138 YGPVWRSLRRNMVQNMLSSNRLKEFGSVRKSAMDKLIERIKSEARDNDGLVWVLQNSRYA 197
Query: 185 VFCLLVFMCFGERVNDEKINDIERVQRTILLNFSRFNILNFWPKVTRILLRKRWDELAKL 244
FC+L+ +CFG + +E I ++++ T +LN + ++ P +T +R + KL
Sbjct: 198 AFCVLLDICFGVEMEEESIEKMDQLM-TAILNAVDPKLHDYLPILTPFNYNER-NRALKL 255
Query: 245 LKDQEDVLLPLIRARKHVKESRLNNINTVVSYVDTLMELELPQ-EKRKLSEDEMISLCSE 303
++ D ++ I R+ K R +++ SY+DTL +L + + + S++++++LCSE
Sbjct: 256 RRELVDFVVGFIEKRR--KAIRTATVSS-FSYLDTLFDLRIIEGSETTPSDEDLVTLCSE 312
Query: 304 FLNGGTDTTSTALQWIMANLVKYPDVQGKLVEEITEVMGGDDAGEKEEVKEEDLHKLRYL 363
FLN GTDTT A++W +A L+ P++Q +L +EI +G V E D+ K+ L
Sbjct: 313 FLNAGTDTTGAAIEWGIAELIANPEIQSRLYDEIKSTVG------DRAVDERDVDKMVLL 366
Query: 364 KCVVLEGLRRHPPGHFVLPHAVSEDVVLNGYLVPKDGTVNFMVAEMGLDPRVWEDPMEFK 423
+ VV E LRRHPP +F L H V+E L+GY +P + F + + DP++W +P +F
Sbjct: 367 QAVVKEILRRHPPTYFTLSHGVTEPTTLSGYNIPVGVNIEFYLPGISEDPKIWSEPKKFD 426
Query: 424 PERFLKD-ETFDITGSKEIKMMPFGAGRRICPGYNLALLHLEYFVANLVWNFDW-KVPEG 481
P+RFL E DITG +KMMPFG GRRICPG +A +H+ +A +V F+W P
Sbjct: 427 PDRFLSGREDADITGVAGVKMMPFGVGRRICPGMGMATVHVHLMIARMVQEFEWLAYPPQ 486
Query: 482 GRVDLSEKQEFTMVMKNPLQAHISPRI 508
+D + K F +VMK PL+A + PR+
Sbjct: 487 SEMDFAGKLVFAVVMKKPLRAMVRPRV 513
>At5g04630 cytochrome P450 - like protein
Length = 509
Score = 334 bits (857), Expect = 6e-92
Identities = 180/508 (35%), Positives = 292/508 (57%), Gaps = 11/508 (2%)
Query: 4 WFIALVSLCIIFLIRATLSSISTTKKTTATLPPGPPHIPIITSIQWLRKSFAQLEPFLRT 63
+F LV++ + L+ K A LPPGP P++ ++ +S Q ++
Sbjct: 10 FFFTLVTILLSCLVYILTRHSHNPK--CANLPPGPKGWPVVGNLLQFARSGKQFFEYVDE 67
Query: 64 LHAKHGPIINLNIGSRPSIFIADRALAHHALVQNSSVFSDRPKALPTNKLMSSNQHNISS 123
+ +GPI L +G R I I+D LAH AL++ + F+ RP PT K+ SS+ + S
Sbjct: 68 MRNIYGPIFTLKMGIRTMIIISDANLAHQALIERGAQFATRPAETPTRKIFSSSDITVHS 127
Query: 124 AFYGPTWRTLRRNLASEMLHPSKIKSFSEIRKWVLQTLINRLKT-ASESEDSVTVVSHFR 182
A YGP WR+LRRN+ ML +++K F IRK + L+ ++K+ A E++ V V+ + R
Sbjct: 128 AMYGPVWRSLRRNMVQNMLCSNRLKEFGSIRKSAIDKLVEKIKSEAKENDGLVWVLRNAR 187
Query: 183 YAVFCLLVFMCFGERVNDEKINDIERVQRTILLNFSRFNILNFWPKVTRILLRKRWDELA 242
+A FC+L+ MCFG ++ +E I ++++ IL I ++ P +T ++R + L
Sbjct: 188 FAAFCILLDMCFGVKMEEESIEKMDQMMTEILTAVDP-RIHDYLPILTPFYFKERKNSLE 246
Query: 243 KLLKDQEDVLLPLIRARKHVKESRLNNINTVVSYVDTLMELELPQEKRKLSEDEMISLCS 302
K + V+ + + R ++ + + +Y+DTL +L + + S++++++LCS
Sbjct: 247 LRRKLVQFVVGFIEKRRLAIRNLGSDKTASSFAYLDTLFDLRVDGRETSPSDEDLVTLCS 306
Query: 303 EFLNGGTDTTSTALQWIMANLVKYPDVQGKLVEEITEVMGGDDAGEKEEVKEEDLHKLRY 362
EFLN GTDTT TA++W +A L+ P +Q +L +EI +G D V+E+DL+K+ +
Sbjct: 307 EFLNAGTDTTGTAIEWGIAELISNPKIQSRLYDEIKSTVGDD-----RTVEEKDLNKMVF 361
Query: 363 LKCVVLEGLRRHPPGHFVLPHAVSEDVVLNGYLVPKDGTVNFMVAEMGLDPRVWEDPMEF 422
L+ V E LRRHPP +F L H V+E L GY +P V F + + DP++W P +F
Sbjct: 362 LQAFVKELLRRHPPTYFTLTHGVTEPTNLAGYDIPVGANVEFYLPGISEDPKIWSKPEKF 421
Query: 423 KPERFLK-DETFDITGSKEIKMMPFGAGRRICPGYNLALLHLEYFVANLVWNFDW-KVPE 480
P+RF+ E D+TG +KMMPFG GRRICPG +A++H+E ++ +V F+W P
Sbjct: 422 DPDRFITGGEDADLTGVAGVKMMPFGIGRRICPGLGMAVVHVELMLSRMVQEFEWSSYPP 481
Query: 481 GGRVDLSEKQEFTMVMKNPLQAHISPRI 508
+VD + K F +VMKNPL+A + R+
Sbjct: 482 ESQVDFTGKLVFAVVMKNPLRARVKARV 509
>At1g11600 cytochrome P450 like protein
Length = 510
Score = 326 bits (835), Expect = 2e-89
Identities = 183/492 (37%), Positives = 285/492 (57%), Gaps = 21/492 (4%)
Query: 27 TKKTTATLPPGPPHIPIITSIQWL---RKSFAQLEPFLRTLHAKHGPIINLNIGSRPSIF 83
+ + + +PPGP P++ ++ + R+ F L +R L K+GPI + +G R I
Sbjct: 28 SSQCSLNIPPGPKGWPLVGNLLQVIFQRRHFVFL---MRDLRKKYGPIFTMQMGQRTMII 84
Query: 84 IADRALAHHALVQNSSVFSDRPKALPTNKLMSSNQHNISSAFYGPTWRTLRRNLASEMLH 143
I D L H ALVQ F+ RP P + S + I+SA YG WRTLRRN +E++
Sbjct: 85 ITDEKLIHEALVQRGPTFASRPPDSPIRLMFSVGKCAINSAEYGSLWRTLRRNFVTELVT 144
Query: 144 PSKIKSFSEIRKWVLQTLINRLKTASESEDSVTVVSHFRYAVFCLLVFMCFGERVNDEKI 203
++K S IR W +Q + R+KT + + V V+S R + +L+ +CFG ++++EKI
Sbjct: 145 APRVKQCSWIRSWAMQNHMKRIKTENVEKGFVEVMSQCRLTICSILICLCFGAKISEEKI 204
Query: 204 NDIERVQRTILLNFSRFNILNFWPKVTRILLRKRWDELAKLLKDQEDVLLPLIRARKHVK 263
+IE V + ++L S + +F P T L R++ E +L K Q + L+PLIR R+
Sbjct: 205 KNIENVLKDVMLITSP-TLPDFLPVFTP-LFRRQVREARELRKTQLECLVPLIRNRRKFV 262
Query: 264 ESRLNNINTVVS-----YVDTLMELELPQEKRKLSEDEMISLCSEFLNGGTDTTSTALQW 318
+++ N +VS YVD+L L L + +L ++E+++LCSE ++ GTDT++T L+W
Sbjct: 263 DAKENPNEEMVSPIGAAYVDSLFRLNLIERGGELGDEEIVTLCSEIVSAGTDTSATTLEW 322
Query: 319 IMANLVKYPDVQGKLVEEITEVMGGDDAGEKEEVKEEDLHKLRYLKCVVLEGLRRHPPGH 378
+ +LV ++Q KL EE+ V+G + V+E+D+ K+ YL+ +V E LRRHPPGH
Sbjct: 323 ALFHLVTDQNIQEKLYEEVVGVVGKNGV-----VEEDDVAKMPYLEAIVKETLRRHPPGH 377
Query: 379 FVLPHAVSEDVVLNGYLVPKDGTVNFMVAEMGLDPRVWEDPMEFKPERFL---KDETFDI 435
F+L HA +D L GY +P V A + +P +W DP +F+PERFL D
Sbjct: 378 FLLSHAAVKDTELGGYDIPAGAYVEIYTAWVTENPDIWSDPGKFRPERFLTGGDGVDADW 437
Query: 436 TGSKEIKMMPFGAGRRICPGYNLALLHLEYFVANLVWNFDWKVPEGGRVDLSEKQEFTMV 495
TG++ + M+PFGAGRRICP ++L +LH+ +A ++ +F W D +E FT+V
Sbjct: 438 TGTRGVTMLPFGAGRRICPAWSLGILHINLMLARMIHSFKWIPVPDSPPDPTETYAFTVV 497
Query: 496 MKNPLQAHISPR 507
MKN L+A I R
Sbjct: 498 MKNSLKAQIRSR 509
>At3g26200 cytochrome P450 like protein
Length = 500
Score = 219 bits (558), Expect = 3e-57
Identities = 141/507 (27%), Positives = 247/507 (47%), Gaps = 21/507 (4%)
Query: 9 VSLCIIFLIRATLSSISTTKKTTATLPPGPPHIPIITSIQWLRKSFAQLEPFLRTLHAKH 68
+SL + L+ L + LPPGP +PII ++ L KS L L +
Sbjct: 3 ISLYFLLLLPLFLIFFKKLSPSKGKLPPGPLGLPIIGNLHQLGKS---LHRSFHKLSQNY 59
Query: 69 GPIINLNIGSRPSIFIADRALAHHALVQNSSVFSDRPKALPTNKLMSSNQHNISSAFYGP 128
GP++ L+ G P + ++ R A L + RPK L KL S N +I A YG
Sbjct: 60 GPVMFLHFGVVPVVVVSTREAAEEVLKTHDLETCTRPK-LTATKLFSYNYKDIGFAQYGD 118
Query: 129 TWRTLRRNLASEMLHPSKIKSFSEIRKWVLQTLINRLKTASESEDSVTVVSHFRYAVFCL 188
WR +R+ E+ K+K+F IR+ + L+N+L ++E+ V + +
Sbjct: 119 DWREMRKLAMLELFSSKKLKAFRYIREEESEVLVNKLSKSAETRTMVDLRKALFSYTASI 178
Query: 189 LVFMCFGERVNDEKINDIERVQRTIL---LNFSRFNILNFWPKVTRILLRK---RWDELA 242
+ + FG+ ++ D+++V+ +L N F +F+P ++ + + EL
Sbjct: 179 VCRLAFGQNFHECDFVDMDKVEDLVLESETNLGSFAFTDFFPAGLGWVIDRISGQHSELH 238
Query: 243 KLLKDQEDVLLPLIRARKHVKESRLNNINTVVSYVDTLMELELPQEKRKLSEDEMISLCS 302
K + +I H+K + + + ++ + ++ E +++ D + + S
Sbjct: 239 KAFARLSNFFQHVID--DHLKPGQSQDHSDIIGVMLDMINKESKVGSFQVTYDHLKGVMS 296
Query: 303 EFLNGGTDTTSTALQWIMANLVKYPDVQGKLVEEITEVMGGDDAGEKEEVKEEDLHKLRY 362
+ G + + + W M L ++P V KL +EI E++G + KE++ E+DL K+ Y
Sbjct: 297 DVFLAGVNAGAITMIWAMTELARHPRVMKKLQQEIREILGDN----KEKITEQDLEKVHY 352
Query: 363 LKCVVLEGLRRHPPGHFVLPHAVSEDVVLNGYLVPKDGTVNFMVAEMGLDPRVWEDPMEF 422
LK V+ E R HPP +LP D+ + GY +PK+ + +G DP WE+P +F
Sbjct: 353 LKLVIEETFRLHPPAPLLLPRETMSDLKIQGYNIPKNTMIEINTYSIGRDPNCWENPNDF 412
Query: 423 KPERFLKDETFDITGSKEIKMMPFGAGRRICPGYNLALLHLEYFVANLVWNFDWKVPEGG 482
PERF+ D + G + +++PFGAGRRICPG + +E + N+++ FDW +P+G
Sbjct: 413 NPERFI-DSPVEYKG-QHYELLPFGAGRRICPGMATGITIVELGLLNVLYFFDWSLPDGM 470
Query: 483 R---VDLSEKQEFTMVMKNPLQAHISP 506
+ +D+ E F + K PL+ +P
Sbjct: 471 KIEDIDMEEAGAFVVAKKVPLELIPTP 497
>At4g12300 flavonoid 3',5'-hydroxylase -like protein
Length = 516
Score = 216 bits (551), Expect = 2e-56
Identities = 143/479 (29%), Positives = 242/479 (49%), Gaps = 15/479 (3%)
Query: 33 TLPPGPPHIPIITSIQWLRKSFAQLEPFLRTLHAKHGPIINLNIGSRPSIFIADRALAHH 92
+LPPGP +PI+ ++ +L L + L HGPI LN+GS+ +I + +LA
Sbjct: 41 SLPPGPRGLPIVGNLPFLDPD---LHTYFANLAQSHGPIFKLNLGSKLTIVVNSPSLARE 97
Query: 93 ALVQNSSVFSDRPKALPTNKLMSSNQHNISSAFYGPTWRTLRRNLASEMLHPSKIKSFSE 152
L FS+R L T + + +I YG WR LR+ ++L + SF E
Sbjct: 98 ILKDQDINFSNRDVPL-TGRAATYGGIDIVWTPYGAEWRQLRKICVLKLLSRKTLDSFYE 156
Query: 153 IRKWVLQTLINRLKTASESEDSVTVVSHFRYAVFCLLVFMCFGERVNDEKINDIERVQRT 212
+R+ ++ L + V V + L + M +G V E++ + +
Sbjct: 157 LRRKEVRERTRYLYEQGRKQSPVKVGDQLFLTMMNLTMNMLWGGSVKAEEMESVGTEFKG 216
Query: 213 ILLNFSRF----NILNFWPKVTRILLRKRWDELAKLLKDQEDVLLPLIRARKHVKESRLN 268
++ +R N+ +F+P + R L+ + ++ + VL I K ++ +
Sbjct: 217 VISEITRLLSEPNVSDFFPWLARFDLQGLVKRMGVCARELDAVLDRAIEQMKPLRGRDDD 276
Query: 269 NINTVVSYVDTLMELELPQEKRKLSEDEMISLCSEFLNGGTDTTSTALQWIMANLVKYPD 328
+ + Y+ L + E E ++ + + +L ++ + GGTDT++ +++ MA L+ P+
Sbjct: 277 EVKDFLQYLMKLKDQEGDSEV-PITINHVKALLTDMVVGGTDTSTNTIEFAMAELMSNPE 335
Query: 329 VQGKLVEEITEVMGGDDAGEKEEVKEEDLHKLRYLKCVVLEGLRRHPPGHFVLPHAVSED 388
+ + EE+ EV+G D+ V+E + +L Y+ ++ E LR HP ++PH +E+
Sbjct: 336 LIKRAQEELDEVVGKDNI-----VEESHITRLPYILAIMKETLRLHPTLPLLVPHRPAEN 390
Query: 389 VVLNGYLVPKDGTVNFMVAEMGLDPRVWEDPMEFKPERFLKDETFDITGSKEIKMMPFGA 448
V+ GY +PKD + V + DP VWE+P EF+PERFL + + D TG+ PFG+
Sbjct: 391 TVVGGYTIPKDTKIFVNVWSIQRDPNVWENPTEFRPERFLDNNSCDFTGA-NYSYFPFGS 449
Query: 449 GRRICPGYNLALLHLEYFVANLVWNFDWKVPEGGRVDLSEKQEFTMVMKNPLQAHISPR 507
GRRIC G LA + Y +A L+ +FDWK+PEG +DL EK + +K PL A PR
Sbjct: 450 GRRICAGVALAERMVLYTLATLLHSFDWKIPEGHVLDLKEKFGIVLKLKIPLVALPIPR 508
>At3g26220 cytochrome P450 monooxygenase (CYP71B3)
Length = 501
Score = 211 bits (537), Expect = 8e-55
Identities = 153/507 (30%), Positives = 252/507 (49%), Gaps = 27/507 (5%)
Query: 9 VSLCIIFLIRATLSSISTTKK---TTATLPPGPPHIPIITSIQWLRKSFAQLEPFLRTLH 65
+S+ + F + S+ KK + LPP PP +PII ++ LR F + L L
Sbjct: 1 MSILLYFFFLPVILSLIFMKKFKDSKRNLPPSPPKLPIIGNLHQLRGLFHRC---LHDLS 57
Query: 66 AKHGPIINLNIGSRPSIFIADRALAHHALVQNSSVFSDRPKALPTNKLMSSNQHNISSAF 125
KHGP++ L +G + I+ + A L + RPK ++K S + +I+ A
Sbjct: 58 KKHGPVLLLRLGFIDMVVISSKEAAEEVLKVHDLECCTRPKTNASSKF-SRDGKDIAFAP 116
Query: 126 YGPTWRTLRRNLASEMLHPSKIKSFSEIRKWVLQTLINRLKTASESEDSVTVVSHFRYAV 185
YG R LR+ K++SF IR+ ++ +LK +++ +++V + Y V
Sbjct: 117 YGEVSRELRKLSLINFFSTQKVRSFRYIREEENDLMVKKLKESAKKKNTVDLSQTLFYLV 176
Query: 186 FCLLVFMCFGER------VNDEKINDIE-RVQRTILLNFSRFNILNFWPKVTRILLRKRW 238
++ FG+R VN EKI ++ VQ+ L+ S +I + R
Sbjct: 177 GSIIFRATFGQRLDQNKHVNKEKIEELMFEVQKVGSLSSS--DIFPAGVGWFMDFVSGRH 234
Query: 239 DELAKLLKDQEDVLLPLIRARKHVKESRLNNINTVVSYVDTLMELELPQEKR---KLSED 295
L K+ + + +L +I E + N + +D+++E QE+ KL+ D
Sbjct: 235 KTLHKVFVEVDTLLNHVIDGHLKNPEDKTNQDRPDI--IDSILETIYKQEQDESFKLTID 292
Query: 296 EMISLCSEFLNGGTDTTSTALQWIMANLVKYPDVQGKLVEEITEVMGGDDAGEKEEVKEE 355
+ + G DT++ + W MA LVK P V K EEI +G +KE ++EE
Sbjct: 293 HLKGIIQNIYLAGVDTSAITMIWAMAELVKNPRVMKKAQEEIRTCIG---IKQKERIEEE 349
Query: 356 DLHKLRYLKCVVLEGLRRHPPGHFVLPHAVSEDVVLNGYLVPKDGTVNFMVAEMGLDPRV 415
D+ KL+YLK V+ E LR HPP +LP D+ + GY +P+ + +G +P +
Sbjct: 350 DVDKLQYLKLVIKETLRLHPPAPLLLPRETMADIKIQGYDIPRKTILLVNAWSIGRNPEL 409
Query: 416 WEDPMEFKPERFLKDETFDITGSKEIKMMPFGAGRRICPGYNLALLHLEYFVANLVWNFD 475
WE+P EF PERF+ D D G+ +M+PFG+GR+ICPG + +E + NL++ FD
Sbjct: 410 WENPEEFNPERFI-DCPMDYKGN-SFEMLPFGSGRKICPGIAFGIATVELGLLNLLYYFD 467
Query: 476 WKVPEGGR-VDLSEKQEFTMVMKNPLQ 501
W++ E + +D+ E + T+V K PL+
Sbjct: 468 WRLAEEDKDIDMEEAGDATIVKKVPLE 494
>At3g26280 cytochrome P450 monooxygenase (CYP71B4)
Length = 504
Score = 210 bits (534), Expect = 2e-54
Identities = 153/506 (30%), Positives = 249/506 (48%), Gaps = 22/506 (4%)
Query: 6 IALVSLCIIFLIRATLSSISTTK--KTTATLPPGPPHIPIITSIQWLRKSFAQLEPFLRT 63
++L+S ++ L+ I T K ++ LPPGP +PII ++ L+ L L
Sbjct: 2 VSLLSFFLLLLVPIFFLLIFTKKIKESKQNLPPGPAKLPIIGNLHQLQ---GLLHKCLHD 58
Query: 64 LHAKHGPIINLNIGSRPSIFIADRALAHHALVQNSSVFSDRPKALPTNKLMSSNQHNISS 123
L KHGP+++L +G P + I+ A AL + RP + +++ S N +I
Sbjct: 59 LSKKHGPVMHLRLGFAPMVVISSSEAAEEALKTHDLECCSRPITM-ASRVFSRNGKDIGF 117
Query: 124 AFYGPTWRTLRRNLASEMLHPSKIKSFSEIRKWVLQTLINRLKTASESEDSVTVVSHFRY 183
YG WR LR+ E K++SF IR+ +I +LK + S+ S +S +
Sbjct: 118 GVYGDEWRELRKLSVREFFSVKKVQSFKYIREEENDLMIKKLKELA-SKQSPVDLSKILF 176
Query: 184 AVFCLLVF-MCFGERVNDEKINDIERVQRTILLNFSR--FNILNFWP----KVTRILLRK 236
+ ++F FG+ D K D E ++ + + S F +F+P K +
Sbjct: 177 GLTASIIFRTAFGQSFFDNKHVDQESIKELMFESLSNMTFRFSDFFPTAGLKWFIGFVSG 236
Query: 237 RWDELAKLLKDQEDVLLPLIRARKHVKESRLNNINTVVSYVDTLMELELPQEKRKLSEDE 296
+ L + ++ D I H K++ + + V + +D +++ E KL+ D
Sbjct: 237 QHKRLYNVF-NRVDTFFNHIVDDHHSKKATQDRPDMVDAILD-MIDNEQQYASFKLTVDH 294
Query: 297 MISLCSEFLNGGTDTTSTALQWIMANLVKYPDVQGKLVEEITEVMGGDDAGEKEEVKEED 356
+ + S + G DT++ L W MA LV+ P V K +EI +G G + EED
Sbjct: 295 LKGVLSNIYHAGIDTSAITLIWAMAELVRNPRVMKKAQDEIRTCIGIKQEG---RIMEED 351
Query: 357 LHKLRYLKCVVLEGLRRHPPGHFVLPHAVSEDVVLNGYLVPKDGTVNFMVAEMGLDPRVW 416
L KL+YLK VV E LR HP +LP D+ + GY +P+ + +G DP W
Sbjct: 352 LDKLQYLKLVVKETLRLHPAAPLLLPRETMADIKIQGYDIPQKRALLVNAWSIGRDPESW 411
Query: 417 EDPMEFKPERFLKDETFDITGSKEIKMMPFGAGRRICPGYNLALLHLEYFVANLVWNFDW 476
++P EF PERF+ D D G +++PFG+GRRICPG +A+ +E + NL++ FDW
Sbjct: 412 KNPEEFNPERFI-DCPVDYKG-HSFELLPFGSGRRICPGIAMAIATIELGLLNLLYFFDW 469
Query: 477 KVPEGGR-VDLSEKQEFTMVMKNPLQ 501
+PE + +D+ E + T+ K PL+
Sbjct: 470 NMPEKKKDMDMEEAGDLTVDKKVPLE 495
>At3g26210 cytochrome P450 like protein
Length = 501
Score = 209 bits (533), Expect = 2e-54
Identities = 141/511 (27%), Positives = 250/511 (48%), Gaps = 29/511 (5%)
Query: 9 VSLCIIFLIRATLSSISTTKKTTAT---LPPGPPHIPIITSIQWLRKSFAQLEPFLRTLH 65
+ LC + L+ L +I T+K+ ++ LPPGPP +PII ++ +L L L
Sbjct: 3 IFLCFLLLLLLLLVTIIFTRKSQSSKLKLPPGPPKLPIIGNLHYLN---GLPHKCLLNLW 59
Query: 66 AKHGPIINLNIGSRPSIFIADRALAHHALVQNSSVFSDRPKALPTNKLMSSNQHNISSAF 125
HGP++ L +G P + I+ A L + RP+ + +K +S N +I A
Sbjct: 60 KIHGPVMQLQLGYVPLVVISSNQAAEEVLKTHDLDCCSRPETI-ASKTISYNFKDIGFAP 118
Query: 126 YGPTWRTLRRNLASEMLHPSKIKSFSEIRKWVLQTLINRLKTASESEDSVTVVSHFRYAV 185
YG WR LR+ E+ K SF IR+ L+ +L ASE + V +
Sbjct: 119 YGEEWRALRKLAVIELFSLKKFNSFRYIREEENDLLVKKLSEASEKQSPVNLKKALFTLS 178
Query: 186 FCLLVFMCFGERVNDEKINDIERVQRTILLNF---SRFNILNFWPKVTRILLRKRWDELA 242
++ + FG+ +++ + D + ++ + ++F NF+P + D++
Sbjct: 179 ASIVCRLAFGQNLHESEFIDEDSMEDLASRSEKIQAKFAFSNFFPGGWIL------DKIT 232
Query: 243 KLLKDQEDVLLPLIRARKHVKESRLNNINTVVS---YVDTLMELELPQEKR---KLSEDE 296
K ++ L V + L V+ VD ++++ Q + KL+ D
Sbjct: 233 GQSKSLNEIFADLDGFFNQVLDDHLKPGRKVLETPDVVDVMIDMMNKQSQDGSFKLTTDH 292
Query: 297 MISLCSEFLNGGTDTTSTALQWIMANLVKYPDVQGKLVEEITEVMGGDDAGEKEEVKEED 356
+ + S+ G +T++T + W M L++ P V K+ +E+ V+G +++ + E+D
Sbjct: 293 IKGIISDIFLAGVNTSATTILWAMTELIRNPRVMKKVQDEVRTVLGE----KRDRITEQD 348
Query: 357 LHKLRYLKCVVLEGLRRHPPGHFVLPHAVSEDVVLNGYLVPKDGTVNFMVAEMGLDPRVW 416
L++L Y K V+ E R HP +LP + + GY +P+ + V +G DP +W
Sbjct: 349 LNQLNYFKLVIKETFRLHPAAPLLLPREAMAKIKIQGYDIPEKTQIMVNVYAIGRDPDLW 408
Query: 417 EDPMEFKPERFLKDETFDITGSKEIKMMPFGAGRRICPGYNLALLHLEYFVANLVWNFDW 476
E+P EFKPERF+ D + D G +++PFG+GRRICPG + + +E + NL++ FDW
Sbjct: 409 ENPEEFKPERFV-DSSVDYRG-LNFELLPFGSGRRICPGMTMGIATVELGLLNLLYFFDW 466
Query: 477 KVPEGGRV-DLSEKQEFTMVMKNPLQAHISP 506
+PEG V D+ ++E +++ + + P
Sbjct: 467 GLPEGRTVKDIDLEEEGAIIIGKKVSLELVP 497
>At3g26230 cytochrome P450, putative
Length = 498
Score = 209 bits (532), Expect = 3e-54
Identities = 153/509 (30%), Positives = 255/509 (50%), Gaps = 34/509 (6%)
Query: 9 VSLCIIFLIRATLSSISTTKKTTATLPPGPPHIPIITSIQWLRKSFAQLEPFLRTLHAKH 68
+S+ + F+ +L I K + LPP P +P+I ++ LR F + L L KH
Sbjct: 1 MSILLYFIALLSLIIIKKIKDSNRNLPPSPLKLPVIGNLYQLRGLFHKC---LHDLSKKH 57
Query: 69 GPIINLNIGSRPSIFIADRALAHHALVQNSSVFSDRPKALPTNKLMSSNQHNISSAFYGP 128
GP++ L +G + I+ A AL + RP T+KL Q +I A YG
Sbjct: 58 GPVLLLRLGFLDMVVISSTEAAEEALKVHDLECCTRPITNVTSKLWRDGQ-DIGLAPYGE 116
Query: 129 TWRTLRRNLASEMLHPSKIKSFSEIRKWVLQTLINRLKTASESEDSVTVVSHFRYAVFCL 188
+ R LR+ + +K++SF IR+ ++ +LK A+ + SV + V +
Sbjct: 117 SLRELRKLSFLKFFSTTKVRSFRYIREEENDLMVKKLKEAALKKSSVDLSQTLFGLVGSI 176
Query: 189 LVFMCFGER------VNDEKINDIE-RVQRTILLNFSRFNILNFWPKVTRILLRKRWDEL 241
+ FG+R VN EKI D+ VQ+ L+ S + +P + D +
Sbjct: 177 IFRSAFGQRFDEGNHVNAEKIEDLMFEVQKLGALSNS-----DLFPGGLGWFV----DFV 227
Query: 242 AKLLKDQEDVLLPLIRARKHVKESRLNNINTVVSY-----VDTLMELELPQEKR---KLS 293
+ K V + + H+ + L N +++ +D+L+++ QE+ KL+
Sbjct: 228 SGHNKKLHKVFVEVDTLLNHIIDDHLKNSIEEITHDRPDIIDSLLDMIRKQEQGDSFKLT 287
Query: 294 EDEMISLCSEFLNGGTDTTSTALQWIMANLVKYPDVQGKLVEEITEVMGGDDAGEKEEVK 353
D + + + G DT++ + W MA LVK P V K+ +EI +G + E+++
Sbjct: 288 IDNLKGIIQDIYLAGVDTSAITMIWAMAELVKNPRVMKKVQDEIRTCIG---IKQNEKIE 344
Query: 354 EEDLHKLRYLKCVVLEGLRRHPPGHFVLPHAVSEDVVLNGYLVPKDGTVNFMVAEMGLDP 413
E+D+ KL+YLK VV E LR HP +LP + + GY +P + V +G DP
Sbjct: 345 EDDVDKLQYLKLVVKETLRLHPAAPLLLPRETMSQIKIQGYNIPSKTILLVNVWSIGRDP 404
Query: 414 RVWEDPMEFKPERFLKDETFDITGSKEIKMMPFGAGRRICPGYNLALLHLEYFVANLVWN 473
+ W++P EF PERF+ D D G+ +M+PFG+GRRICPG A+ +E + NL+++
Sbjct: 405 KHWKNPEEFNPERFI-DCPIDYKGN-SFEMLPFGSGRRICPGIAFAIATVELGLLNLLYH 462
Query: 474 FDWKVPEGGR-VDLSEKQEFTMVMKNPLQ 501
FDW++PE + +D+ E + T++ K PL+
Sbjct: 463 FDWRLPEEDKDLDMEEAGDVTIIKKVPLK 491
>At2g45570 cytochrome P450 (CYP76C2)
Length = 512
Score = 206 bits (525), Expect = 2e-53
Identities = 153/503 (30%), Positives = 250/503 (49%), Gaps = 34/503 (6%)
Query: 15 FLIRATLSSISTTKKTTATLPPGPPHIPIITSIQWLRK----SFAQLEPFLRTLHAKHGP 70
F+I T + +++K + PPGPP +PII +I + + SFA L +GP
Sbjct: 20 FIIFFTTTRPRSSRKVVPS-PPGPPRLPIIGNIHLVGRNPHHSFADLSK-------TYGP 71
Query: 71 IINLNIGSRPSIFIADRALAHHALVQNSSVFSDRPKALPTNKLMSSNQHNISSAFYGPT- 129
I++L GS ++ + A L + S R PTN + S N +S + P+
Sbjct: 72 IMSLKFGSLNTVVVTSPEAAREVLRTYDQILSSRT---PTNSIRSINHDKVSVVWLPPSS 128
Query: 130 --WRTLRRNLASEMLHPSKIKSFSEIRKWVLQTLINRLKTASESEDSVTVVSHFRYAVFC 187
WR LR+ A+++ P +I++ +R+ ++ L++ + +SE E++V +
Sbjct: 129 SRWRLLRKLSATQLFSPQRIEATKTLRENKVKELVSFMSESSEREEAVDISRATFITALN 188
Query: 188 LLVFMCFGERVNDEKINDIERVQRTILLNFSRF---NILNFWPKVTRILLRKRWDELAKL 244
++ + F + + N Q T++ + NF+P + + L+ L
Sbjct: 189 IISNILFSVDLGNYDSNKSGVFQDTVIGVMEAVGNPDAANFFPFLGFLDLQGNRKTLKAC 248
Query: 245 LKDQEDVLLPLIRARKHVKESRLNNINTVVS--YVDTLMELELPQEKRKLSEDEMISLCS 302
+ V I A+ K R N V +VD L++L E +L+ ++++ L
Sbjct: 249 SERLFKVFRGFIDAKLAEKSLRDTNSKDVRERDFVDVLLDLTEGDEA-ELNTNDIVHLLL 307
Query: 303 EFLNGGTDTTSTALQWIMANLVKYPDVQGKLVEEITEVMGGDDAGEKEEVKEEDLHKLRY 362
+ GTDT S+ ++W MA L++ P+ K EI V+G +K V+E D+ L Y
Sbjct: 308 DLFGAGTDTNSSTVEWAMAELLRNPETMVKAQAEIDCVIG-----QKGVVEESDISALPY 362
Query: 363 LKCVVLEGLRRHPPGHFVLPHAVSEDVVLNGYLVPKDGTVNFMVAEMGLDPRVWEDPMEF 422
L+ VV E R HP ++P DV + G++VPKD V V +G DP VWE+ F
Sbjct: 363 LQAVVKETFRLHPAAPLLVPRKAESDVEVLGFMVPKDTQVFVNVWAIGRDPNVWENSSRF 422
Query: 423 KPERFLKDETFDITGSKEIKMMPFGAGRRICPGYNLALLHLEYFVANLVWNFDWKVPEG- 481
KPERFL + D+ G ++ ++ PFGAGRRICPG LA+ + +A+L+++FDWK+P G
Sbjct: 423 KPERFL-GKDIDLRG-RDYELTPFGAGRRICPGLPLAVKTVPLMLASLLYSFDWKLPNGV 480
Query: 482 GRVDLSEKQEF--TMVMKNPLQA 502
G DL + F T+ NPL A
Sbjct: 481 GSEDLDMDETFGLTLHKTNPLHA 503
>At3g44250 cytochrome P450-like protein
Length = 499
Score = 206 bits (523), Expect = 3e-53
Identities = 145/510 (28%), Positives = 240/510 (46%), Gaps = 40/510 (7%)
Query: 9 VSLCIIFLIRATLSSISTTKKTTATLPPGPPHIPIITSIQWLRKSFAQLEPFLRTLHAKH 68
+ LC + L+ +L + LPPGP +PII ++ L K L + ++
Sbjct: 3 IFLCFLLLLPLSLILFKKLLPSKGKLPPGPIGLPIIGNLHQLGKL---LYKSFHKISQEY 59
Query: 69 GPIINLNIGSRPSIFIADRALAHHALVQNSSVFSDRPKALPTNKLMSSNQHNISSAFYGP 128
GP++ L +G P I ++ + A L + RPK T L + N +I A +G
Sbjct: 60 GPVVLLRLGVVPVIVVSSKEGAEEVLKTHDLETCTRPKTAATG-LFTYNFKDIGFAPFGD 118
Query: 129 TWRTLRRNLASEMLHPSKIKSFSEIRKWVLQTLINRL-----KTASESEDSVTVVSHFRY 183
WR +R+ E+ K+KSF IR+ + L+ ++ +T + S D V+ F
Sbjct: 119 DWREMRKITTLELFSVKKLKSFRYIREEESELLVKKISKSVDETQNSSVDLRKVLFSFTA 178
Query: 184 AVFCLLVFMCFGERVNDEKIND--IERVQRTILLNFSRFNILNFWPK---VTRIL----- 233
++ C L F G+ + D +E + N F +F+P + RI
Sbjct: 179 SIICRLAF---GQNFHQCDFVDASLEELVLESEANLGTFAFADFFPGGWLIDRISGQHSR 235
Query: 234 LRKRWDELAKLLKDQEDVLLPLIRARKHVKESRLNNINTVVSYVDTLMELELPQEKRKLS 293
+ K + +L K D H+K + + + +VS + ++ + K++
Sbjct: 236 VNKAFYKLTNFYKHVID---------DHLKTGQPQDHSDIVSVMLDMINKPTKADSFKVT 286
Query: 294 EDEMISLCSEFLNGGTDTTSTALQWIMANLVKYPDVQGKLVEEITEVMGGDDAGEKEEVK 353
D + + S+ G + + + W + L ++P V KL EEI ++G + KE +
Sbjct: 287 YDHLKGVMSDIFLAGVNGGANTMIWTLTELSRHPRVMKKLQEEIRAMLGPN----KERIT 342
Query: 354 EEDLHKLRYLKCVVLEGLRRHPPGHFVLPHAVSEDVVLNGYLVPKDGTVNFMVAEMGLDP 413
EEDL K+ YLK V++E R HPP +LP D+ + GY +PK+ + +G DP
Sbjct: 343 EEDLEKVEYLKLVMVETFRLHPPAPLLLPRLTMSDIKIQGYNIPKNTMIQINTYAIGRDP 402
Query: 414 RVWEDPMEFKPERFLKDETFDITGSKEIKMMPFGAGRRICPGYNLALLHLEYFVANLVWN 473
+ W+ P EF PERFL D D G + +++PFGAGRRICPG + +E + NL++
Sbjct: 403 KYWKQPGEFIPERFL-DSPIDYKG-QHFELLPFGAGRRICPGMATGITMVELGLLNLLYF 460
Query: 474 FDWKVPEG---GRVDLSEKQEFTMVMKNPL 500
FDW +P G +D+ E + F + K PL
Sbjct: 461 FDWSLPNGMTIEDIDMEEDEGFAIAKKVPL 490
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.321 0.137 0.412
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 11,886,498
Number of Sequences: 26719
Number of extensions: 528033
Number of successful extensions: 2607
Number of sequences better than 10.0: 258
Number of HSP's better than 10.0 without gapping: 237
Number of HSP's successfully gapped in prelim test: 21
Number of HSP's that attempted gapping in prelim test: 1514
Number of HSP's gapped (non-prelim): 298
length of query: 508
length of database: 11,318,596
effective HSP length: 104
effective length of query: 404
effective length of database: 8,539,820
effective search space: 3450087280
effective search space used: 3450087280
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.9 bits)
S2: 62 (28.5 bits)
Medicago: description of AC130800.11


