

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC128638.4 - phase: 0
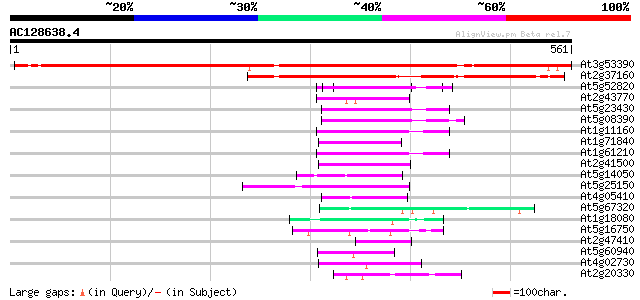
(561 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
At3g53390 putative protein 811 0.0
At2g37160 unknown protein 436 e-122
At5g52820 Notchless protein homolog 54 3e-07
At2g43770 putative splicing factor 50 3e-06
At5g23430 unknown protein 50 3e-06
At5g08390 katanin p80 subunit - like protein 50 4e-06
At1g11160 hypothetical protein 49 1e-05
At1g71840 unknown protein (At1g71840) 48 1e-05
At1g61210 47 3e-05
At2g41500 putative U4/U6 small nuclear ribonucleoprotein 45 8e-05
At5g14050 unknown protein 45 1e-04
At5g25150 transcription initiation factor IID-associated factor-... 45 1e-04
At4g05410 U3 snoRNP-associated -like protein 44 2e-04
At5g67320 unknown protein 44 3e-04
At1g18080 putative guanine nucleotide-binding protein beta sub... 44 3e-04
At5g16750 WD40-repeat protein 43 4e-04
At2g47410 putative WD-40 repeat protein 43 5e-04
At5g60940 cleavage stimulation factor 50K chain 42 7e-04
At4g02730 putative WD-repeat protein 42 7e-04
At2g20330 putative WD-40 repeat protein 42 7e-04
>At3g53390 putative protein
Length = 556
Score = 811 bits (2095), Expect = 0.0
Identities = 411/568 (72%), Positives = 469/568 (82%), Gaps = 26/568 (4%)
Query: 5 TTNGNMTAASSSSSSGPTTTTQSQQQGLKTYFKTPEGRYKLQFDKTHPSGLLQFNHGKTV 64
T+NG ++A SSSSS P QS G+KTYFKTPEG+YKL ++KTH SGLL + HGKTV
Sbjct: 4 TSNGMISAPSSSSS--PAANPQSP--GIKTYFKTPEGKYKLHYEKTHSSGLLHYAHGKTV 59
Query: 65 SMVTLAHLKEKPAPLTPTASSSSFSASSGVRSAAARLLGGSNGNRALSFVGGNGSSKSNG 124
+ VT+A LKE+ AP TPT +SS +SASSG RSA ARLLG NG+RALSFVGGNG KS
Sbjct: 60 TQVTIAQLKERAAPSTPTGTSSGYSASSGFRSATARLLGTGNGSRALSFVGGNGGGKSVS 119
Query: 125 GASRI-GSIGSSSLSSSVANPNFDGKGSYLVFNAGDAILISDLNSQDKDPIKSIHFSNSN 183
+SRI GS +S+ ++S+ N NFDGKG+YLVFN DAI I DLNSQ+KDP+KSIHFSNSN
Sbjct: 120 TSSRISGSFAASNSNTSMTNTNFDGKGTYLVFNVSDAIFICDLNSQEKDPVKSIHFSNSN 179
Query: 184 PVCHAFDQDAKDGHDLLIGLFSGDVYSVSLRQQLQDVGKKIVGAHHYNKDGILNN--SRC 241
P+CHAFD DAKDGHDLLIGL SGDVY+VSLRQQLQDV KK+VGA HYNKDG +NN RC
Sbjct: 180 PMCHAFDPDAKDGHDLLIGLNSGDVYTVSLRQQLQDVSKKLVGALHYNKDGSVNNRQDRC 239
Query: 242 TCISWVPGGDGAFVVAHADGNLYVYEKNKDGAGESSFPILKDQTLFSVAHARYSKSNPIA 301
T ISWVPGGDGAFVVAHADGNLY NKDGA ES+FP ++D T FSV A+YSKSNP+A
Sbjct: 240 TSISWVPGGDGAFVVAHADGNLY----NKDGATESTFPAIRDPTQFSVDKAKYSKSNPVA 295
Query: 302 RWHICQGSINSISFSADGAYLATVGRDGYLRVFDYTKEHLVCGGKSYYGGLLCCAWSMDG 361
RWHICQGSINSI+FS DGA+LATVGRDGYLR+FD+ + LVCGGKSYYG LLCC+WSMDG
Sbjct: 296 RWHICQGSINSIAFSNDGAHLATVGRDGYLRIFDFLTQKLVCGGKSYYGALLCCSWSMDG 355
Query: 362 KYILTGGEDDLVQVWSMEDRKVVAWGEGHNSWVSGVAFDSYWSSPNSNDNGETITYRFGS 421
KYILTGGEDDLVQVWSMEDRKVVAWGEGHNSWVSGVAFDS WSSPNS+ +GE + YRFGS
Sbjct: 356 KYILTGGEDDLVQVWSMEDRKVVAWGEGHNSWVSGVAFDSNWSSPNSDGSGEHVMYRFGS 415
Query: 422 VGQDTQLLLWELEMDEIVVPLRRGPPGGSPTFGSPTFSAGSQSSHWDNAVPLGTLQPAPS 481
VGQDTQLLLW+LEMDEIVVPLRR P GSPT+S GS S+HWDN +P+GTLQPAP
Sbjct: 416 VGQDTQLLLWDLEMDEIVVPLRRAPG------GSPTYSTGS-SAHWDNVIPMGTLQPAPC 468
Query: 482 MRDVPKISPLVAHRVHTEPLSSLIFTQESVLTACREGHIKIWTRPGVAESQPSNSE---- 537
RDVPK+SP++AHRVHTEPLS L+FTQESV+TACREGHIKIWTRP +E+Q ++SE
Sbjct: 469 KRDVPKLSPIIAHRVHTEPLSGLMFTQESVVTACREGHIKIWTRPSESETQTNSSEANPT 528
Query: 538 -TLLATSLKE--KPSLSSK-ISNSIYKQ 561
LL+TS + K SLSSK IS S +KQ
Sbjct: 529 TALLSTSFTKDNKASLSSKIISGSTFKQ 556
>At2g37160 unknown protein
Length = 444
Score = 436 bits (1121), Expect = e-122
Identities = 220/317 (69%), Positives = 251/317 (78%), Gaps = 36/317 (11%)
Query: 238 NSRCTCISWVPGGDGAFVVAHADGNLYVYEKNKDGAGESSFPILKDQTLFSVAHARYSKS 297
NSRCT I+WVPGGDG+FV AHADGNLY NK+GA +SSF ++D T FSV A+YSKS
Sbjct: 150 NSRCTSIAWVPGGDGSFVAAHADGNLY----NKEGATDSSFSAIRDPTQFSVDKAKYSKS 205
Query: 298 NPIARWHICQGSINSISFSADGAYLATVGRDGYLRVFDYTKEHLVCGGKSYYGGLLCCAW 357
NP+ARWHI QG+INSI+FS DGAYLATVGRDGYLR+FD++ + LVCG KSYYG LLCCAW
Sbjct: 206 NPVARWHIGQGAINSIAFSNDGAYLATVGRDGYLRIFDFSTQKLVCGVKSYYGALLCCAW 265
Query: 358 SMDGKYILTGGEDDLVQVWSMEDRKVVAWGEGHNSWVSGVAFDSYWSSPNSNDNGETITY 417
SMDGKY+LTGGEDDLVQVWSMEDRKVVAW EG +GE + Y
Sbjct: 266 SMDGKYLLTGGEDDLVQVWSMEDRKVVAW-EG---------------------SGENVMY 303
Query: 418 RFGSVGQDTQLLLWELEMDEIVVPLRRGPPGGSPTFGSPTFSAGSQSSHWDNAVPLGTLQ 477
RFGSVGQDTQLLLW+LEMDEIVVPLRR PPG GSPT+S GSQS+HWDN VP+GTLQ
Sbjct: 304 RFGSVGQDTQLLLWDLEMDEIVVPLRR-PPG-----GSPTYSTGSQSAHWDNIVPMGTLQ 357
Query: 478 PAPSMRDVPKISPLVAHRVHTEPLSSLIFTQESVLTACREGHIKIWTRPGVAESQPSNSE 537
PAP MRDVPK+SP+VAHRVHTEPLS LIFTQES++TACREGH+KIWTRP ++QPS+S
Sbjct: 358 PAPCMRDVPKLSPVVAHRVHTEPLSGLIFTQESLITACREGHLKIWTRP---DTQPSSSS 414
Query: 538 TLLATSLKEKPSLSSKI 554
KPSL+SK+
Sbjct: 415 E-ATNPTTSKPSLTSKV 430
>At5g52820 Notchless protein homolog
Length = 473
Score = 53.5 bits (127), Expect = 3e-07
Identities = 33/132 (25%), Positives = 67/132 (50%), Gaps = 11/132 (8%)
Query: 313 ISFSADGAYLATVGRDGYLRVFDYTKEHLVCGGKSYYGGLLCCAWSMDGKYILTGGEDDL 372
+SFS DG LA+ D +R++D E + K + +L AWS DGK++++G +
Sbjct: 115 VSFSPDGKQLASGSGDTTVRLWDLYTETPLFTCKGHKNWVLTVAWSPDGKHLVSGSKSGE 174
Query: 373 VQVWSMEDRKVVAWG-EGHNSWVSGVAFDS-YWSSPNSNDNGETITYRFGSVGQDTQLLL 430
+ W+ + ++ GH W++G++++ + SSP RF + +D +
Sbjct: 175 ICCWNPKKGELEGSPLTGHKKWITGISWEPVHLSSP---------CRRFVTSSKDGDARI 225
Query: 431 WELEMDEIVVPL 442
W++ + + ++ L
Sbjct: 226 WDITLKKSIICL 237
Score = 49.7 bits (117), Expect = 4e-06
Identities = 33/126 (26%), Positives = 60/126 (47%), Gaps = 13/126 (10%)
Query: 307 QGSINSISFSADGAYLATVGRDGYLRVFDYTKEHLVCGGKSYYGGLLCCAWSMDGKYILT 366
Q +N + FS DG ++A+ D +R+++ V + + G + +WS D + +L+
Sbjct: 360 QQLVNHVYFSPDGKWIASASFDKSVRLWNGITGQFVTVFRGHVGPVYQVSWSADSRLLLS 419
Query: 367 GGEDDLVQVWSMEDRKVVAWGEGHNSWVSGVAFDSYWSSPNSNDNGETITYRFGSVGQDT 426
G +D +++W + +K+ GH V V WS +GE + S G+D
Sbjct: 420 GSKDSTLKIWEIRTKKLKQDLPGHADEVFAVD----WS-----PDGEKVV----SGGKDR 466
Query: 427 QLLLWE 432
L LW+
Sbjct: 467 VLKLWK 472
Score = 42.7 bits (99), Expect = 5e-04
Identities = 20/78 (25%), Positives = 38/78 (48%), Gaps = 1/78 (1%)
Query: 324 TVGRDGYLRVFDYTKEHLVCGGKSYYGGLLCCAWSMDGKYILTGGEDDLVQVWSMEDRKV 383
T +DG R++D T + + + + C W DG I TG +D +++W K+
Sbjct: 216 TSSKDGDARIWDITLKKSIICLSGHTLAVTCVKWGGDG-IIYTGSQDCTIKMWETTQGKL 274
Query: 384 VAWGEGHNSWVSGVAFDS 401
+ +GH W++ +A +
Sbjct: 275 IRELKGHGHWINSLALST 292
Score = 37.7 bits (86), Expect = 0.017
Identities = 16/48 (33%), Positives = 28/48 (58%)
Query: 352 LLCCAWSMDGKYILTGGEDDLVQVWSMEDRKVVAWGEGHNSWVSGVAF 399
+LC ++S DGK + +G D V++W + + +GH +WV VA+
Sbjct: 112 VLCVSFSPDGKQLASGSGDTTVRLWDLYTETPLFTCKGHKNWVLTVAW 159
>At2g43770 putative splicing factor
Length = 343
Score = 50.4 bits (119), Expect = 3e-06
Identities = 31/101 (30%), Positives = 46/101 (44%), Gaps = 8/101 (7%)
Query: 307 QGSINSISFSADGAYLATVGRDGYLRVFD---YTKEHLVCG-----GKSYYGGLLCCAWS 358
Q +I +S S DG+YL T G D L V+D Y ++ ++ LL C+WS
Sbjct: 222 QDTITGMSLSPDGSYLLTNGMDNKLCVWDMRPYAPQNRCVKIFEGHQHNFEKNLLKCSWS 281
Query: 359 MDGKYILTGGEDDLVQVWSMEDRKVVAWGEGHNSWVSGVAF 399
DG + G D +V +W R+ + GH V+ F
Sbjct: 282 PDGTKVTAGSSDRMVHIWDTTSRRTIYKLPGHTGSVNECVF 322
Score = 37.4 bits (85), Expect = 0.022
Identities = 25/85 (29%), Positives = 39/85 (45%), Gaps = 4/85 (4%)
Query: 310 INSISFSADGAYLATVGRDGYLRVFDYTKEHLVCGGKSYYGGLLCCAWSMDGKYILTGGE 369
I ++SFS + T G D ++V+D K + + + + S DG Y+LT G
Sbjct: 183 ITAVSFSDAADKIFTGGVDNDVKVWDLRKGEATMTLEGHQDTITGMSLSPDGSYLLTNGM 242
Query: 370 DDLVQVWSME----DRKVVAWGEGH 390
D+ + VW M + V EGH
Sbjct: 243 DNKLCVWDMRPYAPQNRCVKIFEGH 267
Score = 33.9 bits (76), Expect = 0.25
Identities = 22/90 (24%), Positives = 42/90 (46%), Gaps = 5/90 (5%)
Query: 309 SINSISFSADGAYLATVGRDGYL---RVFDYTKEHLVCGGKSYYGGLLCCAWSMDGKYIL 365
++ ++ F+ G +A+ D + RV K +V K + +L W+ DG I+
Sbjct: 55 AVYTMKFNPAGTLIASGSHDREIFLWRVHGDCKNFMVL--KGHKNAILDLHWTSDGSQIV 112
Query: 366 TGGEDDLVQVWSMEDRKVVAWGEGHNSWVS 395
+ D V+ W +E K + H+S+V+
Sbjct: 113 SASPDKTVRAWDVETGKQIKKMAEHSSFVN 142
Score = 32.0 bits (71), Expect = 0.94
Identities = 19/70 (27%), Positives = 32/70 (45%), Gaps = 13/70 (18%)
Query: 364 ILTGGEDDLVQVWSMEDRKVVAWGEGHNSWVSGVAFDSYWSSPNSNDNGETITYRFGSVG 423
I TGG D+ V+VW + + EGH ++G++ S D +T G
Sbjct: 195 IFTGGVDNDVKVWDLRKGEATMTLEGHQDTITGMSL--------SPDGSYLLTN-----G 241
Query: 424 QDTQLLLWEL 433
D +L +W++
Sbjct: 242 MDNKLCVWDM 251
>At5g23430 unknown protein
Length = 837
Score = 50.1 bits (118), Expect = 3e-06
Identities = 27/128 (21%), Positives = 60/128 (46%), Gaps = 13/128 (10%)
Query: 312 SISFSADGAYLATVGRDGYLRVFDYTKEHLVCGGKSYYGGLLCCAWSMDGKYILTGGEDD 371
S+ F G + A+ D L+++D K+ + K + G+ ++ DG+++++GGED+
Sbjct: 106 SVDFHPFGEFFASGSLDTNLKIWDIRKKGCIHTYKGHTRGVNVLRFTPDGRWVVSGGEDN 165
Query: 372 LVQVWSMEDRKVVAWGEGHNSWVSGVAFDSYWSSPNSNDNGETITYRFGSVGQDTQLLLW 431
+V+VW + K++ + H + + F + + + D + W
Sbjct: 166 IVKVWDLTAGKLLTEFKSHEGQIQSLDFHPH-------------EFLLATGSADRTVKFW 212
Query: 432 ELEMDEIV 439
+LE E++
Sbjct: 213 DLETFELI 220
Score = 40.8 bits (94), Expect = 0.002
Identities = 22/91 (24%), Positives = 45/91 (49%)
Query: 310 INSISFSADGAYLATVGRDGYLRVFDYTKEHLVCGGKSYYGGLLCCAWSMDGKYILTGGE 369
+N + F+ DG ++ + G D ++V+D T L+ KS+ G + + + TG
Sbjct: 146 VNVLRFTPDGRWVVSGGEDNIVKVWDLTAGKLLTEFKSHEGQIQSLDFHPHEFLLATGSA 205
Query: 370 DDLVQVWSMEDRKVVAWGEGHNSWVSGVAFD 400
D V+ W +E +++ G + V ++F+
Sbjct: 206 DRTVKFWDLETFELIGSGGPETAGVRCLSFN 236
Score = 35.8 bits (81), Expect = 0.065
Identities = 35/164 (21%), Positives = 66/164 (39%), Gaps = 25/164 (15%)
Query: 362 KYILTGGEDDLVQVWSMEDRKVVAWGEGHNSWVSGVAFDSYWSSPNSNDNGETITYRFGS 421
+ ++TGGED V +W++ + GH+S + V FD+ E + +
Sbjct: 30 RVLVTGGEDHKVNLWAIGKPNAILSLYGHSSGIDSVTFDA----------SEVLVAAGAA 79
Query: 422 VGQDTQLLLWELEMDEIVVPLRRGPPGGSPTFGSPTFSAGSQSSHWDNAVPLGTLQPAPS 481
G + LW+LE +IV L G + + G+L
Sbjct: 80 SG---TIKLWDLEEAKIVRTLT----------GHRSNCISVDFHPFGEFFASGSLDTNLK 126
Query: 482 MRDVPKISPLVAHRVHTEPLSSLIFTQES--VLTACREGHIKIW 523
+ D+ K + ++ HT ++ L FT + V++ + +K+W
Sbjct: 127 IWDIRKKGCIHTYKGHTRGVNVLRFTPDGRWVVSGGEDNIVKVW 170
Score = 35.0 bits (79), Expect = 0.11
Identities = 21/73 (28%), Positives = 37/73 (49%), Gaps = 1/73 (1%)
Query: 307 QGSINSISFSADGAYLATVGRDGYLRVFDYTKEHLVCGGKSYYGGLLCCAWSMDGKYILT 366
+G I S+ F LAT D ++ +D L+ G G+ C +++ DGK +L
Sbjct: 185 EGQIQSLDFHPHEFLLATGSADRTVKFWDLETFELIGSGGPETAGVRCLSFNPDGKTVLC 244
Query: 367 GGEDDLVQVWSME 379
G ++ L +++S E
Sbjct: 245 GLQESL-KIFSWE 256
Score = 32.0 bits (71), Expect = 0.94
Identities = 24/112 (21%), Positives = 43/112 (37%), Gaps = 13/112 (11%)
Query: 322 LATVGRDGYLRVFDYTKEHLVCGGKSYYGGLLCCAWSMDGKYILTGGEDDLVQVWSMEDR 381
L T G D + ++ K + + + G+ + + G +++W +E+
Sbjct: 32 LVTGGEDHKVNLWAIGKPNAILSLYGHSSGIDSVTFDASEVLVAAGAASGTIKLWDLEEA 91
Query: 382 KVVAWGEGHNSWVSGVAFDSYWSSPNSNDNGETITYRFGSVGQDTQLLLWEL 433
K+V GH S V F + GE F S DT L +W++
Sbjct: 92 KIVRTLTGHRSNCISVDFHPF---------GEF----FASGSLDTNLKIWDI 130
>At5g08390 katanin p80 subunit - like protein
Length = 823
Score = 49.7 bits (117), Expect = 4e-06
Identities = 32/143 (22%), Positives = 66/143 (45%), Gaps = 19/143 (13%)
Query: 312 SISFSADGAYLATVGRDGYLRVFDYTKEHLVCGGKSYYGGLLCCAWSMDGKYILTGGEDD 371
S++F G + A+ D L+++D K+ + K + G+ ++ DG++I++GGED+
Sbjct: 199 SVNFHPFGEFFASGSLDTNLKIWDIRKKGCIHTYKGHTRGVNVLRFTPDGRWIVSGGEDN 258
Query: 372 LVQVWSMEDRKVVAWGEGHNSWVSGVAFDSYWSSPNSNDNGETITYRFGSVGQDTQLLLW 431
+V+VW + K++ + H + + F + + + D + W
Sbjct: 259 VVKVWDLTAGKLLHEFKSHEGKIQSLDFHPH-------------EFLLATGSADKTVKFW 305
Query: 432 ELEMDEIVVPLRRGPPGGSPTFG 454
+LE E++ GG+ T G
Sbjct: 306 DLETFELI------GSGGTETTG 322
Score = 40.4 bits (93), Expect = 0.003
Identities = 22/91 (24%), Positives = 44/91 (48%)
Query: 310 INSISFSADGAYLATVGRDGYLRVFDYTKEHLVCGGKSYYGGLLCCAWSMDGKYILTGGE 369
+N + F+ DG ++ + G D ++V+D T L+ KS+ G + + + TG
Sbjct: 239 VNVLRFTPDGRWIVSGGEDNVVKVWDLTAGKLLHEFKSHEGKIQSLDFHPHEFLLATGSA 298
Query: 370 DDLVQVWSMEDRKVVAWGEGHNSWVSGVAFD 400
D V+ W +E +++ G + V + F+
Sbjct: 299 DKTVKFWDLETFELIGSGGTETTGVRCLTFN 329
Score = 34.7 bits (78), Expect = 0.14
Identities = 33/164 (20%), Positives = 66/164 (40%), Gaps = 25/164 (15%)
Query: 362 KYILTGGEDDLVQVWSMEDRKVVAWGEGHNSWVSGVAFDSYWSSPNSNDNGETITYRFGS 421
+ ++TGGED V +W++ + GH+S + V FD+ E + +
Sbjct: 123 RVLVTGGEDHKVNLWAIGKPNAILSLYGHSSGIDSVTFDA----------SEGLVAAGAA 172
Query: 422 VGQDTQLLLWELEMDEIVVPLRRGPPGGSPTFGSPTFSAGSQSSHWDNAVPLGTLQPAPS 481
G + LW+LE ++V L G + + G+L
Sbjct: 173 SG---TIKLWDLEEAKVVRTLT----------GHRSNCVSVNFHPFGEFFASGSLDTNLK 219
Query: 482 MRDVPKISPLVAHRVHTEPLSSLIFTQES--VLTACREGHIKIW 523
+ D+ K + ++ HT ++ L FT + +++ + +K+W
Sbjct: 220 IWDIRKKGCIHTYKGHTRGVNVLRFTPDGRWIVSGGEDNVVKVW 263
Score = 34.7 bits (78), Expect = 0.14
Identities = 26/103 (25%), Positives = 42/103 (40%), Gaps = 2/103 (1%)
Query: 307 QGSINSISFSADGAYLATVGRDGYLRVFDYTKEHLVCGGKSYYGGLLCCAWSMDGKYILT 366
+G I S+ F LAT D ++ +D L+ G + G+ C ++ DGK +L
Sbjct: 278 EGKIQSLDFHPHEFLLATGSADKTVKFWDLETFELIGSGGTETTGVRCLTFNPDGKSVLC 337
Query: 367 GGEDDLVQVWSMEDRKVVAWGEGHNSWVSGVAFDSYWSSPNSN 409
G ++ L + M H SG D + NS+
Sbjct: 338 GLQESLKRTEPMSGG--ATQSNSHPEKTSGSGRDQAGLNDNSS 378
Score = 32.3 bits (72), Expect = 0.72
Identities = 25/112 (22%), Positives = 43/112 (38%), Gaps = 13/112 (11%)
Query: 322 LATVGRDGYLRVFDYTKEHLVCGGKSYYGGLLCCAWSMDGKYILTGGEDDLVQVWSMEDR 381
L T G D + ++ K + + + G+ + + G +++W +E+
Sbjct: 125 LVTGGEDHKVNLWAIGKPNAILSLYGHSSGIDSVTFDASEGLVAAGAASGTIKLWDLEEA 184
Query: 382 KVVAWGEGHNSWVSGVAFDSYWSSPNSNDNGETITYRFGSVGQDTQLLLWEL 433
KVV GH S V F + GE F S DT L +W++
Sbjct: 185 KVVRTLTGHRSNCVSVNFHPF---------GEF----FASGSLDTNLKIWDI 223
>At1g11160 hypothetical protein
Length = 974
Score = 48.5 bits (114), Expect = 1e-05
Identities = 29/133 (21%), Positives = 63/133 (46%), Gaps = 13/133 (9%)
Query: 307 QGSINSISFSADGAYLATVGRDGYLRVFDYTKEHLVCGGKSYYGGLLCCAWSMDGKYILT 366
+ + +++ F G +LA+ D LRV+D K+ + K + G+ +S DG+++++
Sbjct: 49 RSNCSAVEFHPFGEFLASGSSDTNLRVWDTRKKGCIQTYKGHTRGISTIEFSPDGRWVVS 108
Query: 367 GGEDDLVQVWSMEDRKVVAWGEGHNSWVSGVAFDSYWSSPNSNDNGETITYRFGSVGQDT 426
GG D++V+VW + K++ + H + + F + + + D
Sbjct: 109 GGLDNVVKVWDLTAGKLLHEFKCHEGPIRSLDF-------------HPLEFLLATGSADR 155
Query: 427 QLLLWELEMDEIV 439
+ W+LE E++
Sbjct: 156 TVKFWDLETFELI 168
Score = 38.5 bits (88), Expect = 0.010
Identities = 35/163 (21%), Positives = 65/163 (39%), Gaps = 27/163 (16%)
Query: 237 NNSRCTCISWVPGGDGAFVVAHADGNLYVYEKNKDGAGESSFPILKDQTLFSVAHARYSK 296
+ S C+ + + P G+ +D NL V++ K G ++ H R
Sbjct: 48 HRSNCSAVEFHPFGE-FLASGSSDTNLRVWDTRKKGCIQTY-----------KGHTR--- 92
Query: 297 SNPIARWHICQGSINSISFSADGAYLATVGRDGYLRVFDYTKEHLVCGGKSYYGGLLCCA 356
I++I FS DG ++ + G D ++V+D T L+ K + G +
Sbjct: 93 ------------GISTIEFSPDGRWVVSGGLDNVVKVWDLTAGKLLHEFKCHEGPIRSLD 140
Query: 357 WSMDGKYILTGGEDDLVQVWSMEDRKVVAWGEGHNSWVSGVAF 399
+ + TG D V+ W +E +++ + V +AF
Sbjct: 141 FHPLEFLLATGSADRTVKFWDLETFELIGTTRPEATGVRAIAF 183
Score = 33.9 bits (76), Expect = 0.25
Identities = 39/170 (22%), Positives = 62/170 (35%), Gaps = 25/170 (14%)
Query: 356 AWSMDGKYILTGGEDDLVQVWSMEDRKVVAWGEGHNSWVSGVAFDSYWSSPNSNDNGETI 415
A++ + +L G ++++W +E+ K+V GH S S V F + GE +
Sbjct: 14 AFNSEEVLVLAGASSGVIKLWDLEESKMVRAFTGHRSNCSAVEFHPF---------GEFL 64
Query: 416 TYRFGSVGQDTQLLLWELEMDEIVVPLRRGPPGGSPTFGSPTFSAGSQSSHWDNAVPLGT 475
S DT L +W+ + + G S SP W V G
Sbjct: 65 ----ASGSSDTNLRVWDTRKKGCIQTYKGHTRGISTIEFSP-------DGRW---VVSGG 110
Query: 476 LQPAPSMRDVPKISPLVAHRVHTEPLSSLIFTQESVL--TACREGHIKIW 523
L + D+ L + H P+ SL F L T + +K W
Sbjct: 111 LDNVVKVWDLTAGKLLHEFKCHEGPIRSLDFHPLEFLLATGSADRTVKFW 160
>At1g71840 unknown protein (At1g71840)
Length = 407
Score = 48.1 bits (113), Expect = 1e-05
Identities = 25/83 (30%), Positives = 41/83 (49%)
Query: 309 SINSISFSADGAYLATVGRDGYLRVFDYTKEHLVCGGKSYYGGLLCCAWSMDGKYILTGG 368
S++ ++FS DG LA+ G DG +++FD + L C G+ W G +L G
Sbjct: 115 SVSCLAFSYDGQLLASGGLDGVVQIFDASSGTLKCVLDGPGAGIEWVRWHPRGHIVLAGS 174
Query: 369 EDDLVQVWSMEDRKVVAWGEGHN 391
ED + +W+ + + GHN
Sbjct: 175 EDCSLWMWNADKEAYLNMFSGHN 197
Score = 35.8 bits (81), Expect = 0.065
Identities = 33/123 (26%), Positives = 55/123 (43%), Gaps = 13/123 (10%)
Query: 315 FSADGAYLATVGRDGYLRVFD---YTKEHLVCGGKSYYGGLLCCAWSMDGKYILTGGEDD 371
F+ DG + T D L V++ H+V G + GL C + + ++G +D
Sbjct: 205 FTPDGKLICTGSDDASLIVWNPKTCESIHIVKGHPYHTEGLTCLDINSNSSLAISGSKDG 264
Query: 372 LVQVWSMEDRKVVAWGEGHNSWVSGVAFDSYWSSPNSNDNGETITYRFGSVGQDTQLLLW 431
V + ++ KVV+ H V V F SP+S TI + G D +L++W
Sbjct: 265 SVHIVNIVTGKVVSSLNSHTDSVECVKF-----SPSS----ATIPLA-ATGGMDKKLIIW 314
Query: 432 ELE 434
+L+
Sbjct: 315 DLQ 317
Score = 30.4 bits (67), Expect = 2.7
Identities = 23/95 (24%), Positives = 38/95 (39%), Gaps = 8/95 (8%)
Query: 309 SINSISFSADGAYL---ATVGRDGYLRVFD--YTKEHLVCGGKSYYGGLLCCAWSMDGKY 363
S+ + FS A + AT G D L ++D ++ +C + G+ W KY
Sbjct: 286 SVECVKFSPSSATIPLAATGGMDKKLIIWDLQHSTPRFIC---EHEEGVTSLTWIGTSKY 342
Query: 364 ILTGGEDDLVQVWSMEDRKVVAWGEGHNSWVSGVA 398
+ TG + V +W V GH V ++
Sbjct: 343 LATGCANGTVSIWDSLLGNCVHTYHGHQDAVQAIS 377
>At1g61210
Length = 282
Score = 47.0 bits (110), Expect = 3e-05
Identities = 26/133 (19%), Positives = 63/133 (46%), Gaps = 13/133 (9%)
Query: 307 QGSINSISFSADGAYLATVGRDGYLRVFDYTKEHLVCGGKSYYGGLLCCAWSMDGKYILT 366
+ + +++ F G +LA+ D L+++D K+ + K + G+ ++ DG+++++
Sbjct: 130 RSNCSAVEFHPFGEFLASGSSDANLKIWDIRKKGCIQTYKGHSRGISTIRFTPDGRWVVS 189
Query: 367 GGEDDLVQVWSMEDRKVVAWGEGHNSWVSGVAFDSYWSSPNSNDNGETITYRFGSVGQDT 426
GG D++V+VW + K++ + H + + F + + + D
Sbjct: 190 GGLDNVVKVWDLTAGKLLHEFKFHEGPIRSLDF-------------HPLEFLLATGSADR 236
Query: 427 QLLLWELEMDEIV 439
+ W+LE E++
Sbjct: 237 TVKFWDLETFELI 249
Score = 31.2 bits (69), Expect = 1.6
Identities = 36/162 (22%), Positives = 58/162 (35%), Gaps = 25/162 (15%)
Query: 364 ILTGGEDDLVQVWSMEDRKVVAWGEGHNSWVSGVAFDSYWSSPNSNDNGETITYRFGSVG 423
+L G ++++W +E+ K+V GH S S V F + GE + S
Sbjct: 103 VLAGASSGVIKLWDVEEAKMVRAFTGHRSNCSAVEFHPF---------GEFL----ASGS 149
Query: 424 QDTQLLLWELEMDEIVVPLRRGPPGGSPTFGSPTFSAGSQSSHWDNAVPLGTLQPAPSMR 483
D L +W++ + + G S +P W V G L +
Sbjct: 150 SDANLKIWDIRKKGCIQTYKGHSRGISTIRFTP-------DGRW---VVSGGLDNVVKVW 199
Query: 484 DVPKISPLVAHRVHTEPLSSLIFTQESVL--TACREGHIKIW 523
D+ L + H P+ SL F L T + +K W
Sbjct: 200 DLTAGKLLHEFKFHEGPIRSLDFHPLEFLLATGSADRTVKFW 241
>At2g41500 putative U4/U6 small nuclear ribonucleoprotein
Length = 554
Score = 45.4 bits (106), Expect = 8e-05
Identities = 25/92 (27%), Positives = 46/92 (49%)
Query: 309 SINSISFSADGAYLATVGRDGYLRVFDYTKEHLVCGGKSYYGGLLCCAWSMDGKYILTGG 368
S+ I+F DGA A+ G D RV+D + + + + +S +G ++ +GG
Sbjct: 383 SVYGIAFQQDGALAASCGLDSLARVWDLRTGRSILVFQGHIKPVFSVNFSPNGYHLASGG 442
Query: 369 EDDLVQVWSMEDRKVVAWGEGHNSWVSGVAFD 400
ED+ ++W + RK + H + VS V ++
Sbjct: 443 EDNQCRIWDLRMRKSLYIIPAHANLVSQVKYE 474
Score = 35.0 bits (79), Expect = 0.11
Identities = 57/262 (21%), Positives = 93/262 (34%), Gaps = 50/262 (19%)
Query: 313 ISFSADGAYLATVGRDGYLRVFDYTKEHLVCGGKSYYGGLLCCAWSMDGKYILTGGEDDL 372
+ FS LAT D +++ T L+ + + L A+ GKY+ T D
Sbjct: 304 VVFSPVDDCLATASADRTAKLWK-TDGTLLQTFEGHLDRLARVAFHPSGKYLGTTSYDKT 362
Query: 373 VQVWSMEDRKVVAWGEGHNSWVSGVAFDS---------------YWS------------- 404
++W + + EGH+ V G+AF W
Sbjct: 363 WRLWDINTGAELLLQEGHSRSVYGIAFQQDGALAASCGLDSLARVWDLRTGRSILVFQGH 422
Query: 405 -----SPNSNDNGETITYRFGSVGQDTQLLLWELEMDEIVVPLRRGPPGGSPTFGSPTFS 459
S N + NG Y S G+D Q +W+L M + + + S P
Sbjct: 423 IKPVFSVNFSPNG----YHLASGGEDNQCRIWDLRMRKSLYIIPAHANLVSQVKYEPQEG 478
Query: 460 AGSQSSHWDNAVPLGTLQPAPSMRDVPKISPLVAHRVHTEPLSSLIFTQES--VLTACRE 517
++ +D V + S RD + L H ++SL T +S + T +
Sbjct: 479 YFLATASYDMKVNIW------SGRDFSLVKSLAGHE---SKVASLDITADSSCIATVSHD 529
Query: 518 GHIKIWTRPGVAESQPSNSETL 539
IK+WT G + + ET+
Sbjct: 530 RTIKLWTSSG-NDDEDEEKETM 550
Score = 33.9 bits (76), Expect = 0.25
Identities = 39/177 (22%), Positives = 78/177 (44%), Gaps = 8/177 (4%)
Query: 262 NLYVYEKNKDGAGESSFPILKDQTLFSVAHARYSKSNPIARWHICQGSINSISFSADGAY 321
++Y +DGA +S + +L V R +S + + HI + S++FS +G +
Sbjct: 383 SVYGIAFQQDGALAASCGL---DSLARVWDLRTGRSILVFQGHI--KPVFSVNFSPNGYH 437
Query: 322 LATVGRDGYLRVFDYTKEHLVCGGKSYYGGLLCCAWS-MDGKYILTGGEDDLVQVWSMED 380
LA+ G D R++D + ++ + + +G ++ T D V +WS D
Sbjct: 438 LASGGEDNQCRIWDLRMRKSLYIIPAHANLVSQVKYEPQEGYFLATASYDMKVNIWSGRD 497
Query: 381 RKVVAWGEGHNSWVSG--VAFDSYWSSPNSNDNGETITYRFGSVGQDTQLLLWELEM 435
+V GH S V+ + DS + S+D + G+ +D + ++++
Sbjct: 498 FSLVKSLAGHESKVASLDITADSSCIATVSHDRTIKLWTSSGNDDEDEEKETMDIDL 554
>At5g14050 unknown protein
Length = 546
Score = 45.1 bits (105), Expect = 1e-04
Identities = 30/106 (28%), Positives = 48/106 (44%), Gaps = 3/106 (2%)
Query: 287 FSVAHARYSKSNPIARWHICQGSINSISFSADGAYLATVGRDGYLRVFDYTKEHLVCGGK 346
F + A++ K P+ + S+ S D +A VG +GY+ + TK + G
Sbjct: 315 FDLEKAKFDKIGPLVGRE--EKSLEYFEVSQDSNTIAFVGNEGYILLVS-TKTKELIGTL 371
Query: 347 SYYGGLLCCAWSMDGKYILTGGEDDLVQVWSMEDRKVVAWGEGHNS 392
G + A+S DGK++L+ G D V VW + K + G S
Sbjct: 372 KMNGSVRSLAFSEDGKHLLSSGGDGQVYVWDLRTMKCLYKGVDEGS 417
>At5g25150 transcription initiation factor IID-associated
factor-like protein
Length = 700
Score = 44.7 bits (104), Expect = 1e-04
Identities = 39/167 (23%), Positives = 70/167 (41%), Gaps = 6/167 (3%)
Query: 233 DGILNNSRCTCISWVPGGDGAFVVAHADGNLYVYEKNKDGAGESSFPILKDQTLFSVAHA 292
D +L++S T I A +V + N V++ G D+T A
Sbjct: 431 DFVLSSSADTTIRLWSTKLNANLVCYKGHNYPVWDAQFSPFGHYFASCSHDRT------A 484
Query: 293 RYSKSNPIARWHICQGSINSISFSADGAYLATVGRDGYLRVFDYTKEHLVCGGKSYYGGL 352
R + I I G ++ + + + Y+AT D +R++D V + +
Sbjct: 485 RIWSMDRIQPLRIMAGHLSDVDWHPNCNYIATGSSDKTVRLWDVQTGECVRIFIGHRSMV 544
Query: 353 LCCAWSMDGKYILTGGEDDLVQVWSMEDRKVVAWGEGHNSWVSGVAF 399
L A S DG+Y+ +G ED + +W + + + GHNS V +++
Sbjct: 545 LSLAMSPDGRYMASGDEDGTIMMWDLSTARCITPLMGHNSCVWSLSY 591
Score = 38.5 bits (88), Expect = 0.010
Identities = 31/128 (24%), Positives = 51/128 (39%), Gaps = 16/128 (12%)
Query: 315 FSADGAYLATVGRDGYLRVFDYTKEHLVCGGKSYYGGLLCCAWSMDGKYILTGGEDDLVQ 374
FS G Y A+ D R++ + + + G L W + YI TG D V+
Sbjct: 468 FSPFGHYFASCSHDRTARIWSMDRIQPL---RIMAGHLSDVDWHPNCNYIATGSSDKTVR 524
Query: 375 VWSMEDRKVVAWGEGHNSWVSGVAFDSYWSSPNSNDNGETITYRFGSVGQDTQLLLWELE 434
+W ++ + V GH S V +A SP+ S +D +++W+L
Sbjct: 525 LWDVQTGECVRIFIGHRSMVLSLAM-----SPDGR--------YMASGDEDGTIMMWDLS 571
Query: 435 MDEIVVPL 442
+ PL
Sbjct: 572 TARCITPL 579
>At4g05410 U3 snoRNP-associated -like protein
Length = 504
Score = 44.3 bits (103), Expect = 2e-04
Identities = 24/87 (27%), Positives = 45/87 (51%), Gaps = 2/87 (2%)
Query: 312 SISFSADGAYLATVGRDGYLRVFDY-TKEHLVCGGKSYYGGLLCCAWSMDGKYILTGGED 370
+++ S+DG YLAT G D ++ ++D T+EH V + + C + + +G D
Sbjct: 227 ALAVSSDGRYLATGGVDRHVHIWDVRTREH-VQAFPGHRNTVSCLCFRYGTSELYSGSFD 285
Query: 371 DLVQVWSMEDRKVVAWGEGHNSWVSGV 397
V+VW++ED+ + GH + +
Sbjct: 286 RTVKVWNVEDKAFITENHGHQGEILAI 312
Score = 38.1 bits (87), Expect = 0.013
Identities = 19/47 (40%), Positives = 26/47 (54%)
Query: 353 LCCAWSMDGKYILTGGEDDLVQVWSMEDRKVVAWGEGHNSWVSGVAF 399
L A S DG+Y+ TGG D V +W + R+ V GH + VS + F
Sbjct: 226 LALAVSSDGRYLATGGVDRHVHIWDVRTREHVQAFPGHRNTVSCLCF 272
>At5g67320 unknown protein
Length = 613
Score = 43.5 bits (101), Expect = 3e-04
Identities = 50/245 (20%), Positives = 88/245 (35%), Gaps = 32/245 (13%)
Query: 310 INSISFSADGAYLATVGRDGYLRVFDYTKEHLVCGGKSYYGGLLCCAWSMDGKYILTGGE 369
+ ++ ++ +G LAT DG R++ E L+ + G + W+ G Y+LTG
Sbjct: 327 VTTLDWNGEGTLLATGSCDGQARIWTLNGE-LISTLSKHKGPIFSLKWNKKGDYLLTGSV 385
Query: 370 DDLVQVWSMEDRKVVAWGEGHN------SWVSGVAFDS-------YWSSPNSNDNGETIT 416
D VW ++ + E H+ W + V+F + Y +T T
Sbjct: 386 DRTAVVWDVKAEEWKQQFEFHSGPTLDVDWRNNVSFATSSTDSMIYLCKIGETRPAKTFT 445
Query: 417 YRFGSV---------------GQDTQLLLWELEMDEIVVPLRRGPPGGSPTFGSPTFSAG 461
G V D+ +W ++ V LR SPT G
Sbjct: 446 GHQGEVNCVKWDPTGSLLASCSDDSTAKIWNIKQSTFVHDLREHTKEIYTIRWSPT-GPG 504
Query: 462 SQSSHWDNAVPLGTLQPAPSMRDVPKISPLVAHRVHTEPLSSLIFTQ--ESVLTACREGH 519
+ + + + + + D L + H EP+ SL F+ E + + +
Sbjct: 505 TNNPNKQLTLASASFDSTVKLWDAELGKMLCSFNGHREPVYSLAFSPNGEYIASGSLDKS 564
Query: 520 IKIWT 524
I IW+
Sbjct: 565 IHIWS 569
Score = 37.4 bits (85), Expect = 0.022
Identities = 20/83 (24%), Positives = 42/83 (50%), Gaps = 5/83 (6%)
Query: 322 LATVGRDGYLRVFDYTKEHLVCGGKSYYGGLLCCAWSMDGKYILTGGEDDLVQVWSMEDR 381
LA+ D ++++D ++C + + A+S +G+YI +G D + +WS+++
Sbjct: 514 LASASFDSTVKLWDAELGKMLCSFNGHREPVYSLAFSPNGEYIASGSLDKSIHIWSIKEG 573
Query: 382 KVVAWGEGHNSWVSGVAFDSYWS 404
K+V G +G F+ W+
Sbjct: 574 KIVKTYTG-----NGGIFEVCWN 591
Score = 29.3 bits (64), Expect = 6.1
Identities = 26/99 (26%), Positives = 38/99 (38%), Gaps = 9/99 (9%)
Query: 355 CAWSMDGKYILTGGEDDLVQVWSMEDRKVVAWGEGHNSWVSGVAFDSYWSSPNSND---- 410
CAWS + +G D ++WS+ + A G N + S+ S D
Sbjct: 271 CAWSPSASLLASGSGDATARIWSIPEGSFKAVHTGRNINALILKHAKGKSNEKSKDVTTL 330
Query: 411 --NGETITYRFGSVGQDTQLLLWELEMDEI-VVPLRRGP 446
NGE GS D Q +W L + I + +GP
Sbjct: 331 DWNGEGTLLATGSC--DGQARIWTLNGELISTLSKHKGP 367
>At1g18080 putative guanine nucleotide-binding protein beta
subunit
Length = 327
Score = 43.5 bits (101), Expect = 3e-04
Identities = 37/157 (23%), Positives = 63/157 (39%), Gaps = 23/157 (14%)
Query: 280 ILKDQTLFSVAHARYSKSNPIARWHICQGSINSISFSADGAYLATVGRDGYLRVFDYTKE 339
+ KD + VA R + + + + S+DG + + DG LR++D
Sbjct: 45 LTKDDKAYGVAQRRLTGHSHF---------VEDVVLSSDGQFALSGSWDGELRLWDLAAG 95
Query: 340 HLVCGGKSYYGGLLCCAWSMDGKYILTGGEDDLVQVWSMEDR---KVVAWGEGHNSWVSG 396
+ +L A+S+D + I++ D +++W+ + GEGH WVS
Sbjct: 96 VSTRRFVGHTKDVLSVAFSLDNRQIVSASRDRTIKLWNTLGECKYTISEGGEGHRDWVSC 155
Query: 397 VAFDSYWSSPNSNDNGETITYRFGSVGQDTQLLLWEL 433
V F SPN T+ S D + +W L
Sbjct: 156 VRF-----SPN------TLQPTIVSASWDKTVKVWNL 181
Score = 37.0 bits (84), Expect = 0.029
Identities = 32/127 (25%), Positives = 61/127 (47%), Gaps = 18/127 (14%)
Query: 312 SISFSADGAYLATVGRDGYLRVFDYTKE---HLVCGGKSYYGGLLCCAWSMDGKY--ILT 366
S++FS D + + RD +++++ E + GG+ + + C +S + I++
Sbjct: 110 SVAFSLDNRQIVSASRDRTIKLWNTLGECKYTISEGGEGHRDWVSCVRFSPNTLQPTIVS 169
Query: 367 GGEDDLVQVWSMEDRKVVAWGEGHNSWVSGVAFDSYWSSPNSNDNGETITYRFGSVGQDT 426
D V+VW++ + K+ + GH +VS VA SP+ + S G+D
Sbjct: 170 ASWDKTVKVWNLSNCKLRSTLAGHTGYVSTVAV-----SPDGS--------LCASGGKDG 216
Query: 427 QLLLWEL 433
+LLW+L
Sbjct: 217 VVLLWDL 223
Score = 32.0 bits (71), Expect = 0.94
Identities = 17/81 (20%), Positives = 39/81 (47%), Gaps = 10/81 (12%)
Query: 308 GSINSISFSADGAYLATVGRDGYLRVFDYTKEHLVCGGKSYY----GGLLCCAWSMDGKY 363
G +++++ S DG+ A+ G+DG + ++D + GK Y ++ +Y
Sbjct: 195 GYVSTVAVSPDGSLCASGGKDGVVLLWDLAE------GKKLYSLEANSVIHALCFSPNRY 248
Query: 364 ILTGGEDDLVQVWSMEDRKVV 384
L + +++W +E + +V
Sbjct: 249 WLCAATEHGIKIWDLESKSIV 269
>At5g16750 WD40-repeat protein
Length = 876
Score = 43.1 bits (100), Expect = 4e-04
Identities = 43/164 (26%), Positives = 72/164 (43%), Gaps = 27/164 (16%)
Query: 283 DQTLFSVAHARYSKS------NPIARWHICQGSINSISFSADGAYLATVGRDGYLRVFDY 336
D+ LFS H+R + I W +G + ++ A G LAT G D + V+D
Sbjct: 72 DKLLFSAGHSRQIRVWDLETLKCIRSWKGHEGPVMGMACHASGGLLATAGADRKVLVWDV 131
Query: 337 TK---EHLVCGGKSYYGGLLCCAWSMDGKYILTGGEDDLVQVWSME----DRKVVAWGEG 389
H G K +L S + +++G +D V+VW + ++K +A E
Sbjct: 132 DGGFCTHYFRGHKGVVSSILFHPDS-NKNILISGSDDATVRVWDLNAKNTEKKCLAIMEK 190
Query: 390 HNSWVSGVAFDSYWSSPNSNDNGETITYRFGSVGQDTQLLLWEL 433
H S V+ +A +++G T+ S G+D + LW+L
Sbjct: 191 HFSAVTSIAL---------SEDGLTLF----SAGRDKVVNLWDL 221
Score = 40.8 bits (94), Expect = 0.002
Identities = 46/210 (21%), Positives = 84/210 (39%), Gaps = 21/210 (10%)
Query: 321 YLATVGRDGYLRVFDYTK---EHLVCGGKSYYGGLLCCAWSMDGKYILTGGEDDLVQVWS 377
+LA +RV+D +++ G K L C S I+TG +D V++W+
Sbjct: 373 FLAVATNLEEVRVYDVATMSCSYVLAGHKEVVLSLDTCVSSSGNVLIVTGSKDKTVRLWN 432
Query: 378 MEDRKVVAWGEGHNSWVSGVAFDSYWSSPNSNDNGETITYRFGSVGQDTQLLLWELE--M 435
+ + G GHN + VAF ++ ++ F S D L +W L+
Sbjct: 433 ATSKSCIGVGTGHNGDILAVAFAK-----------KSFSF-FVSGSGDRTLKVWSLDGIS 480
Query: 436 DEIVVPLRRGPPGGSPTFGSPTFSAGSQSSHWDNAVPLGTLQPAPSMRDVPKISPLVAHR 495
++ P+ S + D+ V G+ S+ +P + +V +
Sbjct: 481 EDSEEPINLKTRSVVAAHDKDINSVAVARN--DSLVCTGSEDRTASIWRLPDLVHVVTLK 538
Query: 496 VHTEPLSSLIFT--QESVLTACREGHIKIW 523
H + S+ F+ + V+TA + +KIW
Sbjct: 539 GHKRRIFSVEFSTVDQCVMTASGDKTVKIW 568
Score = 40.8 bits (94), Expect = 0.002
Identities = 26/103 (25%), Positives = 44/103 (42%), Gaps = 5/103 (4%)
Query: 309 SINSISFSADGAYLATVGRDGYLRVFDYTKEHLVCGGKSYYGGLLCCAWSMDGKYILTGG 368
++ +++ S D L + G +RV+D + K + G ++ A G + T G
Sbjct: 62 TLTALALSPDDKLLFSAGHSRQIRVWDLETLKCIRSWKGHEGPVMGMACHASGGLLATAG 121
Query: 369 EDDLVQVWSMEDRKVVAWGEGHNSWVSGVAFDSYWSSPNSNDN 411
D V VW ++ + GH VS + F P+SN N
Sbjct: 122 ADRKVLVWDVDGGFCTHYFRGHKGVVSSILF-----HPDSNKN 159
Score = 32.7 bits (73), Expect = 0.55
Identities = 28/130 (21%), Positives = 51/130 (38%), Gaps = 13/130 (10%)
Query: 310 INSISFSADGAYLATVGRDGYLRVFDYTKEHLVCGGKSYYGGLLCCAWSMDGKYILTGGE 369
INS++ + + + + T D ++ V K + + +S + ++T
Sbjct: 502 INSVAVARNDSLVCTGSEDRTASIWRLPDLVHVVTLKGHKRRIFSVEFSTVDQCVMTASG 561
Query: 370 DDLVQVWSMEDRKVVAWGEGHNSWVSGVAFDSYWSSPNSNDNGETITYRFGSVGQDTQLL 429
D V++W++ D + EGH S V +F T +F S G D L
Sbjct: 562 DKTVKIWAISDGSCLKTFEGHTSSVLRASF-------------ITDGTQFVSCGADGLLK 608
Query: 430 LWELEMDEIV 439
LW + E +
Sbjct: 609 LWNVNTSECI 618
Score = 32.3 bits (72), Expect = 0.72
Identities = 34/145 (23%), Positives = 62/145 (42%), Gaps = 18/145 (12%)
Query: 307 QGSINSISFSADGA--YLATVGRDGYLRVFDY----TKEHLVCGGKSYYGGLLCCAWSMD 360
+G ++SI F D L + D +RV+D T++ + + ++ + A S D
Sbjct: 144 KGVVSSILFHPDSNKNILISGSDDATVRVWDLNAKNTEKKCLAIMEKHFSAVTSIALSED 203
Query: 361 GKYILTGGEDDLVQVWSMEDRKVVAWG------EGHNSWVSGVAFDSYWSS-----PNSN 409
G + + G D +V +W + D A E + SG F S+ +S
Sbjct: 204 GLTLFSAGRDKVVNLWDLHDYSCKATVATYEVLEAVTTVSSGTPFASFVASLDQKKSKKK 263
Query: 410 DNGETITYRFGSVGQDTQLLLWELE 434
++ TY F +VG+ + +W+ E
Sbjct: 264 ESDSQATY-FITVGERGVVRIWKSE 287
Score = 31.6 bits (70), Expect = 1.2
Identities = 16/68 (23%), Positives = 29/68 (42%)
Query: 309 SINSISFSADGAYLATVGRDGYLRVFDYTKEHLVCGGKSYYGGLLCCAWSMDGKYILTGG 368
S+ SF DG + G DG L++++ + + + A + I TGG
Sbjct: 585 SVLRASFITDGTQFVSCGADGLLKLWNVNTSECIATYDQHEDKVWALAVGKKTEMIATGG 644
Query: 369 EDDLVQVW 376
D ++ +W
Sbjct: 645 GDAVINLW 652
>At2g47410 putative WD-40 repeat protein
Length = 1389
Score = 42.7 bits (99), Expect = 5e-04
Identities = 17/56 (30%), Positives = 31/56 (55%)
Query: 346 KSYYGGLLCCAWSMDGKYILTGGEDDLVQVWSMEDRKVVAWGEGHNSWVSGVAFDS 401
+ + + C + G+Y++TG +D LV++WSME +A GH ++ +A S
Sbjct: 233 RGHRNAVYCAIFDRSGRYVITGSDDRLVKIWSMETALCLASCRGHEGDITDLAVSS 288
Score = 34.3 bits (77), Expect = 0.19
Identities = 17/85 (20%), Positives = 37/85 (43%)
Query: 315 FSADGAYLATVGRDGYLRVFDYTKEHLVCGGKSYYGGLLCCAWSMDGKYILTGGEDDLVQ 374
F G Y+ T D ++++ + + + G + A S + + + D +++
Sbjct: 244 FDRSGRYVITGSDDRLVKIWSMETALCLASCRGHEGDITDLAVSSNNALVASASNDFVIR 303
Query: 375 VWSMEDRKVVAWGEGHNSWVSGVAF 399
VW + D ++ GH V+ +AF
Sbjct: 304 VWRLPDGMPISVLRGHTGAVTAIAF 328
Score = 29.6 bits (65), Expect = 4.6
Identities = 21/88 (23%), Positives = 37/88 (41%), Gaps = 18/88 (20%)
Query: 352 LLCCAWSMDGKYILTGGEDD------LVQVWSMEDRKVVAWGEGHNSWVSGVAFDSYWSS 405
+LCCA++ +G +TG D + VW+ D +V GH+ S D + +
Sbjct: 394 ILCCAYNANGTIFVTGSSDSNARVYCRICVWNAADGSLVHCLTGHSE--SSYVLDVHPFN 451
Query: 406 PNSNDNGETITYRFGSVGQDTQLLLWEL 433
P S G D + ++W++
Sbjct: 452 PRI----------AMSAGYDGKTIIWDI 469
>At5g60940 cleavage stimulation factor 50K chain
Length = 429
Score = 42.4 bits (98), Expect = 7e-04
Identities = 20/79 (25%), Positives = 41/79 (51%), Gaps = 2/79 (2%)
Query: 308 GSINSISFSADGAYLATVGRDGYLRVFDYTKEHLV--CGGKSYYGGLLCCAWSMDGKYIL 365
G+IN + +S+ G+ T +DG +R+FD V G + ++ D +++L
Sbjct: 267 GAINQVRYSSTGSIYITASKDGAIRLFDGVSAKCVRSIGNAHGKSEVTSAVFTKDQRFVL 326
Query: 366 TGGEDDLVQVWSMEDRKVV 384
+ G+D V++W + ++V
Sbjct: 327 SSGKDSTVKLWEIGSGRMV 345
>At4g02730 putative WD-repeat protein
Length = 333
Score = 42.4 bits (98), Expect = 7e-04
Identities = 26/106 (24%), Positives = 53/106 (49%), Gaps = 3/106 (2%)
Query: 309 SINSISFSADGAYLATVGRDGYLRVFDYTKEHLVCGGKSYYGGLLCC--AWSM-DGKYIL 365
+++ FS +G ++ D L++ +Y + + + C A+S+ +GKYI+
Sbjct: 215 AVSFAKFSPNGKFILVATLDSTLKLSNYATGKFLKVYTGHTNKVFCITSAFSVTNGKYIV 274
Query: 366 TGGEDDLVQVWSMEDRKVVAWGEGHNSWVSGVAFDSYWSSPNSNDN 411
+G ED+ V +W ++ R ++ EGH V V+ + +S+ N
Sbjct: 275 SGSEDNCVYLWDLQARNILQRLEGHTDAVISVSCHPVQNEISSSGN 320
Score = 40.4 bits (93), Expect = 0.003
Identities = 40/182 (21%), Positives = 77/182 (41%), Gaps = 28/182 (15%)
Query: 346 KSYYGGLLCCAWSMDGKYILTGGEDDLVQVWSMEDRKVVAWGEGHNSWVSGVAFDSYWSS 405
+ + + C +S DG + + D + +WS + ++ EGH+S +S +A WSS
Sbjct: 40 EGHTAAISCVKFSNDGNLLASASVDKTMILWSATNYSLIHRYEGHSSGISDLA----WSS 95
Query: 406 PNSNDNGETITYRFGSVGQDTQLLLWELEMDEIVVPLRRGPPG--GSPTFGSPTFSAGSQ 463
D+ T S D L +W+ + + RG F P+ S
Sbjct: 96 ----DSHYTC-----SASDDCTLRIWDARSPYECLKVLRGHTNFVFCVNFNPPSNLIVSG 146
Query: 464 SSHWDNAVPLGTLQPAPSMRDVPKISPLVAHRVHTEPLSSLIFTQES--VLTACREGHIK 521
S +D + + ++ +R + + H+ P+SS+ F ++ +++A +G K
Sbjct: 147 S--FDETIRIWEVKTGKCVRMI---------KAHSMPISSVHFNRDGSLIVSASHDGSCK 195
Query: 522 IW 523
IW
Sbjct: 196 IW 197
Score = 38.9 bits (89), Expect = 0.008
Identities = 26/96 (27%), Positives = 46/96 (47%), Gaps = 7/96 (7%)
Query: 309 SINSISFSADGAYLATVGRDGYLRVFDYTKEHLVCGGKSYYGGLLCCAWSMDGKYILTGG 368
+I+ + FS DG LA+ D + ++ T L+ + + G+ AWS D Y +
Sbjct: 45 AISCVKFSNDGNLLASASVDKTMILWSATNYSLIHRYEGHSSGISDLAWSSDSHYTCSAS 104
Query: 369 EDDLVQVWS----MEDRKVVAWGEGHNSWVSGVAFD 400
+D +++W E KV+ GH ++V V F+
Sbjct: 105 DDCTLRIWDARSPYECLKVL---RGHTNFVFCVNFN 137
Score = 37.7 bits (86), Expect = 0.017
Identities = 27/121 (22%), Positives = 57/121 (46%), Gaps = 8/121 (6%)
Query: 283 DQTLFSVAHARYSKSNPIARWHICQGSINSISFSADGAYLATVGRDGYLRVFDYTKEH-- 340
D+T+ + YS I R+ I+ +++S+D Y + D LR++D +
Sbjct: 64 DKTMILWSATNYSL---IHRYEGHSSGISDLAWSSDSHYTCSASDDCTLRIWDARSPYEC 120
Query: 341 -LVCGGKSYYGGLLCCAWSMDGKYILTGGEDDLVQVWSMEDRKVVAWGEGHNSWVSGVAF 399
V G + + + C ++ I++G D+ +++W ++ K V + H+ +S V F
Sbjct: 121 LKVLRGHTNF--VFCVNFNPPSNLIVSGSFDETIRIWEVKTGKCVRMIKAHSMPISSVHF 178
Query: 400 D 400
+
Sbjct: 179 N 179
>At2g20330 putative WD-40 repeat protein
Length = 648
Score = 42.4 bits (98), Expect = 7e-04
Identities = 36/134 (26%), Positives = 55/134 (40%), Gaps = 15/134 (11%)
Query: 324 TVGRDGYLRVFD---YTKEHLVCGGKSYYGG---LLCCAWSMDGKYILTGGEDDLVQVWS 377
T DG LR++D + + V K G + CAW DGK I G D +Q+WS
Sbjct: 296 TSSEDGSLRIWDVNNFLSQTQVIKPKLARPGRVPVTTCAWDRDGKRIAGGVGDGSIQIWS 355
Query: 378 MEDRKVVAWGEGHNSWVSGVAFDSYWSSPNSNDNGETITYRFGSVGQDTQLLLWELEMDE 437
++ WG + +V D S S+D ++ F D L +W+L +
Sbjct: 356 LKP----GWGSRPDIYVGKAHTDDITSVKFSSDGRILLSRSF-----DGSLKVWDLRQMK 406
Query: 438 IVVPLRRGPPGGSP 451
+ + G P P
Sbjct: 407 EALKVFEGLPNYYP 420
Score = 30.8 bits (68), Expect = 2.1
Identities = 25/61 (40%), Positives = 32/61 (51%), Gaps = 3/61 (4%)
Query: 310 INSISFSADGAYLATVGRDGYLRVFD--YTKEHL-VCGGKSYYGGLLCCAWSMDGKYILT 366
I S+ FS+DG L + DG L+V+D KE L V G Y A+S D + ILT
Sbjct: 376 ITSVKFSSDGRILLSRSFDGSLKVWDLRQMKEALKVFEGLPNYYPQTNVAFSPDEQIILT 435
Query: 367 G 367
G
Sbjct: 436 G 436
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.313 0.131 0.389
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 14,288,186
Number of Sequences: 26719
Number of extensions: 705218
Number of successful extensions: 2760
Number of sequences better than 10.0: 193
Number of HSP's better than 10.0 without gapping: 105
Number of HSP's successfully gapped in prelim test: 90
Number of HSP's that attempted gapping in prelim test: 2157
Number of HSP's gapped (non-prelim): 569
length of query: 561
length of database: 11,318,596
effective HSP length: 104
effective length of query: 457
effective length of database: 8,539,820
effective search space: 3902697740
effective search space used: 3902697740
T: 11
A: 40
X1: 16 ( 7.2 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 42 (21.9 bits)
S2: 63 (28.9 bits)
Medicago: description of AC128638.4


