

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC127429.4 + phase: 0
(301 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
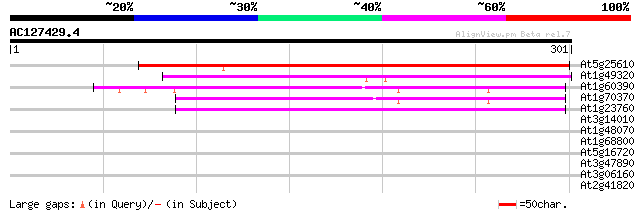
Score E
Sequences producing significant alignments: (bits) Value
At5g25610 dehydration-induced protein RD22 241 3e-64
At1g49320 unknown protein 150 8e-37
At1g60390 122 2e-28
At1g70370 unknown protein 120 9e-28
At1g23760 putative polygalacuronase isoenzyme 1 beta subunit 115 2e-26
At3g14010 unknown protein 32 0.42
At1g48070 unknown protein 29 3.6
At1g68800 hypothetical protein 28 4.6
At5g16720 putative protein 28 6.1
At3g47890 putative protein 28 6.1
At3g06160 hypothetical protein 28 7.9
At2g41820 putative receptor-like protein kinase 28 7.9
>At5g25610 dehydration-induced protein RD22
Length = 392
Score = 241 bits (616), Expect = 3e-64
Identities = 123/236 (52%), Positives = 159/236 (67%), Gaps = 5/236 (2%)
Query: 70 YFYENDANEAELHDNRNLAMFLLEKDLHHGTKFNIQFTKTSDHG--PTFLPGDVANSIPF 127
+ Y A E +LHD+ N A+F LEKDL G + N++F +G FLP A ++PF
Sbjct: 156 FVYNYAAKETQLHDDPNAALFFLEKDLVRGKEMNVRFNAEDGYGGKTAFLPRGEAETVPF 215
Query: 128 SSNKLENILNYFSIKQGSTESEIVKNTISECEAYGIKGEEKLCVTSLESMIDFTTLKLGN 187
S K L FS++ GS E+E++K TI ECEA + GEEK C TSLESM+DF+ KLG
Sbjct: 216 GSEKFSETLKRFSVEAGSEEAEMMKKTIEECEARKVSGEEKYCATSLESMVDFSVSKLGK 275
Query: 188 -NVDTVSTEVNGESG-LQQYVIAN-GVKKMGENNLVVCHKRNYPYAVFYCHKTDATKVYS 244
+V VSTEV ++ +Q+Y IA GVKK+ ++ VVCHK+ YP+AVFYCHK T VY+
Sbjct: 276 YHVRAVSTEVAKKNAPMQKYKIAAAGVKKLSDDKSVVCHKQKYPFAVFYCHKAMMTTVYA 335
Query: 245 VPLEGADGSRVKAVAICHSDTSQWSLKHLAFQVLKVQQGTFPVCHILQQGQVVWFS 300
VPLEG +G R KAVA+CH +TS W+ HLAF+VLKV+ GT PVCH L + VVWFS
Sbjct: 336 VPLEGENGMRAKAVAVCHKNTSAWNPNHLAFKVLKVKPGTVPVCHFLPETHVVWFS 391
Score = 33.1 bits (74), Expect = 0.19
Identities = 14/31 (45%), Positives = 20/31 (64%)
Query: 10 LVATHATLPPELYWKSKLPTTQIPKAITDIL 40
+VA A L PE YW + LP T IP ++ ++L
Sbjct: 16 VVAIAADLTPERYWSTALPNTPIPNSLHNLL 46
>At1g49320 unknown protein
Length = 280
Score = 150 bits (379), Expect = 8e-37
Identities = 86/230 (37%), Positives = 124/230 (53%), Gaps = 11/230 (4%)
Query: 83 DNRNLAMFLLEKDLHHGTKFNIQFTKTS-DHGPTFLPGDVANSIPFSSNKLENILNYFSI 141
D+ +L M+ DL GTK I F K P L A+ IPF+ +KL+ +L++FSI
Sbjct: 51 DDPSLYMYFTLNDLKLGTKLLIYFYKNDLQKLPPLLTRQQADLIPFTKSKLDFLLDHFSI 110
Query: 142 KQGSTESEIVKNTISECEAYGIKGEEKLCVTSLESMIDFTTLKLGNNVD----TVSTEVN 197
+ S + + +K T+ C+A I+GE K C TSLES+ID +G NVD T V
Sbjct: 111 TKDSPQGKAIKETLGHCDAKAIEGEHKFCGTSLESLIDLVKKTMGYNVDLKVMTTKVMVP 170
Query: 198 GES----GLQQYVIANGVKKMGENNLVVCHKRNYPYAVFYCH-KTDATKVYSVPLEGADG 252
++ L Y K++ ++ CH+ YPYAV+YCH ++V+ V L DG
Sbjct: 171 AQNSISYALHNYTFVEAPKELVGIKMLGCHRMPYPYAVYYCHGHKGGSRVFEVNLVTDDG 230
Query: 253 -SRVKAVAICHSDTSQWSLKHLAFQVLKVQQGTFPVCHILQQGQVVWFSK 301
RV A+CH DTS W H+AF+VLK++ + PVCH +VW +K
Sbjct: 231 RQRVVGPAVCHMDTSTWDADHVAFKVLKMEPRSAPVCHFFPLDNIVWVTK 280
>At1g60390
Length = 624
Score = 122 bits (306), Expect = 2e-28
Identities = 86/265 (32%), Positives = 119/265 (44%), Gaps = 13/265 (4%)
Query: 46 GKISVDGKISVN----GKTRIVYDAAPIYF--YENDANEAELHDNRNL--AMFLLEKDLH 97
G+ +V G +S G++ VY + F Y N+ + N+ + F E L
Sbjct: 357 GQDNVGGNVSFKTYGQGQSFKVYTKDGVVFARYSNNVSSNGKTVNKWVEEGKFFREAMLK 416
Query: 98 HGTKFNIQFTKTSDHGPTFLPGDVANSIPFSSNKLENILNYFSIKQGSTESEIVKNTISE 157
GT + K TFLP ++ ++PFSS+ + I F + S+ + I+ + +SE
Sbjct: 417 EGTLMQMPDIKDKMPKRTFLPRNIVKNLPFSSSTIGEIWRVFGAGENSSMAGIISSAVSE 476
Query: 158 CEAYGIKGEEKLCVTSLESMIDFTTLKLGNNVDTVSTEVNGESGLQQYVI--ANGVKKMG 215
CE GE K CV S E MIDF T LG V V T N ++ VI NG+
Sbjct: 477 CERPASHGETKRCVGSAEDMIDFATSVLGRGV-VVRTTENVVGSKKKVVIGKVNGINGGD 535
Query: 216 ENNLVVCHKRNYPYAVFYCHKTDATKVYSVPLEGADGSRV--KAVAICHSDTSQWSLKHL 273
V CH+ YPY ++YCH +VY L VAICH DTS WS H
Sbjct: 536 VTRAVSCHQSLYPYLLYYCHSVPRVRVYETDLLDPKSLEKINHGVAICHIDTSAWSPSHG 595
Query: 274 AFQVLKVQQGTFPVCHILQQGQVVW 298
AF L G VCH + + + W
Sbjct: 596 AFLALGSGPGQIEVCHWIFENDMTW 620
>At1g70370 unknown protein
Length = 626
Score = 120 bits (301), Expect = 9e-28
Identities = 73/213 (34%), Positives = 100/213 (46%), Gaps = 5/213 (2%)
Query: 90 FLLEKDLHHGTKFNIQFTKTSDHGPTFLPGDVANSIPFSSNKLENILNYFSIKQGSTESE 149
F E L GT + K +FLP + +PFS++KL I F + ST
Sbjct: 411 FFRESSLKEGTVIPMPDIKDKMPKRSFLPRSIITKLPFSTSKLGEIKRIFHAVENSTMGG 470
Query: 150 IVKNTISECEAYGIKGEEKLCVTSLESMIDFTTLKLGNNVDTVSTEVNGESGLQQYVI-- 207
I+ + ++ECE GE K CV S E MIDF T LG +V +TE N ++ VI
Sbjct: 471 IITDAVTECERPPSVGETKRCVGSAEDMIDFATSVLGRSVVLRTTE-NVAGSKEKVVIGK 529
Query: 208 ANGVKKMGENNLVVCHKRNYPYAVFYCHKTDATKVYSVPLEGADGSRV--KAVAICHSDT 265
NG+ V CH+ YPY ++YCH +VY L + + +AICH DT
Sbjct: 530 VNGINGGKLTKAVSCHQSLYPYLLYYCHSVPKVRVYEADLLELNSKKKINHGIAICHMDT 589
Query: 266 SQWSLKHLAFQVLKVQQGTFPVCHILQQGQVVW 298
S W H AF L + G VCH + + + W
Sbjct: 590 SSWGPSHGAFLALGSKPGRIEVCHWIFENDMNW 622
>At1g23760 putative polygalacuronase isoenzyme 1 beta subunit
Length = 622
Score = 115 bits (289), Expect = 2e-26
Identities = 71/212 (33%), Positives = 99/212 (46%), Gaps = 3/212 (1%)
Query: 90 FLLEKDLHHGTKFNIQFTKTSDHGPTFLPGDVANSIPFSSNKLENILNYFSIKQGSTESE 149
F E L GT + K +FLP + + +PFS++K+ I F ST
Sbjct: 407 FFRESMLKEGTLIWMPDIKDKMPKRSFLPRSIVSKLPFSTSKIAEIKRVFHANDNSTMEG 466
Query: 150 IVKNTISECEAYGIKGEEKLCVTSLESMIDFTTLKLGNNVDTVSTE-VNGESGLQQYVIA 208
I+ + + ECE E K CV S E MIDF T LG +V +TE V G
Sbjct: 467 IITDAVRECERPPTVSETKRCVGSAEDMIDFATSVLGRSVVLRTTESVAGSKEKVMIGKV 526
Query: 209 NGVKKMGENNLVVCHKRNYPYAVFYCHKTDATKVY-SVPLEGADGSRVK-AVAICHSDTS 266
NG+ V CH+ YPY ++YCH +VY S L+ +++ +AICH DTS
Sbjct: 527 NGINGGRVTKSVSCHQSLYPYLLYYCHSVPKVRVYESDLLDPKSKAKINHGIAICHMDTS 586
Query: 267 QWSLKHLAFQVLKVQQGTFPVCHILQQGQVVW 298
W H AF +L + G VCH + + + W
Sbjct: 587 AWGANHGAFMLLGSRPGQIEVCHWIFENDMNW 618
>At3g14010 unknown protein
Length = 595
Score = 32.0 bits (71), Expect = 0.42
Identities = 19/54 (35%), Positives = 29/54 (53%), Gaps = 2/54 (3%)
Query: 137 NYFSIKQGSTESEIVKNTISECEAYGIKGEEKLCVTSLESMIDFTTLKLGNNVD 190
N F +K+G TE + K S G G K SL+ ++ FTT K+G++V+
Sbjct: 11 NGFPLKRGETEEVLHKTNTSNTVFNGEAGSLKRL--SLDRLVYFTTCKIGHHVE 62
>At1g48070 unknown protein
Length = 114
Score = 28.9 bits (63), Expect = 3.6
Identities = 21/75 (28%), Positives = 35/75 (46%), Gaps = 1/75 (1%)
Query: 17 LPPELYWKSKLPTTQIPKAITDILHPMDDGKISVDGKISVNGKTRIVYDAAPIYFYENDA 76
L E+ W+ L TTQ+P +T DD +D +++ +T + I END
Sbjct: 18 LSNEVEWRLILTTTQLPIVVTVTSSLCDDRCTILDDQLARLSETYVFQTGKEIARLENDL 77
Query: 77 NEAELHD-NRNLAMF 90
+ + D NL++F
Sbjct: 78 SWGAITDIIDNLSLF 92
>At1g68800 hypothetical protein
Length = 317
Score = 28.5 bits (62), Expect = 4.6
Identities = 15/53 (28%), Positives = 29/53 (54%), Gaps = 5/53 (9%)
Query: 124 SIPFSSNKLEN-----ILNYFSIKQGSTESEIVKNTISECEAYGIKGEEKLCV 171
S PFSS+ LE+ ++N F ++Q + +V++ C+ +K + + CV
Sbjct: 22 SFPFSSDFLESFDESFLINQFLLQQQDVAANVVESPWKFCKKLELKKKNEKCV 74
>At5g16720 putative protein
Length = 600
Score = 28.1 bits (61), Expect = 6.1
Identities = 21/88 (23%), Positives = 38/88 (42%), Gaps = 2/88 (2%)
Query: 110 SDHGPTFLPGDVANSIPFSSNKLENILNYFSIKQGSTESEIVKNTISECEAYGIKGEEKL 169
S+ G LP + S KLE I S+ + E E + C + KG++
Sbjct: 511 SEDGSQGLPESDEKNFGSDSEKLEIIKQVDSVYERLQELETDGEFLKNCMSSAKKGDKGT 570
Query: 170 CVTS--LESMIDFTTLKLGNNVDTVSTE 195
+ L+ + D T++L N ++ +T+
Sbjct: 571 DILKDILQHLRDLRTIELTNTIENQTTQ 598
>At3g47890 putative protein
Length = 1528
Score = 28.1 bits (61), Expect = 6.1
Identities = 11/30 (36%), Positives = 19/30 (62%)
Query: 131 KLENILNYFSIKQGSTESEIVKNTISECEA 160
KL ++ ++FS + E+EI+ +S CEA
Sbjct: 306 KLSDLKSHFSASKDGNENEIISEALSFCEA 335
>At3g06160 hypothetical protein
Length = 330
Score = 27.7 bits (60), Expect = 7.9
Identities = 32/120 (26%), Positives = 52/120 (42%), Gaps = 13/120 (10%)
Query: 45 DGKISVDGKISVNG---KTRIVYDAAPIYFYENDANEAELHDNRNLAMFLLEKDLHHGTK 101
DG S + I G +TR V I D N LH N+++ L D HH T
Sbjct: 103 DGHKSYEVSIYGRGDCKETRAVIQVEEISDDTEDDN-VSLHSPSNVSLDSLSNDSHHSTS 161
Query: 102 FNIQFTKTSD---HGPTFLPGDVANS---IPFSSNKLEN--ILNYFSIKQGSTESEIVKN 153
N+ S+ HG + D S +P +S +E+ ++N + +Q S + + ++N
Sbjct: 162 -NVSLRSLSNDSLHGDAEIESDSEYSPENLPSASISVESVEVVNPTTSRQRSYKRKTIEN 220
>At2g41820 putative receptor-like protein kinase
Length = 890
Score = 27.7 bits (60), Expect = 7.9
Identities = 18/70 (25%), Positives = 33/70 (46%), Gaps = 4/70 (5%)
Query: 2 INVVLQLALVATHATLPPELYWKSKLPTTQIP-KAITDILHPMDDGKISVDGKISVNGKT 60
+ + L L+ H +LPPEL KL + + +T + P+ G +S+ I VN
Sbjct: 424 LQIALNLSFNHLHGSLPPELGKLDKLVSLDVSNNLLTGSIPPLLKGMMSL---IEVNFSN 480
Query: 61 RIVYDAAPIY 70
++ P++
Sbjct: 481 NLLNGPVPVF 490
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.317 0.134 0.393
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 6,905,757
Number of Sequences: 26719
Number of extensions: 292460
Number of successful extensions: 672
Number of sequences better than 10.0: 12
Number of HSP's better than 10.0 without gapping: 7
Number of HSP's successfully gapped in prelim test: 5
Number of HSP's that attempted gapping in prelim test: 653
Number of HSP's gapped (non-prelim): 13
length of query: 301
length of database: 11,318,596
effective HSP length: 99
effective length of query: 202
effective length of database: 8,673,415
effective search space: 1752029830
effective search space used: 1752029830
T: 11
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.6 bits)
S2: 60 (27.7 bits)
Medicago: description of AC127429.4


