

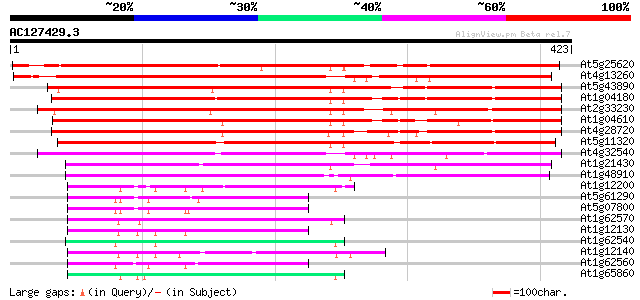
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC127429.3 - phase: 0 /pseudo
(423 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
At5g25620 unknown protein 428 e-120
At4g13260 unknown protein 425 e-119
At5g43890 dimethylaniline monooxygenase-like 376 e-104
At1g04180 putative dimethylaniline monooxygenase 373 e-104
At2g33230 putative flavin-containing monooxygenase 373 e-103
At1g04610 putative dimethylaniline monooxygenase 370 e-103
At4g28720 unknown protein 360 1e-99
At5g11320 putative protein 348 4e-96
At4g32540 dimethylaniline monooxygenase - like protein 331 4e-91
At1g21430 flavin-containing monooxygenases, putative 273 1e-73
At1g48910 hypothetical protein 258 6e-69
At1g12200 unknown protein 78 8e-15
At5g61290 unknown protein 77 2e-14
At5g07800 dimethylaniline monooxygenase-like protein 71 1e-12
At1g62570 similar to glutamate synthase 71 1e-12
At1g12130 hypothetical protein 71 1e-12
At1g62540 unknown protein 70 2e-12
At1g12140 unknown protein 70 2e-12
At1g62560 similar to flavin-containing monooxygenase (sp|P36366)... 69 5e-12
At1g65860 flavin-containing monooxygenase FMO3, putative 69 6e-12
>At5g25620 unknown protein
Length = 417
Score = 428 bits (1101), Expect = e-120
Identities = 234/429 (54%), Positives = 284/429 (65%), Gaps = 38/429 (8%)
Query: 3 CYLRELEGKQAHDPLFIEKMINNNKNSTSSSDRCLWIPGPLIVGAGPSGLAVAAYLKQKG 62
C+ RE+EGK AHD ++ TS C+ + GP+IVGAGPSGLA AA LK++G
Sbjct: 4 CWKREMEGKLAHD----------HRGMTSPRRICV-VTGPVIVGAGPSGLATAACLKERG 52
Query: 63 VPSLILERSNCIASLWKLKTYDRLRLHLPKQVCELPLMEFPSGFPTYPTKQQFIEYLESY 122
+ S++LERSNCIASLW+LKTYDRL LHLPKQ CELP++ FP FPTYPTKQQFIEYLE Y
Sbjct: 53 ITSVLLERSNCIASLWQLKTYDRLHLHLPKQFCELPIIPFPGDFPTYPTKQQFIEYLEDY 112
Query: 123 SKNFDIRPWFNETVMHAEFDATLGFWRVRSEGKAGMVTEFVCRWLIVATGENAEAVVPEI 182
++ FDI+P FN+TV A FD LG WRV S G+ G TE+VCRWL+ ATGENAE VVP
Sbjct: 113 ARRFDIKPEFNQTVESAAFDENLGMWRVTSVGEEG-TTEYVCRWLVAATGENAEPVVPRF 171
Query: 183 EGVDEF--VGSIRHTSLYKSGEEFRGKKVLVVGCGNSGMEVCLDLCNHDAAPSIVVRDSP 240
EG+D+F G ++HT YK+G +F GK+VLVVGCGNSGMEVCLDLCN A PS+VVRD+
Sbjct: 172 EGMDKFAAAGVVKHTCHYKTGGDFAGKRVLVVGCGNSGMEVCLDLCNFGAQPSLVVRDAV 231
Query: 241 ---TKRYAGKINF--------W----VVHVVAQVATRATGGSHPSHCVMVNARQHRTVRF 285
+ G F W +V V +R G ++ + R
Sbjct: 232 HVLPREMLGTSTFGLSMFLLKWLPIRLVDRFLLVVSRFILGD----TTLLGLNRPRLGPL 287
Query: 286 GSASVGSP*TQKTVWKDTGPRCGCPCQD*KRRHQGIKRLKRYTVEFADGSTENFDAIILA 345
++ KT D G D K GI+RLKR+ VEF +G TE FDAIILA
Sbjct: 288 ELKNISG----KTPVLDVGTLAKIKTGDIK-VCSGIRRLKRHEVEFDNGKTERFDAIILA 342
Query: 346 TGYKSNVPYWLKDKGMFSKEDGYPRKPFPNGWKGENGLYAVGFTKRGLLGASMDAKNIAE 405
TGYKSNVP WLK+ MFSK+DG+P + FP GW+GE GLYAVGFTKRG+ GASMDAK IAE
Sbjct: 343 TGYKSNVPSWLKENKMFSKKDGFPIQEFPEGWRGECGLYAVGFTKRGISGASMDAKRIAE 402
Query: 406 DIERCWKAE 414
DI +CWK +
Sbjct: 403 DIHKCWKQD 411
>At4g13260 unknown protein
Length = 415
Score = 425 bits (1092), Expect = e-119
Identities = 234/430 (54%), Positives = 283/430 (65%), Gaps = 56/430 (13%)
Query: 4 YLRELEGKQAHDPLFIEKMINNNKNSTSSSDRCLWIPGPLIVGAGPSGLAVAAYLKQKGV 63
++ E GK+ HDP ++E+ RCL IPGP+IVG+GPSGLA AA LK + +
Sbjct: 3 FVTETLGKRIHDP-YVEET------------RCLMIPGPIIVGSGPSGLATAACLKSRDI 49
Query: 64 PSLILERSNCIASLWKLKTYDRLRLHLPKQVCELPLMEFPSGFPTYPTKQQFIEYLESYS 123
PSLILERS CIASLW+ KTYDRLRLHLPK CELPLM FPS +PTYPTKQQF++YLESY+
Sbjct: 50 PSLILERSTCIASLWQHKTYDRLRLHLPKDFCELPLMPFPSSYPTYPTKQQFVQYLESYA 109
Query: 124 KNFDIRPWFNETVMHAEFDATLGFWRVRSE-GKAGMVTEFVCRWLIVATGENAEAVVPEI 182
++FD++P FN+TV A+FD G WRVR+ GK E+V RWL+VATGENAE V+PEI
Sbjct: 110 EHFDLKPVFNQTVEEAKFDRRCGLWRVRTTGGKKDETMEYVSRWLVVATGENAEEVMPEI 169
Query: 183 EGVDEFVGSIRHTSLYKSGEEFRGKKVLVVGCGNSGMEVCLDLCNHDAAPSIVVRDSPTK 242
+G+ +F G I HTS YKSGE F KK+LVVGCGNSGMEVCLDLCN +A PS+VVRDS
Sbjct: 170 DGIPDFGGPILHTSSYKSGEIFSEKKILVVGCGNSGMEVCLDLCNFNALPSLVVRDS--- 226
Query: 243 RYAGKINFWVVHVVAQ----VATRATGGS----HPSHCVMVNARQHRTVRFGSASVGSP* 294
VHV+ Q ++T S P H V +R +G
Sbjct: 227 ----------VHVLPQEMLGISTFGISTSLLKWFPVHVV-----DRFLLRMSRLVLGDTD 271
Query: 295 TQKTVWKDTGP-----RCG-CPCQD*K----------RRHQGIKRLKRYTVEFADGSTEN 338
V GP +CG P D + + +KR+ Y+ EF DG +N
Sbjct: 272 RLGLVRPKLGPLERKIKCGKTPVLDVGTLAKIRSGHIKVYPELKRVMHYSAEFVDGRVDN 331
Query: 339 FDAIILATGYKSNVPYWLKDKGMFSKEDGYPRKPFPNGWKGENGLYAVGFTKRGLLGASM 398
FDAIILATGYKSNVP WLK MFS++DG+P KPFPNGWKGE+GLYAVGFTK GLLGA++
Sbjct: 332 FDAIILATGYKSNVPMWLKGVNMFSEKDGFPHKPFPNGWKGESGLYAVGFTKLGLLGAAI 391
Query: 399 DAKNIAEDIE 408
DAK IAEDIE
Sbjct: 392 DAKKIAEDIE 401
>At5g43890 dimethylaniline monooxygenase-like
Length = 424
Score = 376 bits (966), Expect = e-104
Identities = 201/408 (49%), Positives = 263/408 (64%), Gaps = 30/408 (7%)
Query: 29 STSSSDR--CLWIPGPLIVGAGPSGLAVAAYLKQKGVPSLILERSNCIASLWKLKTYDRL 86
S SSDR C+W+ GP+IVGAGPSGLA AA L+++GVP ++LER++CIASLW+ +TYDR+
Sbjct: 10 SEDSSDRRRCIWVNGPVIVGAGPSGLATAACLREEGVPFVVLERADCIASLWQKRTYDRI 69
Query: 87 RLHLPKQVCELPLMEFPSGFPTYPTKQQFIEYLESYSKNFDIRPWFNETVMHAEFDATLG 146
+LHLPK+VC+LP M FP +P YPTK+QFIEYLESY+ F+I P FNE V A +D T G
Sbjct: 70 KLHLPKKVCQLPKMPFPEDYPEYPTKRQFIEYLESYANKFEITPQFNECVQSARYDETSG 129
Query: 147 FWRVR--SEGKAGMVTEFVCRWLIVATGENAEAVVPEIEGV-DEFVGSIRHTSLYKSGEE 203
WR++ S +G E++CRWL+VATGENAE VVPEI+G+ EF G + H+ YKSGE+
Sbjct: 130 LWRIKTTSSSSSGSEMEYICRWLVVATGENAEKVVPEIDGLTTEFEGEVIHSCEYKSGEK 189
Query: 204 FRGKKVLVVGCGNSGMEVCLDLCNHDAAPSIVVRDSP---TKRYAGKINF---------- 250
+RGK VLVVGCGNSGMEV LDL NH+A S+VVR S + GK +F
Sbjct: 190 YRGKSVLVVGCGNSGMEVSLDLANHNANASMVVRSSVHVLPREILGKSSFEISMMLMKWF 249
Query: 251 --WVVHVVAQVATRATGGSHPSHCVMVNARQHRTVRFGSASVGSP*TQKTVWKDTGPRCG 308
W+V + + G+ + + ++ S KT D G
Sbjct: 250 PLWLVDKILLILAWLILGNLTKYGLKRPTMGPMELKIVSG--------KTPVLDIGAMEK 301
Query: 309 CPCQD*KRRHQGIKRLKRYTVEFADGSTENFDAIILATGYKSNVPYWLKDKGMFSKEDGY 368
+ GIKR R VE DG + DA++LATGY+SNVP WL++ +FSK +G+
Sbjct: 302 IKSGE-VEIVPGIKRFSRSHVELVDGQRLDLDAVVLATGYRSNVPSWLQENDLFSK-NGF 359
Query: 369 PRKPFPNGWKGENGLYAVGFTKRGLLGASMDAKNIAEDIERCWKAEAK 416
P+ PFPN WKG++GLYA GFT++GL GAS DA NIA+DI W+ E K
Sbjct: 360 PKSPFPNAWKGKSGLYAAGFTRKGLAGASADAVNIAQDIGNVWREETK 407
>At1g04180 putative dimethylaniline monooxygenase
Length = 421
Score = 373 bits (958), Expect = e-104
Identities = 202/401 (50%), Positives = 260/401 (64%), Gaps = 27/401 (6%)
Query: 32 SSDRCLWIPGPLIVGAGPSGLAVAAYLKQKGVPSLILERSNCIASLWKLKTYDRLRLHLP 91
S RC+W+ GP+IVGAGPSGLA AA L +GVP +++ERS+CIASLW+ +TYDRL+LHLP
Sbjct: 15 SERRCVWVNGPVIVGAGPSGLATAACLHDQGVPFVVVERSDCIASLWQKRTYDRLKLHLP 74
Query: 92 KQVCELPLMEFPSGFPTYPTKQQFIEYLESYSKNFDIRPWFNETVMHAEFDATLGFWRVR 151
K+ C+LP M FP +P YPTK+QFI+YLESY+ FDI+P FN++V A FD T G WRVR
Sbjct: 75 KKFCQLPKMPFPDHYPEYPTKRQFIDYLESYANRFDIKPEFNKSVESARFDETSGLWRVR 134
Query: 152 SEGKAGMVTEFVCRWLIVATGENAEAVVPEIEG-VDEFVGSIRHTSLYKSGEEFRGKKVL 210
+ G E++CRWL+VATGENAE VVPEI G + EF G + H YKSGE+FRGK+VL
Sbjct: 135 TTSD-GEEMEYICRWLVVATGENAERVVPEINGLMTEFDGEVIHACEYKSGEKFRGKRVL 193
Query: 211 VVGCGNSGMEVCLDLCNHDAAPSIVVRDSP---TKRYAGKINF------------WVVHV 255
VVGCGNSGMEV LDL NH+A S+VVR S + GK F W+V
Sbjct: 194 VVGCGNSGMEVSLDLANHNAITSMVVRSSVHVLPREIMGKSTFGISVMMMKWLPLWLVDK 253
Query: 256 VAQVATRATGGSHPSHCVMVNARQHRTVRFGSASVGSP*TQKTVWKDTGPRCGCPCQD*K 315
+ + + GS ++ + + G + S T KT D G D
Sbjct: 254 LLLILSWLVLGSLSNYGL-------KRPDIGPMELKSM-TGKTPVLDIGALEKIKSGD-V 304
Query: 316 RRHQGIKRLKRYTVEFADGSTENFDAIILATGYKSNVPYWLKDKGMFSKEDGYPRKPFPN 375
IK+ R+ VE DG + DA++LATGY+SNVP WL++ FSK +G+P+ PFPN
Sbjct: 305 EIVPAIKQFSRHHVELVDGQKLDIDAVVLATGYRSNVPSWLQESEFFSK-NGFPKSPFPN 363
Query: 376 GWKGENGLYAVGFTKRGLLGASMDAKNIAEDIERCWKAEAK 416
WKG++GLYA GFT++GL GAS+DA NIA+DI W+ E K
Sbjct: 364 AWKGKSGLYAAGFTRKGLAGASVDAVNIAQDIGNVWREETK 404
>At2g33230 putative flavin-containing monooxygenase
Length = 431
Score = 373 bits (957), Expect = e-103
Identities = 211/430 (49%), Positives = 259/430 (60%), Gaps = 51/430 (11%)
Query: 22 MINNNKNSTSS-----------SDRCLWIPGPLIVGAGPSGLAVAAYLKQKGVPSLILER 70
M NNN S + S RC+W+ GP+IVGAGPSGLAVAA LK++ VP +ILER
Sbjct: 1 MCNNNNTSCVNISSMLQPEDIFSRRCIWVNGPVIVGAGPSGLAVAADLKRQEVPFVILER 60
Query: 71 SNCIASLWKLKTYDRLRLHLPKQVCELPLMEFPSGFPTYPTKQQFIEYLESYSKNFDIRP 130
+NCIASLW+ +TYDRL+LHLPKQ C+LP + FP P YPTK QFIEYLESY+ +FD+RP
Sbjct: 61 ANCIASLWQNRTYDRLKLHLPKQFCQLPNLPFPEDIPEYPTKYQFIEYLESYATHFDLRP 120
Query: 131 WFNETVMHAEFDATLGFWRV----RSEGKAGMVTEFVCRWLIVATGENAEAVVPEIEGVD 186
FNETV A++D G WRV RSE E++CRWL+VATGENAE VVPE EG++
Sbjct: 121 KFNETVQSAKYDKRFGLWRVQTVLRSELLGYCEFEYICRWLVVATGENAEKVVPEFEGLE 180
Query: 187 EFVGSIRHTSLYKSGEEFRGKKVLVVGCGNSGMEVCLDLCNHDAAPSIVVRDSP---TKR 243
+F G + H YKSGE +RGK+VLVVGCGNSGMEV LDLCNHDA+PS+VVR S +
Sbjct: 181 DFGGDVLHAGDYKSGERYRGKRVLVVGCGNSGMEVSLDLCNHDASPSMVVRSSVHVLPRE 240
Query: 244 YAGKINF------------WVVHVVAQVATRATGGSHPSHCVMVNARQHRTVRFG--SAS 289
GK F W+V V TR G+ T ++G
Sbjct: 241 VLGKSTFELSVTMMKWMPVWLVDKTLLVLTRLLLGN--------------TDKYGLKRPE 286
Query: 290 VGSP*TQKTVWKDTGPRCGCPCQD*KRRHQ---GIKRLKRYTVEFADGSTENFDAIILAT 346
+G + T K G + + GI + VE DG D++ILAT
Sbjct: 287 IGPLELKNTAGKTPVLDIGAISMIKSGKIKIVAGIAKFGPGKVELVDGRVLQIDSVILAT 346
Query: 347 GYKSNVPYWLKDKGMFSKEDGYPRKPFPNGWKGENGLYAVGFTKRGLLGASMDAKNIAED 406
GY+SNVP WLK+ + E G + PFP GWKG+ GLYAVGFT RGL GAS DA ++A D
Sbjct: 347 GYRSNVPSWLKENDL--GEIGIEKNPFPKGWKGKAGLYAVGFTGRGLSGASFDAMSVAHD 404
Query: 407 IERCWKAEAK 416
I WK E K
Sbjct: 405 IANSWKEETK 414
>At1g04610 putative dimethylaniline monooxygenase
Length = 437
Score = 370 bits (950), Expect = e-103
Identities = 200/410 (48%), Positives = 256/410 (61%), Gaps = 43/410 (10%)
Query: 33 SDRCLWIPGPLIVGAGPSGLAVAAYLKQKGVPSLILERSNCIASLWKLKTYDRLRLHLPK 92
S RC+W+ GP+IVGAGPSGLAVAA LK++GVP +ILER+NCIASLW+ +TYDRL+LHLPK
Sbjct: 28 SRRCIWVNGPVIVGAGPSGLAVAAGLKREGVPFIILERANCIASLWQNRTYDRLKLHLPK 87
Query: 93 QVCELPLMEFPSGFPTYPTKQQFIEYLESYSKNFDIRPWFNETVMHAEFDATLGFWRVRS 152
Q C+LP FP FP YPTK QFI+YLESY+ NFDI P FNETV A++D T G WRV++
Sbjct: 88 QFCQLPNYPFPDEFPEYPTKFQFIQYLESYAANFDINPKFNETVQSAKYDETFGLWRVKT 147
Query: 153 EGKAGMV----TEFVCRWLIVATGENAEAVVPEIEGVDEFVGSIRHTSLYKSGEEFRGKK 208
G + E++CRW++VATGENAE VVP+ EG+++F G + H YKSG ++GKK
Sbjct: 148 ISNMGQLGSCEFEYICRWIVVATGENAEKVVPDFEGLEDFGGDVLHAGDYKSGGRYQGKK 207
Query: 209 VLVVGCGNSGMEVCLDLCNHDAAPSIVVRDSP---TKRYAGKINF------------WVV 253
VLVVGCGNSGMEV LDL NH A PS+VVR + + GK F W+
Sbjct: 208 VLVVGCGNSGMEVSLDLYNHGANPSMVVRSAVHVLPREIFGKSTFELGVTMMKYMPVWLA 267
Query: 254 HVVAQVATRATGGSHPSHCVMVNARQHRTVRFGSASVGSP*TQKTVWKDTGPRCGCPCQD 313
R G+ + + + + G + + KT D G
Sbjct: 268 DKTILFLARIILGNTDKYGL-------KRPKIGPLELKNK-EGKTPVLDIGAL------- 312
Query: 314 *KRRHQGIKRLKRYTVEFADGSTE-------NFDAIILATGYKSNVPYWLKDKGMFSKED 366
+ G ++ ++F G E D++ILATGY+SNVP WLKD FS +D
Sbjct: 313 -PKIRSGKIKIVPGIIKFGKGKVELIDGRVLEIDSVILATGYRSNVPSWLKDNDFFS-DD 370
Query: 367 GYPRKPFPNGWKGENGLYAVGFTKRGLLGASMDAKNIAEDIERCWKAEAK 416
G P+ PFPNGWKGE GLYAVGFT++GL GAS+DA ++A DI WK E+K
Sbjct: 371 GIPKNPFPNGWKGEAGLYAVGFTRKGLFGASLDAMSVAHDIANRWKEESK 420
>At4g28720 unknown protein
Length = 426
Score = 360 bits (923), Expect = 1e-99
Identities = 202/410 (49%), Positives = 257/410 (62%), Gaps = 41/410 (10%)
Query: 32 SSDRCLWIPGPLIVGAGPSGLAVAAYLKQKGVPSLILERSNCIASLWKLKTYDRLRLHLP 91
+++RC+W+ GP+IVGAGPSGLA AA L ++ VP ++LER++CIASLW+ +TYDRL+LHLP
Sbjct: 15 TNNRCIWVNGPVIVGAGPSGLATAACLHEQNVPFVVLERADCIASLWQKRTYDRLKLHLP 74
Query: 92 KQVCELPLMEFPSGFPTYPTKQQFIEYLESYSKNFDIRPWFNETVMHAEFDATLGFWRVR 151
KQ C+LP M FP FP YPTK+QFI+YLESY+ F+I P FNE V A FD T G WRV+
Sbjct: 75 KQFCQLPKMPFPEDFPEYPTKRQFIDYLESYATRFEINPKFNECVQTARFDETSGLWRVK 134
Query: 152 SEGKAGMV---TEFVCRWLIVATGENAEAVVPEIEGVDEFVGSIRHTSLYKSGEEFRGKK 208
+ K+ E++CRWL+VATGENAE V+PEI+G+ EF G + H YKSGE+F GKK
Sbjct: 135 TVSKSESTQTEVEYICRWLVVATGENAERVMPEIDGLSEFSGEVIHACDYKSGEKFAGKK 194
Query: 209 VLVVGCGNSGMEVCLDLCNHDAAPSIVVRDS---PTKRYAGKINF------------WVV 253
VLVVGCGNSGMEV LDL NH A PS+VVR S + GK F W+V
Sbjct: 195 VLVVGCGNSGMEVSLDLANHFAKPSMVVRSSLHVMPREVMGKSTFELAMKMLRWFPLWLV 254
Query: 254 HVVAQVATRATGGSHPSHCVMVNARQHRTVR--FGSASVGSP*TQKTVWKDTGP----RC 307
+ V S V+ N ++ R G + S KT D G R
Sbjct: 255 DKILLVL---------SWMVLGNIEKYGLKRPEMGPMELKSV-KGKTPVLDIGAIEKIRL 304
Query: 308 GCPCQD*KRRHQGIKRLKRYTVEFADGSTENFDAIILATGYKSNVPYWLKDKGMFSKEDG 367
G GIKR VE +G + D+++LATGY+SNVPYWL++ F+K +G
Sbjct: 305 GK-----INVVPGIKRFNGNKVELVNGEQLDVDSVVLATGYRSNVPYWLQENEFFAK-NG 358
Query: 368 YPRKPFP-NGWKGENGLYAVGFTKRGLLGASMDAKNIAEDIERCWKAEAK 416
+P+ NGWKG GLYAVGFT++GL GASMDA IA+DI W+ E K
Sbjct: 359 FPKTVADNNGWKGRTGLYAVGFTRKGLSGASMDAVKIAQDIGSVWQLETK 408
>At5g11320 putative protein
Length = 411
Score = 348 bits (892), Expect = 4e-96
Identities = 190/386 (49%), Positives = 240/386 (61%), Gaps = 22/386 (5%)
Query: 37 LWIPGPLIVGAGPSGLAVAAYLKQKGVPSLILERSNCIASLWKLKTYDRLRLHLPKQVCE 96
+++PGP+IVGAGPSGLAVAA L +GVPS+ILER++C+ASLW+ +TYDRL+LHLPK CE
Sbjct: 12 IFVPGPIIVGAGPSGLAVAACLSNRGVPSVILERTDCLASLWQKRTYDRLKLHLPKHFCE 71
Query: 97 LPLMEFPSGFPTYPTKQQFIEYLESYSKNFDIRPWFNETVMHAEFDATLGFWRVRSEGKA 156
LPLM FP FP YP+KQ FI Y+ESY+ F+I+P FN+TV AEFD G W V+++
Sbjct: 72 LPLMPFPKNFPKYPSKQLFISYVESYAARFNIKPVFNQTVEKAEFDDASGLWNVKTQDGV 131
Query: 157 GMVTEFVCRWLIVATGENAEAVVPEIEGVDEFVGSIRHTSLYKSGEEFRGKKVLVVGCGN 216
+ WL+VATGENAE V P I G+ +F G + HTS YKSG F +KVLVVGCGN
Sbjct: 132 -----YTSTWLVVATGENAEPVFPNIPGLKKFTGPVVHTSAYKSGSAFANRKVLVVGCGN 186
Query: 217 SGMEVCLDLCNHDAAPSIVVRDSP---TKRYAGKINF--------WVVHVVAQVATRATG 265
SGMEV LDLC ++A P +VVR+S + + G F W +
Sbjct: 187 SGMEVSLDLCRYNALPHMVVRNSVHVLPRDFFGLSTFGIAMTLLKWFPLKLVDKFLLLLA 246
Query: 266 GSHPSHCVMVNARQHRTVRFGSASVGSP*TQKTVWKDTGPRCGCPCQD*KRRHQGIKRLK 325
S + ++ R+ +T +V T KT D G K Q +K +
Sbjct: 247 NSTLGNTDLLGLRRPKTGPIELKNV----TGKTPVLDVGAISLIRSGQIKVT-QAVKEIT 301
Query: 326 RYTVEFADGSTENFDAIILATGYKSNVPYWLKDKGMFSKEDGYPRKPFPNGWKGENGLYA 385
R +F +G FD+IILATGYKSNVP WLK+ F+KE G P+ PFPNGWKGE GLY
Sbjct: 302 RNGAKFLNGKEIEFDSIILATGYKSNVPDWLKENSFFTKE-GMPKTPFPNGWKGEKGLYT 360
Query: 386 VGFTKRGLLGASMDAKNIAEDIERCW 411
VGFT+RGL G + DA IAEDI W
Sbjct: 361 VGFTRRGLSGTAYDAVKIAEDITDQW 386
>At4g32540 dimethylaniline monooxygenase - like protein
Length = 414
Score = 331 bits (849), Expect = 4e-91
Identities = 193/415 (46%), Positives = 250/415 (59%), Gaps = 38/415 (9%)
Query: 22 MINNNKNSTSSSDRCLWIPGPLIVGAGPSGLAVAAYLKQKGVPSLILERSNCIASLWKLK 81
M ++ N T + + + GP+I+GAGPSGLA +A L +GVPSLILERS+ IASLWK K
Sbjct: 1 MESHPHNKTDQTQHIILVHGPIIIGAGPSGLATSACLSSRGVPSLILERSDSIASLWKSK 60
Query: 82 TYDRLRLHLPKQVCELPLMEFPSGFPTYPTKQQFIEYLESYSKNFDIRPWFNETVMHAEF 141
TYDRLRLHLPK C LPL++FP +P YP+K +F+ YLESY+ +F I P FN+ V +A +
Sbjct: 61 TYDRLRLHLPKHFCRLPLLDFPEYYPKYPSKNEFLAYLESYASHFRIAPRFNKNVQNAAY 120
Query: 142 DATLGFWRVRSEGKAGMVTEFVCRWLIVATGENAEAVVPEIEGVDEFV-GSIRHTSLYKS 200
D++ GFWRV++ TE++ +WLIVATGENA+ PEI G +F G I H S YKS
Sbjct: 121 DSSSGFWRVKTHDN----TEYLSKWLIVATGENADPYFPEIPGRKKFSGGKIVHASEYKS 176
Query: 201 GEEFRGKKVLVVGCGNSGMEVCLDLCNHDAAPSIVVRDSPTKRYAGKINFWVVHVVAQ-- 258
GEEFR +KVLVVGCGNSGME+ LDL H+A+P +VVR++ VHV+ +
Sbjct: 177 GEEFRRQKVLVVGCGNSGMEISLDLVRHNASPHLVVRNT-------------VHVLPREI 223
Query: 259 --VATRATGGS----HPSHCV------MVNARQHRTVRFG--SASVGSP*TQKTVWKDTG 304
V+T G + P V M N T R G G + K
Sbjct: 224 LGVSTFGVGMTLLKCLPLRLVDKFLLLMANLSFGNTDRLGLRRPKTGPLELKNVTGKSPV 283
Query: 305 PRCGCPCQD*KRRHQGIKRLKRYT---VEFADGSTENFDAIILATGYKSNVPYWLKDKGM 361
G Q ++ +K T +F DG ++FD+II ATGYKSNVP WL+ G
Sbjct: 284 LDVGAMSLIRSGMIQIMEGVKEITKKGAKFMDGQEKDFDSIIFATGYKSNVPTWLQG-GD 342
Query: 362 FSKEDGYPRKPFPNGWKGENGLYAVGFTKRGLLGASMDAKNIAEDIERCWKAEAK 416
F +DG P+ PFPNGW+G GLY VGFT+RGLLG + DA IA +I W+ E K
Sbjct: 343 FFTDDGMPKTPFPNGWRGGKGLYTVGFTRRGLLGTASDAVKIAGEIGDQWRDEIK 397
>At1g21430 flavin-containing monooxygenases, putative
Length = 391
Score = 273 bits (699), Expect = 1e-73
Identities = 159/381 (41%), Positives = 225/381 (58%), Gaps = 28/381 (7%)
Query: 43 LIVGAGPSGLAVAAYLKQKGVPSLILERSNCIASLWKLKTYDRLRLHLPKQVCELPLMEF 102
LI+GAGP+GLA +A L + +P++++ER C ASLWK ++YDRL+LHL KQ C+LP M F
Sbjct: 10 LIIGAGPAGLATSACLNRLNIPNIVVERDVCSASLWKRRSYDRLKLHLAKQFCQLPHMPF 69
Query: 103 PSGFPTYPTKQQFIEYLESYSKNFDIRPWFNETVMHAEFDATLGFWRVRSEGK-AGMVTE 161
PS PT+ +K FI YL+ Y+ F++ P +N V A F G W V+ K ++
Sbjct: 70 PSNTPTFVSKLGFINYLDEYATRFNVNPRYNRNVKSAYFKD--GQWIVKVVNKTTALIEV 127
Query: 162 FVCRWLIVATGENAEAVVPEIEG-VDEFVGSIRHTSLYKSGEEFRGKKVLVVGCGNSGME 220
+ ++++ ATGEN E V+PEI G V+ F G H+S YK+GE+F GK VLVVGCGNSGME
Sbjct: 128 YSAKFMVAATGENGEGVIPEIPGLVESFQGKYLHSSEYKNGEKFAGKDVLVVGCGNSGME 187
Query: 221 VCLDLCNHDAAPSIVVRDSP-------TKRYAGKINFWVVHVVAQVATRATGGSHPSHCV 273
+ DL +A SIVVR + + F+ V +V ++ +
Sbjct: 188 IAYDLSKCNANVSIVVRSQVHVLTRCIVRIGMSLLRFFPVKLVDRLC-----------LL 236
Query: 274 MVNARQHRTVRFGSASVGS-P*TQKTV-WKDTGPRCGCPCQD*KRRHQ---GIKRLKRYT 328
+ R T R+G + P K + + GC + + Q IKR++ T
Sbjct: 237 LAELRFRNTSRYGLVRPNNGPFLNKLITGRSATIDVGCVGEIKSGKIQVVTSIKRIEGKT 296
Query: 329 VEFADGSTENFDAIILATGYKSNVPYWLK-DKGMFSKEDGYPRKPFPNGWKGENGLYAVG 387
VEF DG+T+N D+I+ ATGYKS+V WL+ D G E+G P++ FP+ WKG+NGLY+ G
Sbjct: 297 VEFIDGNTKNVDSIVFATGYKSSVSKWLEVDDGDLFNENGMPKREFPDHWKGKNGLYSAG 356
Query: 388 FTKRGLLGASMDAKNIAEDIE 408
F K+GL G S DA+NIA DI+
Sbjct: 357 FGKQGLAGISRDARNIARDID 377
>At1g48910 hypothetical protein
Length = 383
Score = 258 bits (658), Expect = 6e-69
Identities = 149/375 (39%), Positives = 216/375 (56%), Gaps = 17/375 (4%)
Query: 43 LIVGAGPSGLAVAAYLKQKGVPSLILERSNCIASLWKLKTYDRLRLHLPKQVCELPLMEF 102
+IVGAGP+GLA + L Q +P++ILE+ + ASLWK + YDRL+LHL K+ C+LP M
Sbjct: 6 VIVGAGPAGLATSVCLNQHSIPNVILEKEDIYASLWKKRAYDRLKLHLAKEFCQLPFMPH 65
Query: 103 PSGFPTYPTKQQFIEYLESYSKNFDIRPWFNETVMHAEFDATLGFWRVRSEGKAGMVTE- 161
PT+ +K+ F+ YL++Y FDI P +N TV + FD + WRV +E TE
Sbjct: 66 GREVPTFMSKELFVNYLDAYVARFDINPRYNRTVKSSTFDESNNKWRVVAENTVTGETEV 125
Query: 162 FVCRWLIVATGENAEAVVPEIEGVDEFVGSIRHTSLYKSGEEFRGKKVLVVGCGNSGMEV 221
+ +L+VATGEN + +P +EG+D F G I H+S YKSG +F+ K VLVVG GNSGME+
Sbjct: 126 YWSEFLVVATGENGDGNIPMVEGIDTFGGEIMHSSEYKSGRDFKDKNVLVVGGGNSGMEI 185
Query: 222 CLDLCNHDAAPSIVVRDSPTKRYAGKINFWVVHV---------VAQVATRATGGSHPSHC 272
DLCN A +I++R T R+ + V+H+ VA V T T + +
Sbjct: 186 SFDLCNFGANTTILIR---TPRHV--VTKEVIHLGMTLLKYAPVAMVDTLVTTMAKILYG 240
Query: 273 VMVNARQHRTVRFGSASVGSP*TQKTVWKDTGPRCGCPCQD*KRRHQGIKRLKRYTVEFA 332
+ R + A+ T K D G + + + GI + T+ F
Sbjct: 241 DLSKYGLFRPKQGPFAT--KLFTGKAPVIDVGTVEKIRDGEIQVINGGIGSINGKTLTFE 298
Query: 333 DGSTENFDAIILATGYKSNVPYWLKDKGMFSKEDGYPRKPFPNGWKGENGLYAVGFTKRG 392
+G ++FDAI+ ATGYKS+V WL+D K+DG+P+ P P WKGE LY GF+++G
Sbjct: 299 NGHKQDFDAIVFATGYKSSVCNWLEDYEYVMKKDGFPKAPMPKHWKGEKNLYCAGFSRKG 358
Query: 393 LLGASMDAKNIAEDI 407
+ G + DA ++A+DI
Sbjct: 359 IAGGAEDAMSVADDI 373
>At1g12200 unknown protein
Length = 465
Score = 78.2 bits (191), Expect = 8e-15
Identities = 66/257 (25%), Positives = 111/257 (42%), Gaps = 46/257 (17%)
Query: 44 IVGAGPSGLAVAAYLKQKGVPSLILERSNCIASLWKLKT--------------------Y 83
++GAG +GL A L+++G ++LER + I +W + Y
Sbjct: 16 VIGAGAAGLVAARELRREGHSVVVLERGSQIGGVWAYTSQVEPDPLSLDPTRPVVHSSLY 75
Query: 84 DRLRLHLPKQVCELPLMEFPSGFPT-----------YPTKQQFIEYLESYSKNFDIRPW- 131
LR ++P++ + +FP F T +P + + YL ++K FDI
Sbjct: 76 RSLRTNIPREC--MGFTDFP--FATRPHDGSRDPRRHPAHTEVLAYLRDFAKEFDIEEMV 131
Query: 132 -FNETVMHAEFDAT----LGFWRVRSEGKAGMVTEFVCRWLIVATGENAEAVVPEIEGVD 186
F V+ AE A G WRV S G+V E + ++V G E I G+D
Sbjct: 132 RFETEVVKAEQVAAEGEERGKWRVESRSSDGVVDE-IYDAVVVCNGHYTEPRHALITGID 190
Query: 187 EFVGSIRHTSLYKSGEEFRGKKVLVVGCGNSGMEVCLDLCNHDAAPSIVVRDSPTKRY-- 244
+ G H+ Y+ ++F+ + V+V+G SG+++C D+ + R + Y
Sbjct: 191 SWPGKQIHSHNYRVPDQFKDQVVIVIGSSASGVDICRDIAQVAKEVHVSSRSTSPDTYEK 250
Query: 245 -AGKINFWVVHVVAQVA 260
G N W +H Q+A
Sbjct: 251 LTGYENLW-LHSTIQIA 266
>At5g61290 unknown protein
Length = 461
Score = 76.6 bits (187), Expect = 2e-14
Identities = 60/218 (27%), Positives = 103/218 (46%), Gaps = 39/218 (17%)
Query: 44 IVGAGPSGLAVAAYLKQKGVPSLILERSNCIASLW-------------KLKT-------Y 83
++GAGPSGL A LK++G +++E+++ + W K KT Y
Sbjct: 18 VIGAGPSGLVSARELKKEGHKVVVMEQNHDVGGQWLYQPNVDEEDTLGKTKTLKVHSSVY 77
Query: 84 DRLRLHLPKQVCELPLMEFP------SGFPTYPTKQQFIEYLESYSKNFDIRPWFNETVM 137
LRL P++V + +FP +P ++ + YL+ + + F +R V
Sbjct: 78 SSLRLASPREV--MGFSDFPFIAKEGRDSRRFPGHEELLLYLKDFCQVFGLREMIRFNV- 134
Query: 138 HAEF----------DATLGFWRVRSEGKAGMVTEFVCRWLIVATGENAEAVVPEIEGVDE 187
EF D + W V+S K+G V E V ++VA+G + +P I+G+D
Sbjct: 135 RVEFVGMVNEDDDDDDDVKKWMVKSVKKSGEVMEEVFDAVVVASGHYSYPRLPTIKGMDL 194
Query: 188 FVGSIRHTSLYKSGEEFRGKKVLVVGCGNSGMEVCLDL 225
+ H+ +Y+ E F + V+VVGC SG ++ ++L
Sbjct: 195 WKRKQLHSHIYRVPEPFCDEVVVVVGCSMSGQDISIEL 232
>At5g07800 dimethylaniline monooxygenase-like protein
Length = 460
Score = 71.2 bits (173), Expect = 1e-12
Identities = 59/219 (26%), Positives = 104/219 (46%), Gaps = 39/219 (17%)
Query: 44 IVGAGPSGLAVAAYLKQKGVPSLILERSNCIASLW---------------------KLKT 82
++GAGP+GL A L+++G ++LE++ + W +LK
Sbjct: 18 VIGAGPAGLVSARELRKEGHKVVVLEQNEDVGGQWFYQPNVEEEDPLGRSSGSINGELKV 77
Query: 83 ----YDRLRLHLPKQVCELPLMEFP------SGFPTYPTKQQFIEYLESYSKNFDIRPW- 131
Y LRL P+++ + +FP +P ++ YL+ +S+ F +R
Sbjct: 78 HSSIYSSLRLTSPREI--MGYSDFPFLAKKGRDMRRFPGHKELWLYLKDFSEAFGLREMI 135
Query: 132 -FN---ETVMHAEFDATLGFWRVRSEGK-AGMVTEFVCRWLIVATGENAEAVVPEIEGVD 186
FN E V E + + W VRS K +G V E + ++VATG + +P I+G+D
Sbjct: 136 RFNVRVEFVGEKEEEDDVKKWIVRSREKFSGKVMEEIFDAVVVATGHYSHPRLPSIKGMD 195
Query: 187 EFVGSIRHTSLYKSGEEFRGKKVLVVGCGNSGMEVCLDL 225
+ H+ +Y+ + FR + V+VVG SG ++ ++L
Sbjct: 196 SWKRKQIHSHVYRVPDPFRNEVVVVVGNSMSGQDISMEL 234
>At1g62570 similar to glutamate synthase
Length = 461
Score = 71.2 bits (173), Expect = 1e-12
Identities = 54/240 (22%), Positives = 102/240 (42%), Gaps = 31/240 (12%)
Query: 44 IVGAGPSGLAVAAYLKQKGVPSLILERSNCIASLWKL--------------------KTY 83
++GAG +GL A L+++G ++L+R + LW Y
Sbjct: 15 VIGAGAAGLVAARELRREGHTVVVLDREKQVGGLWVYTPETESDELGLDPTRPIVHSSVY 74
Query: 84 DRLRLHLPKQVCE------LPLMEFPS-GFPTYPTKQQFIEYLESYSKNFDIRPWFNETV 136
LR +LP++ +P + PS YP+ ++ + YL+ ++ F+I
Sbjct: 75 KSLRTNLPRECMGYKDFPFVPRGDDPSRDSRRYPSHREVLAYLQDFATEFNIEEMIRFET 134
Query: 137 MHAEFDATLGFWRVRSEGKAGMVTEFVCRWLIVATGENAEAVVPEIEGVDEFVGSIRHTS 196
+ G WRV+S+ G + + +++ G AE + +I G++ + G H+
Sbjct: 135 EVLRVEPVNGKWRVQSKTGGGFSNDEIYDAVVMCCGHFAEPNIAQIPGIESWPGRQTHSH 194
Query: 197 LYKSGEEFRGKKVLVVGCGNSGMEVCLDLCNHDAAPSIVVRDSPT----KRYAGKINFWV 252
Y+ + F+ + V+V+G SG ++ D+ I R S + KR N W+
Sbjct: 195 SYRVPDPFKDEVVVVIGNFASGADISRDISKVAKEVHIASRASKSNTFEKRPVPNNNLWM 254
>At1g12130 hypothetical protein
Length = 470
Score = 70.9 bits (172), Expect = 1e-12
Identities = 51/212 (24%), Positives = 91/212 (42%), Gaps = 30/212 (14%)
Query: 44 IVGAGPSGLAVAAYLKQKGVPSLILERSNCIASLWKL--------------------KTY 83
++GAG +GL A L+++G I ER + LW Y
Sbjct: 15 VIGAGAAGLVAARELRREGHTVTIFERQKQVGGLWVCTPNVEPDLLSIDPDRTVVHSSVY 74
Query: 84 DRLRLHLPKQVC---ELPLMEFPSGFPT----YPTKQQFIEYLESYSKNFDIRPW--FNE 134
LR +LP++ + P + P YP ++ + YL+ ++K F I F
Sbjct: 75 QSLRTNLPRECMGYSDFPFVTRPDDESRDPRRYPDHREVMRYLQDFAKEFKIEEMIRFET 134
Query: 135 TVMHAEFDATLGF-WRVRSEGKAGMVTEFVCRWLIVATGENAEAVVPEIEGVDEFVGSIR 193
V E A WRV+ +G+ E + +++ G E + I G++ + G
Sbjct: 135 EVFRVEPTAENSCKWRVQFRSSSGVSGEDIFDAVVICNGHFTEPRLAHIPGIESWPGKQI 194
Query: 194 HTSLYKSGEEFRGKKVLVVGCGNSGMEVCLDL 225
H+ Y+ + F+G+ V+V+G +SG ++ D+
Sbjct: 195 HSHNYRVSDPFKGQVVIVIGYQSSGSDISRDI 226
>At1g62540 unknown protein
Length = 457
Score = 70.5 bits (171), Expect = 2e-12
Identities = 53/241 (21%), Positives = 96/241 (38%), Gaps = 31/241 (12%)
Query: 43 LIVGAGPSGLAVAAYLKQKGVPSLILERSNCIASLW--------------------KLKT 82
+++GAG +GL A L ++G ++LER + LW
Sbjct: 14 VVIGAGAAGLVAARELSREGHTVVVLEREKEVGGLWIYSPKAESDPLSLDPTRSIVHSSV 73
Query: 83 YDRLRLHLPKQVCELPLMEFPSGFPT-------YPTKQQFIEYLESYSKNFDIRPWFNET 135
Y+ LR +LP++ F F YP+ + + YL+ +++ F++
Sbjct: 74 YESLRTNLPRECMGFTDFPFVPRFDDESRDSRRYPSHMEVLAYLQDFAREFNLEEMVRFE 133
Query: 136 VMHAEFDATLGFWRVRSEGKAGMVTEFVCRWLIVATGENAEAVVPEIEGVDEFVGSIRHT 195
+ + G WRV S+ G+ + + ++V +G E V I G+ + G H+
Sbjct: 134 IEVVRVEPVNGKWRVWSKTSGGVSHDEIFDAVVVCSGHYTEPNVAHIPGIKSWPGKQIHS 193
Query: 196 SLYKSGEEFRGKKVLVVGCGNSGMEVCLDLCNHDAAPSIVVRDSPTKRY----AGKINFW 251
Y+ F + V+V+G SG ++ D+ I R S Y + N W
Sbjct: 194 HNYRVPGPFENEVVVVIGNFASGADISRDIAKVAKEVHIASRASEFDTYEKLPVPRNNLW 253
Query: 252 V 252
+
Sbjct: 254 I 254
>At1g12140 unknown protein
Length = 444
Score = 70.5 bits (171), Expect = 2e-12
Identities = 63/277 (22%), Positives = 118/277 (41%), Gaps = 42/277 (15%)
Query: 44 IVGAGPSGLAVAAYLKQKGVPSLILERSNCIASLWKL--------------------KTY 83
++GAG +GL A L+++ ++ ER + + LW Y
Sbjct: 15 VIGAGAAGLVAARELRRENHTVVVFERDSKVGGLWVYTPNSEPDPLSLDPNRTIVHSSVY 74
Query: 84 DRLRLHLPKQVC---ELPLMEFPSGFPT-----YPTKQQFIEYLESYSKNF---DIRPWF 132
D LR +LP++ + P + P + YP+ ++ + YLE +++ F ++ +
Sbjct: 75 DSLRTNLPRECMGYRDFPFVPRPEDDESRDSRRYPSHREVLAYLEDFAREFKLVEMVRFK 134
Query: 133 NETVMHAEFDATLGFWRVRSEGKAGMVTEFVCRWLIVATGENAEAVVPEIEGVDEFVGSI 192
E V+ D WRV+S+ G+ + + ++V G E V + D + G
Sbjct: 135 TEVVLVEPEDKK---WRVQSKNSDGISKDEIFDAVVVCNGHYTEPRVAHVP--DSWPGKQ 189
Query: 193 RHTSLYKSGEEFRGKKVLVVGCGNSGMEVCLDLCNHDAAPSIVVRDSPTKRYA---GKIN 249
H+ Y+ ++F+ + V+V+G SG ++ D+ I R +P+K Y+ G N
Sbjct: 190 IHSHNYRVPDQFKDQVVVVIGNFASGADISRDITGVAKEVHIASRSNPSKTYSKLPGSNN 249
Query: 250 FWVVHV---VAQVATRATGGSHPSHCVMVNARQHRTV 283
W+ + V Q T + H +N + TV
Sbjct: 250 LWLHSMNGKVVQADTIVHCTGYKYHFPFLNTNGYITV 286
>At1g62560 similar to flavin-containing monooxygenase (sp|P36366);
similar to ESTs gb|R30018, gb|H36886, gb|N37822, and
gb|T88100
Length = 462
Score = 68.9 bits (167), Expect = 5e-12
Identities = 47/213 (22%), Positives = 93/213 (43%), Gaps = 34/213 (15%)
Query: 44 IVGAGPSGLAVAAYLKQKGVPSLILERSNCIASLW--------------------KLKTY 83
++GAGP+GL + L+++G ++ ER + LW Y
Sbjct: 15 VIGAGPAGLITSRELRREGHSVVVFEREKQVGGLWVYTPKSDSDPLSLDPTRSKVHSSIY 74
Query: 84 DRLRLHLPKQVCELPLMEFP---------SGFPTYPTKQQFIEYLESYSKNFDIRPW--F 132
+ LR ++P++ + + +FP YP ++ + Y++ +++ F I F
Sbjct: 75 ESLRTNVPRE--SMGVRDFPFLPRFDDESRDARRYPNHREVLAYIQDFAREFKIEEMIRF 132
Query: 133 NETVMHAEFDATLGFWRVRSEGKAGMVTEFVCRWLIVATGENAEAVVPEIEGVDEFVGSI 192
V+ E G WRV+S+ G + + + ++V G E + I G+ + G
Sbjct: 133 ETEVVRVE-PVDNGNWRVQSKNSGGFLEDEIYDAVVVCNGHYTEPNIAHIPGIKSWPGKQ 191
Query: 193 RHTSLYKSGEEFRGKKVLVVGCGNSGMEVCLDL 225
H+ Y+ + F + V+V+G SG ++ D+
Sbjct: 192 IHSHNYRVPDPFENEVVVVIGNFASGADISRDI 224
>At1g65860 flavin-containing monooxygenase FMO3, putative
Length = 459
Score = 68.6 bits (166), Expect = 6e-12
Identities = 52/240 (21%), Positives = 96/240 (39%), Gaps = 31/240 (12%)
Query: 44 IVGAGPSGLAVAAYLKQKGVPSLILERSNCIASLWKLKT--------------------Y 83
++GAG +GL A L+++G ++ +R + LW + Y
Sbjct: 15 VIGAGAAGLVTARELRREGHTVVVFDREKQVGGLWNYSSKADSDPLSLDTTRTIVHTSIY 74
Query: 84 DRLRLHLPKQVC---ELPLM----EFPSGFPTYPTKQQFIEYLESYSKNFDIRPWFNETV 136
+ LR +LP++ + P + + YP+ ++ + YL+ +++ F I
Sbjct: 75 ESLRTNLPRECMGFTDFPFVPRIHDISRDSRRYPSHREVLAYLQDFAREFKIEEMVRFET 134
Query: 137 MHAEFDATLGFWRVRSEGKAGMVTEFVCRWLIVATGENAEAVVPEIEGVDEFVGSIRHTS 196
+ G W VRS+ G + ++V +G E V I G+ + G H+
Sbjct: 135 EVVCVEPVNGKWSVRSKNSVGFAAHEIFDAVVVCSGHFTEPNVAHIPGIKSWPGKQIHSH 194
Query: 197 LYKSGEEFRGKKVLVVGCGNSGMEVCLDLCNHDAAPSIVVRDSPTKRY----AGKINFWV 252
Y+ F + V+V+G SG ++ D+ I R S + Y + N WV
Sbjct: 195 NYRVPGPFNNEVVVVIGNYASGADISRDIAKVAKEVHIASRASESDTYQKLPVPQNNLWV 254
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.322 0.138 0.442
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 10,317,032
Number of Sequences: 26719
Number of extensions: 461309
Number of successful extensions: 1127
Number of sequences better than 10.0: 50
Number of HSP's better than 10.0 without gapping: 43
Number of HSP's successfully gapped in prelim test: 7
Number of HSP's that attempted gapping in prelim test: 992
Number of HSP's gapped (non-prelim): 89
length of query: 423
length of database: 11,318,596
effective HSP length: 102
effective length of query: 321
effective length of database: 8,593,258
effective search space: 2758435818
effective search space used: 2758435818
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.9 bits)
S2: 61 (28.1 bits)
Medicago: description of AC127429.3


