

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC127429.2 + phase: 2 /pseudo
(500 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
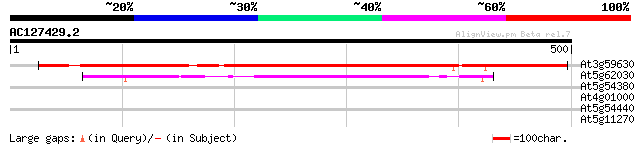
At3g59630 unknown protein 533 e-151
At5g62030 unknown protein (At5g62030) 91 2e-18
At5g54380 receptor-protein kinase-like protein 33 0.37
At4g01000 putative protein 30 4.0
At5g54440 unknown protein 29 5.3
At5g11270 unknown protein 28 9.0
>At3g59630 unknown protein
Length = 491
Score = 533 bits (1372), Expect = e-151
Identities = 275/482 (57%), Positives = 350/482 (72%), Gaps = 29/482 (6%)
Query: 26 QFPDDLLKDSTRVVSVLRKRLRSLRESDTTGSGDEPDIGLFVMADTAYGSCCVDEVGASH 85
QFPD+LLKDST+VVS L+ + R L + + + FVMADT YGSCC+DEVGA H
Sbjct: 29 QFPDELLKDSTKVVSALKSKTRLLTDRE---------VRFFVMADTTYGSCCIDEVGALH 79
Query: 86 INADCVIHYGHTCFSPTTTLPAFFVFGKASIGLADCVESMSQYALTSSKPIMVLYGLEYA 145
I+++CV+HYG TC SPT+ LPAFFVFGKASI ++ CV+ + YA S KPIM+LYGLEYA
Sbjct: 80 IDSECVVHYGQTCLSPTSVLPAFFVFGKASINVSSCVKHLIDYASKSDKPIMILYGLEYA 139
Query: 146 HSIQQVRKALLESSTFCTSDPKSEVHFADVPSPYMFPSKDTKKLNDLQEETCGCGDNSSS 205
H I Q+R+ L S T S++ A+V ++ PSKD + + E ++ SS
Sbjct: 140 HVIPQIREELGLSKT------DSQLSVANVLCSFISPSKDPR---ESMEHPRPYSESDSS 190
Query: 206 DGASGA-AYSIGGLTWKLLEGQSMEDYSLFWIGHDNSAFANVVLRFNACEIVRYDATKSQ 264
D S + +Y +GGLTW L EG +EDY LFWIG D+SAFANVVL FN C+IVRYDA +
Sbjct: 191 DSLSSSRSYRLGGLTWDLPEGSKIEDYLLFWIGSDSSAFANVVLTFNGCDIVRYDAEEDS 250
Query: 265 MVADLSQQRRILKRRYYLVERAKDANIVGILVGTLGVAGYLHIINQMMGLITAAGKKAYT 324
+V + QQRRILKRRYYLVE+AKDANI+GILVGTLGVAGYLH+I+ M LI+AAGKK+Y
Sbjct: 251 LVTEFYQQRRILKRRYYLVEKAKDANIIGILVGTLGVAGYLHMIHHMQALISAAGKKSYI 310
Query: 325 LVMGKPNPAKLANFPECDVFLYVSCAQTALLDSKDYLAPVITPFEATIAFNRGSQWTGAY 384
L MG+PNPAKLANFPECDVF+Y+SCAQTALLDSK++++PVITPFEA +AF+RGS+WTGAY
Sbjct: 311 LAMGRPNPAKLANFPECDVFIYISCAQTALLDSKEFMSPVITPFEANLAFSRGSEWTGAY 370
Query: 385 VMGFGDLINL----PQVQVGNQEEARFSFLKGGYVEDFENQE---NNVEEEREDLALANA 437
+M F D+IN + +G+ EE RFSF +GGYVED + + N E+ E + L A
Sbjct: 371 LMHFQDVINSVKSESEAHIGS-EEPRFSFFQGGYVEDHKTNDQAKNGEEDTGETMTLVQA 429
Query: 438 TEKALQLRDN-CNSIIK-GGAKSGAEFFANRTYQGLNIPDDNTTPQSFVIGRKGRAAGYE 495
EKALQLR N NS+ K AKSG E+F NR Y+GL I +NT P+ +++GR G+A+GY+
Sbjct: 430 AEKALQLRGNDHNSLTKQTAAKSGPEYFLNRVYRGLEINSENTLPEPYIVGRSGKASGYK 489
Query: 496 DE 497
E
Sbjct: 490 HE 491
>At5g62030 unknown protein (At5g62030)
Length = 453
Score = 90.5 bits (223), Expect = 2e-18
Identities = 81/373 (21%), Positives = 159/373 (41%), Gaps = 64/373 (17%)
Query: 66 FVMADTAYGSCCVDEVGASHINADCVIHYGHTCFSP--TTTLPAFFVFGKASIGLADCVE 123
FV+ D YG+CCVD+ A + AD +IHYGH+C P +T +P +VF + I + +
Sbjct: 95 FVLGDVTYGACCVDDFSACALGADLLIHYGHSCLVPIDSTKIPCLYVFVEIQIDVKCLLN 154
Query: 124 SMSQYALTSSKPIMVLYGLEYAHSIQQVRKALLESSTFCTSDPKSEVHFADVPSPYMFPS 183
++ + K I++ +++ +I+ V K LE F P+S+ A
Sbjct: 155 TIHLNLASDVKNIILAGTIQFTSAIRAV-KPELEKQGFNVLIPQSKPLSAG--------- 204
Query: 184 KDTKKLNDLQEETCGCGDNSSSDGASGAAYSIGGLTWKLLEGQSMEDYSLFWIGHDNSAF 243
E GC K+ + +D L ++
Sbjct: 205 -----------EVLGC------------------TAPKVKTVEDCKDQVLVFVADGRFHL 235
Query: 244 ANVVLRFNACEIVRYDATKSQMVADLSQQRRILKRRYYLVERAKDANIVGILVGTLGVAG 303
++ + RYD ++ + + + + R + RA++A GI++GTLG G
Sbjct: 236 EAFMIANPKIKAFRYDPYLGKLFLEEYDHKGMRETRMRAIARAREAKTWGIVLGTLGRQG 295
Query: 304 YLHIINQMMGLITAAGKKAYTLVMGKPNPAKLANFPE-CDVFLYVSCAQTALLDSKDYLA 362
I+ ++ + G + ++M + +P ++A F + D ++ ++C + ++ + +L
Sbjct: 296 NPKILERLEKKMMEKGIDSTVVLMSELSPTRVALFEDSVDAWVQIACPRLSIDWGEAFLK 355
Query: 363 PVITPFEATIAFNRGSQWTGAYVMGFGDLINLPQVQVGNQEEARFSFLKGGYVEDFEN-- 420
P++T FEA IA ++ G+ + N +R + G ED E
Sbjct: 356 PLLTTFEAEIAL--------GFIRGWWE----------NDSSSRVNSSSGCCKEDKETSC 397
Query: 421 --QENNVEEERED 431
+ NV+++++D
Sbjct: 398 ACKNENVKDDKKD 410
>At5g54380 receptor-protein kinase-like protein
Length = 855
Score = 33.1 bits (74), Expect = 0.37
Identities = 23/91 (25%), Positives = 40/91 (43%), Gaps = 7/91 (7%)
Query: 125 MSQYALTSSKPIMVLYGLEYAHSIQQVRKALLESSTFCTSDPKSEVHFADVPSPYMFPSK 184
+++Y + VL+ LEYA +++ AL+E D S H +P M P
Sbjct: 763 LAEYGVDRPSMGDVLWNLEYALQLEETSSALME------PDDNSTNHIPGIPMAPMEPFD 816
Query: 185 DTKKLNDLQEETCGCG-DNSSSDGASGAAYS 214
++ + D G G D+ + D + A +S
Sbjct: 817 NSMSIIDRGGVNSGTGTDDDAEDATTSAVFS 847
>At4g01000 putative protein
Length = 415
Score = 29.6 bits (65), Expect = 4.0
Identities = 27/120 (22%), Positives = 51/120 (42%), Gaps = 6/120 (5%)
Query: 387 GFGDLINLPQVQVGNQEEARFSFLK--GGYVEDFENQENNVEEEREDLALANATEKALQ- 443
GFG L+ ++ G ++ F + G N EN ++E ++ N +KAL+
Sbjct: 84 GFGSLLRGGGMKAGQKKTNNFDACRDMSGRRLRHVNAENRLQEWKDGEEGRNLEKKALEY 143
Query: 444 LRDNCNSIIKGGAKSGAEFFANRTYQGLN---IPDDNTTPQSFVIGRKGRAAGYEDENNK 500
L+ N + +G + + N+ + + + D +SF G++ G E E K
Sbjct: 144 LKKQSNKVKQGVGNGATQKYVNKYKEESDKCILAVDLALNESFKNGKRKAKIGAESEKKK 203
>At5g54440 unknown protein
Length = 1280
Score = 29.3 bits (64), Expect = 5.3
Identities = 29/97 (29%), Positives = 40/97 (40%), Gaps = 14/97 (14%)
Query: 358 KDYLAPVITPFEATIAFNRGSQWTGAYVMGFGDLINLPQVQVGNQEEARFSFLKGGYV-- 415
K A V+ P TI F Q G+YV+G + Q+G SF KGG
Sbjct: 771 KSSAATVLNPGRNTITFALPPQKPGSYVLG------VVTGQIGRLRFRSHSFSKGGPADS 824
Query: 416 EDFENQEN------NVEEEREDLALANATEKALQLRD 446
+DF + E V + R + LA A AL + +
Sbjct: 825 DDFMSYEKPTRPILKVSKPRALVDLAAAVSSALLINE 861
>At5g11270 unknown protein
Length = 354
Score = 28.5 bits (62), Expect = 9.0
Identities = 27/106 (25%), Positives = 44/106 (41%), Gaps = 9/106 (8%)
Query: 363 PVITPFEATIAFNRGSQWTGAYVMGFGDLINLPQVQV-------GNQEEARFSFLKGGYV 415
P++ P + + F RG G + + G +EE F L
Sbjct: 47 PILNPSRSVLVFARGKNRKGFVSSSSSSPKKNKKKSLDGADNGGGEEEEDPFEALFNLLE 106
Query: 416 EDFENQENNVEE--EREDLALANATEKALQLRDNCNSIIKGGAKSG 459
ED +N ++ EE E E ALA+ +AL + D+ + I G+ +G
Sbjct: 107 EDLKNDNSDDEEISEEELEALADELARALGVGDDVDDIDLFGSVTG 152
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.321 0.137 0.412
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 11,393,550
Number of Sequences: 26719
Number of extensions: 495942
Number of successful extensions: 1362
Number of sequences better than 10.0: 6
Number of HSP's better than 10.0 without gapping: 2
Number of HSP's successfully gapped in prelim test: 4
Number of HSP's that attempted gapping in prelim test: 1353
Number of HSP's gapped (non-prelim): 7
length of query: 500
length of database: 11,318,596
effective HSP length: 103
effective length of query: 397
effective length of database: 8,566,539
effective search space: 3400915983
effective search space used: 3400915983
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.9 bits)
S2: 62 (28.5 bits)
Medicago: description of AC127429.2


