

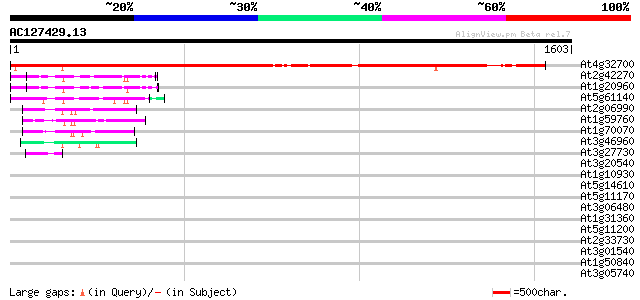
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC127429.13 - phase: 0 /pseudo
(1603 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
At4g32700 putative protein 1779 0.0
At2g42270 ATP-dependent RNA helicase like protein 135 2e-31
At1g20960 putative ATP-dependent RNA helicase 124 6e-28
At5g61140 RNA helicase - like protein 113 1e-24
At2g06990 HUA enhancer 2 (HEN2) 102 2e-21
At1g59760 hypothetical protein 102 2e-21
At1g70070 Similar to Synechocystis antiviral protein (At1g70070) 88 3e-17
At3g46960 putative helicase 82 2e-15
At3g27730 hypothetical protein 47 1e-04
At3g20540 DNA polymerase, putative 42 0.002
At1g10930 DNA helicase isolog 40 0.014
At5g14610 DRH1 DEAD box protein - like 39 0.019
At5g11170 DEAD BOX RNA helicase RH15 - like protein 39 0.024
At3g06480 putative RNA helicase 39 0.024
At1g31360 unknown protein 38 0.041
At5g11200 DEAD BOX RNA helicase RH15 38 0.054
At2g33730 putative U5 small nuclear ribonucleoprotein, an RNA he... 37 0.092
At3g01540 AT3g01540/F4P13_9 37 0.12
At1g50840 unknown protein 37 0.12
At3g05740 putative DNA helicase 35 0.27
>At4g32700 putative protein
Length = 1548
Score = 1779 bits (4607), Expect = 0.0
Identities = 977/1623 (60%), Positives = 1157/1623 (71%), Gaps = 171/1623 (10%)
Query: 1 MAILVLPYVSICTEK----------AEHLEKLLEPLGKHVRSYYGNQGGGSLPKDTSVAV 50
MA+LVLPYVSIC EK AEHLE LLEPLGKHVRSYYGNQGGG+LPKDTSVAV
Sbjct: 1 MALLVLPYVSICAEKVSVVSPSILHAEHLEVLLEPLGKHVRSYYGNQGGGTLPKDTSVAV 60
Query: 51 CTIEKANSLINRLLEEGRLSEMGIIVIDELHMVGDPKRGYLLELMLTKLRYAAGEGISKS 110
CTIEKANSLINRLLEEGRLSE+GIIVIDELHMVGD RGYLLELMLTKLRYAAGEG S+S
Sbjct: 61 CTIEKANSLINRLLEEGRLSELGIIVIDELHMVGDQHRGYLLELMLTKLRYAAGEGSSES 120
Query: 111 SDGEGSGGSSDKNDPAQGLQIVGMSATMPNVAAVADWLQ-----------AALYQTEFRP 159
S GE SG SS K DPA GLQIVGMSATMPNV AVADWLQ AALYQTEFRP
Sbjct: 121 SSGESSGTSSGKADPAHGLQIVGMSATMPNVGAVADWLQVSKTNHNLCVYAALYQTEFRP 180
Query: 160 VPLEEYIKVGNSIYNKSMELSRTIPKGADLGGKDPDHVVELCNEVVQEGQSVLIFCSSRK 219
VPLEEYIKVG++IYNK ME+ RTIPK AD+GGKDPDH+VELCNEVVQEG SVLIFCSSRK
Sbjct: 181 VPLEEYIKVGSTIYNKKMEVVRTIPKAADMGGKDPDHIVELCNEVVQEGNSVLIFCSSRK 240
Query: 220 GCESTARHVAKFLKSFTVDINENNCEFADITSAINSLRKCPAGLDPVLEETFPAGVAFHH 279
GCESTARH++K +K+ V+++ N EF DI SAI++LR+ P+G+DPVLEET P+GVA+HH
Sbjct: 241 GCESTARHISKLIKNVPVNVDGENSEFMDIRSAIDALRRSPSGVDPVLEETLPSGVAYHH 300
Query: 280 AGLTVEEREIVETCYRKGLLRVLTATSTLAAGVNLPARRVIFRQPRIGCDFIDGTRYMQM 339
AGLTVEEREIVETCYRKGL+RVLTATSTLAAGVNLPARRVIFRQP IG DFIDGTRY QM
Sbjct: 301 AGLTVEEREIVETCYRKGLVRVLTATSTLAAGVNLPARRVIFRQPMIGRDFIDGTRYKQM 360
Query: 340 AGRAGRTGIDTKGESVLICKPQELKKVMGLLNESCPPLHSCLSEDLNGMTHAILEVVAGG 399
+GRAGRTGIDTKG+SVLICKP ELK++M LLNE+CPPL SCLSED NGMTHAILEVVAGG
Sbjct: 361 SGRAGRTGIDTKGDSVLICKPGELKRIMALLNETCPPLQSCLSEDKNGMTHAILEVVAGG 420
Query: 400 IVQTANDIH---RKFLEWNEDTKLYSTTPLGRASFGSSLCPEESLIVLADLSRAREGFVL 456
IVQTA DIH RKFLEWNE+TKLY+TTPLGR SFGSSLCPEESLIVL DL RAREG V+
Sbjct: 421 IVQTAKDIHRYVRKFLEWNEETKLYTTTPLGRGSFGSSLCPEESLIVLDDLLRAREGLVM 480
Query: 457 ASDLHLVYLVTPINVDVEPDWESYYERFVKLSPLDQSVGNRVGVTEPFLMRMAHGVPMGS 516
ASDLHLVYLVTPINV VEP+WE YYERF++LSPL+QSVGNRVGV EPFLMRMAHG + +
Sbjct: 481 ASDLHLVYLVTPINVGVEPNWELYYERFMELSPLEQSVGNRVGVVEPFLMRMAHGATVRT 540
Query: 517 -----NKSRWSNNKRQNQHGMSSGIVNSDDQTLRVCKRFYVALILSLLVQETPVGEVCEA 571
+ + + ++HG +S + SD+Q LRVCKRF+VALILS LVQE V EVCEA
Sbjct: 541 LNRPQDVKKNLRGEYDSRHGSTSMKMLSDEQMLRVCKRFFVALILSKLVQEASVTEVCEA 600
Query: 572 FKVARGMVQGLQENAGRFASMVAVFCERLGWHDFEGLVAKFQNRVSFGVRAEVVELTTIP 631
FKVARGMVQ LQENAGRF+SMV+VFCERLGWHD EGLVAKFQNRVSFGVRAE+VELT+IP
Sbjct: 601 FKVARGMVQALQENAGRFSSMVSVFCERLGWHDLEGLVAKFQNRVSFGVRAEIVELTSIP 660
Query: 632 YVKGSRARALYKAGLRTPLAIAEASIPELVKALFESSSWGTEGSAQRSLQFGVAKKIKNG 691
Y+KGSRARALYKAGLRT AIAEASIPE+VKALFESS+W EG+ QR + G+AKKIKNG
Sbjct: 661 YIKGSRARALYKAGLRTSQAIAEASIPEIVKALFESSAWAAEGTGQRRIHLGLAKKIKNG 720
Query: 692 ARKIVLDKAEEARIAAFSAFKSLGYDVPQFAPPISTAVCNSIR----KEVGSSSGSDTAD 747
ARKIVL+KAEEAR AAFSAFKSLG DV + + P+ A +S+ E S GS D
Sbjct: 721 ARKIVLEKAEEARAAAFSAFKSLGLDVNELSKPLPLAPASSLNGQETTERDISRGSVGPD 780
Query: 748 TSHSFIDTNHIDNSNVPA---LEKEKDLIKSSDNGALVSVEGKSDSVMPHSLSTVPVVVP 804
I+ H++ N EK +++ + G VS E S +P+
Sbjct: 781 GLQQSIE-GHMECENFDMDNHREKPSEVLGDATLG--VSSEINLTSRLPN--------FR 829
Query: 805 SINELSMTSGPAKIPDVTTLSVHLQKQNDKSIMHNGCHAQGTGEQDHRGNLASGNMINSS 864
I T+GP+ + +++ + + +++ I Q DH S N +
Sbjct: 830 PIGTAVGTNGPSAVSILSSDTFPIPVYDNREIKPKDNVEQHLTRNDHIP--LSSNKDGTG 887
Query: 865 RKGPINAVSSPGGLDSFLDLWDTVAEFYFDIHYIKRLELHSAAPFEVHGIAICWENSPMY 924
KGP+ A + GG DSFL+LW + EF+FD+HY K +L+S +E+HGIAICW SP+Y
Sbjct: 888 EKGPVTAGNISGGFDSFLELWGSAGEFFFDLHYNKLQDLNSRISYEIHGIAICWNCSPVY 947
Query: 925 YINLPRDILLSGNRKDDGFSLTACSSKQKVSSSNSKQDLMNAMHRWSRISKIIEKKEVRK 984
Y+NL +D+ + A K +V +S++ D++ + RW++ISKI+
Sbjct: 948 YVNLNKDLPNLECVEKQKLIEDAVIGKSEVLASHNMLDVIKS--RWNKISKIMG------ 999
Query: 985 FTWNLKVQIQVLKKPSVSVQRFGSLDTIDKNMHLEVVDNSYILLPPIHVKDAIDMCIVAW 1044
VLK P++S+QR L+ + + + E+VD S++++PP+H IDM IV W
Sbjct: 1000 ---------NVLKSPAISIQRCTRLN-LPEGIRDELVDGSWLMMPPLHTSHTIDMSIVIW 1049
Query: 1045 ILWPDEESSSSPNLDKEVKKRLSPEDAAAANQCGRWRNQMRKAAHNGCCRRVAQTRALCS 1104
ILWPDEE S+PN+DKEVKKRLSPE A AAN+ GRWRNQ+R+ AHNGCCRRVAQTRALCS
Sbjct: 1050 ILWPDEERHSNPNIDKEVKKRLSPEAAEAANRSGRWRNQIRRVAHNGCCRRVAQTRALCS 1109
Query: 1105 VLWKLLVSEKLVEALMEIEIPLVNVLADMELWGIGVDLEGCIQARKLLVKRLKQLEKEAY 1164
LWK+LVSE+L++AL IE+PLVNVLADMELWGIG+D+EGC++AR +L +L+ LEK+A+
Sbjct: 1110 ALWKILVSEELLQALTTIEMPLVNVLADMELWGIGIDIEGCLRARNILRDKLRSLEKKAF 1169
Query: 1165 KLAGMTFSLSMPADIAKVLFEHLKLPIPDGKNKGKNHPSTGKHCLDALRV---------- 1214
+LAGMTFSL PADIA VLF LKLPIP+ ++KGK HPST KHCLD LR
Sbjct: 1170 ELAGMTFSLHNPADIANVLFGQLKLPIPENQSKGKLHPSTDKHCLDLLRYSEWLIAFPYI 1229
Query: 1215 ------------------------------NARD--------------YFVPTQDNWLLL 1230
N RD +FVPTQ+NWLLL
Sbjct: 1230 AFCFFIIKDNRLFELLSASVEHEVEFKLDKNGRDVSSDADRYKINARDFFVPTQENWLLL 1289
Query: 1231 TADYSQIELRLMAHFSKDSTLIELLRKPDGDVFTMIAARWIGCPEVSVGSRQREQTKKMV 1290
TADYSQIELRLMAHFS+DS+LI L +P+GDVFTMIAA+W G E SV R+QTK+++
Sbjct: 1290 TADYSQIELRLMAHFSRDSSLISKLSQPEGDVFTMIAAKWTGKAEDSVSPHDRDQTKRLI 1349
Query: 1291 YGILYGMGPNSLAEQMDCTSDEASERISNFKSTFPGVASWLHEAVAFCRSKG*G--DLVP 1348
YGILYGMG N LAEQ++CTSDEA E+I +FKS+FP V SWL+E ++FC+ KG G L P
Sbjct: 1350 YGILYGMGANRLAEQLECTSDEAKEKIRSFKSSFPAVTSWLNETISFCQEKGWGCYRLFP 1409
Query: 1349 FLHFVYAFL-LNLNYLWAVCNLIMCKYIHDPDM*RLSREGNDFCQK*NLAVVQKNQKLRG 1407
L + A L + LN+++ YI Q L+G
Sbjct: 1410 LLFVLNAVLCVPLNHIFI--------YI---------------------------QTLKG 1434
Query: 1408 KLSIPFVRCNTLFSFSSKSGYVSLQAVSIHVNIVFLPSTSQGSAADIIKIAMIKIYSEIF 1467
+ F + + QAV+ S QGSAADIIKIAMI IYS I
Sbjct: 1435 RRRF---LSKIKFGNAKEKSKAQRQAVN---------SMCQGSAADIIKIAMINIYSAIA 1482
Query: 1468 PGVDKLDSSSSVTAKFEMLRDRCRILLQVHDELVLEVDPSVVKEAALLLQTSMENAVSLL 1527
VD SSSS +F ML+ RCRILLQVHDELVLEVDPS VK AA+LLQTSMENAVSLL
Sbjct: 1483 EDVDTAASSSSSETRFHMLKGRCRILLQVHDELVLEVDPSYVKLAAMLLQTSMENAVSLL 1542
Query: 1528 GLY 1530
G++
Sbjct: 1543 GMF 1545
>At2g42270 ATP-dependent RNA helicase like protein
Length = 2172
Score = 135 bits (340), Expect = 2e-31
Identities = 129/440 (29%), Positives = 193/440 (43%), Gaps = 73/440 (16%)
Query: 3 ILVLPYVSICTEKAEHLEKLLEPLGKHVRSYYGNQG-GGSLPKDTSVAVCTIEKANSLIN 61
+ V P ++ E + L + L+ G V+ G+Q G K+T + V T EK + +
Sbjct: 564 VYVAPMKALVAEVVDSLSQRLKDFGVTVKELSGDQSLTGQEIKETQIIVTTPEKWDIITR 623
Query: 62 RLLEEGRLSEMGIIVIDELHMVGDPKRGYLLELMLTKLRYAAGEGISKSSDGEGSGGSSD 121
+ + + +++IDE+H++ D + L ++ LR
Sbjct: 624 KSGDRTYTQLVRLLIIDEIHLLDDNRGPVLESIVARTLRQI------------------- 664
Query: 122 KNDPAQGLQIVGMSATMPNVAAVADWLQAAL------YQTEFRPVPL-EEYIKVGNSIYN 174
+ +++VG+SAT+PN VA +L+ L + +RPVPL ++YI +
Sbjct: 665 -ESTKEHIRLVGLSATLPNCDDVASFLRVDLKNGLFIFDRSYRPVPLGQQYIGINVKKPL 723
Query: 175 KSMELSRTI--PKGADLGGKDPDHVVELCNEVVQEGQSVLIFCSSRKGCESTARHVAKFL 232
+ +L I K + GK VLIF SRK TAR +
Sbjct: 724 RRFQLMNDICYQKVVAVAGK----------------HQVLIFVHSRKETAKTARAIRD-- 765
Query: 233 KSFTVDINENNCEFADITSAINSLRKCPAGL--DPVLEETFPAGVAFHHAGLTVEEREIV 290
T N+ F S + KC AGL + L+E P G A HHAGLT +REIV
Sbjct: 766 ---TAMANDTLSRFLKEDSQSREILKCLAGLLKNNDLKELLPYGFAIHHAGLTRTDREIV 822
Query: 291 ETCYRKGLLRVLTATSTLAAGVNLPARRVIFRQPRI------GCDFIDGTRYMQMAGRAG 344
E +R G L+VL +T+TLA GVNLPA VI + ++ + MQM GRAG
Sbjct: 823 ENQFRWGNLQVLISTATLAWGVNLPAHTVIIKGTQVYNPERGEWMELSPLDVMQMIGRAG 882
Query: 345 RTGIDTKGESVLICKPQELKKVMGLLNESCP---PLHSCLSEDLNGMTHAILEVVAGGIV 401
R D +GE ++I +L+ + L+NE P S L++ LN E+V G I
Sbjct: 883 RPQYDQQGEGIIITGYSKLQYYLRLMNEQLPIESQFISKLADQLNA------EIVLGTIQ 936
Query: 402 QTANDIHRKFLEWNEDTKLY 421
H W T LY
Sbjct: 937 NAREACH-----WLGYTYLY 951
Score = 75.9 bits (185), Expect = 2e-13
Identities = 87/390 (22%), Positives = 169/390 (43%), Gaps = 75/390 (19%)
Query: 48 VAVCTIEKANSLINRLLEEGRLSEMGIIVIDELHMVGDPKRGYLLELMLTKLRYAAGEGI 107
+ + T EK ++L R + + ++ + ++DELH++G + G +LE++++++RY + +
Sbjct: 1450 IIISTPEKWDALSRRWKQRKYIQQVSLFIVDELHLIGG-QGGQVLEVIVSRMRYISSQVG 1508
Query: 108 SKSSDGEGSGGSSDKNDPAQGLQIVGMSATMPNVAAVADWLQAAL-----YQTEFRPVPL 162
+K ++IV +S ++ N + +W+ A+ + RPVPL
Sbjct: 1509 NK-------------------IRIVALSTSLANAKDLGEWIGASSCGVFNFPPNVRPVPL 1549
Query: 163 EEYIKVGNSIYNKSMELSRTIPKGADLGGKDPDHVVELCNEVVQEGQS---VLIFCSSRK 219
E +I + + ++ + T P +VQ ++ ++F +RK
Sbjct: 1550 EIHIHGVDILSFEARMQAMTKP---------------TYTAIVQHAKNKKPAIVFVPTRK 1594
Query: 220 GCESTARHVAKFLKSFTVDINENNCEFADITSAINSLRKCPAGL----DPVLEETFPAGV 275
TA + + + +N + D + +L + L + L+ET G+
Sbjct: 1595 HVRLTAVDLIAY-------SHMDNMKSPDF--LLGNLEELEPFLIQICEETLKETLRHGI 1645
Query: 276 AFHHAGLTVEEREIVETCYRKGLLRVLTATSTLAAGVNLPARRVIFRQPRIGCDFIDG-- 333
+ H GL+ ++EIV + G ++V +S+L G L A V+ +G F DG
Sbjct: 1646 GYLHEGLSNLDQEIVTQLFEAGRIQVCVMSSSLCWGTPLKAHLVVV----MGTHFYDGRE 1701
Query: 334 --------TRYMQMAGRAGRTGIDTKGESVLICKPQELKKVMGLLNESCPPLHSCLSEDL 385
+ +QM GR R +D G+ V+ C + L E+ P+ S L L
Sbjct: 1702 NSHSDYPISNLLQMMGRGSRPLLDDAGKCVIFCHAPRKEYYKKFLYEAL-PVESHLQHFL 1760
Query: 386 NGMTHAILEVVAGGIVQTANDIHRKFLEWN 415
+ +A EVVA I + + +L W+
Sbjct: 1761 HDNFNA--EVVARVIENKQDAV--DYLTWS 1786
>At1g20960 putative ATP-dependent RNA helicase
Length = 2171
Score = 124 bits (310), Expect = 6e-28
Identities = 126/441 (28%), Positives = 192/441 (42%), Gaps = 75/441 (17%)
Query: 3 ILVLPYVSICTEKAEHLEKLLEPLGKHVRSYYGNQG-GGSLPKDTSVAVCTIEKANSLIN 61
+ V P ++ E +L L+ G VR G+Q G ++T + V T EK + +
Sbjct: 563 VYVAPMKALVAEVVGNLSNRLKDYGVIVRELSGDQSLTGREIEETQIIVTTPEKWDIITR 622
Query: 62 RLLEEGRLSEMGIIVIDELHMVGDPKRGYLLELMLTKLRYAAGEGISKSSDGEGSGGSSD 121
+ + + +++IDE+H++ D + L ++ LR
Sbjct: 623 KSGDRTYTQLVRLLIIDEIHLLHDNRGPVLESIVARTLRQI------------------- 663
Query: 122 KNDPAQGLQIVGMSATMPNVAAVADWLQAAL------YQTEFRPVPL-EEYIKVGNSIYN 174
+ +++VG+SAT+PN VA +L+ L + +RPVPL ++YI +
Sbjct: 664 -ETTKENIRLVGLSATLPNYEDVALFLRVDLKKGLFKFDRSYRPVPLHQQYIGISVKKPL 722
Query: 175 KSMELSRTIPKGADLGGKDPDHVVELCNEVVQEG---QSVLIFCSSRKGCESTARHVAKF 231
+ +L +LC + V G VLIF SRK TAR +
Sbjct: 723 QRFQLMN-----------------DLCYQKVLAGAGKHQVLIFVHSRKETSKTARAIRD- 764
Query: 232 LKSFTVDINENNCEFADITSAINSLRKCPAGL--DPVLEETFPAGVAFHHAGLTVEEREI 289
T N+ F S + + + L++ P G A HHAGL+ +REI
Sbjct: 765 ----TAMANDTLSRFLKEDSVTRDVLHSHEDIVKNSDLKDILPYGFAIHHAGLSRGDREI 820
Query: 290 VETCYRKGLLRVLTATSTLAAGVNLPARRVIFR-----QPRIGCDF-IDGTRYMQMAGRA 343
VET + +G ++VL +T+TLA GVNLPA VI + P G + MQM GRA
Sbjct: 821 VETLFSQGHVQVLVSTATLAWGVNLPAHTVIIKGTQVYNPEKGAWMELSPLDVMQMLGRA 880
Query: 344 GRTGIDTKGESVLICKPQELKKVMGLLNESCP---PLHSCLSEDLNGMTHAILEVVAGGI 400
GR D GE ++I EL+ + L+NE P S L++ LN E+V G
Sbjct: 881 GRPQYDQHGEGIIITGYSELQYYLSLMNEQLPIESQFISKLADQLNA------EIVL-GT 933
Query: 401 VQTANDIHRKFLEWNEDTKLY 421
VQ A R+ W T LY
Sbjct: 934 VQNA----REACHWLGYTYLY 950
Score = 82.4 bits (202), Expect = 2e-15
Identities = 95/402 (23%), Positives = 177/402 (43%), Gaps = 82/402 (20%)
Query: 48 VAVCTIEKANSLINRLLEEGRLSEMGIIVIDELHMVGDPKRGYLLELMLTKLRYAAGEGI 107
+ + T EK ++L R + + ++ + ++DELH++G + G +LE++++++RY + + I
Sbjct: 1449 IIISTPEKWDALSRRWKQRKYVQQVSLFIVDELHLIGG-QHGPVLEVIVSRMRYISSQVI 1507
Query: 108 SKSSDGEGSGGSSDKNDPAQGLQIVGMSATMPNVAAVADWLQAAL-----YQTEFRPVPL 162
+K ++IV +S ++ N + +W+ A+ + RPVPL
Sbjct: 1508 NK-------------------IRIVALSTSLANAKDLGEWIGASSHGLFNFPPGVRPVPL 1548
Query: 163 EEYIK-VGNSIYNKSMELSRTIPKGADLGGKDPDHVVELCNEVVQEGQS---VLIFCSSR 218
E +I+ V S + M+ + T P +VQ ++ ++F +R
Sbjct: 1549 EIHIQGVDISSFEARMQ-AMTKP---------------TYTAIVQHAKNKKPAIVFVPTR 1592
Query: 219 KGCESTARHVAKFLKSFTVDINENNCEF-----ADITSAINSLRKCPAGLDPVLEETFPA 273
K TA L +++ N + +F ++ + +R + L+ET
Sbjct: 1593 KHVRLTAVD----LMAYSHMDNPQSPDFLLGKLEELDPFVEQIR------EETLKETLCH 1642
Query: 274 GVAFHHAGLTVEEREIVETCYRKGLLRVLTATSTLAAGVNLPARRVIFRQPRIGCDFIDG 333
G+ + H GL+ ++EIV + G ++V +S+L G L A V+ +G + DG
Sbjct: 1643 GIGYLHEGLSSLDQEIVTQLFEAGRIQVCVMSSSLCWGTPLTAHLVVV----MGTQYYDG 1698
Query: 334 TR----------YMQMAGRAGRTGIDTKGESVLICKPQELKKVMGLLNESCPPLHSCLSE 383
+QM GRA R +D G+ V+ C + L E+ P+ S L
Sbjct: 1699 RENSHSDYPVPDLLQMMGRASRPLLDNAGKCVIFCHAPRKEYYKKFLYEAF-PVESQLQH 1757
Query: 384 DLNGMTHAILEVVAGGIVQTANDIHRKFLEWNEDTKLYSTTP 425
L+ +A EVVAG I + + +L W T +Y P
Sbjct: 1758 FLHDNFNA--EVVAGVIENKQDAV--DYLTW---TFMYRRLP 1792
>At5g61140 RNA helicase - like protein
Length = 2139
Score = 113 bits (282), Expect = 1e-24
Identities = 106/418 (25%), Positives = 180/418 (42%), Gaps = 65/418 (15%)
Query: 3 ILVLPYVSICTEKAEHLEKLLEPLGKHVRSYYGN-QGGGSLPKDTSVAVCTIEKANSLIN 61
+ V P ++ E + L PL V+ G+ Q + ++T + V T EK + +
Sbjct: 565 VYVAPMKALAAEVTSAFSRRLAPLNMVVKELTGDMQLTKTELEETQMIVTTPEKWDVITR 624
Query: 62 RLLEEGRLSEMGIIVIDELHMVGDPKRGYLLELMLTKLRYAAGEGISKSSDGEGSGGSSD 121
+ + + +++IDE+H++ D + + L+ LR
Sbjct: 625 KSSDMSMSMLVKLLIIDEVHLLNDDRGAVIEALVARTLRQVESTQTM------------- 671
Query: 122 KNDPAQGLQIVGMSATMPNVAAVADWLQAAL------YQTEFRPVPL-EEYIKVGNSIYN 174
++IVG+SAT+P+ VA +L+ + + +RPVPL ++YI + +
Sbjct: 672 -------IRIVGLSATLPSYLQVAQFLRVNTDTGLFYFDSSYRPVPLAQQYIGITEHNFA 724
Query: 175 KSMELSRTIPKGADLGGKDPDHVVELCNEVVQEGQSVLIFCSSRKGCESTARHVAKFLKS 234
EL I + + +++G +IF SRK TA + +
Sbjct: 725 ARNELLNEI-------------CYKKVVDSIKQGHQAMIFVHSRKDTSKTAEKLVDLARQ 771
Query: 235 F-TVDI--NENNCEFADITSAINSLRKCPAGLDPVLEETFPAGVAFHHAGLTVEEREIVE 291
+ T+D+ NE + +F + + R + L + F AG HHAG+ +R + E
Sbjct: 772 YETLDLFTNETHPQFQLMKKDVMKSR------NKDLVKFFEAGFGIHHAGMLRSDRTLTE 825
Query: 292 TCYRKGLLRVLTATSTLAAGVNLPARRVIFRQPRI------GCDFIDGTRYMQMAGRAGR 345
+ GLL+VL T+TLA GVNLPA V+ + ++ G + MQ+ GRAGR
Sbjct: 826 RLFSDGLLKVLVCTATLAWGVNLPAHTVVIKGTQLYDAKAGGWKDLGMLDVMQIFGRAGR 885
Query: 346 TGIDTKGESVLICKPQELKKVMGLLNESCP---PLHSCLSEDLNGMTHAILEVVAGGI 400
D GE ++I +L + LL P S L ++LN EVV G +
Sbjct: 886 PQFDKSGEGIIITSHDKLAYYLRLLTSQLPIESQFISSLKDNLNA------EVVLGTV 937
Score = 85.5 bits (210), Expect = 2e-16
Identities = 120/500 (24%), Positives = 192/500 (38%), Gaps = 125/500 (25%)
Query: 3 ILVLPYVSICTEKAEHLEK-LLEPLGKHVRSYYGNQGGGSLPK-DTSVAVCTIEKANSLI 60
+ + P +I E+ +K L+ PLGK + G+ + + + T EK + +
Sbjct: 1394 VYIAPLKAIVRERMNDWKKHLVAPLGKEMVEMTGDYTPDLVALLSADIIISTPEKWDGIS 1453
Query: 61 NRLLEEGRLSEMGIIVIDELHMVGDPKRGYLLEL---------MLTKLRYAAGEGISKSS 111
+ ++G++++DE+H++G RG +LEL +L K Y G ++ +
Sbjct: 1454 RNWHTRSYVKKVGLVILDEIHLLG-ADRGPILELYHDTCNVSMVLVKSLYLFGSALANAG 1512
Query: 112 DGEGSGGSSDKNDPAQGLQIVGMSATMPNVAAVADWLQAAL-----YQTEFRPVPLEEYI 166
D +ADWL ++ RPVP+E +I
Sbjct: 1513 D-------------------------------LADWLGVGEIGLFNFKPSVRPVPIEVHI 1541
Query: 167 KVGNSIYNKSMELSRTIPKGADLGGKDPDHVVELCNEVVQEGQSVLIFCSSRKGCESTAR 226
+ Y S P A + P + VLIF SSR+ TA
Sbjct: 1542 QGYPGKYYCPRMNSMNKPAYAAICTHSPT-------------KPVLIFVSSRRQTRLTAL 1588
Query: 227 HVA--KFLKSFTVDINENNCEFADITSAINSLRKCPAGL-DPVLEETFPAGVAFHHAGLT 283
+ KF S +E+ +F ++ L+ + + D L T G+ HHAGL
Sbjct: 1589 DLIQMKFAAS-----DEHPRQFLSVSE--EDLQMVLSQITDQNLRHTLQFGIGLHHAGLN 1641
Query: 284 VEEREIVETCYRKGL--------------------------------LRVLTATSTLAAG 311
+R VE + L+VL +TSTLA G
Sbjct: 1642 DHDRSAVEELFTNNKIQAISSTYSLCCLYRIHGYTFTLRLMMESMLPLQVLVSTSTLAWG 1701
Query: 312 VNLPARRVIFRQPRIGCDFIDG----------TRYMQMAGRAGRTGIDTKGESVLICKPQ 361
VNLPA VI + G ++ DG T +QM GRAGR D G++V++
Sbjct: 1702 VNLPAHLVIIK----GTEYFDGKTKRYVDFPLTEILQMMGRAGRPQFDQHGKAVILVHEP 1757
Query: 362 ELKKVMGLLNESCPPLHSCLSEDLNGMTHAILEVVAGGIVQTANDIHRKFLEWNEDTKLY 421
+ L E P+ S L E L+ H E+V+G I + +H +L W T L+
Sbjct: 1758 KKSFYKKFLYEPF-PVESSLKEKLH--DHFNAEIVSGTIGNKEDAVH--YLTW---TYLF 1809
Query: 422 STTPLGRASFGSSLCPEESL 441
A +G +E++
Sbjct: 1810 RRLMANPAYYGLEGTQDETI 1829
>At2g06990 HUA enhancer 2 (HEN2)
Length = 995
Score = 102 bits (253), Expect = 2e-21
Identities = 99/372 (26%), Positives = 161/372 (42%), Gaps = 83/372 (22%)
Query: 38 GGGSLPKDTSVAVCTIEKANSLINRLLEEGRLSEMGIIVIDELHMVGDPKRGYLLELMLT 97
G +L + S V T E +++ R E L E+ ++ DE+H + D +RG + E +
Sbjct: 156 GDVTLSPNASCLVMTTEILRAMLYRGSEV--LKEVAWVIFDEIHYMKDRERGVVWEESII 213
Query: 98 KLRYAAGEGISKSSDGEGSGGSSDKNDPAQGLQIVGMSATMPNVAAVADWL------QAA 151
L A +++V +SATM N A+W+
Sbjct: 214 FLPPA--------------------------IKMVFLSATMSNATEFAEWICYLHKQPCH 247
Query: 152 LYQTEFRPVPLEEYI--KVGNSIY-----------NKSMELSRTIPK------------- 185
+ T+FRP PL+ Y G +Y + +++ T PK
Sbjct: 248 VVYTDFRPTPLQHYAFPMGGGGLYLVVDDNEQFREDSFVKMQDTFPKPKSNDGKKSANGK 307
Query: 186 ----GADLGGKDPDHVVELCNEVVQEG--QSVLIFCSSRKGCESTARHVAKFLKSFTVDI 239
GA GG D V +++ E + V+IF SR+ CE A ++K +D
Sbjct: 308 SGGRGAKGGGGPGDSDVYKIVKMIMERKFEPVIIFSFSRRECEQHALSMSK------LDF 361
Query: 240 NENNCEFADITSAINSLRKCPAGLD---PVLEETFPA---GVAFHHAGLTVEEREIVETC 293
N + E + N+ +C D P +E P G+A HH+GL +E+VE
Sbjct: 362 NTDE-EKEVVEQVFNNAMQCLNEEDRSLPAIELMLPLLQRGIAVHHSGLLPVIKELVELL 420
Query: 294 YRKGLLRVLTATSTLAAGVNLPARRVIFRQPR----IGCDFIDGTRYMQMAGRAGRTGID 349
+++GL++ L AT T A G+N+PA+ V+F + +I Y+QM+GRAGR G D
Sbjct: 421 FQEGLVKALFATETFAMGLNMPAKTVVFTAVKKWDGDSHRYIGSGEYIQMSGRAGRRGKD 480
Query: 350 TKGESVLICKPQ 361
+G +++ Q
Sbjct: 481 ERGICIIMIDEQ 492
>At1g59760 hypothetical protein
Length = 988
Score = 102 bits (253), Expect = 2e-21
Identities = 106/397 (26%), Positives = 179/397 (44%), Gaps = 84/397 (21%)
Query: 38 GGGSLPKDTSVAVCTIEKANSLINRLLEEGRLSEMGIIVIDELHMVGDPKRGYLLELMLT 97
G ++ + S V T E S+ + E R E+ I+ DE+H + D +RG + E +
Sbjct: 142 GDVTIDPNASCLVMTTEILRSMQYKGSEIMR--EVAWIIFDEVHYMRDSERGVVWEESIV 199
Query: 98 KLRYAAGEGISKSSDGEGSGGSSDKNDPAQGLQIVGMSATMPNVAAVADWLQAALYQ--- 154
+ KN + V +SAT+PN ADW+ Q
Sbjct: 200 M---------------------APKNS-----RFVFLSATVPNAKEFADWVAKVHQQPCH 233
Query: 155 ---TEFRPVPLEEYI--KVGNSIY------------NKSMELSRTIP------------- 184
T++RP PL+ Y+ GN +Y + L+ +P
Sbjct: 234 IVYTDYRPTPLQHYVFPAGGNGLYLVVDEKSKFHEDSFQKSLNALVPTNESDKKRDNGKF 293
Query: 185 -KGADLG--GKDPDHVVELCNEVVQ-EGQSVLIFCSSRKGCESTARHVAKFLKSFTVDIN 240
KG +G G++ D + +L ++Q + V++F S+K CE+ A ++K + + + +
Sbjct: 294 QKGLVIGKLGEESD-IFKLVKMIIQRQYDPVILFSFSKKECEALAMQMSKMVLNSDDEKD 352
Query: 241 ENNCEFADITSAINSL----RKCP--AGLDPVLEETFPAGVAFHHAGLTVEEREIVETCY 294
FA SAI+ L +K P + + P+L+ G+ HH+GL +E++E +
Sbjct: 353 AVETIFA---SAIDMLSDDDKKLPQVSNILPILKR----GIGVHHSGLLPILKEVIEILF 405
Query: 295 RKGLLRVLTATSTLAAGVNLPARRVIFRQPR--IGCDF--IDGTRYMQMAGRAGRTGIDT 350
++GL++ L AT T + G+N+PA+ V+F R G F + Y+QM+GRAGR GID
Sbjct: 406 QEGLIKCLFATETFSIGLNMPAKTVVFTNVRKFDGDKFRWLSSGEYIQMSGRAGRRGIDK 465
Query: 351 KGESVL-ICKPQELKKVMGLLNESCPPLHSCLSEDLN 386
+G +L + + E +L S L+S N
Sbjct: 466 RGICILMVDEKMEPAVAKSMLKGSADSLNSAFHLSYN 502
>At1g70070 Similar to Synechocystis antiviral protein (At1g70070)
Length = 1171
Score = 88.2 bits (217), Expect = 3e-17
Identities = 94/387 (24%), Positives = 165/387 (42%), Gaps = 100/387 (25%)
Query: 38 GGGSLPKDTSVAVCTIEKANSLINR---LLEEGR-LSEMGIIVIDELHMVGDPKRGYLLE 93
G ++ KD + + T E +++ + + G L + IV+DE+H + D RG + E
Sbjct: 231 GDSAINKDAQIVIMTTEILRNMLYQSVGMASSGTGLFHVDAIVLDEVHYLSDISRGTVWE 290
Query: 94 LMLTKLRYAAGEGISKSSDGEGSGGSSDKNDPAQGLQIVGMSATMPNVAAVADWLQAALY 153
++ Y E +Q++ +SAT+ N +A W+
Sbjct: 291 EIVI---YCPKE-----------------------VQLICLSATVANPDELAGWIGEIHG 324
Query: 154 QTEF-----RPVPLEEYIKVGNSIY----------NKSMELS------------------ 180
+TE RPVPL Y +S+ N+ + L+
Sbjct: 325 KTELVTSTRRPVPLTWYFSTKHSLLPLLDEKGINVNRKLSLNYLQLSASEARFRDDDDGY 384
Query: 181 ---RTIPKGADLGGKDPDHVVELC---NEVVQ----------------EGQSVLI---FC 215
R+ +G D + +V + NE+ + +G+++L F
Sbjct: 385 RKRRSKKRGGDTSYNNLVNVTDYPLSKNEINKIRRSQVPQISDTLWHLQGKNMLPAIWFI 444
Query: 216 SSRKGCESTARHVAKFLKSFTVDINENNCEFADITSAINSLRKC-PAGLDPVLEETFPAG 274
+R+GC++ ++V F ++CE +++ A+ R P + E+ G
Sbjct: 445 FNRRGCDAAVQYVENFQLL-------DDCEKSEVELALKKFRVLYPDAVRESAEKGLLRG 497
Query: 275 VAFHHAGLTVEEREIVETCYRKGLLRVLTATSTLAAGVNLPARRVIFR--QPRIGCDFID 332
+A HHAG + +E +++GL++V+ AT TLAAG+N+PAR + + G + I+
Sbjct: 498 IAAHHAGCLPLWKSFIEELFQRGLVKVVFATETLAAGINMPARTAVISSLSKKAGNERIE 557
Query: 333 --GTRYMQMAGRAGRTGIDTKGESVLI 357
QMAGRAGR GID KG +VL+
Sbjct: 558 LGPNELYQMAGRAGRRGIDEKGYTVLV 584
>At3g46960 putative helicase
Length = 1347
Score = 82.0 bits (201), Expect = 2e-15
Identities = 100/412 (24%), Positives = 158/412 (38%), Gaps = 108/412 (26%)
Query: 31 RSYYGNQGGGSLPKDTSV---AVCTIEKANSLINRLLEEGR--LSEMGIIVIDELHMVGD 85
R + G G L D S+ A C I ++ +L G + ++ ++ DE+H V D
Sbjct: 412 RDFCGKFDVGLLTGDVSIRPEASCLI-MTTEILRSMLYRGADIIRDIEWVIFDEVHYVND 470
Query: 86 PKRGYLLELMLTKLRYAAGEGISKSSDGEGSGGSSDKNDPAQGLQIVGMSATMPNVAAVA 145
+RG + E ++ L + + V +SAT+PN A
Sbjct: 471 VERGVVWEEVIIML--------------------------PRHINFVLLSATVPNTFEFA 504
Query: 146 DWL------QAALYQTEFRPVPLEEYIKVGNSIY---NKSMELSRTIPKGADLGGKDPDH 196
DW+ + + T RPVPLE + +Y + LS+ I D K +
Sbjct: 505 DWIGRTKQKEIRVTGTTKRPVPLEHCLFYSGELYKVCENEVFLSKGIKDAKDSQKKKNSN 564
Query: 197 VV------ELCNEVVQEGQSVLIFCSSRKGCESTARHVAKFLKSFTVDINENNCEF---- 246
V ++ + Q+G + +G ++ V KS ++NN F
Sbjct: 565 AVSVAPKQQMGSSAHQDGSKSQKHEAHSRGKQNKHSSVKDVGKSSYSGNSQNNGAFRRSA 624
Query: 247 --------------------------------------ADITS-------------AINS 255
D+TS A +
Sbjct: 625 ASNWLLLINKLSKMSLLPVVVFCFSKNYCDRCADALTGTDLTSSSEKSEIRVFCDKAFSR 684
Query: 256 LRKCPAGLDPVL--EETFPAGVAFHHAGLTVEEREIVETCYRKGLLRVLTATSTLAAGVN 313
L+ L VL + G+ HHAGL +E+VE + +G+++VL +T T A GVN
Sbjct: 685 LKGSDRNLPQVLRLQSLLHRGIGVHHAGLLPIVKEVVEMLFCRGVIKVLFSTETFAMGVN 744
Query: 314 LPARRVIFRQPR--IGCDF--IDGTRYMQMAGRAGRTGIDTKGESVLICKPQ 361
PAR V+F R G +F + Y QMAGRAGR G+D G V++C+ +
Sbjct: 745 APARTVVFDALRKFDGKEFRQLLPGEYTQMAGRAGRRGLDKTGTVVVMCRDE 796
>At3g27730 hypothetical protein
Length = 1113
Score = 46.6 bits (109), Expect = 1e-04
Identities = 28/109 (25%), Positives = 60/109 (54%), Gaps = 17/109 (15%)
Query: 44 KDTSVAVCTIEKANSLINRLLEEGRL---SEMGIIVIDELHMVGDPKRGYLLELMLTKLR 100
+D + + T EK +++ + G L S++ +++IDE+H++ DP RG LE ++++L+
Sbjct: 128 QDADIILTTPEKFDAVSRYRVTSGGLGFFSDIALVLIDEVHLLNDP-RGAALEAIVSRLK 186
Query: 101 YAAGEGISKSSDGEGSGGSSDKNDPAQGLQIVGMSATMPNVAAVADWLQ 149
+ +SS ++++ +SAT+PN+ +A+WL+
Sbjct: 187 ILSSNHELRSS-------------TLASVRLLAVSATIPNIEDLAEWLK 222
>At3g20540 DNA polymerase, putative
Length = 1160
Score = 42.4 bits (98), Expect = 0.002
Identities = 30/122 (24%), Positives = 56/122 (45%), Gaps = 2/122 (1%)
Query: 1209 LDALRVNARDYFVPTQDNWLLLTADYSQIELRLMAHFSKDSTLIELLRKPDGDVFTMIAA 1268
L+ R R F+ + N L++ ADY Q+ELR++AH + ++ E GD + A
Sbjct: 782 LEKDRYKIRQAFIASPGNSLIV-ADYGQLELRILAHLASCESMKEAF-IAGGDFHSRTAM 839
Query: 1269 RWIGCPEVSVGSRQREQTKKMVYGILYGMGPNSLAEQMDCTSDEASERISNFKSTFPGVA 1328
+V + +R + K + + I YG L+ + +EA + ++ + + V
Sbjct: 840 NMYPHIREAVENGERRKAKMLNFSIAYGKTAIGLSRDWKVSREEAQDTVNLWYNDRQEVR 899
Query: 1329 SW 1330
W
Sbjct: 900 KW 901
Score = 35.0 bits (79), Expect = 0.35
Identities = 31/106 (29%), Positives = 50/106 (46%), Gaps = 19/106 (17%)
Query: 1399 VQKNQKLRGKLSIPFVRCNTLFSFSSKSGYVSLQAVSIHVNIVFLPSTSQGSAADIIKIA 1458
V+K Q+LR K +I TL + K +A H+ + + QGSAAD+ A
Sbjct: 898 VRKWQELRKKEAIQKGYVLTLLGRARKFPEYRSRAQKNHIERAAINTPVQGSAADVAMCA 957
Query: 1459 MIKIYSEIFPGVDKLDSSSSVTAKFEMLRD-RCRILLQVHDELVLE 1503
M++I + + L++ ++LLQVHDE++LE
Sbjct: 958 MLEISNN------------------QRLKELGWKLLLQVHDEVILE 985
>At1g10930 DNA helicase isolog
Length = 665
Score = 39.7 bits (91), Expect = 0.014
Identities = 25/81 (30%), Positives = 39/81 (47%), Gaps = 8/81 (9%)
Query: 269 ETFPAGVAFHHAGLTVEEREIVETCYRKGLLRVLTATSTLAAGVNLPARRVIFRQ--PRI 326
+ F AF+H + E+R ++T + K + ++ AT G+N P R + P+
Sbjct: 365 QEFGHKAAFYHGSMEPEQRAFIQTQWSKDEINIICATVAFGMGINKPDVRFVIHHSLPK- 423
Query: 327 GCDFIDGTRYMQMAGRAGRTG 347
I+G Y Q GRAGR G
Sbjct: 424 ---SIEG--YHQECGRAGRDG 439
>At5g14610 DRH1 DEAD box protein - like
Length = 713
Score = 39.3 bits (90), Expect = 0.019
Identities = 42/178 (23%), Positives = 67/178 (37%), Gaps = 55/178 (30%)
Query: 206 QEGQSVLIFCSSRKGCESTARHVAKFLKSFTVDINENNCEFADITSAINSLRKCPAGLDP 265
+ G ++IFCS+++ C+ AR++ +
Sbjct: 472 EPGSKIIIFCSTKRMCDQLARNLTR----------------------------------- 496
Query: 266 VLEETFPAGVAFHHAGLTVEEREIVETCYRKGLLRVLTATSTLAAGVNLPARRVIFRQPR 325
TF G A H + ER+ V +R G VL AT A G+++ RV+
Sbjct: 497 ----TF--GAAAIHGDKSQAERDDVLNQFRSGRTPVLVATDVAARGLDVKDIRVV----- 545
Query: 326 IGCDFIDGTR-YMQMAGRAGRTGIDTKGESVLICKPQELK------KVMGLLNESCPP 376
+ DF +G Y+ GR GR G G + Q+ K K++ N+ PP
Sbjct: 546 VNYDFPNGVEDYVHRIGRTGRAG--ATGLAYTFFGDQDAKHASDLIKILEGANQKVPP 601
>At5g11170 DEAD BOX RNA helicase RH15 - like protein
Length = 427
Score = 38.9 bits (89), Expect = 0.024
Identities = 37/177 (20%), Positives = 77/177 (42%), Gaps = 13/177 (7%)
Query: 200 LCNEVVQEGQSVLIFCSSRKGCESTARHVAKFLK-SFTVDINE--NNCEFADITSAINSL 256
+C + +Q+ + + ++ +H K + T +N+ + +F + + S+
Sbjct: 239 VCKKFMQDPMEIYVDDEAKLTLHGLVQHYIKLSEMEKTRKLNDLLDALDFNQVVIFVKSV 298
Query: 257 RKCPAGLDPVLEETFPAGVAFHHAGLTVEEREIVETCYRKGLLRVLTATSTLAAGVNLPA 316
+ ++E FP+ H+G++ EER +++G R+L AT + G+++
Sbjct: 299 SRAAELNKLLVECNFPSICI--HSGMSQEERLTRYKSFKEGHKRILVATDLVGRGIDIER 356
Query: 317 RRVIFRQPRIGCDFIDGT-RYMQMAGRAGRTGIDTKGESVLICKPQELKKVMGLLNE 372
++ I D D Y+ GRAGR G TKG ++ +V+ + E
Sbjct: 357 VNIV-----INYDMPDSADTYLHRVGRAGRFG--TKGLAITFVASASDSEVLNQVQE 406
>At3g06480 putative RNA helicase
Length = 1088
Score = 38.9 bits (89), Expect = 0.024
Identities = 44/178 (24%), Positives = 62/178 (34%), Gaps = 55/178 (30%)
Query: 206 QEGQSVLIFCSSRKGCESTARHVAKFLKSFTVDINENNCEFADITSAINSLRKCPAGLDP 265
+ G V+IFCS+++ C+ AR V +
Sbjct: 678 ERGSKVIIFCSTKRLCDHLARSVGRHF--------------------------------- 704
Query: 266 VLEETFPAGVAFHHAGLTVEEREIVETCYRKGLLRVLTATSTLAAGVNLPARRVIFRQPR 325
G H T ER+ V +R G VL AT A G+++ RV+
Sbjct: 705 --------GAVVIHGDKTQGERDWVLNQFRSGKSCVLIATDVAARGLDIKDIRVV----- 751
Query: 326 IGCDFIDGTR-YMQMAGRAGRTGIDTKGESVLICKPQELK------KVMGLLNESCPP 376
I DF G Y+ GR GR G G + Q+ K KV+ N+ PP
Sbjct: 752 INYDFPTGVEDYVHRIGRTGRAG--ATGVAFTFFTEQDWKYAPDLIKVLEGANQQVPP 807
>At1g31360 unknown protein
Length = 705
Score = 38.1 bits (87), Expect = 0.041
Identities = 36/157 (22%), Positives = 54/157 (33%), Gaps = 44/157 (28%)
Query: 195 DHVVELCNEVVQEGQSVLIFCSSRKGCESTARHVAKFLKSFTVDINENNCEFADITSAIN 254
D + E E +S +++C SRK CE A D+ E AD
Sbjct: 300 DEIAEFIRESYSNNESGIVYCFSRKECEQIAG-----------DLRERGIS-AD------ 341
Query: 255 SLRKCPAGLDPVLEETFPAGVAFHHAGLTVEEREIVETCYRKGLLRVLTATSTLAAGVNL 314
++HA + RE V + K L+V+ T G+N
Sbjct: 342 ----------------------YYHADMDANMREKVHMRWSKNKLQVIVGTVAFGMGINK 379
Query: 315 PARRVIFRQPRIGCDFIDGTRYMQMAGRAGRTGIDTK 351
P R + Y Q +GRAGR G+ ++
Sbjct: 380 PDVRFVIHHSLSK----SMETYYQESGRAGRDGLPSE 412
>At5g11200 DEAD BOX RNA helicase RH15
Length = 427
Score = 37.7 bits (86), Expect = 0.054
Identities = 26/95 (27%), Positives = 45/95 (47%), Gaps = 8/95 (8%)
Query: 279 HAGLTVEEREIVETCYRKGLLRVLTATSTLAAGVNLPARRVIFRQPRIGCDFIDGT-RYM 337
H+G++ EER +++G R+L AT + G+++ ++ I D D Y+
Sbjct: 319 HSGMSQEERLTRYKSFKEGHKRILVATDLVGRGIDIERVNIV-----INYDMPDSADTYL 373
Query: 338 QMAGRAGRTGIDTKGESVLICKPQELKKVMGLLNE 372
GRAGR G TKG ++ +V+ + E
Sbjct: 374 HRVGRAGRFG--TKGLAITFVASASDSEVLNQVQE 406
>At2g33730 putative U5 small nuclear ribonucleoprotein, an RNA
helicase
Length = 733
Score = 37.0 bits (84), Expect = 0.092
Identities = 58/249 (23%), Positives = 95/249 (37%), Gaps = 37/249 (14%)
Query: 139 PNVAAVADWLQAALYQTEFRPVPLEEYIKVGNSIYNKSMELSRTIPKGADLGGKDPDHVV 198
P VA V D + ++ + E L+E IY + S T+P G V
Sbjct: 482 PQVAGVLDAMPSSNLKPENEEEELDE-----KKIYRTTYMFSATMPPG-----------V 525
Query: 199 ELCNEVVQEGQSVLIFCSSRKGCESTARHVAKFLKSFTV-DINENNCEFADITSAI--NS 255
E V+ ++ K + ++HV +S + + E + T+ + N+
Sbjct: 526 ERLARKYLRNPVVVTIGTAGKTTDLISQHVIMMKESEKFFRLQKLLDELGEKTAIVFVNT 585
Query: 256 LRKCPAGLDPVLEETFPAG--VAFHHAGLTVEEREIVETCYRKGLLRVLTATSTLAAGVN 313
+ C D + + AG V H G + E+REI +R VL AT + G++
Sbjct: 586 KKNC----DSIAKNLDKAGYRVTTLHGGKSQEQREISLEGFRAKRYNVLVATDVVGRGID 641
Query: 314 LP--ARRVIFRQPRIGCDFIDGTRYMQMAGRAGRTGIDTKGESVLICKPQE----LKKVM 367
+P A + + P+ Y GR GR G S L E LK+++
Sbjct: 642 IPDVAHVINYDMPK------HIEMYTHRIGRTGRAGKSGVATSFLTLHDTEVFYDLKQML 695
Query: 368 GLLNESCPP 376
N + PP
Sbjct: 696 VQSNSAVPP 704
>At3g01540 AT3g01540/F4P13_9
Length = 619
Score = 36.6 bits (83), Expect = 0.12
Identities = 39/178 (21%), Positives = 64/178 (35%), Gaps = 55/178 (30%)
Query: 206 QEGQSVLIFCSSRKGCESTARHVAKFLKSFTVDINENNCEFADITSAINSLRKCPAGLDP 265
+ G V+IFCS+++ C+ R++ +
Sbjct: 401 EPGSKVIIFCSTKRMCDQLTRNLTRQF--------------------------------- 427
Query: 266 VLEETFPAGVAFHHAGLTVEEREIVETCYRKGLLRVLTATSTLAAGVNLPARRVIFRQPR 325
G A H + ER+ V +R G VL AT A G+++ R +
Sbjct: 428 --------GAAAIHGDKSQPERDNVLNQFRSGRTPVLVATDVAARGLDVKDIRAV----- 474
Query: 326 IGCDFIDGTR-YMQMAGRAGRTGIDTKGESVLICKPQELK------KVMGLLNESCPP 376
+ DF +G Y+ GR GR G G++ Q+ K K++ N+ PP
Sbjct: 475 VNYDFPNGVEDYVHRIGRTGRAG--ATGQAFTFFGDQDSKHASDLIKILEGANQRVPP 530
>At1g50840 unknown protein
Length = 1050
Score = 36.6 bits (83), Expect = 0.12
Identities = 31/105 (29%), Positives = 50/105 (47%), Gaps = 17/105 (16%)
Query: 1399 VQKNQKLRGKLSIPFVRCNTLFSFSSKSGYVSLQAVSIHVNIVFLPSTSQGSAADIIKIA 1458
V+K Q++R K +I TL S + +A H+ + + QGSAAD+ A
Sbjct: 918 VRKWQEMRKKEAIEDGYVLTLLGRSRRFPASKSRAQRNHIQRAAINTPVQGSAADVAMCA 977
Query: 1459 MIKIYSEIFPGVDKLDSSSSVTAKFEMLRDRCRILLQVHDELVLE 1503
M++I S+ + + L R+LLQ+HDE++LE
Sbjct: 978 MLEI---------------SINQQLKKL--GWRLLLQIHDEVILE 1005
Score = 33.9 bits (76), Expect = 0.78
Identities = 34/148 (22%), Positives = 57/148 (37%), Gaps = 28/148 (18%)
Query: 1209 LDALRVNARDYFVPTQDNWLLLTADYSQIELRLMAHFSKDSTLIELLRKPDGDVFTMIAA 1268
L+ R R FV + N L++ ADY Q+ELR++AH + +++E K GD + A
Sbjct: 776 LEKDRYKIRKAFVASPGNTLVV-ADYGQLELRILAHLTGCKSMMEAF-KAGGDFHSRTAM 833
Query: 1269 RWIGCPEVSVGSRQ--------------------------REQTKKMVYGILYGMGPNSL 1302
+V + Q R + K + + I YG L
Sbjct: 834 NMYPHVREAVENGQVILEWHPEPGEDKPPVPLLKDAFGSERRKAKMLNFSIAYGKTAVGL 893
Query: 1303 AEQMDCTSDEASERISNFKSTFPGVASW 1330
+ ++ EA E + + + V W
Sbjct: 894 SRDWKVSTKEAQETVDLWYNDRQEVRKW 921
>At3g05740 putative DNA helicase
Length = 624
Score = 35.4 bits (80), Expect = 0.27
Identities = 23/104 (22%), Positives = 49/104 (47%), Gaps = 6/104 (5%)
Query: 267 LEETFPAGVAFHHAGLTVEEREIVETCYRKGLLRVLTATSTLAAGVNLPARRVIFRQPRI 326
L E ++HAG+ ++R V+ ++ G +R++ AT G++ R +
Sbjct: 465 LNEKCKVKTVYYHAGVPAKQRVDVQRKWQTGEVRIVCATIAFGMGIDKADVRFVIHNTLS 524
Query: 327 GCDFIDGTRYMQMAGRAGRTGIDTKGESVLICKPQELKKVMGLL 370
Y Q +GRAGR G+ + + + + + ++ +V+ +L
Sbjct: 525 KA----VESYYQESGRAGRDGL--QAQCICLYQKKDFSRVVCML 562
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.320 0.136 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 35,567,127
Number of Sequences: 26719
Number of extensions: 1537896
Number of successful extensions: 4075
Number of sequences better than 10.0: 37
Number of HSP's better than 10.0 without gapping: 11
Number of HSP's successfully gapped in prelim test: 26
Number of HSP's that attempted gapping in prelim test: 3984
Number of HSP's gapped (non-prelim): 79
length of query: 1603
length of database: 11,318,596
effective HSP length: 113
effective length of query: 1490
effective length of database: 8,299,349
effective search space: 12366030010
effective search space used: 12366030010
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.8 bits)
S2: 67 (30.4 bits)
Medicago: description of AC127429.13


