

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC127428.4 + phase: 0
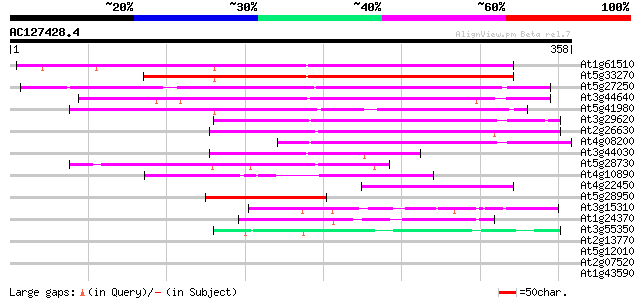
(358 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
At1g61510 hypothetical protein 211 5e-55
At5g33270 putative protein 201 5e-52
At5g27250 putative protein 196 1e-50
At3g44640 putative protein 181 7e-46
At5g41980 unknown protein 177 6e-45
At3g29620 hypothetical protein 169 3e-42
At2g26630 En/Spm-like transposon protein 165 4e-41
At4g08200 putative protein 119 2e-27
At3g44030 putative protein 102 2e-22
At5g28730 putative protein 97 2e-20
At4g10890 putative protein 82 6e-16
At4g22450 hypothetical protein 79 3e-15
At5g28950 putative protein 71 8e-13
At3g15310 unknown protein 46 4e-05
At1g24370 hypothetical protein 45 5e-05
At3g55350 unknown protein 43 2e-04
At2g13770 hypothetical protein 39 0.006
At5g12010 putative protein 38 0.007
At2g07520 pseudogene 34 0.11
At1g43590 hypothetical protein 34 0.11
>At1g61510 hypothetical protein
Length = 608
Score = 211 bits (537), Expect = 5e-55
Identities = 136/372 (36%), Positives = 187/372 (49%), Gaps = 56/372 (15%)
Query: 5 KEQLEHNTQEEEDDD---TFEESYILAALLGE-YATKYLCKEPCRTSELTGHAW--VQEI 58
K+ + +EDDD EE + AL+ E Y K + E G W VQ +
Sbjct: 205 KQSTSQSVMLQEDDDMDLVLEEENNMYALVKELYGGKNVNTERQLVRINRGGGWRRVQRL 264
Query: 59 LQGNPTRCYEMFRMEKHIFHKLCHELVE-HDLKSSKHMGVEEMVAMFLVVVGHGVGNRMI 117
+ + C+++ RM + F LC L E + L+ S ++ +EE VAMF+ +V + R+I
Sbjct: 265 MVESKVECFDILRMHQSTFRTLCKILSEQYKLEESCNIYLEESVAMFIEMVAQDLTVRVI 324
Query: 118 QERFQHSGETVR-----------------------------------------------V 130
ER+QHS ETV+
Sbjct: 325 AERYQHSLETVKRKLDEVLSALLKLAADIVKPTRDDVTGISPFLVNDKRYMPYFIDCIGA 384
Query: 131 IDGTHVSCVVSASEQPRFIGRKGYPTQNIMAVCDWNMCFTFVLAGWEGTAHDARVFDKAL 190
+DGTHVS + + R+ GRK T NI+AVC+++M F G G AHD +V
Sbjct: 385 LDGTHVSVRPPSGDVERYRGRKSEATMNILAVCNFSMKFNIAYVGVPGRAHDTKVLTYC- 443
Query: 191 TTANLNFPHPPQGKYYLVDSGYPTPIGYIGPYRCERYHLPEFRRSSGFENHNEVFNYYHS 250
T +FPHPP GKYYLVDSGYPT GY+GP+R RYHL F R N E+FN HS
Sbjct: 444 ATHEASFPHPPAGKYYLVDSGYPTRSGYLGPHRRTRYHLELFNRGGPPTNSRELFNRRHS 503
Query: 251 SLRCTIERTFGVWKNRFAIL-RSMPKFKYETQVHIVVATMAIHNFIRRSAEMDVDFNLYE 309
SLR IERTFGVWK ++ IL R PK++ + + IV +TMA+HN+IR S + D DF +E
Sbjct: 504 SLRSVIERTFGVWKAKWRILDRKHPKYEVKKWIKIVTSTMALHNYIRDSQQEDSDFRHWE 563
Query: 310 DENTVIHHDDDH 321
+ H D++
Sbjct: 564 IVESYEQHGDEN 575
>At5g33270 putative protein
Length = 343
Score = 201 bits (511), Expect = 5e-52
Identities = 110/248 (44%), Positives = 151/248 (60%), Gaps = 13/248 (5%)
Query: 86 EHDLKSSKHMGVEEMVAMFLVVVGHGVGNRMIQERFQHSGETVR-----------VIDGT 134
++ L+ S ++ +EE VAMF+ +V + R+I ER+QHS ETV+ +DGT
Sbjct: 64 QYKLEESCNIYLEESVAMFIEMVAQDLTVRVIAERYQHSLETVKRKLDEVLNCIGALDGT 123
Query: 135 HVSCVVSASEQPRFIGRKGYPTQNIMAVCDWNMCFTFVLAGWEGTAHDARVFDKALTTAN 194
HVS + + R+ GRK T NI+A+C+++M FT+ G G AHD +V T
Sbjct: 124 HVSVRPPSGDVERYRGRKSEATMNILALCNFSMKFTYAYVGVPGRAHDTKVLTYC-ATHE 182
Query: 195 LNFPHPPQGKYYLVDSGYPTPIGYIGPYRCERYHLPEFRRSSGFENHNEVFNYYHSSLRC 254
+FPHPP GKYYLVDSGYPT GY+GP+R RYHL F R N E+FN HSSLR
Sbjct: 183 ASFPHPPAGKYYLVDSGYPTRSGYLGPHRRTRYHLELFNRGGPPTNSRELFNRRHSSLRS 242
Query: 255 TIERTFGVWKNRFAIL-RSMPKFKYETQVHIVVATMAIHNFIRRSAEMDVDFNLYEDENT 313
IERTFGVWK ++ IL R K++ + + IV +TMA+HN+IR S + D DF +E +
Sbjct: 243 VIERTFGVWKAKWRILDRKHLKYEVKKWIKIVTSTMALHNYIRDSQQEDSDFRHWEIVES 302
Query: 314 VIHHDDDH 321
H D++
Sbjct: 303 YEQHGDEN 310
>At5g27250 putative protein
Length = 348
Score = 196 bits (499), Expect = 1e-50
Identities = 114/341 (33%), Positives = 186/341 (54%), Gaps = 16/341 (4%)
Query: 8 LEHNTQEEEDDDTFEESYILAALLGEYATKYLCKEPCRTSELTGHAWVQEILQGNPTRCY 67
+E N EE+D D ++ + Y ++ + + R GHA++Q+ L+ +P
Sbjct: 1 MERNLCEEDDSDVDIMLLMVVGSVDAYESQ-IPRPMRRQITKKGHAYIQKALKDDPIHFR 59
Query: 68 EMFRMEKHIFHKLCHEL-VEHDLKSSKHMGVEEMVAMFLVVVGHGVGNRMIQERFQHSG- 125
+++RM +F +LCH L ++ +S KH + G G ++ + H
Sbjct: 60 QLYRMNPEVFAELCHLLQMKTGSQSFKHACTN--------IDGQGYKYSALKNKQNHKIL 111
Query: 126 ETVRVIDGTHVSCVVSASEQPRFIGRKGYPTQNIMAVCDWNMCFTFVLAGWEGTAHDARV 185
++ IDGTH++ +V E+ + RKG +QN++A C++++ F +VL+GWEG+AHD++V
Sbjct: 112 SLLQAIDGTHINAMVQGPEKASYRNRKGVISQNVLAACNFDLEFIYVLSGWEGSAHDSKV 171
Query: 186 FDKALTTANLNFPHPPQGKYYLVDSGYPTPIGYIGPYRCERYHLPEFR-RSSGFENHNEV 244
ALT N P+GKYYL D G+P ++ P R RYHL +FR N NE+
Sbjct: 172 LQDALTRRT-NRLQVPEGKYYLADCGFPNRRNFLAPLRSTRYHLQDFRGEGRDPTNQNEL 230
Query: 245 FNYYHSSLRCTIERTFGVWKNRFAILRSMPKFKYETQVHIVVATMAIHNFIRRSAEMDVD 304
FN H+SLR IER FG++K+RF I +S P F ++TQ IV++ A+HNF+R+ D
Sbjct: 231 FNLRHASLRNVIERIFGIFKSRFLIFKSAPPFSFKTQAEIVLSCAALHNFLRQKCRSDEF 290
Query: 305 FNLYEDENTVIHHDDDHRSTNLNQSQSFNVASSSEMDHARN 345
+ EDE V D+ ++++ N + E +HAR+
Sbjct: 291 SSDEEDETDV---DNANQNSEENGGEENVETQEQEREHARH 328
>At3g44640 putative protein
Length = 377
Score = 181 bits (458), Expect = 7e-46
Identities = 102/319 (31%), Positives = 173/319 (53%), Gaps = 25/319 (7%)
Query: 45 RTSELTGHAWVQEILQGNPTRCYEMFRMEKHIFHKLCHELVEHDLKSS------KHMGVE 98
R+ G+ ++Q L NP +++RME +FHKLC ++ ++ S KH +
Sbjct: 50 RSVSRVGYNYIQTALTVNPAHFRQLYRMEPEVFHKLCDVILLYNGHISEIKLFYKHKFHK 109
Query: 99 EMVAMFLVV--------VGHGVGNRMIQERFQHSGETVRVIDGTHVSCVVSASEQPRFIG 150
+ A+ V V R + + + + IDGTH+ +V + F
Sbjct: 110 ALKALAFVAPNWMAKPEVRVPAKIRESTSFYPYFKDCIGAIDGTHIFAMVPTCDAASFRN 169
Query: 151 RKGYPTQNIMAVCDWNMCFTFVLAGWEGTAHDARVFDKALTTANLNFPHPPQGKYYLVDS 210
RKG+ +QN++A C++++ F ++L+GW+G+AHD++V + AL T N N P+GK+YLVD
Sbjct: 170 RKGFISQNVLAACNFDLQFIYILSGWKGSAHDSKVLNDAL-TRNTNRLQVPEGKFYLVDC 228
Query: 211 GYPTPIGYIGPYRCERYHLPEFR-RSSGFENHNEVFNYYHSSLRCTIERTFGVWKNRFAI 269
GY ++ P+R RYHL +FR + + NE+FN H+SLR IER FG++K+RF I
Sbjct: 229 GYANRRKFLAPFRRTRYHLQDFRGQGRDPKTQNELFNLRHASLRNVIERIFGIFKSRFLI 288
Query: 270 LRSMPKFKYETQVHIVVATMAIHNFIR---RSAEMDVDFNLYEDENTVIHHDDDHRSTNL 326
+S P + ++TQ +V+A +HNF+ RS E + ++ E+ + DD+ N
Sbjct: 289 FKSAPPYPFKTQTELVLACAGLHNFLHQECRSDEFPPETSINEE------NSDDNEENNY 342
Query: 327 NQSQSFNVASSSEMDHARN 345
++ + S + ++A N
Sbjct: 343 EENGDVGILESQQREYANN 361
>At5g41980 unknown protein
Length = 374
Score = 177 bits (450), Expect = 6e-45
Identities = 111/332 (33%), Positives = 163/332 (48%), Gaps = 52/332 (15%)
Query: 39 LCKEPCRTSELTGHAWVQEILQGNPTRCYEMFRMEKHIFHKLCHELVEHDL-KSSKHMGV 97
L KE + S G+ +V +IL G +C+E FRM+K +F+KLC L L + + + +
Sbjct: 15 LPKEVSKISISDGNKFVYQILNGPNEQCFENFRMDKPVFYKLCDLLQTRGLLRHTNRIKI 74
Query: 98 EEMVAMFLVVVGHGVGNRMIQERFQHSGETVR---------------------------- 129
E +A+FL ++GH + R +QE F +SGET+
Sbjct: 75 EAQLAIFLFIIGHNLRTRAVQELFCYSGETISRHFNNVLNAVIAISKDFFQPNSNSDTLE 134
Query: 130 -----------VIDGTHVSCVVSASEQPRFIGRKGYPTQNIMAVCDWNMCFTFVLAGWEG 178
V+D H+ +V EQ F G TQN++A +++ F +VLAGWEG
Sbjct: 135 NDDPYFKDCVGVVDSFHIPVMVGVDEQGPFRNGNGLLTQNVLAASSFDLRFNYVLAGWEG 194
Query: 179 TAHDARVFDKALTTANLNFPHPPQGKYYLVDSGYPTPIGYIGPYRCERYHLPEFRRSSGF 238
+A D +V + ALT N PQGKYY+VD+ YP G+I PY ++
Sbjct: 195 SASDQQVLNAALTRRNKL--QVPQGKYYIVDNKYPNLPGFIAPYHGVS--------TNSR 244
Query: 239 ENHNEVFNYYHSSLRCTIERTFGVWKNRFAILRSMPKFKYETQVHIVVATMAIHNFIRRS 298
E E+FN H L I RTFG K RF IL S P + +TQV +V+A A+HN++R
Sbjct: 245 EEAKEMFNERHKLLHRAIHRTFGALKERFPILLSAPPYPLQTQVKLVIAACALHNYVRLE 304
Query: 299 AEMDVDFNLYEDENTVIHHDDDHRSTNLNQSQ 330
D+ F ++E+E +D R L + Q
Sbjct: 305 KPDDLVFRMFEEETLAEAGED--REVALEEEQ 334
>At3g29620 hypothetical protein
Length = 222
Score = 169 bits (427), Expect = 3e-42
Identities = 89/221 (40%), Positives = 130/221 (58%), Gaps = 8/221 (3%)
Query: 131 IDGTHVSCVVSASEQPRFIGRKGYPTQNIMAVCDWNMCFTFVLAGWEGTAHDARVFDKAL 190
+DGTHV +VS + R+ RK + NI+AVC+++M FT++ G G+AHD +V A+
Sbjct: 1 MDGTHVPAMVSGRDHQRYWNRKSICSMNILAVCNFDMLFTYIYVGVFGSAHDTKVLSLAM 60
Query: 191 TTANLNFPHPPQGKYYLVDSGYPTPIGYIGPYRCERYHLPEFRRSSGFENHNEVFNYYHS 250
+ NFPHP GKYYLVDSGY GY+GP+R YH +F+ + NH E FN+ HS
Sbjct: 61 E-GDPNFPHPHIGKYYLVDSGYALRRGYLGPFRQTWYHHNQFQNQAPPNNHKEKFNWRHS 119
Query: 251 SLRCTIERTFGVWKNRFAILRSMPKFKYETQVHIVVATMAIHNFIRRSAEMDVDFNLYED 310
LRC IERTFGVWK ++ I++ + T I+VATMA+HNF+ + D+DF++
Sbjct: 120 LLRCVIERTFGVWKGKWRIMQDRAWYNIVTTRKIMVATMALHNFVWKLGIPDLDFDIDWM 179
Query: 311 ENTVIHHDDDHRSTNLNQSQSFNVASSSEMDHARNSIRDQI 351
+ D+DH T ++ ++ + H RD+I
Sbjct: 180 Q------DNDHHPTLDDEDETVEQDMTGSRQH-MEGFRDEI 213
>At2g26630 En/Spm-like transposon protein
Length = 292
Score = 165 bits (417), Expect = 4e-41
Identities = 94/238 (39%), Positives = 126/238 (52%), Gaps = 15/238 (6%)
Query: 128 VRVIDGTHVSCVVSASEQPRFIGRKGYPTQNIMAVCDWNMCFTFVLAGWEGTAHDARVFD 187
V +DGTHV + ++ GRK PT N++A+C+++M F + G G AHD +V +
Sbjct: 45 VGALDGTHVPVRPPSQTAKKYKGRKLEPTMNVLAICNFDMKFIYAYVGVPGRAHDTKVLN 104
Query: 188 KALTTANLNFPHPPQGKYYLVDSGYPTPIGYIGPYRCERYHLPEFRRSSGFENHNEVFNY 247
T F HPP GKYYLVDSGYPT GY+GP+R RYHL +F R E+FN
Sbjct: 105 YCATNEPY-FSHPPNGKYYLVDSGYPTRTGYLGPHRRMRYHLGQFGRGGPPVTARELFNR 163
Query: 248 YHSSLRCTIERTFGVWKNRFAIL-RSMPKFKYETQVHIVVATMAIHNFIRRSAEMDVDFN 306
HS LR IERTFGVWK ++ I+ R PK+ + IV ATMA+HNFIR S D DF
Sbjct: 164 KHSGLRSVIERTFGVWKAKWRIVDRKHPKYGLAKWIKIVTATMALHNFIRDSHREDHDFL 223
Query: 307 LY-------------EDENTVIHHDDDHRSTNLNQSQSFNVASSSEMDHARNSIRDQI 351
+ E+E ++ + + +VA D A ++RD I
Sbjct: 224 QWQSIEEYHVDDEEEEEEEEGEEEEEGEEEEGEGEEEEEHVAYEPTGDRAMEALRDNI 281
>At4g08200 putative protein
Length = 202
Score = 119 bits (299), Expect = 2e-27
Identities = 69/187 (36%), Positives = 98/187 (51%), Gaps = 7/187 (3%)
Query: 172 VLAGWEGTAHDARVFDKALTTANLNFPHPPQGKYYLVDSGYPTPIGYIGPYRCERYHLPE 231
+ G G AHDA+V A+ + NF HPP KYYLVDSGY GY+ YR +YH
Sbjct: 21 IYVGVLGYAHDAKVLALAMQ-GDPNFSHPPISKYYLVDSGYGLHRGYLISYRQSQYHPSH 79
Query: 232 FRRSSGFENHNEVFNYYHSSLRCTIERTFGVWKNRFAILRSMPKFKYETQVHIVVATMAI 291
F+ + N+ E FN HSSLR ERTF VWK ++ I+ + ++ T +VV TMA+
Sbjct: 80 FQNQAPPNNYKEKFNRLHSSLRLVTERTFRVWKGKWKIMHNRARYDVRTTKKLVVETMAL 139
Query: 292 HNFIRRSAEMDVDFNLYEDENTVIHHDDDHRSTNLNQSQSFNVASSSEMDHARNSIRDQI 351
HNF+R+S +D DF ++ DD+H+ + + + + + IRD I
Sbjct: 140 HNFVRKSNILDPDFEANWEQ------DDNHQPSLKEEVEVQDDGQIFDSRQYMEGIRDDI 193
Query: 352 IAYKLNN 358
NN
Sbjct: 194 AMNLWNN 200
>At3g44030 putative protein
Length = 775
Score = 102 bits (255), Expect = 2e-22
Identities = 59/140 (42%), Positives = 75/140 (53%), Gaps = 6/140 (4%)
Query: 128 VRVIDGTHVSCVVSASEQPRFIGRKGYPTQNIMAVCDWNMCFTFVLAGWEGTAHDARVFD 187
V +DGTHV V Q + R + NIMA+CD NM FT++ G G+ HD V
Sbjct: 637 VGAMDGTHVCVKVKPELQGMYWNRHDNASLNIMAICDLNMLFTYIWNGVPGSCHDTVVLQ 696
Query: 188 KALTTANLNFPHPPQGKYYLVDSGYPTPIGYIGPYRCE-----RYHLPEFRRSSGFENHN 242
A ++ FP PP KYYLVDS YP G++ YR RYH+ +F N +
Sbjct: 697 IA-QQSDSEFPLPPSEKYYLVDSCYPNKQGFLALYRSSRNRVVRYHMSQFYPGPPPRNKH 755
Query: 243 EVFNYYHSSLRCTIERTFGV 262
E+FN H+SLR IERT GV
Sbjct: 756 ELFNQCHTSLRSVIERTVGV 775
>At5g28730 putative protein
Length = 296
Score = 96.7 bits (239), Expect = 2e-20
Identities = 71/234 (30%), Positives = 105/234 (44%), Gaps = 36/234 (15%)
Query: 39 LCKEPCRTSELTGHAWVQEILQGNPTRCYEMFRMEKHIFHKLCHELV-EHDLKSSKHMGV 97
L +E E H + N C + RM F +LC L ++ L+SS ++ +
Sbjct: 2 LLEEEANMEECIAHQ-----IYSNEVSCQTLIRMSSEAFTQLCEILHGKYGLQSSTNISL 56
Query: 98 EEMVAMFLVVVGHGVGNRMIQERFQHSGETV--------RVIDGTHVSCVVSAS-EQPRF 148
+E VA+FL++ R I RF H+ ET+ + ++ V + E+ R
Sbjct: 57 DESVAIFLIICASNDTQRDIALRFGHAQETIWRKFHDVLKAMERLAVEYIRPRKVEELRA 116
Query: 149 IGRK---------------GYPTQNIMAVCDWNMCFTFVLAGWEGTAHDARVFDKALTTA 193
I + G + N++A+CD +M FT+ G G+ HDARV A++
Sbjct: 117 ISNRLQDDTRYWPFLMDLLGIASFNVLAICDLDMLFTYCFVGMAGSTHDARVLSAAISDD 176
Query: 194 NLNFPHPPQGKYYLVDSGYPTPIGYIGPYRCERYHLPE-----FRRSSGFENHN 242
L F PP KYYLVDSGY GY+ PYR E + F + FE HN
Sbjct: 177 PL-FHVPPDSKYYLVDSGYANKRGYLAPYRREHREAQDIISNNFLTVNLFETHN 229
>At4g10890 putative protein
Length = 380
Score = 81.6 bits (200), Expect = 6e-16
Identities = 61/185 (32%), Positives = 88/185 (46%), Gaps = 31/185 (16%)
Query: 87 HDLKSSKHMGVEEMVAMFLVVVGHGVGNRMIQERFQHSGETV-RVIDGTHVSCVVSASEQ 145
+ L+ +++ +EE VAMFL V R I R+Q S V R ID + + A++
Sbjct: 6 YGLQELENVYLEESVAMFLEEVDKNRTVRDIVARYQQSLNVVKRKIDDVLSALLKFAADT 65
Query: 146 PRFIGRKGYPTQNIMAVCDWNMCFTFVLAGWEGTAHDARVFDKALTTANLNFPHPPQGKY 205
+ ++ + T+ ++ C N F+ PHP KY
Sbjct: 66 LK--SQESHDTK-VLKYCARNESFS---------------------------PHPSNRKY 95
Query: 206 YLVDSGYPTPIGYIGPYRCERYHLPEFRRSSGFENHNEVFNYYHSSLRCTIERTFGVWKN 265
YLV+S YPT GY+GP+R YHL +F R E+FN H LR I+RTFGVWK
Sbjct: 96 YLVNSVYPTTTGYLGPHRRILYHLGQFGRGGPPVTVQELFNRKHLDLRSVIDRTFGVWKA 155
Query: 266 RFAIL 270
++ IL
Sbjct: 156 KWRIL 160
>At4g22450 hypothetical protein
Length = 457
Score = 79.3 bits (194), Expect = 3e-15
Identities = 43/98 (43%), Positives = 58/98 (58%), Gaps = 1/98 (1%)
Query: 225 ERYHLPEFRRSSGFENHNEVFNYYHSSLRCTIERTFGVWKNRFAIL-RSMPKFKYETQVH 283
E L F R N E+FN HSSLR IERTFGVWK ++ IL R PK++ + +
Sbjct: 327 ENVSLMLFNRGGPPTNSRELFNRRHSSLRSVIERTFGVWKAKWRILDRKHPKYEVKKWIK 386
Query: 284 IVVATMAIHNFIRRSAEMDVDFNLYEDENTVIHHDDDH 321
IV +TMA+HN+IR S + D DF +E + H D++
Sbjct: 387 IVTSTMALHNYIRDSQQEDSDFRHWEIVESYEQHGDEN 424
>At5g28950 putative protein
Length = 148
Score = 71.2 bits (173), Expect = 8e-13
Identities = 32/77 (41%), Positives = 51/77 (65%)
Query: 126 ETVRVIDGTHVSCVVSASEQPRFIGRKGYPTQNIMAVCDWNMCFTFVLAGWEGTAHDARV 185
+ V ID TH+ +VS + P F RKG +QN++A C++++ F +VL+GWEG+AHD++V
Sbjct: 24 DCVGAIDDTHIFAMVSQKKMPSFRNRKGDISQNMLAACNFDVEFMYVLSGWEGSAHDSKV 83
Query: 186 FDKALTTANLNFPHPPQ 202
+ ALT + P P +
Sbjct: 84 LNDALTRNSNRLPVPEE 100
>At3g15310 unknown protein
Length = 415
Score = 45.8 bits (107), Expect = 4e-05
Identities = 56/212 (26%), Positives = 94/212 (43%), Gaps = 30/212 (14%)
Query: 153 GYPTQNIMAVCDWNMCFTFVLAGWEGTAHDARV------FDKALTTANLNFPHPPQGK-- 204
G PT + A+ ++ V G GT +D + FD L N + G+
Sbjct: 215 GKPTIVLEAIASQDLWIWHVFFGPPGTLNDINILDRSPIFDDILQGRAPNVKYKVNGREY 274
Query: 205 ---YYLVDSGYPTPIGYIGPYRCERYHLPEFRRSSGFENHNEVFNYYHSSLRCTIERTFG 261
YYL D YP +I R LP+ R+++ F H E R +ER FG
Sbjct: 275 HLAYYLTDGIYPKWATFIQSIR-----LPQNRKATLFATHQEAD-------RKDVERAFG 322
Query: 262 VWKNRFAILRSMPKFKYETQV--HIVVATMAIHNFIRRSAEMDVDFNLYEDENTVIHHDD 319
V + RF I+++ P ++ + +I+ A + +HN I E D +N+ D + + +
Sbjct: 323 VLQARFHIIKN-PALVWDKEKIGNIMKACIILHNMIVED-ERD-GYNIQFDVSEFL-QVE 378
Query: 320 DHRSTNLNQSQSFNVASSSE-MDHARNSIRDQ 350
+++ ++ S S + + E M RN +RDQ
Sbjct: 379 GNQTPQVDLSYSTGMPLNIENMMGMRNQLRDQ 410
>At1g24370 hypothetical protein
Length = 413
Score = 45.4 bits (106), Expect = 5e-05
Identities = 45/175 (25%), Positives = 72/175 (40%), Gaps = 25/175 (14%)
Query: 147 RFIGRKGYPTQNIMAVCDWNMCFTFVLAGWEGTAHDARVFDKALTTANLNFPHPPQGKY- 205
++ GR PT + AV D+++ G G+ +D V + + ANL P Y
Sbjct: 92 QYAGRSRSPTIILEAVADYDLWIWHAYFGLPGSNNDINVLEASHLFANLAEGTAPPASYV 151
Query: 206 ----------YLVDSGYPTPIGYIGPYRCERYHLPEFRRSSGFENHNEVFNYYHSSLRCT 255
YL D YP + + H P + F E + R
Sbjct: 152 INEKPYNMSYYLADGIYPKWSTLV-----QTIHDPRGPKKKLFAMKQE-------ACRKD 199
Query: 256 IERTFGVWKNRFAILRSMPKFKYETQVH-IVVATMAIHNFIRRSAEMDVDFNLYE 309
+ER FGV ++RFAI+ + +T +H I+ + + +HN I E D+D + E
Sbjct: 200 VERAFGVLQSRFAIVAGPSRLWNKTVLHDIMTSCIIMHNMIIED-ERDIDAPIEE 253
>At3g55350 unknown protein
Length = 406
Score = 43.1 bits (100), Expect = 2e-04
Identities = 54/237 (22%), Positives = 92/237 (38%), Gaps = 44/237 (18%)
Query: 131 IDGTHVSCVVSASEQPRFI---GRKGYPTQNIMAVCDWNMCFTFVLAGWEGTAHDARVF- 186
ID TH+ + A E + G K + + + AV D +M F V+AGW G+ +D V
Sbjct: 187 IDITHIVMNLPAVEPSNKVWLDGEKNF-SMTLQAVVDPDMRFLDVIAGWPGSLNDDVVLK 245
Query: 187 ----------DKALTTANLNFPHPPQGKYYLV-DSGYPTPIGYIGPYRCERYHLPEFRRS 235
K L L + + Y+V DSG+P + PY+ + LP+
Sbjct: 246 NSGFYKLVEKGKRLNGEKLPLSERTELREYIVGDSGFPLLPWLLTPYQGKPTSLPQTE-- 303
Query: 236 SGFENHNEVFNYYHSSLRCTIERTFGVWKNRFAILRSMPKFKYETQV-HIVVATMAIHNF 294
FN HS + K+R+ I+ + ++ I+ +HN
Sbjct: 304 ---------FNKRHSEATKAAQMALSKLKDRWRIINGVMWMPDRNRLPRIIFVCCLLHNI 354
Query: 295 IRRSAEMDVDFNLYEDENTVIHHDDDHRSTNLNQSQSFNVASSSEMDHARNSIRDQI 351
I +D++ +D+ HD ++R S D A + +RD++
Sbjct: 355 I-----IDMEDQTLDDQPLSQQHDMNYRQ-----------RSCKLADEASSVLRDEL 395
>At2g13770 hypothetical protein
Length = 244
Score = 38.5 bits (88), Expect = 0.006
Identities = 46/180 (25%), Positives = 76/180 (41%), Gaps = 18/180 (10%)
Query: 152 KGYPTQNIMAVCDWNMCFTFVLAGWEGTAHDARVFDKALTTANLNFPHPPQGKYYLVDSG 211
+G+P AV D+++ G G+ + V + + ANL P Y + +G
Sbjct: 66 RGFPE----AVADYDLWIWHAYFGLPGSNNGINVLEASHLFANLAEGTAPPASYVI--NG 119
Query: 212 YPTPIGYI---GPYRCERYHLPEFRRSSGFENHNEVFNYYHSSLRCTIERTFGVWKNRFA 268
P +GY G Y + G + ++F + R +ER FGV + RFA
Sbjct: 120 KPYNMGYYLADGIYPKWSTLVQTIHDPRGPKK--KLFAMKQEACRKDVERAFGVLQLRFA 177
Query: 269 ILRSMPKFKYETQVH-IVVATMAIHNFIRRSAEMDVDFNLYED-----ENTVIHHDDDHR 322
I+ + +T +H I+ + + +HN I E D+D + E E + DDD R
Sbjct: 178 IVAGPSRLWNKTVLHDIMTSCIIMHNMIIED-ERDIDAPIEERVEVPIEEVEMTGDDDTR 236
>At5g12010 putative protein
Length = 502
Score = 38.1 bits (87), Expect = 0.007
Identities = 41/196 (20%), Positives = 73/196 (36%), Gaps = 21/196 (10%)
Query: 115 RMIQERFQHSGETVRVIDG---THVSCVVSASEQPRFIGRKGYPTQN-------IMAVCD 164
R I+ERF+ V+ TH+ + + ++ I AV +
Sbjct: 270 RNIRERFESVSGIPNVVGSMYTTHIPIIAPKISVASYFNKRHTERNQKTSYSITIQAVVN 329
Query: 165 WNMCFTFVLAGWEGTAHDARVFDKALTTANLNFPHPPQGKYYLVDSGYPTPIGYIGPYRC 224
FT + GW G+ D +V +K+L N +G + G+P + PY
Sbjct: 330 PKGVFTDLCIGWPGSMPDDKVLEKSLLYQRANNGGLLKGMWVAGGPGHPLLDWVLVPYT- 388
Query: 225 ERYHLPEFRRSSGFENHNEVFNYYHSSLRCTIERTFGVWKNRFAILRSMPKFKYETQVHI 284
+++ + H FN S ++ + FG K R+A L+ + K + +
Sbjct: 389 --------QQNLTWTQH--AFNEKMSEVQGVAKEAFGRLKGRWACLQKRTEVKLQDLPTV 438
Query: 285 VVATMAIHNFIRRSAE 300
+ A +HN E
Sbjct: 439 LGACCVLHNICEMREE 454
>At2g07520 pseudogene
Length = 222
Score = 34.3 bits (77), Expect = 0.11
Identities = 45/216 (20%), Positives = 80/216 (36%), Gaps = 39/216 (18%)
Query: 150 GRKGYPTQNIMAVCDWNMCFTFVLAGWEGTAHDARVFDKALTTANLNFPHPPQGK----- 204
G KG T + AV ++ G T +D D++ ++ + P+
Sbjct: 29 GDKGTSTISFEAVASHDLWIWHTFFGCASTLNDINFLDRSPVFDDIEQGNTPRVNFFVYQ 88
Query: 205 ------YYLVDSGYPTPIGYIGPYRCERYHLPEFRRSSGFENHNEVFNYYHSSLRCTIER 258
YYL D YP+ ++ R LP+ +++F R IER
Sbjct: 89 RQYNMAYYLADGIYPSYPTFVKSIR-----LPQ-------SEPDKLFVQLQEGCRKDIER 136
Query: 259 TFGVWKNRFAILRSMPKFKYETQVHIVVATMAIHNFIRRSAEMDVDFNLYEDENTVIHHD 318
FGV RF I+ ++ + I++ M + N A+ D+
Sbjct: 137 AFGVLHARFKIIWNIADLAIIMRSCIILHNMIVENERDTYAQHWTDY------------- 183
Query: 319 DDHRSTNLNQSQSFN---VASSSEMDHARNSIRDQI 351
D + + + Q F+ + + + HAR+ +RD I
Sbjct: 184 DQSKESGSSTPQPFSTEVLHAFANHVHARSKLRDSI 219
>At1g43590 hypothetical protein
Length = 168
Score = 34.3 bits (77), Expect = 0.11
Identities = 27/92 (29%), Positives = 45/92 (48%), Gaps = 13/92 (14%)
Query: 205 YYLVDSGYPTPIGYIGPYRCERYHLPEFRRSSGFENHNEVFNYYHSSLRCTIERTFGVWK 264
YYL D YP +I + LP+ ++S F + E + R +ER FGV +
Sbjct: 14 YYLTDEIYPKWATFI-----QSISLPQDEKASLFATNQE-------ACRKDVERAFGVLQ 61
Query: 265 NRFAILRSMPKFKYETQV-HIVVATMAIHNFI 295
RFAI++ + ++ +I+ A + +HN I
Sbjct: 62 ARFAIVKHPALIWDKIKIGNIMRACIILHNMI 93
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.322 0.136 0.422
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 8,571,625
Number of Sequences: 26719
Number of extensions: 371104
Number of successful extensions: 1097
Number of sequences better than 10.0: 30
Number of HSP's better than 10.0 without gapping: 16
Number of HSP's successfully gapped in prelim test: 14
Number of HSP's that attempted gapping in prelim test: 1050
Number of HSP's gapped (non-prelim): 37
length of query: 358
length of database: 11,318,596
effective HSP length: 100
effective length of query: 258
effective length of database: 8,646,696
effective search space: 2230847568
effective search space used: 2230847568
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.9 bits)
S2: 61 (28.1 bits)
Medicago: description of AC127428.4


