

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC127169.5 + phase: 0 /pseudo
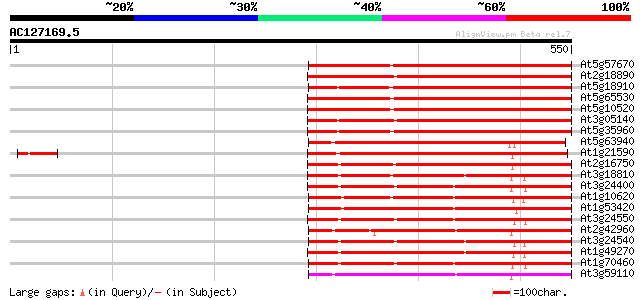
(550 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
At5g57670 putative protein 394 e-110
At2g18890 putative protein kinase 330 2e-90
At5g18910 protein kinase - like protein 306 1e-83
At5g65530 unknown protein 302 3e-82
At5g10520 Pto kinase interactor - like protein 300 1e-81
At3g05140 putative serine/threonine protein kinase 295 4e-80
At5g35960 serine/threonine protein kinase-like 294 8e-80
At5g63940 unknown protein 221 6e-58
At1g21590 unknown protein 218 5e-57
At2g16750 putative protein kinase 217 2e-56
At3g18810 protein kinase, putative 211 6e-55
At3g24400 protein kinase, putative 209 4e-54
At1g10620 putative serine/threonine protein kinase emb|CAA18823.1 208 7e-54
At1g53420 hypothetical protein, 5' partial 207 9e-54
At3g24550 protein kinase, putative 206 2e-53
At2g42960 putative protein kinase 205 5e-53
At3g24540 protein kinase, putative 205 6e-53
At1g49270 hypothetical protein 205 6e-53
At1g70460 putative protein kinase 204 8e-53
At3g59110 receptor-like protein kinase 204 1e-52
>At5g57670 putative protein
Length = 416
Score = 394 bits (1011), Expect = e-110
Identities = 185/257 (71%), Positives = 219/257 (84%), Gaps = 2/257 (0%)
Query: 294 GGYSEVYKGDLCNGETIAVKRLAKDNKDPNKEKEFLMELGIIGHVCHPNTASLLGYCFEN 353
GGYSEVY+GDL +G IAVKRLAK++ D NKEKEFL ELGII HV HPNTA LLG C E
Sbjct: 113 GGYSEVYRGDLWDGRRIAVKRLAKESGDMNKEKEFLTELGIISHVSHPNTALLLGCCVEK 172
Query: 354 GLYLIFNYSQNGNLSTALHGYGKAGNSLDWPIRYKIAIGVARGLHYLHKCCKHRIIHRDI 413
GLYL+F +S+NG L +ALH SLDWP+RYKIA+GVARGLHYLHK C HRIIHRDI
Sbjct: 173 GLYLVFRFSENGTLYSALHE--NENGSLDWPVRYKIAVGVARGLHYLHKRCNHRIIHRDI 230
Query: 414 KASNVLLGPDYEPQITDFGLAKWLPNKWTHHAVIPVEGTFGYLAPEVFMHGIVDEKTDIF 473
K+SNVLLGPDYEPQITDFGLAKWLPNKWTHHAVIPVEGTFGYLAPE M G +DEKTDI+
Sbjct: 231 KSSNVLLGPDYEPQITDFGLAKWLPNKWTHHAVIPVEGTFGYLAPESLMQGTIDEKTDIY 290
Query: 474 AFGVLLLEIVTGRRPVDSSKQNILLWAKPLMESGNIAELADPRMEGRYDVEQLYRVVLTA 533
AFG+LLLEI+TGRRPV+ ++++ILLWAKP ME+GN +EL DP+++ +YD +Q+ ++VLTA
Sbjct: 291 AFGILLLEIITGRRPVNPTQKHILLWAKPAMETGNTSELVDPKLQDKYDDQQMNKLVLTA 350
Query: 534 SYCVRQTAIWRPAMTEV 550
S+CV+Q+ I RP MT+V
Sbjct: 351 SHCVQQSPILRPTMTQV 367
>At2g18890 putative protein kinase
Length = 392
Score = 330 bits (845), Expect = 2e-90
Identities = 153/260 (58%), Positives = 205/260 (78%), Gaps = 4/260 (1%)
Query: 293 RGGYSEVYKGDLC-NGETIAVKRLAKDNKDPNK-EKEFLMELGIIGHVCHPNTASLLGYC 350
RGG++EVYKG L NGE IAVKR+ + +D + EKEFLME+G IGHV HPN SLLG C
Sbjct: 76 RGGFAEVYKGILGKNGEEIAVKRITRGGRDDERREKEFLMEIGTIGHVSHPNVLSLLGCC 135
Query: 351 FENGLYLIFNYSQNGNLSTALHGYGKAGNSLDWPIRYKIAIGVARGLHYLHKCCKHRIIH 410
+NGLYL+F +S G+L++ LH +A L+W RYKIAIG A+GLHYLHK C+ RIIH
Sbjct: 136 IDNGLYLVFIFSSRGSLASLLHDLNQA--PLEWETRYKIAIGTAKGLHYLHKGCQRRIIH 193
Query: 411 RDIKASNVLLGPDYEPQITDFGLAKWLPNKWTHHAVIPVEGTFGYLAPEVFMHGIVDEKT 470
RDIK+SNVLL D+EPQI+DFGLAKWLP++W+HH++ P+EGTFG+LAPE + HGIVDEKT
Sbjct: 194 RDIKSSNVLLNQDFEPQISDFGLAKWLPSQWSHHSIAPIEGTFGHLAPEYYTHGIVDEKT 253
Query: 471 DIFAFGVLLLEIVTGRRPVDSSKQNILLWAKPLMESGNIAELADPRMEGRYDVEQLYRVV 530
D+FAFGV LLE+++G++PVD+S Q++ WAK +++ G I +L DPR+ +D++QL+R+
Sbjct: 254 DVFAFGVFLLELISGKKPVDASHQSLHSWAKLIIKDGEIEKLVDPRIGEEFDLQQLHRIA 313
Query: 531 LTASYCVRQTAIWRPAMTEV 550
AS C+R +++ RP+M EV
Sbjct: 314 FAASLCIRSSSLCRPSMIEV 333
>At5g18910 protein kinase - like protein
Length = 511
Score = 306 bits (785), Expect = 1e-83
Identities = 141/257 (54%), Positives = 198/257 (76%), Gaps = 4/257 (1%)
Query: 294 GGYSEVYKGDLCNGETIAVKRLAKDNKDPNKEKEFLMELGIIGHVCHPNTASLLGYCFEN 353
GGY+EVYKG + +G+ +A+K+L + + + ++L ELGII HV HPN A L+GYC E
Sbjct: 201 GGYAEVYKGQMADGQIVAIKKLTRGSAE-EMTMDYLSELGIIVHVDHPNIAKLIGYCVEG 259
Query: 354 GLYLIFNYSQNGNLSTALHGYGKAGNSLDWPIRYKIAIGVARGLHYLHKCCKHRIIHRDI 413
G++L+ S NG+L++ L+ +A L+W +RYK+A+G A GL+YLH+ C+ RIIH+DI
Sbjct: 260 GMHLVLELSPNGSLASLLY---EAKEKLNWSMRYKVAMGTAEGLYYLHEGCQRRIIHKDI 316
Query: 414 KASNVLLGPDYEPQITDFGLAKWLPNKWTHHAVIPVEGTFGYLAPEVFMHGIVDEKTDIF 473
KASN+LL ++E QI+DFGLAKWLP++WTHH V VEGTFGYL PE FMHGIVDEKTD++
Sbjct: 317 KASNILLTQNFEAQISDFGLAKWLPDQWTHHTVSKVEGTFGYLPPEFFMHGIVDEKTDVY 376
Query: 474 AFGVLLLEIVTGRRPVDSSKQNILLWAKPLMESGNIAELADPRMEGRYDVEQLYRVVLTA 533
A+GVLLLE++TGR+ +DSS+ +I++WAKPL++ I +L DP +E YDVE+L R+V A
Sbjct: 377 AYGVLLLELITGRQALDSSQHSIVMWAKPLIKENKIKQLVDPILEDDYDVEELDRLVFIA 436
Query: 534 SYCVRQTAIWRPAMTEV 550
S C+ QT++ RP M++V
Sbjct: 437 SLCIHQTSMNRPQMSQV 453
>At5g65530 unknown protein
Length = 456
Score = 302 bits (774), Expect = 3e-82
Identities = 145/260 (55%), Positives = 190/260 (72%), Gaps = 5/260 (1%)
Query: 293 RGGYSEVYKGDLCNGETIAVKRLAKDNKDPNKE-KEFLMELGIIGHVCHPNTASLLGYCF 351
+GG++EVYKG L +GET+A+K+L + K+ + +FL ELGII HV HPN A L G+
Sbjct: 152 KGGHAEVYKGVLPDGETVAIKKLTRHAKEVEERVSDFLSELGIIAHVNHPNAARLRGFSC 211
Query: 352 ENGLYLIFNYSQNGNLSTALHGYGKAGNSLDWPIRYKIAIGVARGLHYLHKCCKHRIIHR 411
+ GL+ + YS +G+L++ L G + LDW RYK+A+G+A GL YLH C RIIHR
Sbjct: 212 DRGLHFVLEYSSHGSLASLLFG---SEECLDWKKRYKVAMGIADGLSYLHNDCPRRIIHR 268
Query: 412 DIKASNVLLGPDYEPQITDFGLAKWLPNKWTHHAVIPVEGTFGYLAPEVFMHGIVDEKTD 471
DIKASN+LL DYE QI+DFGLAKWLP W HH V P+EGTFGYLAPE FMHGIVDEKTD
Sbjct: 269 DIKASNILLSQDYEAQISDFGLAKWLPEHWPHHIVFPIEGTFGYLAPEYFMHGIVDEKTD 328
Query: 472 IFAFGVLLLEIVTGRRPVDS-SKQNILLWAKPLMESGNIAELADPRMEGRYDVEQLYRVV 530
+FAFGVLLLEI+TGRR VD+ S+Q+I++WAKPL+E N+ E+ DP++ +D ++ RV+
Sbjct: 329 VFAFGVLLLEIITGRRAVDTDSRQSIVMWAKPLLEKNNMEEIVDPQLGNDFDETEMKRVM 388
Query: 531 LTASYCVRQTAIWRPAMTEV 550
TAS C+ + RP M +
Sbjct: 389 QTASMCIHHVSTMRPDMNRL 408
>At5g10520 Pto kinase interactor - like protein
Length = 467
Score = 300 bits (768), Expect = 1e-81
Identities = 144/260 (55%), Positives = 187/260 (71%), Gaps = 5/260 (1%)
Query: 293 RGGYSEVYKGDLCNGETIAVKRLAKDNKDPNKE-KEFLMELGIIGHVCHPNTASLLGYCF 351
+GG++EVYKG L NGET+A+K+L K+ + +FL ELGII HV HPN A L G+
Sbjct: 161 KGGHAEVYKGVLINGETVAIKKLMSHAKEEEERVSDFLSELGIIAHVNHPNAARLRGFSS 220
Query: 352 ENGLYLIFNYSQNGNLSTALHGYGKAGNSLDWPIRYKIAIGVARGLHYLHKCCKHRIIHR 411
+ GL+ + Y+ G+L++ L G + L+W IRYK+A+G+A GL YLH C RIIHR
Sbjct: 221 DRGLHFVLEYAPYGSLASMLFG---SEECLEWKIRYKVALGIADGLSYLHNACPRRIIHR 277
Query: 412 DIKASNVLLGPDYEPQITDFGLAKWLPNKWTHHAVIPVEGTFGYLAPEVFMHGIVDEKTD 471
DIKASN+LL DYE QI+DFGLAKWLP W HH V P+EGTFGYLAPE FMHGIVDEK D
Sbjct: 278 DIKASNILLNHDYEAQISDFGLAKWLPENWPHHVVFPIEGTFGYLAPEYFMHGIVDEKID 337
Query: 472 IFAFGVLLLEIVTGRRPVD-SSKQNILLWAKPLMESGNIAELADPRMEGRYDVEQLYRVV 530
+FAFGVLLLEI+T RR VD +S+Q+I+ WAKP +E ++ ++ DPR+ ++ ++ RV+
Sbjct: 338 VFAFGVLLLEIITSRRAVDTASRQSIVAWAKPFLEKNSMEDIVDPRLGNMFNPTEMQRVM 397
Query: 531 LTASYCVRQTAIWRPAMTEV 550
LTAS CV A RP MT +
Sbjct: 398 LTASMCVHHIAAMRPDMTRL 417
>At3g05140 putative serine/threonine protein kinase
Length = 460
Score = 295 bits (755), Expect = 4e-80
Identities = 146/258 (56%), Positives = 184/258 (70%), Gaps = 3/258 (1%)
Query: 293 RGGYSEVYKGDLCNGETIAVKRLAKDNKDPNKEKEFLMELGIIGHVCHPNTASLLGYCFE 352
RGGY++VY+G L G+ IAVKRL K D + EFL ELGII HV HPNTA +G C E
Sbjct: 151 RGGYADVYQGILPEGKLIAVKRLTKGTPD-EQTAEFLSELGIIAHVDHPNTAKFIGCCIE 209
Query: 353 NGLYLIFNYSQNGNLSTALHGYGKAGNSLDWPIRYKIAIGVARGLHYLHKCCKHRIIHRD 412
G++L+F S G+L + LHG K L W RY +A+G A GL YLH+ C+ RIIHRD
Sbjct: 210 GGMHLVFRLSPLGSLGSLLHGPSKY--KLTWSRRYNVALGTADGLVYLHEGCQRRIIHRD 267
Query: 413 IKASNVLLGPDYEPQITDFGLAKWLPNKWTHHAVIPVEGTFGYLAPEVFMHGIVDEKTDI 472
IKA N+LL D++PQI DFGLAKWLP + THH V EGTFGY APE FMHGIVDEKTD+
Sbjct: 268 IKADNILLTEDFQPQICDFGLAKWLPKQLTHHNVSKFEGTFGYFAPEYFMHGIVDEKTDV 327
Query: 473 FAFGVLLLEIVTGRRPVDSSKQNILLWAKPLMESGNIAELADPRMEGRYDVEQLYRVVLT 532
FAFGVLLLE++TG +D S+Q+++LWAKPL+E I EL DP + Y+ E+L R+ T
Sbjct: 328 FAFGVLLLELITGHPALDESQQSLVLWAKPLLERKAIKELVDPSLGDEYNREELIRLTST 387
Query: 533 ASYCVRQTAIWRPAMTEV 550
AS C+ Q+++ RP M++V
Sbjct: 388 ASLCIDQSSLLRPRMSQV 405
>At5g35960 serine/threonine protein kinase-like
Length = 429
Score = 294 bits (753), Expect = 8e-80
Identities = 144/258 (55%), Positives = 187/258 (71%), Gaps = 4/258 (1%)
Query: 293 RGGYSEVYKGDLCNGETIAVKRLAKDNKDPNKEKEFLMELGIIGHVCHPNTASLLGYCFE 352
+GGY+EVYKG L NG+ +A+KRL + N + +FL E+GI+ HV HPN A LLGY E
Sbjct: 142 KGGYAEVYKGMLPNGQMVAIKRLMRGNSE-EIIVDFLSEMGIMAHVNHPNIAKLLGYGVE 200
Query: 353 NGLYLIFNYSQNGNLSTALHGYGKAGNSLDWPIRYKIAIGVARGLHYLHKCCKHRIIHRD 412
G++L+ S +G+L++ L+ + + W IRYKIA+GVA GL YLH+ C RIIHRD
Sbjct: 201 GGMHLVLELSPHGSLASMLYS---SKEKMKWSIRYKIALGVAEGLVYLHRGCHRRIIHRD 257
Query: 413 IKASNVLLGPDYEPQITDFGLAKWLPNKWTHHAVIPVEGTFGYLAPEVFMHGIVDEKTDI 472
IKA+N+LL D+ PQI DFGLAKWLP WTHH V EGTFGYLAPE HGIVDEKTD+
Sbjct: 258 IKAANILLTHDFSPQICDFGLAKWLPENWTHHIVSKFEGTFGYLAPEYLTHGIVDEKTDV 317
Query: 473 FAFGVLLLEIVTGRRPVDSSKQNILLWAKPLMESGNIAELADPRMEGRYDVEQLYRVVLT 532
FA GVLLLE+VTGRR +D SKQ+++LWAKPLM+ I EL DP + G Y+ Q+ V+L
Sbjct: 318 FALGVLLLELVTGRRALDYSKQSLVLWAKPLMKKNKIRELIDPSLAGEYEWRQIKLVLLA 377
Query: 533 ASYCVRQTAIWRPAMTEV 550
A+ ++Q++I RP M++V
Sbjct: 378 AALSIQQSSIERPEMSQV 395
>At5g63940 unknown protein
Length = 705
Score = 221 bits (564), Expect = 6e-58
Identities = 116/257 (45%), Positives = 165/257 (64%), Gaps = 8/257 (3%)
Query: 294 GGYSEVYKGDLCNGETIAVKRLAKDNKDPNKEKEFLMELGIIGHVCHPNTASLLGYCFEN 353
GG S VY+GDL +G +AVK L + KEF++E+ +I V H N SL G+CFEN
Sbjct: 371 GGNSYVYRGDLPDGRELAVKIL---KPCLDVLKEFILEIEVITSVHHKNIVSLFGFCFEN 427
Query: 354 G-LYLIFNYSQNGNLSTALHGYGKAGNSLDWPIRYKIAIGVARGLHYLHKCCKHRIIHRD 412
L L+++Y G+L LHG K W RYK+A+GVA L YLH +IHRD
Sbjct: 428 NNLMLVYDYLPRGSLEENLHGNRKDAKKFGWMERYKVAVGVAEALDYLHNTHDPEVIHRD 487
Query: 413 IKASNVLLGPDYEPQITDFGLAKWLPNKWTHHAVIPVEGTFGYLAPEVFMHGIVDEKTDI 472
+K+SNVLL D+EPQ++DFG A + H A + GTFGYLAPE FMHG V +K D+
Sbjct: 488 VKSSNVLLADDFEPQLSDFGFASLASSTSQHVAGGDIAGTFGYLAPEYFMHGKVTDKIDV 547
Query: 473 FAFGVLLLEIVTGRRP--VDSSK--QNILLWAKPLMESGNIAELADPRMEGRYDVEQLYR 528
+AFGV+LLE+++GR+P VD SK ++++LWA P+++SG A+L DP +E + + +
Sbjct: 548 YAFGVVLLELISGRKPICVDQSKGQESLVLWANPILDSGKFAQLLDPSLENDNSNDLIEK 607
Query: 529 VVLTASYCVRQTAIWRP 545
++L A+ C+++T RP
Sbjct: 608 LLLAATLCIKRTPHDRP 624
Score = 33.9 bits (76), Expect = 0.24
Identities = 16/39 (41%), Positives = 28/39 (71%), Gaps = 1/39 (2%)
Query: 6 SSTILIGLSLDPNDSKELLSWAIRVLANPNDTIVAVHVL 44
S+ +++G+ D S+E+L+W++ +A P D IVA+HVL
Sbjct: 19 SALLIVGVKPD-EWSREVLTWSLVNVARPGDRIVALHVL 56
>At1g21590 unknown protein
Length = 756
Score = 218 bits (556), Expect = 5e-57
Identities = 110/260 (42%), Positives = 165/260 (63%), Gaps = 8/260 (3%)
Query: 293 RGGYSEVYKGDLCNGETIAVKRLAKDNKDPNKEKEFLMELGIIGHVCHPNTASLLGYCFE 352
+GG S V++G L NG +AVK L + K+F+ E+ II + H N SLLGYCFE
Sbjct: 417 KGGSSRVFRGYLPNGREVAVKILKRTEC---VLKDFVAEIDIITTLHHKNVISLLGYCFE 473
Query: 353 NG-LYLIFNYSQNGNLSTALHGYGKAGNSLDWPIRYKIAIGVARGLHYLHKCCKHRIIHR 411
N L L++NY G+L LHG K + W RYK+A+G+A L YLH +IHR
Sbjct: 474 NNNLLLVYNYLSRGSLEENLHGNKKDLVAFRWNERYKVAVGIAEALDYLHNDAPQPVIHR 533
Query: 412 DIKASNVLLGPDYEPQITDFGLAKWLPNKWTHHAVIPVEGTFGYLAPEVFMHGIVDEKTD 471
D+K+SN+LL D+EPQ++DFGLAKW T V GTFGYLAPE FM+G ++ K D
Sbjct: 534 DVKSSNILLSDDFEPQLSDFGLAKWASESTTQIICSDVAGTFGYLAPEYFMYGKMNNKID 593
Query: 472 IFAFGVLLLEIVTGRRPVDS----SKQNILLWAKPLMESGNIAELADPRMEGRYDVEQLY 527
++A+GV+LLE+++GR+PV+S ++ ++++WAKP+++ ++L D ++ + +Q+
Sbjct: 594 VYAYGVVLLELLSGRKPVNSESPKAQDSLVMWAKPILDDKEYSQLLDSSLQDDNNSDQME 653
Query: 528 RVVLTASYCVRQTAIWRPAM 547
++ L A+ C+R RP M
Sbjct: 654 KMALAATLCIRHNPQTRPTM 673
Score = 44.7 bits (104), Expect = 1e-04
Identities = 19/40 (47%), Positives = 29/40 (72%), Gaps = 1/40 (2%)
Query: 8 TILIGLSLDPNDSKELLSWAIRVLANPNDTIVAVHVLGEE 47
T+++G+ D + S ELL WA+ +A P DT++A+HVLG E
Sbjct: 13 TVMVGVKFDES-SNELLDWALVKVAEPGDTVIALHVLGNE 51
>At2g16750 putative protein kinase
Length = 474
Score = 217 bits (552), Expect = 2e-56
Identities = 112/264 (42%), Positives = 169/264 (63%), Gaps = 10/264 (3%)
Query: 293 RGGYSEVYKGDLCNGETIAVKRLAKDNKDPNKEKEFLMELGIIGHVCHPNTASLLGYCFE 352
+GG +EVYKG L +G+ +AVK L K+ KEF+ E+ I+ + H N + L+G C
Sbjct: 139 KGGCNEVYKGFLEDGKGVAVKILKPSVKEA--VKEFVHEVSIVSSLSHSNISPLIGVCVH 196
Query: 353 -NGLYLIFNYSQNGNLSTALH-GYGKAGNSLDWPIRYKIAIGVARGLHYLHKCCKHRIIH 410
N L ++N S G+L L GK + L W R KIAIG+ L YLH C + +IH
Sbjct: 197 YNDLISVYNLSSKGSLEETLQVSVGK--HVLRWEERLKIAIGLGEALDYLHNQCSNPVIH 254
Query: 411 RDIKASNVLLGPDYEPQITDFGLAKWLPNKWTHHAVIPVEGTFGYLAPEVFMHGIVDEKT 470
RD+K+SNVLL ++EPQ++DFGL+ W + V GTFGYLAPE FM+G V +K
Sbjct: 255 RDVKSSNVLLSDEFEPQLSDFGLSMWGSKSCRYTIQRDVVGTFGYLAPEYFMYGKVSDKV 314
Query: 471 DIFAFGVLLLEIVTGRRPVDS----SKQNILLWAKPLMESGNIAELADPRMEGRYDVEQL 526
D++AFGV+LLE+++GR + S ++++++WAKP++E GN EL DP + G +D +Q
Sbjct: 315 DVYAFGVVLLELISGRTSISSDSPRGQESLVMWAKPMIEKGNAKELLDPNIAGTFDEDQF 374
Query: 527 YRVVLTASYCVRQTAIWRPAMTEV 550
+++VL A++C+ + A +RP + E+
Sbjct: 375 HKMVLAATHCLTRAATYRPNIKEI 398
>At3g18810 protein kinase, putative
Length = 700
Score = 211 bits (538), Expect = 6e-55
Identities = 118/266 (44%), Positives = 169/266 (63%), Gaps = 13/266 (4%)
Query: 293 RGGYSEVYKGDLCNGETIAVKRLAKDNKDPNKEKEFLMELGIIGHVCHPNTASLLGYCFE 352
+GG+ V+KG L NG+ IAVK L + E+EF E+ II V H SL+GYC
Sbjct: 345 QGGFGYVHKGILPNGKEIAVKSLKAGSGQG--EREFQAEVDIISRVHHRFLVSLVGYCIA 402
Query: 353 NGL-YLIFNYSQNGNLSTALHGYGKAGNSLDWPIRYKIAIGVARGLHYLHKCCKHRIIHR 411
G L++ + N L LHG K+G LDWP R KIA+G A+GL YLH+ C RIIHR
Sbjct: 403 GGQRMLVYEFLPNDTLEFHLHG--KSGKVLDWPTRLKIALGSAKGLAYLHEDCHPRIIHR 460
Query: 412 DIKASNVLLGPDYEPQITDFGLAKWLPNKWTHHAVIPVEGTFGYLAPEVFMHGIVDEKTD 471
DIKASN+LL +E ++ DFGLAK + TH + + GTFGYLAPE G + +++D
Sbjct: 461 DIKASNILLDESFEAKVADFGLAKLSQDNVTHVST-RIMGTFGYLAPEYASSGKLTDRSD 519
Query: 472 IFAFGVLLLEIVTGRRPVD---SSKQNILLWAKPL----MESGNIAELADPRMEGRYDVE 524
+F+FGV+LLE+VTGRRPVD + +++ WA+P+ + G+ +EL DPR+E +Y+
Sbjct: 520 VFSFGVMLLELVTGRRPVDLTGEMEDSLVDWARPICLNAAQDGDYSELVDPRLENQYEPH 579
Query: 525 QLYRVVLTASYCVRQTAIWRPAMTEV 550
++ ++V A+ VR +A RP M+++
Sbjct: 580 EMAQMVACAAAAVRHSARRRPKMSQI 605
>At3g24400 protein kinase, putative
Length = 694
Score = 209 bits (531), Expect = 4e-54
Identities = 114/267 (42%), Positives = 168/267 (62%), Gaps = 14/267 (5%)
Query: 293 RGGYSEVYKGDLCNGETIAVKRLAKDNKDPNKEKEFLMELGIIGHVCHPNTASLLGYCFE 352
+GG+ V+KG L NG+ +AVK+L + + E+EF E+GII V H + +L+GYC
Sbjct: 339 QGGFGYVFKGMLRNGKEVAVKQLKEGSSQG--EREFQAEVGIISRVHHRHLVALVGYCIA 396
Query: 353 NGL-YLIFNYSQNGNLSTALHGYGKAGNSLDWPIRYKIAIGVARGLHYLHKCCKHRIIHR 411
+ L++ + N L LHG G+ +++W R KIA+G A+GL YLH+ C +IIHR
Sbjct: 397 DAQRLLVYEFVPNNTLEFHLHGKGRP--TMEWSSRLKIAVGSAKGLSYLHENCNPKIIHR 454
Query: 412 DIKASNVLLGPDYEPQITDFGLAKWLPNKWTHHAVIPVEGTFGYLAPEVFMHGIVDEKTD 471
DIKASN+L+ +E ++ DFGLAK + + H V GTFGYLAPE G + EK+D
Sbjct: 455 DIKASNILIDFKFEAKVADFGLAK-IASDTNTHVSTRVMGTFGYLAPEYASSGKLTEKSD 513
Query: 472 IFAFGVLLLEIVTGRRPVD----SSKQNILLWAKPLM----ESGNIAELADPRMEGRYDV 523
+F+FGV+LLE++TGRRP+D + +++ WA+PL+ E GN + D ++ YD
Sbjct: 514 VFSFGVVLLELITGRRPIDVNNVHADNSLVDWARPLLNQVSELGNFEVVVDKKLNNEYDK 573
Query: 524 EQLYRVVLTASYCVRQTAIWRPAMTEV 550
E++ R+V A+ CVR TA RP M +V
Sbjct: 574 EEMARMVACAAACVRSTAPRRPRMDQV 600
>At1g10620 putative serine/threonine protein kinase emb|CAA18823.1
Length = 718
Score = 208 bits (529), Expect = 7e-54
Identities = 115/266 (43%), Positives = 172/266 (64%), Gaps = 14/266 (5%)
Query: 294 GGYSEVYKGDLCNGETIAVKRLAKDNKDPNKEKEFLMELGIIGHVCHPNTASLLGYCF-E 352
GG+ VYKG L G+ +A+K+L + + +E F E+ II V H + SL+GYC E
Sbjct: 379 GGFGCVYKGILFEGKPVAIKQLKSVSAEGYRE--FKAEVEIISRVHHRHLVSLVGYCISE 436
Query: 353 NGLYLIFNYSQNGNLSTALHGYGKAGNSLDWPIRYKIAIGVARGLHYLHKCCKHRIIHRD 412
+LI+ + N L LHG K L+W R +IAIG A+GL YLH+ C +IIHRD
Sbjct: 437 QHRFLIYEFVPNNTLDYHLHG--KNLPVLEWSRRVRIAIGAAKGLAYLHEDCHPKIIHRD 494
Query: 413 IKASNVLLGPDYEPQITDFGLAKWLPNKWTHHAVIPVEGTFGYLAPEVFMHGIVDEKTDI 472
IK+SN+LL ++E Q+ DFGLA+ L + H V GTFGYLAPE G + +++D+
Sbjct: 495 IKSSNILLDDEFEAQVADFGLAR-LNDTAQSHISTRVMGTFGYLAPEYASSGKLTDRSDV 553
Query: 473 FAFGVLLLEIVTGRRPVDSS----KQNILLWAKP----LMESGNIAELADPRMEGRYDVE 524
F+FGV+LLE++TGR+PVD+S +++++ WA+P +E G+I+E+ DPR+E Y
Sbjct: 554 FSFGVVLLELITGRKPVDTSQPLGEESLVEWARPRLIEAIEKGDISEVVDPRLENDYVES 613
Query: 525 QLYRVVLTASYCVRQTAIWRPAMTEV 550
++Y+++ TA+ CVR +A+ RP M +V
Sbjct: 614 EVYKMIETAASCVRHSALKRPRMVQV 639
>At1g53420 hypothetical protein, 5' partial
Length = 856
Score = 207 bits (528), Expect = 9e-54
Identities = 115/262 (43%), Positives = 162/262 (60%), Gaps = 8/262 (3%)
Query: 294 GGYSEVYKGDLCNGETIAVKRLAKDNKDPNKEKEFLMELGIIGHVCHPNTASLLGYCFEN 353
GG+ VYKG L +G IAVK+L+ +K N+E FL E+G+I + HPN L G C E
Sbjct: 536 GGFGPVYKGKLFDGTIIAVKQLSTGSKQGNRE--FLNEIGMISALHHPNLVKLYGCCVEG 593
Query: 354 G-LYLIFNYSQNGNLSTALHGYGKAGNSLDWPIRYKIAIGVARGLHYLHKCCKHRIIHRD 412
G L L++ + +N +L+ AL G + LDWP R KI IGVARGL YLH+ + +I+HRD
Sbjct: 594 GQLLLVYEFVENNSLARALFGPQETQLRLDWPTRRKICIGVARGLAYLHEESRLKIVHRD 653
Query: 413 IKASNVLLGPDYEPQITDFGLAKWLPNKWTHHAVIPVEGTFGYLAPEVFMHGIVDEKTDI 472
IKA+NVLL P+I+DFGLAK L + + H + GTFGY+APE M G + +K D+
Sbjct: 654 IKATNVLLDKQLNPKISDFGLAK-LDEEDSTHISTRIAGTFGYMAPEYAMRGHLTDKADV 712
Query: 473 FAFGVLLLEIVTGR-RPVDSSKQN---ILLWAKPLMESGNIAELADPRMEGRYDVEQLYR 528
++FG++ LEIV GR ++ SK N ++ W + L E N+ EL DPR+ Y+ E+
Sbjct: 713 YSFGIVALEIVHGRSNKIERSKNNTFYLIDWVEVLREKNNLLELVDPRLGSEYNREEAMT 772
Query: 529 VVLTASYCVRQTAIWRPAMTEV 550
++ A C RP+M+EV
Sbjct: 773 MIQIAIMCTSSEPCERPSMSEV 794
>At3g24550 protein kinase, putative
Length = 652
Score = 206 bits (525), Expect = 2e-53
Identities = 115/267 (43%), Positives = 169/267 (63%), Gaps = 14/267 (5%)
Query: 293 RGGYSEVYKGDLCNGETIAVKRLAKDNKDPNKEKEFLMELGIIGHVCHPNTASLLGYCFE 352
+GG+ V+KG L +G+ +AVK+L + E+EF E+ II V H + SL+GYC
Sbjct: 288 QGGFGYVHKGILPSGKEVAVKQLKAGSGQG--EREFQAEVEIISRVHHRHLVSLIGYCMA 345
Query: 353 N-GLYLIFNYSQNGNLSTALHGYGKAGNSLDWPIRYKIAIGVARGLHYLHKCCKHRIIHR 411
L++ + N NL LHG G+ +++W R KIA+G A+GL YLH+ C +IIHR
Sbjct: 346 GVQRLLVYEFVPNNNLEFHLHGKGRP--TMEWSTRLKIALGSAKGLSYLHEDCNPKIIHR 403
Query: 412 DIKASNVLLGPDYEPQITDFGLAKWLPNKWTHHAVIPVEGTFGYLAPEVFMHGIVDEKTD 471
DIKASN+L+ +E ++ DFGLAK + + H V GTFGYLAPE G + EK+D
Sbjct: 404 DIKASNILIDFKFEAKVADFGLAK-IASDTNTHVSTRVMGTFGYLAPEYAASGKLTEKSD 462
Query: 472 IFAFGVLLLEIVTGRRPVDSSK----QNILLWAKPLM----ESGNIAELADPRMEGRYDV 523
+F+FGV+LLE++TGRRPVD++ +++ WA+PL+ E G+ LAD +M YD
Sbjct: 463 VFSFGVVLLELITGRRPVDANNVYVDDSLVDWARPLLNRASEEGDFEGLADSKMGNEYDR 522
Query: 524 EQLYRVVLTASYCVRQTAIWRPAMTEV 550
E++ R+V A+ CVR +A RP M+++
Sbjct: 523 EEMARMVACAAACVRHSARRRPRMSQI 549
>At2g42960 putative protein kinase
Length = 494
Score = 205 bits (522), Expect = 5e-53
Identities = 114/263 (43%), Positives = 162/263 (61%), Gaps = 10/263 (3%)
Query: 294 GGYSEVYKGDLCNGETIAVKRLAKDNKDPNKEKEFLMELGIIGHVCHPNTASLLGYCFEN 353
GGY VY+G L NG +AVK+L N EKEF +E+ IGHV H N LLGYC E
Sbjct: 192 GGYGVVYRGKLVNGTEVAVKKLL--NNLGQAEKEFRVEVEAIGHVRHKNLVRLLGYCIE- 248
Query: 354 GLY--LIFNYSQNGNLSTALHGYGKAGNSLDWPIRYKIAIGVARGLHYLHKCCKHRIIHR 411
G++ L++ Y +GNL LHG + +L W R KI G A+ L YLH+ + +++HR
Sbjct: 249 GVHRMLVYEYVNSGNLEQWLHGAMRQHGNLTWEARMKIITGTAQALAYLHEAIEPKVVHR 308
Query: 412 DIKASNVLLGPDYEPQITDFGLAKWLPNKWTHHAVIPVEGTFGYLAPEVFMHGIVDEKTD 471
DIKASN+L+ ++ +++DFGLAK L + H V GTFGY+APE G+++EK+D
Sbjct: 309 DIKASNILIDDEFNAKLSDFGLAKLL-DSGESHITTRVMGTFGYVAPEYANTGLLNEKSD 367
Query: 472 IFAFGVLLLEIVTGRRPVD----SSKQNILLWAKPLMESGNIAELADPRMEGRYDVEQLY 527
I++FGVLLLE +TGR PVD +++ N++ W K ++ + E+ DPR+E R L
Sbjct: 368 IYSFGVLLLEAITGRDPVDYGRPANEVNLVEWLKMMVGTRRAEEVVDPRLEPRPSKSALK 427
Query: 528 RVVLTASYCVRQTAIWRPAMTEV 550
R +L + CV A RP M++V
Sbjct: 428 RALLVSLRCVDPEAEKRPRMSQV 450
>At3g24540 protein kinase, putative
Length = 509
Score = 205 bits (521), Expect = 6e-53
Identities = 115/266 (43%), Positives = 165/266 (61%), Gaps = 14/266 (5%)
Query: 294 GGYSEVYKGDLCNGETIAVKRLAKDNKDPNKEKEFLMELGIIGHVCHPNTASLLGYCFEN 353
GG+ VYKG L NG +AVK+L + EKEF E+ II + H N SL+GYC
Sbjct: 188 GGFGFVYKGILNNGNEVAVKQLKVGSAQG--EKEFQAEVNIISQIHHRNLVSLVGYCIAG 245
Query: 354 GL-YLIFNYSQNGNLSTALHGYGKAGNSLDWPIRYKIAIGVARGLHYLHKCCKHRIIHRD 412
L++ + N L LHG G+ +++W +R KIA+ ++GL YLH+ C +IIHRD
Sbjct: 246 AQRLLVYEFVPNNTLEFHLHGKGRP--TMEWSLRLKIAVSSSKGLSYLHENCNPKIIHRD 303
Query: 413 IKASNVLLGPDYEPQITDFGLAKWLPNKWTHHAVIPVEGTFGYLAPEVFMHGIVDEKTDI 472
IKA+N+L+ +E ++ DFGLAK + TH + V GTFGYLAPE G + EK+D+
Sbjct: 304 IKAANILIDFKFEAKVADFGLAKIALDTNTHVST-RVMGTFGYLAPEYAASGKLTEKSDV 362
Query: 473 FAFGVLLLEIVTGRRPVDSSK----QNILLWAKPL----MESGNIAELADPRMEGRYDVE 524
++FGV+LLE++TGRRPVD++ +++ WA+PL +E N LAD ++ YD E
Sbjct: 363 YSFGVVLLELITGRRPVDANNVYADDSLVDWARPLLVQALEESNFEGLADIKLNNEYDRE 422
Query: 525 QLYRVVLTASYCVRQTAIWRPAMTEV 550
++ R+V A+ CVR TA RP M +V
Sbjct: 423 EMARMVACAAACVRYTARRRPRMDQV 448
>At1g49270 hypothetical protein
Length = 699
Score = 205 bits (521), Expect = 6e-53
Identities = 116/265 (43%), Positives = 165/265 (61%), Gaps = 10/265 (3%)
Query: 293 RGGYSEVYKGDLCNGETIAVKRLAKDNKDPNKEKEFLMELGIIGHVCHPNTASLLGYCFE 352
+GG+ V+KG L NG+ IAVK L + E+EF E+ II V H + SL+GYC
Sbjct: 344 QGGFGYVHKGILPNGKEIAVKSLKAGSGQG--EREFQAEVEIISRVHHRHLVSLVGYCSN 401
Query: 353 NGLYLIFNYSQNGNLSTALHGYGKAGNSLDWPIRYKIAIGVARGLHYLHKCCKHRIIHRD 412
G + Y N + H +GK+G +DWP R KIA+G A+GL YLH+ C +IIHRD
Sbjct: 402 AGGQRLLVYEFLPNDTLEFHLHGKSGTVMDWPTRLKIALGSAKGLAYLHEDCHPKIIHRD 461
Query: 413 IKASNVLLGPDYEPQITDFGLAKWLPNKWTHHAVIPVEGTFGYLAPEVFMHGIVDEKTDI 472
IKASN+LL ++E ++ DFGLAK + TH + V GTFGYLAPE G + EK+D+
Sbjct: 462 IKASNILLDHNFEAKVADFGLAKLSQDNNTHVST-RVMGTFGYLAPEYASSGKLTEKSDV 520
Query: 473 FAFGVLLLEIVTGRRPVDSS---KQNILLWAKPL----MESGNIAELADPRMEGRYDVEQ 525
F+FGV+LLE++TGR PVD S + +++ WA+PL + G EL DP +E +Y+ +
Sbjct: 521 FSFGVMLLELITGRGPVDLSGDMEDSLVDWARPLCMRVAQDGEYGELVDPFLEHQYEPYE 580
Query: 526 LYRVVLTASYCVRQTAIWRPAMTEV 550
+ R+V A+ VR + RP M+++
Sbjct: 581 MARMVACAAAAVRHSGRRRPKMSQI 605
>At1g70460 putative protein kinase
Length = 710
Score = 204 bits (520), Expect = 8e-53
Identities = 114/266 (42%), Positives = 170/266 (63%), Gaps = 14/266 (5%)
Query: 294 GGYSEVYKGDLCNGETIAVKRLAKDNKDPNKEKEFLMELGIIGHVCHPNTASLLGYCF-E 352
GG+ VYKG L +G+ +AVK+L + ++EF E+ II V H + SL+GYC +
Sbjct: 362 GGFGCVYKGKLNDGKLVAVKQLKVGSGQG--DREFKAEVEIISRVHHRHLVSLVGYCIAD 419
Query: 353 NGLYLIFNYSQNGNLSTALHGYGKAGNSLDWPIRYKIAIGVARGLHYLHKCCKHRIIHRD 412
+ LI+ Y N L LHG G+ L+W R +IAIG A+GL YLH+ C +IIHRD
Sbjct: 420 SERLLIYEYVPNQTLEHHLHGKGRP--VLEWARRVRIAIGSAKGLAYLHEDCHPKIIHRD 477
Query: 413 IKASNVLLGPDYEPQITDFGLAKWLPNKWTHHAVIPVEGTFGYLAPEVFMHGIVDEKTDI 472
IK++N+LL ++E Q+ DFGLAK L + H V GTFGYLAPE G + +++D+
Sbjct: 478 IKSANILLDDEFEAQVADFGLAK-LNDSTQTHVSTRVMGTFGYLAPEYAQSGKLTDRSDV 536
Query: 473 FAFGVLLLEIVTGRRPVDS----SKQNILLWAKPLM----ESGNIAELADPRMEGRYDVE 524
F+FGV+LLE++TGR+PVD +++++ WA+PL+ E+G+ +EL D R+E Y
Sbjct: 537 FSFGVVLLELITGRKPVDQYQPLGEESLVEWARPLLHKAIETGDFSELVDRRLEKHYVEN 596
Query: 525 QLYRVVLTASYCVRQTAIWRPAMTEV 550
+++R++ TA+ CVR + RP M +V
Sbjct: 597 EVFRMIETAAACVRHSGPKRPRMVQV 622
>At3g59110 receptor-like protein kinase
Length = 512
Score = 204 bits (518), Expect = 1e-52
Identities = 114/262 (43%), Positives = 158/262 (59%), Gaps = 8/262 (3%)
Query: 294 GGYSEVYKGDLCNGETIAVKRLAKDNKDPNKEKEFLMELGIIGHVCHPNTASLLGYCFEN 353
GGY VYKG L NG +AVK+L N EKEF +E+ IGHV H N LLGYC E
Sbjct: 199 GGYGVVYKGRLINGNDVAVKKLL--NNLGQAEKEFRVEVEAIGHVRHKNLVRLLGYCIEG 256
Query: 354 -GLYLIFNYSQNGNLSTALHGYGKAGNSLDWPIRYKIAIGVARGLHYLHKCCKHRIIHRD 412
L++ Y +GNL LHG ++L W R KI +G A+ L YLH+ + +++HRD
Sbjct: 257 VNRMLVYEYVNSGNLEQWLHGAMGKQSTLTWEARMKILVGTAQALAYLHEAIEPKVVHRD 316
Query: 413 IKASNVLLGPDYEPQITDFGLAKWLPNKWTHHAVIPVEGTFGYLAPEVFMHGIVDEKTDI 472
IKASN+L+ D+ +++DFGLAK L + H V GTFGY+APE G+++EK+DI
Sbjct: 317 IKASNILIDDDFNAKLSDFGLAKLL-DSGESHITTRVMGTFGYVAPEYANTGLLNEKSDI 375
Query: 473 FAFGVLLLEIVTGRRPVD----SSKQNILLWAKPLMESGNIAELADPRMEGRYDVEQLYR 528
++FGVLLLE +TGR PVD +++ N++ W K ++ + E+ D R+E L R
Sbjct: 376 YSFGVLLLETITGRDPVDYERPANEVNLVEWLKMMVGTRRAEEVVDSRIEPPPATRALKR 435
Query: 529 VVLTASYCVRQTAIWRPAMTEV 550
+L A CV A RP M++V
Sbjct: 436 ALLVALRCVDPEAQKRPKMSQV 457
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.337 0.149 0.491
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 12,091,409
Number of Sequences: 26719
Number of extensions: 510384
Number of successful extensions: 5096
Number of sequences better than 10.0: 1009
Number of HSP's better than 10.0 without gapping: 829
Number of HSP's successfully gapped in prelim test: 180
Number of HSP's that attempted gapping in prelim test: 1680
Number of HSP's gapped (non-prelim): 1195
length of query: 550
length of database: 11,318,596
effective HSP length: 104
effective length of query: 446
effective length of database: 8,539,820
effective search space: 3808759720
effective search space used: 3808759720
T: 11
A: 40
X1: 15 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 39 (21.7 bits)
S2: 63 (28.9 bits)
Medicago: description of AC127169.5


