

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC127021.9 + phase: 0
(540 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
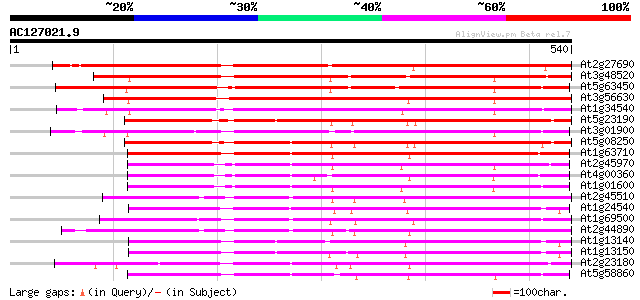
Score E
Sequences producing significant alignments: (bits) Value
At2g27690 putative cytochrome P450 588 e-168
At3g48520 cytochrome P450-like protein 422 e-118
At5g63450 cytochrome P450-like protein 404 e-113
At3g56630 cytochrome P450-like protein 376 e-104
At1g34540 cytochrome p450, putative 367 e-102
At5g23190 cytochrome P450-like protein 355 4e-98
At3g01900 putative cytochrome P450 354 8e-98
At5g08250 cytochrome P450-like protein 338 3e-93
At1g63710 cytochrome P450 like protein 323 1e-88
At2g45970 putative cytochrome P450 316 2e-86
At4g00360 probable cytochrome P450 310 2e-84
At1g01600 unknown protein 308 4e-84
At2g45510 putative cytochrome P450 304 7e-83
At1g24540 putative cytochrome P450 304 9e-83
At1g69500 unknown protein 303 1e-82
At2g44890 putative cytochrome P450 300 1e-81
At1g13140 cytochrome P450 monooxygenase like protein 297 9e-81
At1g13150 putative cytochrome P450 monooxygenase 297 1e-80
At2g23180 putative cytochrome P450 294 7e-80
At5g58860 cytochrome P450 CYP86A1 287 9e-78
>At2g27690 putative cytochrome P450
Length = 495
Score = 588 bits (1516), Expect = e-168
Identities = 298/504 (59%), Positives = 376/504 (74%), Gaps = 19/504 (3%)
Query: 42 TFTFLFFSFTLLFSFFSFLLFISRIKPWCNCNTCKTFLTMSWSNKFVNLVDYYTHLLQES 101
+FT + F F ++FS F LLF+ +++ +CNC C +LT SW F+NL D+YTHLL+ S
Sbjct: 6 SFTIVSFFFIIIFSLFH-LLFLQKLR-YCNCEICHAYLTSSWKKDFINLSDWYTHLLRRS 63
Query: 102 PTGTIHVHVLGNTITSNPENVEYILKTNFNNYPKGKQFSTILGDLLGRGIFNVDGHSWKF 161
PT TI VHVL + IT+NP NVE+ILKTNF+NYPKGKQFS ILGDLLGRGIFN DG +W+F
Sbjct: 64 PTSTIKVHVLNSVITANPSNVEHILKTNFHNYPKGKQFSVILGDLLGRGIFNSDGDTWRF 123
Query: 162 QRKMASLELGSVVIRSYAMELVIEEIKTRLLPLIASVAEKKTASKADNTSEDVLLDMQDI 221
QRK+ASLELGSV +R +A E+V EI+TRLLP++ S ++ + +LD+QD+
Sbjct: 124 QRKLASLELGSVSVRVFAHEIVKTEIETRLLPILTSFSDNPGS----------VLDLQDV 173
Query: 222 LRRFSFDNICKFSFGLDPCCLVPSLPVSNLANAFDLSSTLSAQRALTASPLIWKMKRFFN 281
RRFSFD I K SFG DP CL P+S A AFD +S LSA+RAL PL+WK KR
Sbjct: 174 FRRFSFDTISKLSFGFDPDCLRLPFPISEFAVAFDTASLLSAKRALAPFPLLWKTKRLLR 233
Query: 282 IGSEKKLKEAIKIVNDLANEMIKQRREIENGVESRKDLLSRFMGALNSHDDEYLRDIVVS 341
IGSEKKL+E+I ++N LA ++IKQRR G+ + DL+SRFM + DDEYLRDIVVS
Sbjct: 234 IGSEKKLQESINVINRLAGDLIKQRR--LTGLMGKNDLISRFMAVVAEDDDEYLRDIVVS 291
Query: 342 FLLAGRDTVASALTGFFILLSKNPKVEEKIRVELDRVMNPNQECAT--FEQTREMHYLNG 399
FLLAGRDTVA+ LTGFF LL+++P+VE +IR ELDRVM + T ++ REM YL+
Sbjct: 292 FLLAGRDTVAAGLTGFFWLLTRHPEVENRIREELDRVMGTGFDSVTARCDEMREMDYLHA 351
Query: 400 AIHESMRLFPPVQFDSKFALEDDVLPDGTFIKKGSRVTYHPYAMGRMENIWGPDCLEFKP 459
+++ESMRLFPPVQFDSKFAL DDVL DGTF+ G+RVTYH YAMGRM+ IWGPD EFKP
Sbjct: 352 SLYESMRLFPPVQFDSKFALNDDVLSDGTFVNSGTRVTYHAYAMGRMDRIWGPDYEEFKP 411
Query: 460 ERWL-KDGVFVPKCPFKYPVFQAGSRVCLGKELAIVEMKSVVASLVKRFDVRVVGP--NQ 516
ERWL +G F P+ P KYPVFQAG+RVC+GKE+AI+EMKS+ ++++RF+ RV P +
Sbjct: 412 ERWLDNEGKFRPENPVKYPVFQAGARVCIGKEMAIMEMKSIAVAIIRRFETRVASPETTE 471
Query: 517 EPQFAPGLTASFRGGLPVKIYERT 540
+FAPGLTA+ GGLPV I ER+
Sbjct: 472 TLRFAPGLTATVNGGLPVMIQERS 495
>At3g48520 cytochrome P450-like protein
Length = 506
Score = 422 bits (1084), Expect = e-118
Identities = 225/472 (47%), Positives = 317/472 (66%), Gaps = 29/472 (6%)
Query: 81 MSWSNKFVNLVDYYTHLLQESPTGTIHVHVLGNT---ITSNPENVEYILKTNFNNYPKGK 137
+S++ L+ +YT LL+ SP+ TI V +LGN IT+NP NVEYILKTNF N+PKGK
Sbjct: 46 LSFNKNRHRLLQWYTELLRLSPSQTILVPLLGNRRTIITTNPLNVEYILKTNFFNFPKGK 105
Query: 138 QFSTILGDLLGRGIFNVDGHSWKFQRKMASLELGSVVIRSYAMELVIEEIKTRLLPLIAS 197
F+ +LGDLLG GIFNVDGHSW QRK+AS E + +RS+A E++ +E++ RL+P++++
Sbjct: 106 PFTDLLGDLLGGGIFNVDGHSWSSQRKLASHEFSTRSLRSFAFEVLKDEVENRLVPVLST 165
Query: 198 VAEKKTASKADNTSEDVLLDMQDILRRFSFDNICKFSFGLDPCCLVPSLPVSNLANAFDL 257
A+ T +D+QD+L+RF+FD +CK S G DP CL + PV+ L AFD
Sbjct: 166 AADVGTT-----------VDLQDVLKRFAFDVVCKVSLGWDPDCLDLTRPVNPLVEAFDT 214
Query: 258 SSTLSAQRALTASPLIWKMKRFFNIGSEKKLKEAIKIVNDLANEMIKQRR---EIENGVE 314
++ +SA+RA +WK KR N+GSE+KL+EAI+ V+ L +E+++ ++ EI G E
Sbjct: 215 AAEISARRATEPIYAVWKTKRVLNVGSERKLREAIRTVHVLVSEIVRAKKKSLEIGTGAE 274
Query: 315 SRKDLLSRFMGALNSHDDEYLRDIVVSFLLAGRDTVASALTGFFILLSKNPKVEEKIRVE 374
+++DLLSRF+ A H+ E +RD+V+SF++AGRDT ++A+T F LL++N VE KI E
Sbjct: 275 AKQDLLSRFLAA--GHNGEAVRDMVISFIMAGRDTTSAAMTWLFWLLTENDDVERKILEE 332
Query: 375 LDRVMNPNQECATFEQTREMHYLNGAIHESMRLFPPVQFDSKFALEDDVLPDGTFIKKGS 434
+D +++ FE +EM Y + E+MRL+PPV +DSK A DDVLPDGT +K+G
Sbjct: 333 VDPLVSLG---LGFEDLKEMAYTKACLCEAMRLYPPVSWDSKHAANDDVLPDGTRVKRGD 389
Query: 435 RVTYHPYAMGRMENIWGPDCLEFKPERWLKD------GVFVPKCPFKYPVFQAGSRVCLG 488
+VTY PY MGRME +WG D EF P RW V P P+K+PVFQAG RVC+G
Sbjct: 390 KVTYFPYGMGRMETLWGTDSEEFNPNRWFDSEPGSTRPVLKPISPYKFPVFQAGPRVCVG 449
Query: 489 KELAIVEMKSVVASLVKRFDVRVVGPNQEPQFAPGLTASFRGGLPVKIYERT 540
KE+A ++MK VV S++ RF++ V ++ P F P LTA GGL VKI R+
Sbjct: 450 KEMAFMQMKYVVGSVLSRFEIVPVNKDR-PVFVPLLTAHMAGGLKVKIKRRS 500
>At5g63450 cytochrome P450-like protein
Length = 510
Score = 404 bits (1037), Expect = e-113
Identities = 231/508 (45%), Positives = 312/508 (60%), Gaps = 30/508 (5%)
Query: 45 FLFFSFTLLFSFFSFLLFISRIKPWCNCNTCKTFLTMSWSNKFVNLVDYYTHLLQESPTG 104
F F L+FSF + L P + +S++ L+ +YT LL+ SP+
Sbjct: 12 FPIIGFVLIFSFPTKTLKAKTASPSNPTSYQLIGSILSFNKNRHRLLQWYTDLLRLSPSQ 71
Query: 105 TIHVHVL---GNTITSNPENVEYILKTNFNNYPKGKQFSTILGDLLGRGIFNVDGHSWKF 161
TI V +L IT+NPENVE+ILKTNF N+PKGK F+ +LGDLLG GIFN DG W
Sbjct: 72 TITVDLLFGRRTIITANPENVEHILKTNFYNFPKGKPFTDLLGDLLGGGIFNSDGELWSS 131
Query: 162 QRKMASLELGSVVIRSYAMELVIEEIKTRLLPLIASVAEKKTASKADNTSEDVLLDMQDI 221
QRK+AS E +R + E++ EE++ RL+P+++S + E V D Q++
Sbjct: 132 QRKLASHEFTMRSLREFTFEILREEVQNRLIPVLSSAVD---------CGETV--DFQEV 180
Query: 222 LRRFSFDNICKFSFGLDPCCLVPSLPVSNLANAFDLSSTLSAQRALTASPLIWKMKRFFN 281
L+RF+FD +CK S G DP CL + PV L AFD+++ +SA+RA +WK+KRF N
Sbjct: 181 LKRFAFDVVCKVSLGWDPDCLDLTRPVPELVKAFDVAAEISARRATEPVYAVWKVKRFLN 240
Query: 282 IGSEKKLKEAIKIVNDLANEMIKQRR---EIENGVESRKDLLSRFMGALNSHDDEYLRDI 338
+GSEK+L+EAIK V+ +E+I+ ++ +I V ++DLLSRF+ A H +E +RD
Sbjct: 241 VGSEKRLREAIKTVHLSVSEIIRAKKKSLDIGGDVSDKQDLLSRFLAA--GHGEEAVRDS 298
Query: 339 VVSFLLAGRDTVASALTGFFILLSKNPKVEEKIRVELDRVMNPNQECATFEQTREMHYLN 398
V+SF++AGRDT ++A+T F LLS+N VE KI LD + N FE REM Y
Sbjct: 299 VISFIMAGRDTTSAAMTWLFWLLSQNDDVETKI---LDELRNKGSLGLGFEDLREMSYTK 355
Query: 399 GAIHESMRLFPPVQFDSKFALEDDVLPDGTFIKKGSRVTYHPYAMGRMENIWGPDCLEFK 458
+ E+MRL+PPV +DSK A DD+LPDGT +KKG +VTY PY MGRME +WG D EFK
Sbjct: 356 ACLCEAMRLYPPVAWDSKHAANDDILPDGTPLKKGDKVTYFPYGMGRMEKVWGKDWDEFK 415
Query: 459 PERWLKD-------GVFVPKCPFKYPVFQAGSRVCLGKELAIVEMKSVVASLVKRFDVRV 511
P RW ++ V FK+PVFQAG RVC+GKE+A +MK VV S++ RF +
Sbjct: 416 PNRWFEEEPSYGTKPVLKSVSSFKFPVFQAGPRVCIGKEMAFTQMKYVVGSVLSRFKIIP 475
Query: 512 VGPNQEPQFAPGLTASFRGGLPVKIYER 539
V N P F P LTA GGL VKI R
Sbjct: 476 V-CNNRPVFVPLLTAHMAGGLKVKIKRR 502
>At3g56630 cytochrome P450-like protein
Length = 499
Score = 376 bits (966), Expect = e-104
Identities = 205/460 (44%), Positives = 289/460 (62%), Gaps = 21/460 (4%)
Query: 91 VDYYTHLLQESPTGTIHVHVLGN---TITSNPENVEYILKTNFNNYPKGKQFSTILGDLL 147
+D+ L PT T G +T+NP NVEY+LKT F ++PKG++F +IL D L
Sbjct: 51 LDWTVETLSRCPTQTAIFRRPGKLQFVMTANPANVEYMLKTKFESFPKGERFISILEDFL 110
Query: 148 GRGIFNVDGHSWKFQRKMASLELGSVVIRSYAMELVIEEIKTRLLPLIASVAEKKTASKA 207
GRGIFN DG W QRK AS E + +R + M V EI TRL+P++A A
Sbjct: 111 GRGIFNSDGEMWWKQRKTASYEFSTKSLRDFVMSNVTVEINTRLVPVLAEAA-------- 162
Query: 208 DNTSEDVLLDMQDILRRFSFDNICKFSFGLDPCCLVPSLPVS-NLANAFDLSSTLSAQRA 266
+ L+D+QDIL RF+FDNICK +F +D CL N AF+ ++T+ +QR
Sbjct: 163 ---TNGKLIDLQDILERFAFDNICKLAFNVDSACLGDDGAAGVNFMQAFETAATIISQRF 219
Query: 267 LTASPLIWKMKRFFNIGSEKKLKEAIKIVNDLANEMIKQRREIENGVESRKDLLSRFMGA 326
+ WK+K+ NIGSE+ L+E+I IV+ A+E+++ R E + ++DLLSRF+
Sbjct: 220 QSVISYSWKIKKKLNIGSERVLRESIMIVHKFADEIVRNRIEQGKVSDHKEDLLSRFISK 279
Query: 327 LNSHDDEYLRDIVVSFLLAGRDTVASALTGFFILLSKNPKVEEKIRVELDRVMNPN---- 382
+ E LRDIV+SF+LAGRDT +SAL+ FF LLS +P+V++KI EL+ +
Sbjct: 280 EEMNSPEILRDIVISFILAGRDTTSSALSWFFWLLSMHPEVKDKILQELNSIRERTGKRI 339
Query: 383 QECATFEQTREMHYLNGAIHESMRLFPPVQFDSKFALEDDVLPDGTFIKKGSRVTYHPYA 442
E FE + M+YL+ AI ES+RL+PPV D+ ED+VLPDGTFI K ++Y+ YA
Sbjct: 340 GEVYGFEDLKLMNYLHAAITESLRLYPPVPVDTMSCAEDNVLPDGTFIGKDWGISYNAYA 399
Query: 443 MGRMENIWGPDCLEFKPERWLKD--GVFVPKCPFKYPVFQAGSRVCLGKELAIVEMKSVV 500
MGRME+IWG DC F PERW+ + G F + P+K+P F AG R+CLGKE+A ++MKS+V
Sbjct: 400 MGRMESIWGKDCDRFDPERWIDETNGGFRGENPYKFPAFHAGPRMCLGKEMAYIQMKSIV 459
Query: 501 ASLVKRFDVRVVGPNQEPQFAPGLTASFRGGLPVKIYERT 540
A++++RF V V G + P+ +T RGGL V++ ER+
Sbjct: 460 AAVLERFVVEVPGKKERPEILMSVTLRIRGGLNVRVQERS 499
>At1g34540 cytochrome p450, putative
Length = 498
Score = 367 bits (943), Expect = e-102
Identities = 213/512 (41%), Positives = 305/512 (58%), Gaps = 34/512 (6%)
Query: 46 LFFSFTLLFSFFSFLLFISRIKPWCNCNTCKTFLTMSWSNKFVNLV-------DYYTHLL 98
L F F L F F +F ++ ++ F + F LV D+ L
Sbjct: 4 LIFIFLLCFPISIFFIFFTK-----KSSSEFGFKSYPIVGSFPGLVNNRHRFLDWTVETL 58
Query: 99 QESPTGTIHVHVLGNT---ITSNPENVEYILKTNFNNYPKGKQFSTILGDLLGRGIFNVD 155
PT T G +T+NP NVEY+LKT F ++PKG+QF+++L D LG GIFN D
Sbjct: 59 SRCPTQTAIFRRPGKQQLIMTANPSNVEYMLKTKFESFPKGQQFTSVLEDFLGHGIFNSD 118
Query: 156 GHSWKFQRKMASLELGSVVIRSYAMELVIEEIKTRLLPLIASVAEKKTASKADNTSEDVL 215
G W QRK AS E + +R + M V EI TRL+P++ A T K L
Sbjct: 119 GDMWWKQRKTASYEFSTKSLRDFVMSNVTVEINTRLVPVLVEAA---TTGK--------L 167
Query: 216 LDMQDILRRFSFDNICKFSFGLDPCCLVPSLPVS-NLANAFDLSSTLSAQRALTASPLIW 274
+D+QDIL RF+FDNICK +F +D CL V N AF+ ++T+ +QR + + W
Sbjct: 168 IDLQDILERFAFDNICKLAFNVDCACLGHDGAVGVNFMRAFETAATIISQRFRSVASCAW 227
Query: 275 KMKRFFNIGSEKKLKEAIKIVNDLANEMIKQRREIENGVESRKDLLSRFMGALNSHDDEY 334
++K+ NIGSE+ L+E+I V+ A+E+++ R + + ++DLLSRF+ + E
Sbjct: 228 RIKKKLNIGSERVLRESIATVHKFADEIVRNRIDQGRSSDHKEDLLSRFISKEEMNSPEI 287
Query: 335 LRDIVVSFLLAGRDTVASALTGFFILLSKNPKVEEKIRVELD----RVMNPNQECATFEQ 390
LRDIV+SF+LAGRDT +SAL+ FF LLS +P+VE+KI EL+ R E FE
Sbjct: 288 LRDIVISFILAGRDTTSSALSWFFWLLSMHPEVEDKILQELNSIRARTGKRIGEVYGFEH 347
Query: 391 TREMHYLNGAIHESMRLFPPVQFDSKFALEDDVLPDGTFIKKGSRVTYHPYAMGRMENIW 450
+ M+YL+ AI ES+RL+PPV D K ED+VLPDGTF+ KG +TY+ +AMGRME+IW
Sbjct: 348 LKMMNYLHAAITESLRLYPPVPVDIKSCAEDNVLPDGTFVGKGWAITYNIFAMGRMESIW 407
Query: 451 GPDCLEFKPERWLKD--GVFVPKCPFKYPVFQAGSRVCLGKELAIVEMKSVVASLVKRFD 508
G DC F PERW+ + G F + P K+P F AG R+C+GK++A ++MKS+VA++++RF
Sbjct: 408 GKDCDRFDPERWIDETNGCFRGEDPSKFPAFHAGPRMCVGKDMAYIQMKSIVAAVLERFV 467
Query: 509 VRVVGPNQEPQFAPGLTASFRGGLPVKIYERT 540
V V G + P+ +T +GGL ++ ER+
Sbjct: 468 VEVPG-KERPEILLSMTLRIKGGLFARVQERS 498
>At5g23190 cytochrome P450-like protein
Length = 559
Score = 355 bits (911), Expect = 4e-98
Identities = 199/449 (44%), Positives = 280/449 (62%), Gaps = 33/449 (7%)
Query: 111 LGNTITSNPENVEYILKTNFNNYPKGKQFSTILGDLLGRGIFNVDGHSWKFQRKMASLEL 170
L +TIT +P NVE++LK F+ +PKG F L DLLG GIFN D +W+ QRK AS+E
Sbjct: 107 LNSTITCDPRNVEHLLKNRFSVFPKGSYFRDNLRDLLGDGIFNADDETWQRQRKTASIEF 166
Query: 171 GSVVIRSYAMELVIEEIKTRLLPLIASVAEKKTASKADNTSEDVLLDMQDILRRFSFDNI 230
S R + + E + RLLP++ +T+ K+ + +D+QD+L R +FDN+
Sbjct: 167 HSAKFRQLTTQSLFELVHKRLLPVL------ETSVKSSSP-----IDLQDVLLRLTFDNV 215
Query: 231 CKFSFGLDPCCLVPSLPVSNLANAFDLSSTLSAQRALTASPLIWKMKRFFNIGSEKKLKE 290
C +FG+DP CL P PV A AF+ +T +A +WK R+ +IG+EKKLKE
Sbjct: 216 CMIAFGVDPGCLGPDQPVIPFAKAFE-DATEAAVVRFVMPTCVWKFMRYLDIGTEKKLKE 274
Query: 291 AIKIVNDLANEMIKQRRE---IENGVESRKDLLSRFMGALN----SHDDEYLRDIVVSFL 343
+IK V+D A+E+I+ R++ +E R DLL+ FMG + S D++LRDI V+F+
Sbjct: 275 SIKGVDDFADEVIRTRKKELSLEGETTKRSDLLTVFMGLRDEKGESFSDKFLRDICVNFI 334
Query: 344 LAGRDTVASALTGFFILLSKNPKVEEKIRVELDRVMNP-----NQECATFE------QTR 392
LAGRDT + AL+ FF LL KNP+VEEKI VE+ +++ N E + +E + +
Sbjct: 335 LAGRDTSSVALSWFFWLLEKNPEVEEKIMVEMCKILRQRDDHGNAEKSDYEPVFGPEEIK 394
Query: 393 EMHYLNGAIHESMRLFPPVQFDSKFALEDDVLPDGTFIKKGSRVTYHPYAMGRMENIWGP 452
+M YL A+ E++RL+P V D K EDDV PDGT +KKG +V Y YAMGRME IWG
Sbjct: 395 KMDYLQAALSEALRLYPSVPVDHKEVQEDDVFPDGTMLKKGDKVIYAIYAMGRMEAIWGK 454
Query: 453 DCLEFKPERWLKDGVFVPKCPFKYPVFQAGSRVCLGKELAIVEMKSVVASLVKRFDVRVV 512
DCLEF+PERWL+DG F+ + +K+ F G R+CLGK+ A +MKS A++V R+ V+VV
Sbjct: 455 DCLEFRPERWLRDGRFMSESAYKFTAFNGGPRLCLGKDFAYYQMKSTAAAIVYRYKVKVV 514
Query: 513 -GPNQEPQFAPGLTASFRGGLPVKIYERT 540
G EP+ A LT + GL V + R+
Sbjct: 515 NGHKVEPKLA--LTMYMKHGLMVNLINRS 541
>At3g01900 putative cytochrome P450
Length = 496
Score = 354 bits (908), Expect = 8e-98
Identities = 203/513 (39%), Positives = 296/513 (57%), Gaps = 38/513 (7%)
Query: 40 STTFTFLFFSFTLLFSFFSFLLFISRIKPWCNCNTCKTFLTMSWSNKFVN----LVDYYT 95
++TF L LL S +++ C +T KT+ + F L+D+YT
Sbjct: 3 ASTFILLLVLVLLLVSAGKHVIYS------CRNSTPKTYPVIGCLISFYTNRNRLLDWYT 56
Query: 96 HLLQESPTGTIHVHVLG---NTITSNPENVEYILKTNFNNYPKGKQFSTILGDLLGRGIF 152
LL ESP+ T+ + L +T+NP NVEYILKTNF+NYPKGK F+ ILGD LG GIF
Sbjct: 57 ELLTESPSRTVVIRRLAARRTVVTANPSNVEYILKTNFDNYPKGKPFTEILGDFLGNGIF 116
Query: 153 NVDGHSWKFQRKMASLELGSVVIRSYAMELVIEEIKTRLLPLIASVAEKKTASKADNTSE 212
NVDG+ W QR++A+ + +R Y + ++ E++ LL + + AE
Sbjct: 117 NVDGNLWLKQRRLATHDFTPKSLREY-VTVLRNEVEKELLAFLNAAAEDSQP-------- 167
Query: 213 DVLLDMQDILRRFSFDNICKFSFGLDPCCLVPSLPVSNLANAFDLSSTLSAQRALTASPL 272
D+Q++LRRF+F+ +C G+D C L PS PVS AF +S +SA R
Sbjct: 168 ---FDLQELLRRFTFNIVCIVFLGIDRCTLNPSSPVSEFDRAFQTASAVSAGRGSAPLSF 224
Query: 273 IWKMKRFFNIGSEKKLKEAIKIVNDLANEMIKQRREIENGVESRKDLLSRFMGALNSHDD 332
+WK KR GSEK+L++A+ V++ +E+I+ ++ + +D LSR + A S D
Sbjct: 225 VWKFKRLVGFGSEKELRKAVGEVHNCVDEIIRDKKR----KPANQDFLSRLIVAGES--D 278
Query: 333 EYLRDIVVSFLLAGRDTVASALTGFFILLSKNPKVEEKIRVELDRVMNPNQECATFEQTR 392
E +RD+V+S ++AGRDT ++ T F L++ + + E + E+ V +E +
Sbjct: 279 ETVRDMVISIIMAGRDTTSAVATRLFWLITGHEETEHDLVSEIRSVKEEITGGFDYESLK 338
Query: 393 EMHYLNGAIHESMRLFPPVQFDSKFALEDDVLPDGTFIKKGSRVTYHPYAMGRMENIWGP 452
++ L + E MRL+PPV +DSK AL DD LPDGT ++ G RVTY PY MGRME +WG
Sbjct: 339 KLSLLKACLCEVMRLYPPVPWDSKHALTDDRLPDGTLVRAGDRVTYFPYGMGRMEELWGE 398
Query: 453 DCLEFKPERWLKD------GVFVPKCPFKYPVFQAGSRVCLGKELAIVEMKSVVASLVKR 506
D EFKP RW + V PFK+PVFQAG RVCLG+E+A V+MK +VAS++ R
Sbjct: 399 DWDEFKPNRWAESYDKTCCRVLKKVNPFKFPVFQAGPRVCLGEEMAYVQMKYIVASILDR 458
Query: 507 FDVRVVGPNQEPQFAPGLTASFRGGLPVKIYER 539
F++ + P +P F P LTA GG+ V+++ R
Sbjct: 459 FEIEPI-PTDKPDFVPMLTAHMAGGMQVRVHRR 490
>At5g08250 cytochrome P450-like protein
Length = 550
Score = 338 bits (868), Expect = 3e-93
Identities = 191/452 (42%), Positives = 274/452 (60%), Gaps = 40/452 (8%)
Query: 111 LGNTITSNPENVEYILKTNFNNYPKGKQFSTILGDLLGRGIFNVDGHSWKFQRKMASLEL 170
L +T +P NVE++LKT F+ YPKG F + DLLG GIFN D +W+ QRK AS+E
Sbjct: 108 LNCVVTCDPRNVEHLLKTRFSIYPKGSYFRETMQDLLGDGIFNTDDGTWQRQRKAASVEF 167
Query: 171 GSVVIRSYAMELVIEEIKTRLLPLIASVAEKKTASKADNTSEDVLLDMQDILRRFSFDNI 230
S R + + E + RLLP++ +T+ K +D+QDIL R +FDN+
Sbjct: 168 HSAKFRQLTSQSLHELVHNRLLPVL------ETSGK---------IDLQDILLRLTFDNV 212
Query: 231 CKFSFGLDPCCLVPSLPVSNLANAFDLSSTLSAQRALTASPLIWKMKRFFNIGSEKKLKE 290
C +FG+DP CL P LP A AF+ ++ + R + +WK+ R N+G+EKKLKE
Sbjct: 213 CMIAFGVDPGCLSPKLPEIPFAKAFEDATEATVVRFVMPK-FVWKLMRSLNLGTEKKLKE 271
Query: 291 AIKIVNDLANEMIKQRRE---IENGVESRKDLLSRFMGALNSH----DDEYLRDIVVSFL 343
+I V+D A E+I+ R++ +E + R DLL+ FMG + + D++LRDI V+F+
Sbjct: 272 SINGVDDFAEEVIRTRKKEMSLETEIAKRPDLLTIFMGLRDENGQKFSDKFLRDICVNFI 331
Query: 344 LAGRDTVASALTGFFILLSKNPKVEEKIRVELDRVMNP---------NQECATF---EQT 391
LAGRDT + AL+ FF L+ KNP+VEEKI + + +++ N E E+
Sbjct: 332 LAGRDTSSVALSWFFWLIEKNPEVEEKIMMGICKILEQRVDHGDTKKNMEYEPVFRPEEI 391
Query: 392 REMHYLNGAIHESMRLFPPVQFDSKFALEDDVLPDGTFIKKGSRVTYHPYAMGRMENIWG 451
++M YL A+ E++RL+P V D K LEDDV PDGT +KKG +V Y YAMGRME IWG
Sbjct: 392 KKMDYLQAALSETLRLYPSVPVDHKEVLEDDVFPDGTKLKKGEKVIYAIYAMGRMETIWG 451
Query: 452 PDCLEFKPERWLKDGVFVPKCPFKYPVFQAGSRVCLGKELAIVEMKSVVASLVKRFDVRV 511
DC EFKPERWL+DG ++ + +K+ F G R+CLGK+ A +M+ V A+++ R+ VRV
Sbjct: 452 KDCREFKPERWLRDGRYMSESAYKFTAFNGGPRLCLGKDFAYYQMRYVAAAIIYRYKVRV 511
Query: 512 ---VGPNQEPQFAPGLTASFRGGLPVKIYERT 540
G EP+ A LT + GL V + +R+
Sbjct: 512 DDKGGHKVEPKMA--LTMYMKHGLKVNMVKRS 541
>At1g63710 cytochrome P450 like protein
Length = 523
Score = 323 bits (829), Expect = 1e-88
Identities = 179/439 (40%), Positives = 269/439 (60%), Gaps = 26/439 (5%)
Query: 114 TITSNPENVEYILKTNFNNYPKGKQFSTILGDLLGRGIFNVDGHSWKFQRKMASLELGSV 173
T+T +P+N+E+ILKT F+NYPKG + ++ DLLG GIFN DG +W+FQRK A+LE +
Sbjct: 82 TVTCDPKNLEHILKTRFDNYPKGPSWQSVFHDLLGDGIFNSDGDTWRFQRKTAALEFTTR 141
Query: 174 VIRSYAMELVIEEIKTRLLPLIASVAEKKTASKADNTSEDVLLDMQDILRRFSFDNICKF 233
+R V IK RL+P++ S + +D+QD+L R +FDNIC
Sbjct: 142 TLRQAMARWVDRAIKNRLVPILESARSRAEP-----------IDLQDVLLRLTFDNICGL 190
Query: 234 SFGLDPCCLVPSLPVSNLANAFDLSSTLSAQRALTASPLIWKMKRFFNIGSEKKLKEAIK 293
+FG DP L P P + A AFD ++ + QR + IWK++++ +G E + +I
Sbjct: 191 TFGKDPRTLSPEFPENGFAVAFDGATEATLQRFIMPE-FIWKIRKWLRLGLEDDMSRSIS 249
Query: 294 IVNDLANEMIKQRR-EI--ENGVESRKD-LLSRFMGALNSHDDEYLRDIVVSFLLAGRDT 349
V++ +E+I R+ E+ + ESR D LLSRFM S+ D+YL+ + ++F+LAGRDT
Sbjct: 250 HVDNYLSEIINTRKLELLGQQQDESRHDDLLSRFMKKKESYSDKYLKYVALNFILAGRDT 309
Query: 350 VASALTGFFILLSKNPKVEEKIRVELDRVMNPNQ---------ECATFEQTREMHYLNGA 400
+ A++ FF L+S NP+VEEKI E+ ++ + E TF++ ++ YL A
Sbjct: 310 SSVAMSWFFWLVSLNPRVEEKIINEICTILIKTRDTNVSKWTDEPLTFDEIDQLVYLKAA 369
Query: 401 IHESMRLFPPVQFDSKFALEDDVLPDGTFIKKGSRVTYHPYAMGRMENIWGPDCLEFKPE 460
+ E++RL+P V DSKF + +DVLPDGTF+ GS VTY Y++GRM+ IWG DCLEFKPE
Sbjct: 370 LSETLRLYPSVPEDSKFVVANDVLPDGTFVPSGSNVTYSIYSVGRMKFIWGEDCLEFKPE 429
Query: 461 RWLKDGVFVPKCPFKYPVFQAGSRVCLGKELAIVEMKSVVASLVKRFDVRVVGPNQEPQF 520
RWL++ +K+ F AG R+CLGK+LA ++MKS+ AS++ R + V P +
Sbjct: 430 RWLEESRDEKCNQYKFVAFNAGPRICLGKDLAYLQMKSITASILLRHRL-TVAPGHRVEQ 488
Query: 521 APGLTASFRGGLPVKIYER 539
LT + GL + +++R
Sbjct: 489 KMSLTLFMKFGLKMDVHKR 507
>At2g45970 putative cytochrome P450
Length = 537
Score = 316 bits (810), Expect = 2e-86
Identities = 180/442 (40%), Positives = 265/442 (59%), Gaps = 29/442 (6%)
Query: 114 TITSNPENVEYILKTNFNNYPKGKQFSTILGDLLGRGIFNVDGHSWKFQRKMASLELGSV 173
T+T +P N+E+ILK F+NYPKG + + DLLG+GIFN DG +W FQRK A+LE +
Sbjct: 82 TVTCDPRNLEHILKNRFDNYPKGPTWQAVFHDLLGQGIFNSDGDTWLFQRKTAALEFTTR 141
Query: 174 VIRSYAMELVIEEIKTRLLPLIASVAEKKTASKADNTSEDVLLDMQDILRRFSFDNICKF 233
+R V IK R LP++ + A SE + D+QD+L R +FDNIC
Sbjct: 142 TLRQAMARWVNRAIKLRFLPILEN---------ARLGSEPI--DLQDLLLRLTFDNICGL 190
Query: 234 SFGLDPCCLVPSLPVSNLANAFDLSSTLSAQRALTASPLIWKMKRFFNIGSEKKLKEAIK 293
+FG DP P LPV+ A AFD ++ S QR + ++WK KR+ +G E L ++
Sbjct: 191 TFGKDPRTCAPGLPVNTFAVAFDRATEASLQRFILPE-ILWKFKRWLRLGLEVSLTRSLV 249
Query: 294 IVNDLANEMIKQRREI----ENGVESRKDLLSRFMGALNSHDDEYLRDIVVSFLLAGRDT 349
V++ +E+I R+E N + DLLSRF+ S+ DE L+ + ++F+LAGRDT
Sbjct: 250 QVDNYLSEIITTRKEEMMTQHNNGKHHDDLLSRFIKKKESYSDETLQRVALNFILAGRDT 309
Query: 350 VASALTGFFILLSKNPKVEEKIRVEL---------DRVMNPNQECATFEQTREMHYLNGA 400
+ AL+ FF L++++P +E+KI E+ D V E + E+ + +L A
Sbjct: 310 SSVALSWFFWLITQHPAIEDKILREICTVLVETRGDDVALWTDEPLSCEELDRLVFLKAA 369
Query: 401 IHESMRLFPPVQFDSKFALEDDVLPDGTFIKKGSRVTYHPYAMGRMENIWGPDCLEFKPE 460
+ E++RL+P V DSK A++DDVLPDGTF+ GS +TY Y+ GRM++ WG DCLEFKPE
Sbjct: 370 LSETLRLYPSVPEDSKRAVKDDVLPDGTFVPAGSSITYSIYSAGRMKSTWGEDCLEFKPE 429
Query: 461 RWLKD---GVFVPKCPFKYPVFQAGSRVCLGKELAIVEMKSVVASLVKRFDVRVVGPNQE 517
RW+ G F+ PFK+ F AG R+CLGK+LA ++MKS+ ++++ R + VV ++
Sbjct: 430 RWISQSDGGRFINHDPFKFVAFNAGPRICLGKDLAYLQMKSIASAVLLRHRLTVVTGHKV 489
Query: 518 PQFAPGLTASFRGGLPVKIYER 539
Q LT + GL V ++ER
Sbjct: 490 EQ-KMSLTLFMKYGLLVNVHER 510
>At4g00360 probable cytochrome P450
Length = 553
Score = 310 bits (793), Expect = 2e-84
Identities = 173/446 (38%), Positives = 263/446 (58%), Gaps = 36/446 (8%)
Query: 114 TITSNPENVEYILKTNFNNYPKGKQFSTILGDLLGRGIFNVDGHSWKFQRKMASLELGSV 173
T+T +P+N+E++LKT F+NYPKG + + D LG+GIFN DG +W FQRK A+LE +
Sbjct: 82 TVTCDPKNIEHMLKTRFDNYPKGPTWQAVFHDFLGQGIFNSDGDTWLFQRKTAALEFTTR 141
Query: 174 VIRSYAMELVIEEIKTRLLPLIASVAEKKTASKADNTSEDVLLDMQDILRRFSFDNICKF 233
+R V IK R P++ + A N E V D+QD++ R +FDNIC
Sbjct: 142 TLRQAMGRWVNRGIKLRFCPILET---------AQNNYEPV--DLQDLILRLTFDNICGL 190
Query: 234 SFGLDPCCLVPSLPVSNLANAFDLSSTLSAQRALTASPLIWKMKRFFNIGSEKKLKEAI- 292
+FG D P LP + A+AFD ++ S QR + +W++K++ +G E L ++
Sbjct: 191 AFGKDTRTCAPGLPENGFASAFDRATEASLQRFILPE-FLWRLKKWLGLGLEVSLSRSLG 249
Query: 293 -------KIVNDLANEMIKQRREIENGVESRKDLLSRFMGALN-SHDDEYLRDIVVSFLL 344
++N E++ QR E+GV+ DLLSRFM + S+ + +LR + ++F+L
Sbjct: 250 EIDGYLDAVINTRKQELLSQR---ESGVQRHDDLLSRFMKKKDQSYSETFLRHVALNFIL 306
Query: 345 AGRDTVASALTGFFILLSKNPKVEEKIRVELDRVMNPNQ---------ECATFEQTREMH 395
AGRDT + AL+ FF L++ +P VE+KI E+ V+ + E F++ +
Sbjct: 307 AGRDTSSVALSWFFWLITTHPTVEDKIVREICSVLIETRGTDVSSWTAEPLEFDEVDRLV 366
Query: 396 YLNGAIHESMRLFPPVQFDSKFALEDDVLPDGTFIKKGSRVTYHPYAMGRMENIWGPDCL 455
YL A+ E++RL+P V DSK + DD+LPDGTF+ GS VTY YA GRM++ WG DCL
Sbjct: 367 YLKAALSETLRLYPSVPEDSKHVVNDDILPDGTFVPAGSSVTYSIYAAGRMKSTWGEDCL 426
Query: 456 EFKPERWLK--DGVFVPKCPFKYPVFQAGSRVCLGKELAIVEMKSVVASLVKRFDVRVVG 513
EFKPERW+ DG FV +++ F AG R+CLGK+LA ++MK++ A+++ R + V
Sbjct: 427 EFKPERWISPDDGKFVNHDQYRFVAFNAGPRICLGKDLAYLQMKTIAAAVLLRHRL-TVA 485
Query: 514 PNQEPQFAPGLTASFRGGLPVKIYER 539
P + + LT + GL V +++R
Sbjct: 486 PGHKVEQKMSLTLFMKNGLLVNVHKR 511
>At1g01600 unknown protein
Length = 554
Score = 308 bits (790), Expect = 4e-84
Identities = 176/445 (39%), Positives = 265/445 (59%), Gaps = 32/445 (7%)
Query: 114 TITSNPENVEYILKTNFNNYPKGKQFSTILGDLLGRGIFNVDGHSWKFQRKMASLELGSV 173
T+T +P+N+E++LKT F+NYPKG + ++ DLLG+GIFN DG +W FQRK A+LE +
Sbjct: 82 TVTCDPKNLEHMLKTRFDNYPKGPTWQSVFHDLLGQGIFNSDGDTWLFQRKTAALEFTTR 141
Query: 174 VIRSYAMELVIEEIKTRLLPLIASVAEKKTASKADNTSEDVLLDMQDILRRFSFDNICKF 233
+R V IK R P++A+ A + +E V D+QD++ R +FDNIC
Sbjct: 142 TLRQAMGRWVNRGIKLRFCPILAT---------AQDNAEPV--DLQDLILRLTFDNICGL 190
Query: 234 SFGLDPCCLVPSLPVSNLANAFDLSSTLSAQRALTASPLIWKMKRFFNIGSEKKLKEAIK 293
+FG D P LP + A+AFD ++ S QR + +WK+K++ +G E L ++
Sbjct: 191 AFGKDTRTCAPGLPENGFASAFDRATEASLQRFIIPK-FMWKLKKWLGLGLEVSLSRSLG 249
Query: 294 IVNDLANEMIKQRREI-----ENGVESRKD-LLSRFM-GALNSHDDEYLRDIVVSFLLAG 346
+++ +I R++ E+G R D LLSRFM S+ D +L+ + ++F+LAG
Sbjct: 250 EIDEYLAAVINTRKQELMSQQESGTHQRHDDLLSRFMMKKTESYSDTFLQHVALNFILAG 309
Query: 347 RDTVASALTGFFILLSKNPKVEEKIRVEL----------DRVMNPNQECATFEQTREMHY 396
RDT + AL+ FF L++ +P VE+KI E+ D V + +E F++ + Y
Sbjct: 310 RDTSSVALSWFFWLITMHPTVEDKIVREICSVLIETRGTDDVASWTEEPLGFDEIDRLVY 369
Query: 397 LNGAIHESMRLFPPVQFDSKFALEDDVLPDGTFIKKGSRVTYHPYAMGRMENIWGPDCLE 456
L AI E++RL+P V DSK DDVLPDGTF+ GS VTY YA GRM++ WG DCLE
Sbjct: 370 LKAAISETLRLYPSVPEDSKHVENDDVLPDGTFVPAGSSVTYSIYAAGRMKSTWGEDCLE 429
Query: 457 FKPERWLK--DGVFVPKCPFKYPVFQAGSRVCLGKELAIVEMKSVVASLVKRFDVRVVGP 514
F PERW+ DG F+ +++ F AG R+CLGK+LA ++MK++ A+++ R + VV P
Sbjct: 430 FNPERWISPIDGKFINHDQYRFVAFNAGPRICLGKDLAYLQMKTIAAAVLLRHRLTVV-P 488
Query: 515 NQEPQFAPGLTASFRGGLPVKIYER 539
+ + LT + GL V +Y+R
Sbjct: 489 GHKVEQKMSLTLFMKNGLLVNLYKR 513
>At2g45510 putative cytochrome P450
Length = 511
Score = 304 bits (779), Expect = 7e-83
Identities = 182/468 (38%), Positives = 259/468 (54%), Gaps = 32/468 (6%)
Query: 90 LVDYYTHLLQESPTGTIHVHVLGNTITSNPENVEYILKTNFNNYPKGKQFSTILGDLLGR 149
L DY T + +E PT +T++P NVE+ILKT F+NY KG + DLLG
Sbjct: 57 LYDYETEIAREKPTYRFLSPGQSEILTADPRNVEHILKTRFDNYSKGHSSRENMADLLGH 116
Query: 150 GIFNVDGHSWKFQRKMASLELGSVVIRSYAMELVIEEIKTRLLPLIASVAEKKTASKADN 209
GIF VDG W+ QRK++S E + V+R ++ + + L+ V+E + KA
Sbjct: 117 GIFAVDGEKWRQQRKLSSFEFSTRVLRDFSCSV----FRRNASKLVGFVSEFALSGKA-- 170
Query: 210 TSEDVLLDMQDILRRFSFDNICKFSFGLDPCCLVP-SLPVSNLANAFDLSSTLSAQRALT 268
D QD+L R + D+I K FG++ CL S AFD + ++ R +
Sbjct: 171 ------FDAQDLLMRCTLDSIFKVGFGVELKCLDGFSKEGQEFMEAFDEGNVATSSRFID 224
Query: 269 ASPLIWKMKRFFNIGSEKKLKEAIKIVNDLANEMIKQRREI---ENGVESRKDLLSRFMG 325
+WK+K FFNIGS+ KLK++I ++ +I +R+ E R+D+LSRF+
Sbjct: 225 P---LWKLKWFFNIGSQSKLKKSIATIDKFVYSLITTKRKELAKEQNTVVREDILSRFLV 281
Query: 326 ALNSH----DDEYLRDIVVSFLLAGRDTVASALTGFFILLSKNPKVEEKIRVELDRV--- 378
+D+YLRDI+++F++AG+DT A+ L+ F +L KNP V+EKI E+ V
Sbjct: 282 ESEKDPENMNDKYLRDIILNFMIAGKDTTAALLSWFLYMLCKNPLVQEKIVQEIRDVTFS 341
Query: 379 ------MNPNQECATFEQTREMHYLNGAIHESMRLFPPVQFDSKFALEDDVLPDGTFIKK 432
+N E E EMHYL+ A+ E++RL+PPV D + A DDVLPDG + K
Sbjct: 342 HEKTTDVNGFVESINEEALDEMHYLHAALSETLRLYPPVPVDMRCAENDDVLPDGHRVSK 401
Query: 433 GSRVTYHPYAMGRMENIWGPDCLEFKPERWLKDGVFVPKCPFKYPVFQAGSRVCLGKELA 492
G + Y YAMGRM IWG D EFKPERWLKDG+F P+ PFK+ F AG R+CLGK+ A
Sbjct: 402 GDNIYYIAYAMGRMTYIWGQDAEEFKPERWLKDGLFQPESPFKFISFHAGPRICLGKDFA 461
Query: 493 IVEMKSVVASLVKRFDVRVVGPNQEPQFAPGLTASFRGGLPVKIYERT 540
+MK V +L+ F ++ N + + LT GGL + RT
Sbjct: 462 YRQMKIVSMALLHFFRFKMADENSKVYYKRMLTLHVDGGLHLCAIPRT 509
>At1g24540 putative cytochrome P450
Length = 522
Score = 304 bits (778), Expect = 9e-83
Identities = 176/445 (39%), Positives = 256/445 (56%), Gaps = 35/445 (7%)
Query: 115 ITSNPENVEYILKTNFNNYPKGKQFSTILGDLLGRGIFNVDGHSWKFQRKMASLELGSVV 174
+T++P NVE+ILKTNF NYPKG + DLL GIFN D WK +R++A E+ S
Sbjct: 91 MTADPANVEHILKTNFKNYPKGAFYRERFRDLLEDGIFNADDELWKEERRVAKTEMHSSR 150
Query: 175 IRSYAMELVIEEIKTRLLPLIASVAEKKTASKADNTSEDVLLDMQDILRRFSFDNICKFS 234
+ + + + +L+PL+ +++ K + D+QD+L RF+FDNIC +
Sbjct: 151 FLEHTFTTMRDLVDQKLVPLMENLSTSKR-----------VFDLQDLLLRFTFDNICISA 199
Query: 235 FGLDPCCLVPSLPVSNLANAFDLSSTLSAQRALTASPLIWKMKRFFNIGSEKKLKEAIKI 294
FG+ P L LP A AF+ ++ + R L P +WK RF IG E+KL A++I
Sbjct: 200 FGVYPGSLETGLPEIPFAKAFEDATEYTLARFLIP-PFVWKPMRFLGIGYERKLNNAVRI 258
Query: 295 VNDLANEMIKQRREIEN---GVESRKDLLSRFMGAL----------NSHDDEYLRDIVVS 341
V+ AN+ +++RR + DLLSR M N D+Y R+ S
Sbjct: 259 VHAFANKTVRERRNKMRKLGNLNDYADLLSRLMQREYEKEEDTTRGNYFSDKYFREFCTS 318
Query: 342 FLLAGRDTVASALTGFFILLSKNPKVEEKIRVELDRVMNP-----NQECATFEQTREMHY 396
F++AGRDT + AL FF L+ K+P+VE++I E+ + ++ E REM Y
Sbjct: 319 FIIAGRDTTSVALVWFFWLVQKHPEVEKRILREIREIKRKLTTQETEDQFEAEDFREMVY 378
Query: 397 LNGAIHESMRLFPPVQFDSKFALEDDVLPDGTFIKKGSRVTYHPYAMGRMENIWGPDCLE 456
L A+ ES+RL+P V + K ALEDDVLPDGT +KKG+R+ Y Y+MGR+E+IWG D E
Sbjct: 379 LQAALTESLRLYPSVPMEMKQALEDDVLPDGTRVKKGARIHYSVYSMGRIESIWGKDWEE 438
Query: 457 FKPERWLKDGVFVPKCPFKYPVFQAGSRVCLGKELAIVEMKSVVASLVKRFDVRVVGPNQ 516
FKPERW+K+G V + FKY VF G R+C+GK+ A +MK V A+++ R+ V+VV Q
Sbjct: 439 FKPERWIKEGRIVSEDQFKYVVFNGGPRLCVGKKFAYTQMKMVAAAILMRYSVKVV---Q 495
Query: 517 EPQFAPGLTAS--FRGGLPVKIYER 539
+ P LT + + G+ V + R
Sbjct: 496 GQEIVPKLTTTLYMKNGMNVMLQPR 520
>At1g69500 unknown protein
Length = 524
Score = 303 bits (777), Expect = 1e-82
Identities = 180/497 (36%), Positives = 268/497 (53%), Gaps = 59/497 (11%)
Query: 87 FVNLVDYYTHLLQESPTGTIHVHVLGNTITSNPENVEYILKTNFNNYPKGKQFSTILGDL 146
F + D+ L S T + + T ++P NVEY+LKTNF+NYPKG+ + + + L
Sbjct: 44 FDRMHDWLVEYLYNSRTVVVPMPFTTYTYIADPINVEYVLKTNFSNYPKGETYHSYMEVL 103
Query: 147 LGRGIFNVDGHSWKFQRKMASLELGSVVIRSYAMELVIEEIKTRLLPLIASVAEKKTASK 206
LG GIFN DG W+ QRK AS E S +R ++ +V +E +L +++ + K+
Sbjct: 104 LGDGIFNSDGELWRKQRKTASFEFASKNLRDFST-VVFKEYSLKLFTILSQASFKEQQ-- 160
Query: 207 ADNTSEDVLLDMQDILRRFSFDNICKFSFGLDPCCLVPSLPVSNLANAFDLSSTLSAQRA 266
+DMQ++L R + D+ICK FG++ L P LP ++ A AFD ++ + R
Sbjct: 161 ---------VDMQELLMRMTLDSICKVGFGVEIGTLAPELPENHFAKAFDTANIIVTLRF 211
Query: 267 LTASPLIWKMKRFFNIGSEKKLKEAIKIVNDLANEMIKQRR-------------EIENGV 313
+ +WKMK+F NIGSE L ++IK+VND +I++R+ N
Sbjct: 212 IDP---LWKMKKFLNIGSEALLGKSIKVVNDFTYSVIRRRKAELLEAQISPTNNNNNNNN 268
Query: 314 ESRKDLLSRFMGALNSHD----DEYLRDIVVSFLLAGRDTVASALTGFFILLSKNPKVEE 369
+ + D+LSRF+ + D ++ LRDIV++F++AGRDT A+ LT ++ N V E
Sbjct: 269 KVKHDILSRFIEISDDPDSKETEKSLRDIVLNFVIAGRDTTATTLTWAIYMIMMNENVAE 328
Query: 370 KIRVELDRVMNPNQECAT--------------------------FEQTREMHYLNGAIHE 403
K+ EL + + E ++ ++HYL+ I E
Sbjct: 329 KLYSELQELEKESAEATNTSLHQYDTEDFNSFNEKVTEFAGLLNYDSLGKLHYLHAVITE 388
Query: 404 SMRLFPPVQFDSKFALEDDVLPDGTFIKKGSRVTYHPYAMGRMENIWGPDCLEFKPERWL 463
++RL+P V D K LEDD+LP+GT +K G VTY PY+MGRME WG D FKPERWL
Sbjct: 389 TLRLYPAVPQDPKGVLEDDMLPNGTKVKAGGMVTYVPYSMGRMEYNWGSDAALFKPERWL 448
Query: 464 KDGVFVPKCPFKYPVFQAGSRVCLGKELAIVEMKSVVASLVKRFDVRVVGPNQEPQFAPG 523
KDGVF PFK+ FQAG R+CLGK+ A ++MK +A L + + +V PN ++
Sbjct: 449 KDGVFQNASPFKFTAFQAGPRICLGKDSAYLQMKMAMAILCRFYKFHLV-PNHPVKYRMM 507
Query: 524 LTASFRGGLPVKIYERT 540
S GL V + R+
Sbjct: 508 TILSMAHGLKVTVSRRS 524
>At2g44890 putative cytochrome P450
Length = 490
Score = 300 bits (768), Expect = 1e-81
Identities = 188/509 (36%), Positives = 273/509 (52%), Gaps = 46/509 (9%)
Query: 51 TLLFSFFSFLLFISRIKPWCNCNTCKTFLTMSWSNKFVNLVDYYTHLLQESPTGTIHVHV 110
T +F SF L++ T + F S ++K L DY T + + PT
Sbjct: 7 TTIFILLSFALYL----------TIRIFTGKSRNDKSHKLYDYETEIARTKPTFRFLSPG 56
Query: 111 LGNTITSNPENVEYILKTNFNNYPKGKQFSTILGDLLGRGIFNVDGHSWKFQRKMASLEL 170
T++P NVE+ILKT F+NY KG + L DLLG GIF VDG WK QRK+ S E
Sbjct: 57 QSEIFTADPRNVEHILKTRFHNYSKGPVGTVNLADLLGHGIFAVDGEKWKQQRKLVSFEF 116
Query: 171 GSVVIRSYAMELVIEEIKTRLLPLIASVAEKKTASKADNTSEDVLLDMQDILRRFSFDNI 230
+ V+R+++ + +T L+ +AE + K+ D QD+L + + D+I
Sbjct: 117 STRVLRNFSYSV----FRTSASKLVGFIAEFALSGKS--------FDFQDMLMKCTLDSI 164
Query: 231 CKFSFGLDPCCLVP-SLPVSNLANAFDLSSTLSAQRALTASPLIWKMKRFFNIGSEKKLK 289
K FG++ CL S AFD + ++ R WK+K F NIGSE +LK
Sbjct: 165 FKVGFGVELGCLDGFSKEGEEFMKAFDEGNGATSSRVTDP---FWKLKCFLNIGSESRLK 221
Query: 290 EAIKIVNDLANEMIKQRREI---ENGVESRKDLLSRFMGALNSH------DDEYLRDIVV 340
++I I++ +I +R+ E R+D+LS+F+ L S +D+YLRDI++
Sbjct: 222 KSIAIIDKFVYSLITTKRKELSKEQNTSVREDILSKFL--LESEKDPENMNDKYLRDIIL 279
Query: 341 SFLLAGRDTVASALTGFFILLSKNPKVEEKIRVELDRVMNPNQ---------ECATFEQT 391
+ ++AG+DT A++L+ F +L KNP V+EKI E+ V + ++ E T E
Sbjct: 280 NVMVAGKDTTAASLSWFLYMLCKNPLVQEKIVQEIRDVTSSHEKTTDVNGFIESVTEEAL 339
Query: 392 REMHYLNGAIHESMRLFPPVQFDSKFALEDDVLPDGTFIKKGSRVTYHPYAMGRMENIWG 451
+M YL+ A+ E+MRL+PPV + A DDVLPDG + KG + Y YAMGRM IWG
Sbjct: 340 AQMQYLHAALSETMRLYPPVPEHMRCAENDDVLPDGHRVSKGDNIYYISYAMGRMTYIWG 399
Query: 452 PDCLEFKPERWLKDGVFVPKCPFKYPVFQAGSRVCLGKELAIVEMKSVVASLVKRFDVRV 511
D EFKPERWLKDGVF P+ FK+ F AG R+C+GK+ A +MK V +L+ F ++
Sbjct: 400 QDAEEFKPERWLKDGVFQPESQFKFISFHAGPRICIGKDFAYRQMKIVSMALLHFFRFKM 459
Query: 512 VGPNQEPQFAPGLTASFRGGLPVKIYERT 540
N + + LT GGL + RT
Sbjct: 460 ADENSKVSYKKMLTLHVDGGLHLCAIPRT 488
>At1g13140 cytochrome P450 monooxygenase like protein
Length = 519
Score = 297 bits (761), Expect = 9e-81
Identities = 169/436 (38%), Positives = 247/436 (55%), Gaps = 29/436 (6%)
Query: 115 ITSNPENVEYILKTNFNNYPKGKQFSTILGDLLGRGIFNVDGHSWKFQRKMASLELGSVV 174
+T P NVEY+LKTNF N+PKG F DLL GIFN D SWK QR++ E+ S
Sbjct: 94 VTCVPANVEYMLKTNFKNFPKGAFFKERFNDLLEDGIFNADAESWKEQRRIIITEMHSTR 153
Query: 175 IRSYAMELVIEEIKTRLLPLIASVAEKKTASKADNTSEDVLLDMQDILRRFSFDNICKFS 234
++ + + ++ +LL ++ S A + A D+QD+L R +FDNIC
Sbjct: 154 FVEHSFQTTQDLVRKKLLKVMESFARSQEA-----------FDLQDVLLRLTFDNICIAG 202
Query: 235 FGLDPCCLVPSLPVSNLANAFDLSSTLSAQRALTASPLIWKMKRFFNIGSEKKLKEAIKI 294
G DP L LP+ A AF+ +T S P IWK +FF+IG EK L++A+ +
Sbjct: 203 LGDDPGTLDSDLPLVPFAQAFE-EATESTMFRFMIPPFIWKPLKFFDIGYEKGLRKAVDV 261
Query: 295 VNDLANEMIKQRREIENGVESRKDLLSRFMGALNSHDDEYLRDIVVSFLLAGRDTVASAL 354
L+ + I + +K+ + + ++ R SF+LAGRDT + AL
Sbjct: 262 SMSLSTRWLW----IVSASSKKKEQSHKTTDEKDPSTIKFFRQFCTSFILAGRDTSSVAL 317
Query: 355 TGFFILLSKNPKVEEKIRVELDRVM--------NPNQECATFEQTREMHYLNGAIHESMR 406
T FF ++ K+P+VE KI E+ ++ + N+ T ++ +M YL A+ E+MR
Sbjct: 318 TWFFWVIQKHPEVENKIIREISEILRQRGDSPTSKNESLFTVKELNDMVYLQAALSETMR 377
Query: 407 LFPPVQFDSKFALEDDVLPDGTFIKKGSRVTYHPYAMGRMENIWGPDCLEFKPERWLKDG 466
L+PP+ + K A+EDDV PDGTFI+KGSRV + YAMGRME+IWG DC FKPERW++ G
Sbjct: 378 LYPPIPMEMKQAIEDDVFPDGTFIRKGSRVYFATYAMGRMESIWGKDCESFKPERWIQSG 437
Query: 467 VFVPKCPFKYPVFQAGSRVCLGKELAIVEMKSVVASLVKRFDVRVVGPNQEPQFAPGLTA 526
FV FKY VF AG R+CLGK A ++MK++ AS++ R+ ++V ++ P +T
Sbjct: 438 NFVNDDQFKYVVFNAGPRLCLGKTFAYLQMKTIAASVLSRYSIKVA---KDHVVVPRVTT 494
Query: 527 S--FRGGLPVKIYERT 540
+ R GL V I ++
Sbjct: 495 TLYMRHGLKVTISSKS 510
>At1g13150 putative cytochrome P450 monooxygenase
Length = 529
Score = 297 bits (760), Expect = 1e-80
Identities = 175/448 (39%), Positives = 254/448 (56%), Gaps = 38/448 (8%)
Query: 115 ITSNPENVEYILKTNFNNYPKGKQFSTILGDLLGRGIFNVDGHSWKFQRKMASLELGSVV 174
+T P NVEY+LKTNF N+PKG F + DLL GIFN D SWK QR++ E+ S
Sbjct: 87 VTCVPANVEYMLKTNFKNFPKGTFFKSRFNDLLEEGIFNADDDSWKEQRRIIITEMHSTG 146
Query: 175 IRSYAMELVIEEIKTRLLPLIASVAEKKTASKADNTSEDVLLDMQDILRRFSFDNICKFS 234
++ + ++ +LL ++ S A+ + A D+QD+ R +FD IC
Sbjct: 147 FVEHSFQTTQHLVRKKLLKVMESFAKSQEA-----------FDLQDVFLRLTFDIICLAG 195
Query: 235 FGLDPCCLVPSLPVSNLANAFDLSSTLSAQRALTASPLIWKMKRFFNIGSEKKLKEAIKI 294
G DP L LP A AF+ +T S P IWK RF + G EK L+ A+ +
Sbjct: 196 LGADPETLAVDLPQVPFAKAFE-EATESTLFRFMIPPFIWKPMRFLDTGYEKGLRIAVGV 254
Query: 295 VNDLANEMIKQR---REIENGVESRKDLLSRFMGALNSHDDE---------YLRDIVVSF 342
V+ ++MI R + E +++R D+LSR + + SH E + R SF
Sbjct: 255 VHGFVDKMIVDRICELKEEETLDNRSDVLSRII-QIESHKRENEIDPSTIRFFRQFCTSF 313
Query: 343 LLAGRDTVASALTGFFILLSKNPKVEEKIRVELDRVM--------NPNQECATFEQTREM 394
+LAGRDT + AL+ F ++ K+P+VE KI E+ ++ + N+ T ++ M
Sbjct: 314 ILAGRDTSSVALSWFCWVIQKHPEVENKIICEIREILRQRGDSPTSKNESLFTVKELNNM 373
Query: 395 HYLNGAIHESMRLFPPVQFDSKFALEDDVLPDGTFIKKGSRVTYHPYAMGRMENIWGPDC 454
YL A+ E++RLFPP+ + K A+EDDVLPDGTF++KGSRV + YAMGRME+IWG DC
Sbjct: 374 VYLQAALSETLRLFPPIPMEMKQAIEDDVLPDGTFVRKGSRVYFSIYAMGRMESIWGKDC 433
Query: 455 LEFKPERWLKDGVFVPKCPFKYPVFQAGSRVCLGKELAIVEMKSVVASLVKRFDVRVVGP 514
F+PERW++ G FV FKY VF AG R+C+GK A ++MK + AS++ R+ ++VV
Sbjct: 434 EIFRPERWIQAGKFVSDDQFKYVVFNAGPRLCIGKTFAYLQMKMIAASVLLRYSIKVV-- 491
Query: 515 NQEPQFAPGLTAS--FRGGLPVKIYERT 540
Q+ AP +T + + GL V I R+
Sbjct: 492 -QDHVIAPRVTTNLYMKYGLKVTITPRS 518
>At2g23180 putative cytochrome P450
Length = 516
Score = 294 bits (753), Expect = 7e-80
Identities = 186/524 (35%), Positives = 286/524 (54%), Gaps = 42/524 (8%)
Query: 44 TFLFFSFTLLFSFFSFLLFISRIKPWCNCNTCKTFLTM--SWSNKFVNLVDYYTHLLQES 101
T L S +LLF F + F+ KP + T FL M + + D+ T LL+ S
Sbjct: 5 TLLEVSISLLFFSFLYGYFLISKKPHRSFLTNWPFLGMLPGLLVEIPRVYDFVTELLEAS 64
Query: 102 ----PTGTIHVHVLGNTITSNPENVEYILKTNFNNYPKGKQFSTILGDLLGRGIFNVDGH 157
P L IT +P N+ +I+ +NF NYPKG +F I D+LG GIFN D
Sbjct: 65 NLTYPFKGPCFGGLDMLITVDPANIHHIMSSNFANYPKGTEFKKIF-DVLGDGIFNADSE 123
Query: 158 SWKFQRKMASLELGSVVIRSYAMELVIEEIKTRLLPLIASVAEKKTASKADNTSEDVLLD 217
WK RK A + + + + ++ +++ L+PL+ VAEKK ++D
Sbjct: 124 LWKDLRKSAQSMMTHQDFQRFTLRTIMSKLEKGLVPLLDYVAEKKQ-----------VVD 172
Query: 218 MQDILRRFSFDNICKFSFGLDPCCLVPSLPVSNLANAFDLSSTLSAQRALTASPLIWKMK 277
+QD+ +RF+FD + G+DP CL +P A A D + R + ++WKM+
Sbjct: 173 LQDVFQRFTFDTSFVLATGVDPGCLSTEMPQIEFARALDEAEEAIFFRHVKPE-IVWKMQ 231
Query: 278 RFFNIGSEKKLKEAIKIVNDLANEMIKQRR-EIENGV----ESRKDLLSRFMGA------ 326
RF G E K+K+A + + ++ I +R EI NGV S KDLL +M
Sbjct: 232 RFIGFGDELKMKKAHSTFDRVCSKCIASKRDEITNGVINIDSSSKDLLMCYMNVDTICHT 291
Query: 327 -----LNSHDDEYLRDIVVSFLLAGRDTVASALTGFFILLSKNPKVEEKIRVELDRVMNP 381
LN DD++LRD+++SF+LAGRDT +SALT FF LLSKNPK KIR E++ ++P
Sbjct: 292 TKYKLLNPSDDKFLRDMILSFMLAGRDTTSSALTWFFWLLSKNPKAITKIRQEINTQLSP 351
Query: 382 NQ---ECATFEQTREMHYLNGAIHESMRLFPPVQFDSKFALEDDVLPDGTFIKKGSRVTY 438
+ ++ ++ Y++GA+ E++RL+PPV F K + DVLP G + S++ +
Sbjct: 352 RTNDFDSFNAQELNKLVYVHGALCEALRLYPPVPFQHKSPTKSDVLPSGHRVDASSKIVF 411
Query: 439 HPYAMGRMENIWGPDCLEFKPERWLKD-GVFVPKCPFKYPVFQAGSRVCLGKELAIVEMK 497
Y++GRM+++WG D EFKPERW+ + G + FK+ F AG R CLGKE+A+ +MK
Sbjct: 412 CLYSLGRMKSVWGEDASEFKPERWISESGRLIHVPSFKFLSFNAGPRTCLGKEVAMTQMK 471
Query: 498 SVVASLVKRFDVRVV-GPNQEPQFAPGLTASFRGGLPVKIYERT 540
+V +++ ++++VV G EP P + + GL V + +R+
Sbjct: 472 TVAVKIIQNYEIKVVEGHKIEP--VPSIILHMKHGLKVTVTKRS 513
>At5g58860 cytochrome P450 CYP86A1
Length = 513
Score = 287 bits (735), Expect = 9e-78
Identities = 172/442 (38%), Positives = 253/442 (56%), Gaps = 34/442 (7%)
Query: 114 TITSNPENVEYILKTNFNNYPKGKQFSTILGDLLGRGIFNVDGHSWKFQRKMASLELGSV 173
T+T +P+NVE+ILKT F+NYPKG + DLLG+GIFN DG +W QRK A+LE +
Sbjct: 85 TVTCHPKNVEHILKTRFDNYPKGPMWRAAFHDLLGQGIFNSDGDTWLMQRKTAALEFTTR 144
Query: 174 VIRSYAMELVIEEIKTRLLPLIASVAEKKTASKADNTSEDVLLDMQDILRRFSFDNICKF 233
+R V IK RL ++ + +D+QD+ R +FDNIC
Sbjct: 145 TLRQAMARWVNGTIKNRLWLILDRAVQNNKP-----------VDLQDLFLRLTFDNICGL 193
Query: 234 SFGLDPCCLVPSLPVSNLANAFDLSSTLSAQRALTASPLIWKMKRFFNIGSEKKLKEAIK 293
+FG DP L LP + + AFD ++ + +R L +W++++ IGSE KLK++++
Sbjct: 194 TFGKDPETLSLDLPDNPFSVAFDTATEATLKRLLYTG-FLWRIQKAMGIGSEDKLKKSLE 252
Query: 294 IVNDLANEMIKQRREIENGVESRKDLLSRFMGALNSHDD----EYLRDIVVSFLLAGRDT 349
+V N+ I R+ + DLLSRF+ + + + + L+ I ++F+LAGRDT
Sbjct: 253 VVETYMNDAIDARKNSPSD-----DLLSRFLKKRDVNGNVLPTDVLQRIALNFVLAGRDT 307
Query: 350 VASALTGFFILLSKNPKVEEKIRVELDRVMNPN---------QECATFEQTREMHYLNGA 400
+ AL+ FF L+ N +VE KI EL V+ +E F++ + YL A
Sbjct: 308 SSVALSWFFWLVMNNREVETKIVNELSMVLKETRGNDQEKWTEEPLEFDEADRLVYLKAA 367
Query: 401 IHESMRLFPPVQFDSKFALEDDVLPDGTFIKKGSRVTYHPYAMGRMENIWGPDCLEFKPE 460
+ E++RL+P V D K+ ++DDVLPDGTF+ +GS VTY Y++GRM+ IWG DCLEF+PE
Sbjct: 368 LAETLRLYPSVPQDFKYVVDDDVLPDGTFVPRGSTVTYSIYSIGRMKTIWGEDCLEFRPE 427
Query: 461 RWL-KDG--VFVPKCPFKYPVFQAGSRVCLGKELAIVEMKSVVASLVKRFDVRVVGPNQE 517
RWL DG PK +K+ F AG R CLGK+LA +MKSV ++++ R+ V V P
Sbjct: 428 RWLTADGERFETPKDGYKFVAFNAGPRTCLGKDLAYNQMKSVASAVLLRYRVFPV-PGHR 486
Query: 518 PQFAPGLTASFRGGLPVKIYER 539
+ LT + GL V + R
Sbjct: 487 VEQKMSLTLFMKNGLRVYLQPR 508
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.323 0.138 0.410
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 11,864,593
Number of Sequences: 26719
Number of extensions: 505526
Number of successful extensions: 1999
Number of sequences better than 10.0: 249
Number of HSP's better than 10.0 without gapping: 186
Number of HSP's successfully gapped in prelim test: 63
Number of HSP's that attempted gapping in prelim test: 1351
Number of HSP's gapped (non-prelim): 269
length of query: 540
length of database: 11,318,596
effective HSP length: 104
effective length of query: 436
effective length of database: 8,539,820
effective search space: 3723361520
effective search space used: 3723361520
T: 11
A: 40
X1: 16 ( 7.5 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (22.0 bits)
S2: 62 (28.5 bits)
Medicago: description of AC127021.9


