

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC127021.10 + phase: 0 /pseudo/partial
(264 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
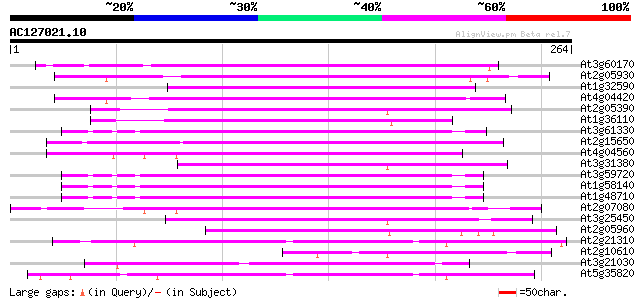
Score E
Sequences producing significant alignments: (bits) Value
At3g60170 putative protein 106 1e-23
At2g05930 copia-like retroelement pol polyprotein 88 5e-18
At1g32590 hypothetical protein, 5' partial 84 8e-17
At4g04420 putative transposon protein 84 1e-16
At2g05390 putative retroelement pol polyprotein 79 2e-15
At1g36110 hypothetical protein 75 4e-14
At3g61330 copia-type polyprotein 73 1e-13
At2g15650 putative retroelement pol polyprotein 73 1e-13
At4g04560 putative transposon protein 73 2e-13
At3g31380 hypothetical protein 72 3e-13
At3g59720 copia-type reverse transcriptase-like protein 72 4e-13
At1g58140 hypothetical protein 72 4e-13
At1g48710 hypothetical protein 72 4e-13
At2g07080 putative gag-protease polyprotein 71 7e-13
At3g25450 hypothetical protein 69 2e-12
At2g05960 putative retroelement pol polyprotein 58 5e-09
At2g21310 putative retroelement pol polyprotein 56 2e-08
At2g10610 pseudogene 55 5e-08
At3g21030 hypothetical protein 54 1e-07
At5g35820 copia-like retrotransposable element 52 4e-07
>At3g60170 putative protein
Length = 1339
Score = 106 bits (264), Expect = 1e-23
Identities = 70/221 (31%), Positives = 121/221 (54%), Gaps = 11/221 (4%)
Query: 13 PKFNGDPEEFSWWKTNMYSFIMGLDEELWDILEDGVDDLDLDEEGAAIDRRIHTPAQKKL 72
P+F+G + +W M +F+ ELW ++E+G+ + + + +R A ++
Sbjct: 13 PRFDG---YYDFWSMTMENFLRS--RELWRLVEEGIPAIVVGTTPVSEAQR---SAVEEA 64
Query: 73 YKKHHKIRGIIVASIPRTEYMKMSDKSTAKAMFASLCANFEGSKKVKEAKALMLVHQYEL 132
K K++ + +I R + DKST+KA++ S+ ++GS KVK A+ L ++EL
Sbjct: 65 KLKDLKVKNFLFQAIDREILETILDKSTSKAIWESMKKKYQGSTKVKRAQLQALRKEFEL 124
Query: 133 FRMKDDESIEEMYSRFQTLVSGLQILKKSYVVSDHVSKILRSLPSRWRPKVTAIEEAKDL 192
MK+ E I+ R T+V+ ++ + S VSKILRSL ++ V +IEE+ DL
Sbjct: 125 LAMKEGEKIDTFLGRTLTVVNKMKTNGEVMEQSTIVSKILRSLTPKFNYVVCSIEESNDL 184
Query: 193 NTLSVEDLVSSLKVHEMSLNEHETSKKSKSIA---LPSKGK 230
+TLS+++L SL VHE LN H +++ + PS+G+
Sbjct: 185 STLSIDELHGSLLVHEQRLNGHVQEEQALKVTHEERPSQGR 225
>At2g05930 copia-like retroelement pol polyprotein
Length = 916
Score = 87.8 bits (216), Expect = 5e-18
Identities = 69/268 (25%), Positives = 117/268 (42%), Gaps = 47/268 (17%)
Query: 22 FSWWKTNMYSFIMGLDEELWDIL-----------EDGVDDLDLDEEGAAIDRRIHTPAQK 70
+ WK M + I GL +E W EDG D L +++ + T +
Sbjct: 33 YGHWKVKMRALIRGLGKEAWIATSIGWKAPVIKGEDGEDVLKTEDQWNDAEEAKATANSR 92
Query: 71 KLYKKHHKIRGIIVASIPRTEYMKMSDKSTAKAMFASLCANFEGSKKVKEAKALMLVHQY 130
L +I S+ + ++ ++ + +AK + L +EG+ VK ++ ML Q+
Sbjct: 93 AL--------SLIFNSVNQNQFKQIQNCESAKEAWDKLAKAYEGTSSVKRSRIDMLASQF 144
Query: 131 ELFRMKDDESIEEMYSRFQTLVSGLQILKKSYVVSDHVSKILRSLPSRWRPKVTAIEEAK 190
E M++ E+IEE + + S L K Y V K+LR LPSR+ K TA+ +
Sbjct: 145 ENLTMEETENIEEFSGKISAIASEAHNLGKKYKDKKLVKKLLRCLPSRFESKRTAMGTSL 204
Query: 191 DLNTLSVEDLVSSLKVHEMSLNEHE----------TSKKSKSI--------------ALP 226
D N++ E++V + +E+ + + S K K + +
Sbjct: 205 DTNSIDFEEVVGMFQAYELEITSGKGGYGHIKAECPSLKRKDLKCSECKGLGHIKFDCVG 264
Query: 227 SKGKTSKSSKAYKASESEEESPDGDSDE 254
SK K +S +SESE +S DGDS++
Sbjct: 265 SKSKPDRSC----SSESESDSNDGDSED 288
>At1g32590 hypothetical protein, 5' partial
Length = 1263
Score = 84.0 bits (206), Expect = 8e-17
Identities = 46/145 (31%), Positives = 83/145 (56%)
Query: 75 KHHKIRGIIVASIPRTEYMKMSDKSTAKAMFASLCANFEGSKKVKEAKALMLVHQYELFR 134
K HK++ + ASI +T + K T+K ++ S+ ++G+ +V+ A+ L +E+
Sbjct: 24 KDHKVKNYLFASIDKTILKTILQKETSKDLWESMKRKYQGNDRVQSAQLQRLRRSFEVLE 83
Query: 135 MKDDESIEEMYSRFQTLVSGLQILKKSYVVSDHVSKILRSLPSRWRPKVTAIEEAKDLNT 194
MK E+I +SR + + ++ L + S V KILR+L ++ V AIEE+ ++
Sbjct: 84 MKIGETITGYFSRVMEITNDMRNLGEDMPDSKVVEKILRTLVEKFTYVVCAIEESNNIKE 143
Query: 195 LSVEDLVSSLKVHEMSLNEHETSKK 219
L+V+ L SSL VHE +L+ H+ ++
Sbjct: 144 LTVDGLQSSLMVHEQNLSRHDVEER 168
>At4g04420 putative transposon protein
Length = 1008
Score = 83.6 bits (205), Expect = 1e-16
Identities = 57/223 (25%), Positives = 104/223 (46%), Gaps = 20/223 (8%)
Query: 22 FSWWKTNMYSFIMGLDEELWDIL-----------EDGVDDLDLDEEGAAIDRRIHTPAQK 70
+ WK M + I GL +E W EDG D L ++ A++
Sbjct: 21 YGHWKVKMRALIRGLGKEAWIATSIGWKAPVIKGEDGEDVLKTKDQW--------NDAEE 72
Query: 71 KLYKKHHKIRGIIVASIPRTEYMKMSDKSTAKAMFASLCANFEGSKKVKEAKALMLVHQY 130
K + + +I + + ++ ++ + +AK + L +EG+ VK ++ ML Q+
Sbjct: 73 AKAKANSRALSLIFNFVNQNQFKRIQNCESAKEAWDKLAKAYEGTSSVKRSRIDMLASQF 132
Query: 131 ELFRMKDDESIEEMYSRFQTLVSGLQILKKSYVVSDHVSKILRSLPSRWRPKVTAIEEAK 190
E M++ E+IEE + + S L K Y V K+LR LPSR+ K TA+ +
Sbjct: 133 ENLSMEETENIEEFSGKISAIASEAHNLGKKYKDKKLVKKLLRCLPSRFESKRTAMGTSL 192
Query: 191 DLNTLSVEDLVSSLKVHEMSLNEHETSKKSKSIALPSKGKTSK 233
D +++ E++V L+ +E+ + + SK +AL + K ++
Sbjct: 193 DTDSIDFEEVVGMLQAYELEITSGK-GGYSKGLALAASAKKNE 234
Score = 28.9 bits (63), Expect = 3.0
Identities = 19/52 (36%), Positives = 28/52 (53%), Gaps = 4/52 (7%)
Query: 203 SLKVHEMSLNEHETSKKSKSIALPSKGKTSKSSKAYKASESEEESPDGDSDE 254
SLK ++ +E +K + SK K KS +SESE +S DGDS++
Sbjct: 297 SLKRKDLKCSECNGLGHTKFDCVGSKSKPDKSC----SSESESDSNDGDSED 344
>At2g05390 putative retroelement pol polyprotein
Length = 1307
Score = 79.0 bits (193), Expect = 2e-15
Identities = 47/199 (23%), Positives = 100/199 (49%), Gaps = 23/199 (11%)
Query: 39 ELWDILEDGVDDLDLDEEGAAIDRRIHTPAQKKLYKKHHKIRGIIVASIPRTEYMKMSDK 98
++W+ ++ G DD++ K+ R ++ S+P + +++
Sbjct: 9 KVWETIDPGSDDME----------------------KNDMARALLFQSVPESTILQVGKH 46
Query: 99 STAKAMFASLCANFEGSKKVKEAKALMLVHQYELFRMKDDESIEEMYSRFQTLVSGLQIL 158
T+KAM+ ++ G+++VKEAK L+ +++ MKD+E+I+E R + + + L
Sbjct: 47 KTSKAMWEAIKTRNLGAERVKEAKLQTLMAEFDRLNMKDNETIDEFVGRISEISTKSESL 106
Query: 159 KKSYVVSDHVSKILRSLP-SRWRPKVTAIEEAKDLNTLSVEDLVSSLKVHEMSLNEHETS 217
+ S V K L+SLP ++ + A+E+ DLNT ED+V +K +E + + + S
Sbjct: 107 GEEIEESKIVKKFLKSLPRKKYIHIIAALEQILDLNTTGFEDIVGRMKTYEDRVCDEDDS 166
Query: 218 KKSKSIALPSKGKTSKSSK 236
+ + + + ++S ++
Sbjct: 167 PEEQGKLMYANSESSYDTR 185
>At1g36110 hypothetical protein
Length = 745
Score = 75.1 bits (183), Expect = 4e-14
Identities = 48/171 (28%), Positives = 84/171 (49%), Gaps = 23/171 (13%)
Query: 39 ELWDILEDGVDDLDLDEEGAAIDRRIHTPAQKKLYKKHHKIRGIIVASIPRTEYMKMSDK 98
++W+ +E G+DD K+ RG+I SIP + +++
Sbjct: 42 KMWETIEPGIDD----------------------GYKNTMARGLIFQSIPESLTLQVGTL 79
Query: 99 STAKAMFASLCANFEGSKKVKEAKALMLVHQYELFRMKDDESIEEMYSRFQTLVSGLQIL 158
+TAK ++ S+ + G+ +VKEA+ L+ ++E +MK+ E I+ R L + L
Sbjct: 80 ATAKLVWDSIKTRYVGADRVKEARLQTLMAEFEKMKMKESEKIDVFAGRLAELATRSDAL 139
Query: 159 KKSYVVSDHVSKILRSLPSR-WRPKVTAIEEAKDLNTLSVEDLVSSLKVHE 208
+ S V K L +LP R + + ++E+ DLN S ED+V +KV+E
Sbjct: 140 GSNIETSKLVKKFLNALPLRKYIHIIASLEQVLDLNNTSFEDIVGRIKVYE 190
>At3g61330 copia-type polyprotein
Length = 1352
Score = 73.2 bits (178), Expect = 1e-13
Identities = 51/200 (25%), Positives = 100/200 (49%), Gaps = 12/200 (6%)
Query: 25 WKTNMYSFIMGLDEELWDILEDGVDDLDLDEEGAAIDRRIHTPAQKKLYKKHHKIRGIIV 84
W M + + D +W+I+E G ++ + EG+ + + K+ K +I
Sbjct: 21 WSLRMKAILGAHD--VWEIVEKGF--IEPENEGSL--SQTQKDGLRDSRKRDKKALCLIY 74
Query: 85 ASIPRTEYMKMSDKSTAKAMFASLCANFEGSKKVKEAKALMLVHQYELFRMKDDESIEEM 144
+ + K+ + ++AK + L +++G+ +VK+ + L ++E +MK+ E + +
Sbjct: 75 QGLDEDTFEKVVEATSAKEAWEKLRTSYKGADQVKKVRLQTLRGEFEALQMKEGELVSDY 134
Query: 145 YSRFQTLVSGLQILKKSYVVSDHVSKILRSLPSRWRPKVTAIEEAKDLNTLSVEDLVSSL 204
+SR T+ + L+ + + K+LRSL ++ VT IEE KDL +++E L+ SL
Sbjct: 135 FSRVLTVTNNLKRNGEKLDDVRIMEKVLRSLDLKFEHIVTVIEETKDLEAMTIEQLLGSL 194
Query: 205 KVHEMSLNEHETSKKSKSIA 224
+ +E E KK + IA
Sbjct: 195 QAYE------EKKKKKEDIA 208
>At2g15650 putative retroelement pol polyprotein
Length = 1347
Score = 73.2 bits (178), Expect = 1e-13
Identities = 50/215 (23%), Positives = 104/215 (48%), Gaps = 3/215 (1%)
Query: 18 DPEEFSWWKTNMYSFIMGLDEELWDILEDGVDDLDLDEEGAAIDRRIHTPAQKKLYKKHH 77
D E++ +W M + +LW ++E+GV + E R T ++ +
Sbjct: 13 DGEKYDFWSIKMATIFR--TRKLWSVVEEGVPVEPVQAEETPETARAKTLREEAVTNDTM 70
Query: 78 KIRGIIVASIPRTEYMKMSDKSTAKAMFASLCANFEGSKKVKEAKALMLVHQYELFRMKD 137
++ I+ ++ + +++ S++K + L ++GS +V+ K L +YE +M D
Sbjct: 71 ALQ-ILQTAVTDQIFSRIAAASSSKEAWDVLKDEYQGSPQVRLVKLQSLRREYENLKMYD 129
Query: 138 DESIEEMYSRFQTLVSGLQILKKSYVVSDHVSKILRSLPSRWRPKVTAIEEAKDLNTLSV 197
+++I+ + L L + + + KIL SLP+++ V+ +E+ +DL+ L++
Sbjct: 130 NDNIKTFTDKLIVLEIQLTYHGEKKTNTQLIQKILISLPAKFDSIVSVLEQTRDLDALTM 189
Query: 198 EDLVSSLKVHEMSLNEHETSKKSKSIALPSKGKTS 232
+L+ LK E + E S K + + SKG+ S
Sbjct: 190 SELLGILKAQEARVTAREESTKEGAFYVRSKGRES 224
>At4g04560 putative transposon protein
Length = 590
Score = 72.8 bits (177), Expect = 2e-13
Identities = 46/199 (23%), Positives = 97/199 (48%), Gaps = 3/199 (1%)
Query: 18 DPEEFSWWKTNMYSFIMGLDEELWDILEDG-VDDLDLDEEGAAIDR-RIHTPAQKKLYKK 75
D E + +WK + I +D + W +EDG + D + + + R A +K
Sbjct: 349 DAEHYGYWKVLIKRSIQSIDMDAWFAVEDGWMPPTTKDAKRDIVSKSRTEWIADEKTAAN 408
Query: 76 HH-KIRGIIVASIPRTEYMKMSDKSTAKAMFASLCANFEGSKKVKEAKALMLVHQYELFR 134
H+ + +I S+ R ++ ++ +AK ++ L +FE + VK + ML ++E
Sbjct: 409 HNSQALSVIFGSLLRNKFTQVQGCLSAKEVWEILQVSFECTNNVKRTRLDMLASEFENLT 468
Query: 135 MKDDESIEEMYSRFQTLVSGLQILKKSYVVSDHVSKILRSLPSRWRPKVTAIEEAKDLNT 194
M+ +ES+++ + ++ +L K+Y V K LRSLP +++ +AI+ + + +
Sbjct: 469 MEAEESVDDFNGKLSSITQEAVVLGKTYKDKKMVKKFLRSLPDKFQSHKSAIDVSLNSDQ 528
Query: 195 LSVEDLVSSLKVHEMSLNE 213
L + +V ++ ++ E
Sbjct: 529 LKFDQVVGMMQAYDTDKEE 547
Score = 38.5 bits (88), Expect = 0.004
Identities = 43/186 (23%), Positives = 81/186 (43%), Gaps = 31/186 (16%)
Query: 13 PKFNGDPEEFSWWKTNMYSFIMGLDEELWDILEDGVDDLDLDEEGAAIDRRIHTP--AQK 70
P FN E + +W+ M + ++LW+I+++GV + + H+P Q+
Sbjct: 8 PIFN--KENYGFWRIKMKTIFQ--TKKLWEIVDEGVPKPPAEGD--------HSPEAVQQ 55
Query: 71 KLYKKHHKIRGIIVASIPRTEYMKMSDKSTAKAMFASLCANFEGSKKVKEAKALMLVHQY 130
K + ++ + I +T +SD + AS AL +Y
Sbjct: 56 KTRCEAASLKDLTALQILQTA---VSDSIFPRIAPAS--------------SALGKPWEY 98
Query: 131 ELFRMKDDESIEEMYSRFQTLVSGLQILKKSYVVSDHVSKILRSLPSRWRPKVTAIEEAK 190
E +MK+ ++I ++ + + L++ + V KIL SLP R+ V +++ K
Sbjct: 99 ENLKMKESDNINTFMTKLIEMGNQLRVHGEEKSDYQIVQKILISLPKRFDIIVAMMKQTK 158
Query: 191 DLNTLS 196
DL +LS
Sbjct: 159 DLTSLS 164
>At3g31380 hypothetical protein
Length = 262
Score = 72.0 bits (175), Expect = 3e-13
Identities = 46/156 (29%), Positives = 79/156 (50%), Gaps = 1/156 (0%)
Query: 80 RGIIVASIPRTEYMKMSDKSTAKAMFASLCANFEGSKKVKEAKALMLVHQYELFRMKDDE 139
RG++V SIP +++ + TAK ++ S+ G+++VKEA+ L+ +E +MK+ E
Sbjct: 3 RGLLVQSIPEAFTLQVGNLQTAKEVWDSIKTRHVGAERVKEARVQTLMADFEKMKMKEAE 62
Query: 140 SIEEMYSRFQTLVSGLQILKKSYVVSDHVSKILRSLP-SRWRPKVTAIEEAKDLNTLSVE 198
I++ R L + L + V V K L SLP ++ + ++E+ DLN + E
Sbjct: 63 KIDDFAGRLSELSTKSAPLGVNIEVPKLVKKFLNSLPRKKYIHIIASLEQVLDLNNTTFE 122
Query: 199 DLVSSLKVHEMSLNEHETSKKSKSIALPSKGKTSKS 234
D+V +KV+E + + E L SKS
Sbjct: 123 DIVGCMKVNEERVYDPEEETNEDQNKLMYTSSDSKS 158
>At3g59720 copia-type reverse transcriptase-like protein
Length = 1272
Score = 71.6 bits (174), Expect = 4e-13
Identities = 50/199 (25%), Positives = 99/199 (49%), Gaps = 12/199 (6%)
Query: 25 WKTNMYSFIMGLDEELWDILEDGVDDLDLDEEGAAIDRRIHTPAQKKLYKKHHKIRGIIV 84
W M + + D +W+I+E G ++ + EG+ + + K+ K +I
Sbjct: 21 WSLRMKAILGAHD--VWEIVEKGF--IEPENEGSL--SQTQKDGLRDSRKRDKKALCLIY 74
Query: 85 ASIPRTEYMKMSDKSTAKAMFASLCANFEGSKKVKEAKALMLVHQYELFRMKDDESIEEM 144
+ + K+ + ++AK + L +++G+ +VK+ + L ++E +MK+ E + +
Sbjct: 75 QGLDEDTFEKVVEATSAKEAWEKLRTSYKGADQVKKVRLQTLRGEFEALQMKEGELVSDY 134
Query: 145 YSRFQTLVSGLQILKKSYVVSDHVSKILRSLPSRWRPKVTAIEEAKDLNTLSVEDLVSSL 204
+SR T+ + L+ + + K+LRSL ++ VT IEE KDL +++E L+ SL
Sbjct: 135 FSRVLTVTNNLKRNGEKLDDVRIMEKVLRSLDLKFEHIVTVIEETKDLEAMTIEQLLGSL 194
Query: 205 KVHEMSLNEHETSKKSKSI 223
+ +E E KK + I
Sbjct: 195 QAYE------EKKKKKEDI 207
>At1g58140 hypothetical protein
Length = 1320
Score = 71.6 bits (174), Expect = 4e-13
Identities = 50/199 (25%), Positives = 99/199 (49%), Gaps = 12/199 (6%)
Query: 25 WKTNMYSFIMGLDEELWDILEDGVDDLDLDEEGAAIDRRIHTPAQKKLYKKHHKIRGIIV 84
W M + + D +W+I+E G ++ + EG+ + + K+ K +I
Sbjct: 21 WSLRMKAILGAHD--VWEIVEKGF--IEPENEGSL--SQTQKDGLRDSRKRDKKALCLIY 74
Query: 85 ASIPRTEYMKMSDKSTAKAMFASLCANFEGSKKVKEAKALMLVHQYELFRMKDDESIEEM 144
+ + K+ + ++AK + L +++G+ +VK+ + L ++E +MK+ E + +
Sbjct: 75 QGLDEDTFEKVVEATSAKEAWEKLRTSYKGADQVKKVRLQTLRGEFEALQMKEGELVSDY 134
Query: 145 YSRFQTLVSGLQILKKSYVVSDHVSKILRSLPSRWRPKVTAIEEAKDLNTLSVEDLVSSL 204
+SR T+ + L+ + + K+LRSL ++ VT IEE KDL +++E L+ SL
Sbjct: 135 FSRVLTVTNNLKRNGEKLDDVRIMEKVLRSLDLKFEHIVTVIEETKDLEAMTIEQLLGSL 194
Query: 205 KVHEMSLNEHETSKKSKSI 223
+ +E E KK + I
Sbjct: 195 QAYE------EKKKKKEDI 207
>At1g48710 hypothetical protein
Length = 1352
Score = 71.6 bits (174), Expect = 4e-13
Identities = 50/199 (25%), Positives = 99/199 (49%), Gaps = 12/199 (6%)
Query: 25 WKTNMYSFIMGLDEELWDILEDGVDDLDLDEEGAAIDRRIHTPAQKKLYKKHHKIRGIIV 84
W M + + D +W+I+E G ++ + EG+ + + K+ K +I
Sbjct: 21 WSLRMKAILGAHD--VWEIVEKGF--IEPENEGSL--SQTQKDGLRDSRKRDKKALCLIY 74
Query: 85 ASIPRTEYMKMSDKSTAKAMFASLCANFEGSKKVKEAKALMLVHQYELFRMKDDESIEEM 144
+ + K+ + ++AK + L +++G+ +VK+ + L ++E +MK+ E + +
Sbjct: 75 QGLDEDTFEKVVEATSAKEAWEKLRTSYKGADQVKKVRLQTLRGEFEALQMKEGELVSDY 134
Query: 145 YSRFQTLVSGLQILKKSYVVSDHVSKILRSLPSRWRPKVTAIEEAKDLNTLSVEDLVSSL 204
+SR T+ + L+ + + K+LRSL ++ VT IEE KDL +++E L+ SL
Sbjct: 135 FSRVLTVTNNLKRNGEKLDDVRIMEKVLRSLDLKFEHIVTVIEETKDLEAMTIEQLLGSL 194
Query: 205 KVHEMSLNEHETSKKSKSI 223
+ +E E KK + I
Sbjct: 195 QAYE------EKKKKKEDI 207
>At2g07080 putative gag-protease polyprotein
Length = 627
Score = 70.9 bits (172), Expect = 7e-13
Identities = 62/252 (24%), Positives = 109/252 (42%), Gaps = 33/252 (13%)
Query: 1 DTADKKTDSGKAPKFNGDPEEFSWWKTNMYSFIMGLDEELWDILEDGVDDLDLDEEGAAI 60
+ D T +GK D + + +WK M I G E+G I
Sbjct: 2 EPTDVSTGTGKVLLL--DTKRYGYWKVCMTQIIRG------------------QEDGFKI 41
Query: 61 DR-RIHTPAQKKLYKKHH-KIRGIIVASIPRTEYMKMSDKSTAKAMFASLCANFEGSKKV 118
+ + + A++KL K + + I + E+ + +AK + +L + EG+ V
Sbjct: 42 TKPKANWTAEEKLQSKFNARAMKAIFNGVDEDEFKLIQGCKSAKQAWDTLQKSHEGTSSV 101
Query: 119 KEAKALMLVHQYELFRMKDDESIEEMYSRFQTLVSGLQILKKSYVVSDHVSKILRSLPSR 178
K + + Q+E +M+ DE I + S+ L + +++ K+Y V K+LR LP +
Sbjct: 102 KRTRLDHIATQFEYLKMEPDEKIVKFSSKISALANEAEVMGKTYKDQKLVKKLLRCLPPK 161
Query: 179 WRPKVTAIEEAKDLNTLSVEDLVSSLKVHEMSLNEHETSKKSKSIALPSKGKTSKSSKAY 238
+ + A + + +S DLV LK+ EM ++ + K SK+IA A
Sbjct: 162 FAAHKAVMRVAGNTDKISFVDLVGMLKLEEMKADQDKV-KPSKNIAF----------NAD 210
Query: 239 KASESEEESPDG 250
+ SE +E DG
Sbjct: 211 QGSEQFQEIKDG 222
>At3g25450 hypothetical protein
Length = 1343
Score = 69.3 bits (168), Expect = 2e-12
Identities = 44/174 (25%), Positives = 89/174 (50%), Gaps = 6/174 (3%)
Query: 74 KKHHKIRGIIVASIPRTEYMKMSDKSTAKAMFASLCANFEGSKKVKEAKALMLVHQYELF 133
+K+ R ++ SIP + +++ + T+ A++ ++ + G+++VKEA+ L+ +++
Sbjct: 58 EKNDMARALLFQSIPESLILQVGKQKTSSAVWEAIKSRNLGAERVKEARLQTLMAEFDKL 117
Query: 134 RMKDDESIEEMYSRFQTLVSGLQILKKSYVVSDHVSKILRSLP-SRWRPKVTAIEEAKDL 192
+MKD E+I++ R + + L + S V K L+SLP ++ V A+E+ DL
Sbjct: 118 KMKDSETIDDYVGRISEITTKAAALGEDIEESKIVKKFLKSLPRKKYIHIVAALEQVLDL 177
Query: 193 NTLSVEDLVSSLKVHEMSLNEHETSKKSKSIALPSKGKTSKSSKAYKASESEEE 246
T + ED+ +K +E + + + S + + K T + E EEE
Sbjct: 178 KTTTFEDIAGRIKTYEDRVWDDDDSHEDQ-----GKLMTEVEEEVVDDLEEEEE 226
>At2g05960 putative retroelement pol polyprotein
Length = 1200
Score = 58.2 bits (139), Expect = 5e-09
Identities = 48/196 (24%), Positives = 91/196 (45%), Gaps = 31/196 (15%)
Query: 93 MKMSDKSTAKAMFASLCANFEGSKKVKEAKALMLVHQYELFRMKDDESIEEMYSRFQTLV 152
+++ T+KA++ + + G+++VKEAK L+ +++ +MKD+E+I+E R +
Sbjct: 35 LQVGKLKTSKAVWDKIQSRNLGAERVKEAKLKTLMAEFDKLKMKDNETIDECAGRLSEIS 94
Query: 153 SGLQILKKSYVVSDHVSKILRSLPS-RWRPKVTAIEEAKDLNTLSVEDLVSSLKVHEMSL 211
+ L + + V K L+SLP+ ++ V A+E+ DL + +D+V +K +E +
Sbjct: 95 TKSTSLGEDIEETKVVKKFLKSLPTKKYIHIVAALEQVLDLKNTTFKDIVGRIKTYEDKI 154
Query: 212 --------NEHETSKK-------------SKSIALP---------SKGKTSKSSKAYKAS 241
E E +K S+ I L + + K K
Sbjct: 155 WVLITCLKKEAEEEEKSVVGVEAEEELVISREITLRLLVIAVINYASNCPDRLLKLIKLQ 214
Query: 242 ESEEESPDGDSDEDQS 257
E ++E+ D D DE +S
Sbjct: 215 ERQQEAEDDDDDEVES 230
>At2g21310 putative retroelement pol polyprotein
Length = 838
Score = 56.2 bits (134), Expect = 2e-08
Identities = 56/247 (22%), Positives = 107/247 (42%), Gaps = 15/247 (6%)
Query: 21 EFSWWKTNMYSFIMGLDEELWDILEDGVDDLDLDEEG--AAIDRRIHTPAQKKLYKKHHK 78
++ WK + + I EL +LE +D ++EE A D + K L +K K
Sbjct: 16 DYVLWKEKLLAHI-----ELLGLLEGLEEDEAIEEEESTAETDSLLTKTEDKVLKEKRGK 70
Query: 79 IRGIIVASIPRTEYMKMSDKSTAKAMFASLCANFEGSKKVKEAKALMLVHQYELFRMKDD 138
R ++ S+ K+ + TA M L F ++ Y +M D
Sbjct: 71 ARSTVILSLGNHVLRKVIKEKTAAGMIRVLDKLFMAKSLPNRIYLKQRLYGY---KMSDS 127
Query: 139 ESIEEMYSRFQTLVSGLQILKKSYVVSDHVSKILRSLPSRWRPKVTAIEEAKDLNTLSVE 198
+IEE + F L+S L+ +K S D +L SLP ++ ++ K TL+++
Sbjct: 128 MTIEENVNDFFKLISDLENVKVSVPDEDQAIVLLMSLPKQFDQLKDTLKYGK--TTLALD 185
Query: 199 DLVSSL--KVHEMSLNEHETSKKSKSIALPSKGKTSKSSKAYKASESEEESPDGDSDEDQ 256
++ ++ KV E+ + S ++ + +G++ K K+ + ++S+ S +
Sbjct: 186 EITGAIRSKVLELGASGKMLKNSSDALFVQDRGRSEKRDKSSERNKSQSRSKSREKKVCW 245
Query: 257 SC-KDGY 262
C K+G+
Sbjct: 246 VCGKEGH 252
>At2g10610 pseudogene
Length = 531
Score = 54.7 bits (130), Expect = 5e-08
Identities = 39/131 (29%), Positives = 68/131 (51%), Gaps = 11/131 (8%)
Query: 129 QYELFRMKDDESIEE---MYSRFQTLVSGLQILKKSYVVSDHVSKILRSLP-SRWRPKVT 184
++ +MKD I++ M S + L ++ + S V K L+S+P R+ V
Sbjct: 3 EFHRLKMKDSHKIDDFAGMLSEISVKSAALGVIIEE---SRLVKKFLKSVPRKRYIHIVA 59
Query: 185 AIEEAKDLNTLSVEDLVSSLKVHEMSLNEHETSKKSKSIALPSKGKTSKSSKAYKASESE 244
A+E+ DLN ++ +D+V LKV+E + + + ++ +S L S +SK ++ E
Sbjct: 60 ALEQVLDLNAITFDDIVGRLKVYEERVYDEDDPEEDQSKLLYSSMDSSKR----ESEGEE 115
Query: 245 EESPDGDSDED 255
EE P G+ DED
Sbjct: 116 EEDPIGEEDED 126
>At3g21030 hypothetical protein
Length = 411
Score = 53.5 bits (127), Expect = 1e-07
Identities = 45/182 (24%), Positives = 86/182 (46%), Gaps = 8/182 (4%)
Query: 36 LDEELWDILEDGVD-DLDLDEEGAAIDRRIHTPAQKKLYKKHHKIRGIIVASIPRTEYMK 94
++ +WD++++GV + + E AA + + + L K +K I+ +S+P + + K
Sbjct: 27 VENGVWDVVQNGVSPNPTTNPELAATIKAVDLAQWRNLVVKDNKALKIMQSSLPDSVFRK 86
Query: 95 MSDKSTAKAMFASLCANFEGSKKVKEAKALMLVHQYELFRMKDDESIEEMYSRFQTLVSG 154
++AK ++ L + KEAK L Q+E M + E ++ R + ++
Sbjct: 87 TISIASAKELWDLL----KKGNDTKEAKLRRLEKQFENLMMYEGERMKLYLKRLEKIIEE 142
Query: 155 LQILKKSYVVSDHVSKILRSLPSRWRPKVTAIEEAKDLNTLSVEDLVSSLKVHEMSLNEH 214
L +L ++K+L SLP + V ++E L L+ DL LK EM ++
Sbjct: 143 LFVLGNPISDDKVIAKLLTSLPRSYDDSVPVLKEFMTLPELTHRDL---LKAFEMFGSDP 199
Query: 215 ET 216
+T
Sbjct: 200 KT 201
>At5g35820 copia-like retrotransposable element
Length = 1342
Score = 51.6 bits (122), Expect = 4e-07
Identities = 57/250 (22%), Positives = 104/250 (40%), Gaps = 19/250 (7%)
Query: 9 SGKAP--KFNGDPEEFSWWKT-----NMYSFIMGLDEELWDILEDGVDDLDLDEEGAAID 61
SG+A KF+GD + W + M + GL EE ++ED ++ +G D
Sbjct: 3 SGRAEVEKFDGDGDYILWKEKLLAHMEMLGLLEGLGEEEEAVVEDSTTEIS---DGGNQD 59
Query: 62 RRIHTPA--QKKLYKKHHKIRGIIVASIPRTEYMKMSDKSTAKAMFASLCANFEGSKKVK 119
T K L +K K R I+ S+ K+ + TA M L F
Sbjct: 60 PETATSKLEDKILKEKRGKARSTIILSLGNNVLRKVIKQKTAAGMIKVLDQLFMAKSLPN 119
Query: 120 EAKALMLVHQYELFRMKDDESIEEMYSRFQTLVSGLQILKKSYVVSDHVSKILRSLPSRW 179
++ Y +M ++ ++EE + F L+S L+ +K D +L SLP ++
Sbjct: 120 RIYLKQRLYGY---KMSENMTMEENVNDFFKLISDLENVKVVVPDEDQAIVLLMSLPRQF 176
Query: 180 RPKVTAIEEAKDLNTLSVEDLVSSL--KVHEMSLNEHETSKKSKSIALPSKGKTSKSSKA 237
++ K TL +E++ S++ K+ E+ + S + + +G++ K
Sbjct: 177 DQLKETLKYCK--TTLHLEEITSAIRSKILELGASGKLLKNNSDGLFVQDRGRSETRGKG 234
Query: 238 YKASESEEES 247
++S +S
Sbjct: 235 PNKNKSRSKS 244
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.310 0.127 0.353
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 6,079,891
Number of Sequences: 26719
Number of extensions: 256616
Number of successful extensions: 1453
Number of sequences better than 10.0: 137
Number of HSP's better than 10.0 without gapping: 54
Number of HSP's successfully gapped in prelim test: 83
Number of HSP's that attempted gapping in prelim test: 1318
Number of HSP's gapped (non-prelim): 185
length of query: 264
length of database: 11,318,596
effective HSP length: 97
effective length of query: 167
effective length of database: 8,726,853
effective search space: 1457384451
effective search space used: 1457384451
T: 11
A: 40
X1: 16 ( 7.2 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 42 (21.8 bits)
S2: 59 (27.3 bits)
Medicago: description of AC127021.10


