

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC127019.17 + phase: 0 /pseudo
(170 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
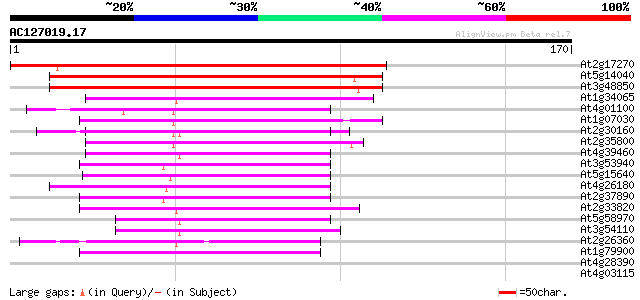
Score E
Sequences producing significant alignments: (bits) Value
At2g17270 putative mitochondrial phosphate translocator protein 159 7e-40
At5g14040 mitochondrial phosphate translocator 114 3e-26
At3g48850 mitochondrial phosphate transporter 109 8e-25
At1g34065 allinase like protein 57 6e-09
At4g01100 putative carrier protein 50 8e-07
At1g07030 unknown protein 47 4e-06
At2g30160 putative mitochondrial carrier protein 47 5e-06
At2g35800 putative mitochondrial carrier protein 47 7e-06
At4g39460 mitochondrial carrier like protein 44 6e-05
At3g53940 unknown protein 43 7e-05
At5g15640 putative mitochondrial carrier protein 42 1e-04
At4g26180 putative mitochondrial carrier protein 42 1e-04
At2g37890 mitochondrial carrier like protein 42 2e-04
At2g33820 mitochondrial basic amino acid carrier (BAC1) 42 2e-04
At5g58970 uncoupling protein AtUCP2 40 6e-04
At3g54110 uncoupling protein (ucp/PUMP) 40 6e-04
At2g26360 mitochondrial carrier like protein 40 6e-04
At1g79900 putative carnitine/acylcarnitine translocase 40 6e-04
At4g28390 ADP,ATP carrier-like protein 39 0.001
At4g03115 putative protein 39 0.002
>At2g17270 putative mitochondrial phosphate translocator protein
Length = 309
Score = 159 bits (402), Expect = 7e-40
Identities = 74/115 (64%), Positives = 93/115 (80%), Gaps = 1/115 (0%)
Query: 1 MAAMEGGISQELT-PRYYALCTIGGMLSAGTTHLATTPLDVLKVNMQVHPIKYYSISSCF 59
M ++ + +EL+ P +Y +CT+GGMLSAGTTHLA TPLDVLKVNMQV+P+KY SI S F
Sbjct: 1 MTRVKSKLDEELSSPWFYTVCTMGGMLSAGTTHLAITPLDVLKVNMQVNPVKYNSIPSGF 60
Query: 60 TTLLREQGPSVLWKGWTGKFFGYGAQGGCRFGLYEYFKGVYSNVLVDQHRSLVYF 114
+TLLRE G S LW+GW+GK GYG QGGCRFGLYEYFK +YS+VL + +R+ +YF
Sbjct: 61 STLLREHGHSYLWRGWSGKLLGYGVQGGCRFGLYEYFKTLYSDVLPNHNRTSIYF 115
>At5g14040 mitochondrial phosphate translocator
Length = 375
Score = 114 bits (285), Expect = 3e-26
Identities = 52/105 (49%), Positives = 70/105 (66%), Gaps = 4/105 (3%)
Query: 13 TPRYYALCTIGGMLSAGTTHLATTPLDVLKVNMQVHPIKYYSISSCFTTLLREQGPSVLW 72
+P +YA CT GG+LS G TH+ TPLD++K NMQ+ P KY SISS F LL+EQG +
Sbjct: 74 SPAFYAACTFGGILSCGLTHMTVTPLDLVKCNMQIDPAKYKSISSGFGILLKEQGVKGFF 133
Query: 73 KGWTGKFFGYGAQGGCRFGLYEYFKGVYSNV----LVDQHRSLVY 113
+GW GY AQG C+FG YEYFK YS++ ++++L+Y
Sbjct: 134 RGWVPTLLGYSAQGACKFGFYEYFKKTYSDLAGPEYTAKYKTLIY 178
>At3g48850 mitochondrial phosphate transporter
Length = 363
Score = 109 bits (272), Expect = 8e-25
Identities = 49/105 (46%), Positives = 71/105 (66%), Gaps = 4/105 (3%)
Query: 13 TPRYYALCTIGGMLSAGTTHLATTPLDVLKVNMQVHPIKYYSISSCFTTLLREQGPSVLW 72
+P Y+A CT+ GMLS G TH A TPLDV+K NMQ+ P+KY +I+S F T ++EQG
Sbjct: 63 SPAYFAACTVAGMLSCGITHTAITPLDVIKCNMQIDPLKYKNITSAFKTTIKEQGLKGFT 122
Query: 73 KGWTGKFFGYGAQGGCRFGLYEYFKGVYSNVL----VDQHRSLVY 113
+GW+ GY AQG ++GLYEY K YS+++ ++++L+Y
Sbjct: 123 RGWSPTLLGYSAQGAFKYGLYEYAKKYYSDIVGPEYAAKYKTLIY 167
>At1g34065 allinase like protein
Length = 327
Score = 56.6 bits (135), Expect = 6e-09
Identities = 30/89 (33%), Positives = 43/89 (47%), Gaps = 2/89 (2%)
Query: 24 GMLSAGTTHLATTPLDVLKVNMQVHP--IKYYSISSCFTTLLREQGPSVLWKGWTGKFFG 81
G + T + TTPLDV+K + V +Y +S C T++RE+G S LWKG +
Sbjct: 239 GAFAGAVTGVLTTPLDVIKTRLMVQGSGTQYKGVSDCIKTIIREEGSSALWKGMGPRVLW 298
Query: 82 YGAQGGCRFGLYEYFKGVYSNVLVDQHRS 110
G G FG+ E K + S H +
Sbjct: 299 IGIGGSIFFGVLEKTKQILSERSQKSHNA 327
>At4g01100 putative carrier protein
Length = 352
Score = 49.7 bits (117), Expect = 8e-07
Identities = 31/97 (31%), Positives = 48/97 (48%), Gaps = 9/97 (9%)
Query: 6 GGISQELTPRYYALCTIGGMLSAGTTHL-ATTPLDVLKVNMQVH----PIKYYSISSCFT 60
G + +LTP L +G +AG + AT P+D+++ + V P +Y I+
Sbjct: 133 GNENAQLTP----LLRLGAGATAGIIAMSATYPMDMVRGRLTVQTANSPYQYRGIAHALA 188
Query: 61 TLLREQGPSVLWKGWTGKFFGYGAQGGCRFGLYEYFK 97
T+LRE+GP L++GW G G F +YE K
Sbjct: 189 TVLREEGPRALYRGWLPSVIGVVPYVGLNFSVYESLK 225
Score = 28.1 bits (61), Expect = 2.4
Identities = 17/54 (31%), Positives = 27/54 (49%), Gaps = 3/54 (5%)
Query: 24 GMLSAGTTHLATTPLDVLKVNMQV---HPIKYYSISSCFTTLLREQGPSVLWKG 74
G ++ G + A PL+ +K+ +QV H IKY + R +G L+KG
Sbjct: 45 GGVAGGVSRTAVAPLERMKILLQVQNPHNIKYSGTVQGLKHIWRTEGLRGLFKG 98
>At1g07030 unknown protein
Length = 326
Score = 47.4 bits (111), Expect = 4e-06
Identities = 28/95 (29%), Positives = 46/95 (47%), Gaps = 5/95 (5%)
Query: 22 IGGMLSAGTTHLATTPLDVLKVNMQVH---PIKYYSISSCFTTLLREQGPSVLWKGWTGK 78
I G ++ H+A P+D +K +MQ P+K I F ++++++GPS L++G
Sbjct: 41 IAGSIAGSVEHMAMFPVDTIKTHMQALRPCPLKPVGIREAFRSIIQKEGPSALYRGIWAM 100
Query: 79 FFGYGAQGGCRFGLYEYFKGVYSNVLVDQHRSLVY 113
G G F YE K S DQ+ S+ +
Sbjct: 101 GLGAGPAHAVYFSFYEVSKKYLS--AGDQNNSVAH 133
Score = 37.0 bits (84), Expect = 0.005
Identities = 23/91 (25%), Positives = 38/91 (41%), Gaps = 6/91 (6%)
Query: 24 GMLSAGTTHLATTPLDVLKVNMQVHPI------KYYSISSCFTTLLREQGPSVLWKGWTG 77
G + G TTPLDV+K +Q + SIS T++++ G L +GW
Sbjct: 234 GAAAGGLAAAVTTPLDVVKTQLQCQGVCGCDRFTSSSISHVLRTIVKKDGYRGLLRGWLP 293
Query: 78 KFFGYGAQGGCRFGLYEYFKGVYSNVLVDQH 108
+ + + YE K + + VD +
Sbjct: 294 RMLFHAPAAAICWSTYEGVKSFFQDFNVDSN 324
Score = 28.9 bits (63), Expect = 1.4
Identities = 16/76 (21%), Positives = 31/76 (40%)
Query: 22 IGGMLSAGTTHLATTPLDVLKVNMQVHPIKYYSISSCFTTLLREQGPSVLWKGWTGKFFG 81
+ G+ + ++ TP+D++K +Q+ Y + C +LRE+G + +
Sbjct: 135 MSGVFATISSDAVFTPMDMVKQRLQMGEGTYKGVWDCVKRVLREEGIGAFYASYRTTVLM 194
Query: 82 YGAQGGCRFGLYEYFK 97
F YE K
Sbjct: 195 NAPFTAVHFATYEAAK 210
>At2g30160 putative mitochondrial carrier protein
Length = 331
Score = 47.0 bits (110), Expect = 5e-06
Identities = 27/92 (29%), Positives = 44/92 (47%), Gaps = 4/92 (4%)
Query: 9 SQELTPRYYALCTIGGMLSAGTTHLATTPLDVLKVNMQVH---PIKYYSISSCFTTLLRE 65
+Q T +++ L + G ++ H+A P+D +K +MQ PIK I F ++++
Sbjct: 31 AQNTTLKFWQLM-VAGSIAGSVEHMAMFPVDTVKTHMQALRSCPIKPIGIRQAFRSIIKT 89
Query: 66 QGPSVLWKGWTGKFFGYGAQGGCRFGLYEYFK 97
GPS L++G G G F YE K
Sbjct: 90 DGPSALYRGIWAMGLGAGPAHAVYFSFYEVSK 121
Score = 39.7 bits (91), Expect = 8e-04
Identities = 23/86 (26%), Positives = 38/86 (43%), Gaps = 6/86 (6%)
Query: 24 GMLSAGTTHLATTPLDVLKVNMQVHPI------KYYSISSCFTTLLREQGPSVLWKGWTG 77
G + G TTPLDV+K +Q + K SIS F T++++ G L +GW
Sbjct: 239 GAAAGGLAAAVTTPLDVVKTQLQCQGVCGCDRFKSSSISDVFRTIVKKDGYRGLARGWLP 298
Query: 78 KFFGYGAQGGCRFGLYEYFKGVYSNV 103
+ + + YE K + ++
Sbjct: 299 RMLFHAPAAAICWSTYETVKSFFQDL 324
Score = 29.6 bits (65), Expect = 0.82
Identities = 14/54 (25%), Positives = 26/54 (47%)
Query: 14 PRYYALCTIGGMLSAGTTHLATTPLDVLKVNMQVHPIKYYSISSCFTTLLREQG 67
P A I G+ + ++ TP+D++K +Q+ Y + C + RE+G
Sbjct: 129 PNNSAAHAISGVFATISSDAVFTPMDMVKQRLQIGNGTYKGVWDCIKRVTREEG 182
>At2g35800 putative mitochondrial carrier protein
Length = 823
Score = 46.6 bits (109), Expect = 7e-06
Identities = 31/89 (34%), Positives = 41/89 (45%), Gaps = 5/89 (5%)
Query: 24 GMLSAGTTHLATTPLDVLKVNMQVH-PIKYYSISSCFTTLLREQGPSVLWKGWTGKFFGY 82
G +S G + TTP DV+K M P + S+S ++LR +GP L+KG +FF
Sbjct: 728 GAVSGGIAAVVTTPFDVMKTRMMTATPGRPISMSMVVVSILRNEGPLGLFKGAVPRFFWV 787
Query: 83 GAQGGCRFGLYEYFKGVYSN----VLVDQ 107
G F YE K VL DQ
Sbjct: 788 APLGAMNFAGYELAKKAMQKNEDAVLADQ 816
>At4g39460 mitochondrial carrier like protein
Length = 325
Score = 43.5 bits (101), Expect = 6e-05
Identities = 26/76 (34%), Positives = 35/76 (45%), Gaps = 2/76 (2%)
Query: 24 GMLSAGTTHLATTPLDVLKVNMQVHPI--KYYSISSCFTTLLREQGPSVLWKGWTGKFFG 81
G + T TTPLDV+K + V +Y I C T++RE+G L KG +
Sbjct: 233 GAFAGALTGAVTTPLDVIKTRLMVQGSAKQYQGIVDCVQTIVREEGAPALLKGIGPRVLW 292
Query: 82 YGAQGGCRFGLYEYFK 97
G G FG+ E K
Sbjct: 293 IGIGGSIFFGVLESTK 308
>At3g53940 unknown protein
Length = 365
Score = 43.1 bits (100), Expect = 7e-05
Identities = 26/78 (33%), Positives = 37/78 (47%), Gaps = 2/78 (2%)
Query: 22 IGGMLSAGTTHLATTPLDVLKVNM--QVHPIKYYSISSCFTTLLREQGPSVLWKGWTGKF 79
+ G L+ T AT PLD+++ + Q + I Y + F T+ RE+G L+KG
Sbjct: 181 VSGGLAGLTAASATYPLDLVRTRLSAQRNSIYYQGVGHAFRTICREEGILGLYKGLGATL 240
Query: 80 FGYGAQGGCRFGLYEYFK 97
G G F YE FK
Sbjct: 241 LGVGPSLAISFAAYETFK 258
Score = 26.2 bits (56), Expect = 9.1
Identities = 22/93 (23%), Positives = 42/93 (44%), Gaps = 7/93 (7%)
Query: 18 ALCTIG-GMLSAGTTHLATTPLDVLKVNMQVH------PIKYYSISSCFTTLLREQGPSV 70
A+ ++G G LS + AT PLD+++ MQ+ + + F + + +G
Sbjct: 271 AVVSLGCGSLSGIVSSTATFPLDLVRRRMQLEGAGGRARVYTTGLFGTFKHIFKTEGMRG 330
Query: 71 LWKGWTGKFFGYGAQGGCRFGLYEYFKGVYSNV 103
L++G +++ G F +E K + S V
Sbjct: 331 LYRGIIPEYYKVVPGVGIAFMTFEELKKLLSTV 363
>At5g15640 putative mitochondrial carrier protein
Length = 323
Score = 42.4 bits (98), Expect = 1e-04
Identities = 26/77 (33%), Positives = 34/77 (43%), Gaps = 2/77 (2%)
Query: 23 GGMLSAGTTHLATTPLDVLKVNMQV--HPIKYYSISSCFTTLLREQGPSVLWKGWTGKFF 80
GG+++ T TTPLD +K +QV H S LL E G ++G +FF
Sbjct: 240 GGIIAGATASSITTPLDTIKTRLQVMGHQENRPSAKQVVKKLLAEDGWKGFYRGLGPRFF 299
Query: 81 GYGAQGGCRFGLYEYFK 97
A G YEY K
Sbjct: 300 SMSAWGTSMILTYEYLK 316
>At4g26180 putative mitochondrial carrier protein
Length = 325
Score = 42.4 bits (98), Expect = 1e-04
Identities = 28/94 (29%), Positives = 39/94 (40%), Gaps = 9/94 (9%)
Query: 13 TPRYYALCTIGGMLSAGTTHLATTPLDVLKVNMQ---------VHPIKYYSISSCFTTLL 63
T R L + G + GT L T PLD+++ + V I Y I CF+
Sbjct: 109 TTRGPLLDLVAGSFAGGTAVLFTYPLDLVRTKLAYQTQVKAIPVEQIIYRGIVDCFSRTY 168
Query: 64 REQGPSVLWKGWTGKFFGYGAQGGCRFGLYEYFK 97
RE G L++G +G G +F YE K
Sbjct: 169 RESGARGLYRGVAPSLYGIFPYAGLKFYFYEEMK 202
>At2g37890 mitochondrial carrier like protein
Length = 337
Score = 41.6 bits (96), Expect = 2e-04
Identities = 26/78 (33%), Positives = 36/78 (45%), Gaps = 2/78 (2%)
Query: 22 IGGMLSAGTTHLATTPLDVLKVNM--QVHPIKYYSISSCFTTLLREQGPSVLWKGWTGKF 79
+ G L+ T AT PLD+++ + Q + I Y I F T+ RE+G L+KG
Sbjct: 153 VSGGLAGITAATATYPLDLVRTRLAAQRNAIYYQGIEHTFRTICREEGILGLYKGLGATL 212
Query: 80 FGYGAQGGCRFGLYEYFK 97
G G F YE K
Sbjct: 213 LGVGPSLAINFAAYESMK 230
>At2g33820 mitochondrial basic amino acid carrier (BAC1)
Length = 311
Score = 41.6 bits (96), Expect = 2e-04
Identities = 24/90 (26%), Positives = 40/90 (43%), Gaps = 5/90 (5%)
Query: 22 IGGMLSAGTTHLATTPLDVLKVNMQVHP-----IKYYSISSCFTTLLREQGPSVLWKGWT 76
+ GM++ T P D +KV +Q H ++Y + C + +L+ +G L++G T
Sbjct: 19 VAGMMAGLATVAVGHPFDTVKVKLQKHNTDVQGLRYKNGLHCASRILQTEGVKGLYRGAT 78
Query: 77 GKFFGYGAQGGCRFGLYEYFKGVYSNVLVD 106
F G + FG+Y K L D
Sbjct: 79 SSFMGMAFESSLMFGIYSQAKLFLRGTLPD 108
>At5g58970 uncoupling protein AtUCP2
Length = 305
Score = 40.0 bits (92), Expect = 6e-04
Identities = 27/75 (36%), Positives = 31/75 (41%), Gaps = 10/75 (13%)
Query: 33 LATTPLDVLKVNMQVHPI----------KYYSISSCFTTLLREQGPSVLWKGWTGKFFGY 82
L T PLD KV +Q+ KY T+ RE+G S LWKG
Sbjct: 28 LCTIPLDTAKVRLQLQRKIPTGDGENLPKYRGSIGTLATIAREEGISGLWKGVIAGLHRQ 87
Query: 83 GAQGGCRFGLYEYFK 97
GG R GLYE K
Sbjct: 88 CIYGGLRIGLYEPVK 102
Score = 28.1 bits (61), Expect = 2.4
Identities = 17/65 (26%), Positives = 28/65 (42%), Gaps = 1/65 (1%)
Query: 36 TPLDVLKVNMQVHPIKYYSISSCFTTLLREQGPSVLWKGWTGKFFGYGAQGGCRFGLYEY 95
+P+DV+K M + Y + CF ++ +G +KG+ F G F E
Sbjct: 235 SPIDVVKSRM-MGDSTYRNTVDCFIKTMKTEGIMAFYKGFLPNFTRLGTWNAIMFLTLEQ 293
Query: 96 FKGVY 100
K V+
Sbjct: 294 VKKVF 298
>At3g54110 uncoupling protein (ucp/PUMP)
Length = 306
Score = 40.0 bits (92), Expect = 6e-04
Identities = 25/77 (32%), Positives = 33/77 (42%), Gaps = 9/77 (11%)
Query: 33 LATTPLDVLKVNMQVHPI---------KYYSISSCFTTLLREQGPSVLWKGWTGKFFGYG 83
+ T PLD KV +Q+ KY + T+ RE+G LWKG
Sbjct: 27 VCTIPLDTAKVRLQLQKSALAGDVTLPKYRGLLGTVGTIAREEGLRSLWKGVVPGLHRQC 86
Query: 84 AQGGCRFGLYEYFKGVY 100
GG R G+YE K +Y
Sbjct: 87 LFGGLRIGMYEPVKNLY 103
Score = 31.2 bits (69), Expect = 0.28
Identities = 15/49 (30%), Positives = 22/49 (44%)
Query: 36 TPLDVLKVNMQVHPIKYYSISSCFTTLLREQGPSVLWKGWTGKFFGYGA 84
+P+DV+K M Y CF L+ GP +KG+ F G+
Sbjct: 233 SPVDVVKSRMMGDSGAYKGTIDCFVKTLKSDGPMAFYKGFIPNFGRLGS 281
>At2g26360 mitochondrial carrier like protein
Length = 369
Score = 40.0 bits (92), Expect = 6e-04
Identities = 28/93 (30%), Positives = 45/93 (48%), Gaps = 6/93 (6%)
Query: 4 MEGGISQELTPRYYALCTIGGMLSAGTTHLATTPLDVLKVNMQVHP--IKYYSISSCFTT 61
+E + +EL P + A+ G LS G T + TTP DV+K M P ++ + + + +
Sbjct: 264 VERQLGRELEP-WEAIAV--GALSGGFTAVLTTPFDVIKTRMMTAPQGVELSMLMAAY-S 319
Query: 62 LLREQGPSVLWKGWTGKFFGYGAQGGCRFGLYE 94
+L +GP +KG +FF G YE
Sbjct: 320 ILTHEGPLAFYKGAVPRFFWTAPLGALNLAGYE 352
>At1g79900 putative carnitine/acylcarnitine translocase
Length = 296
Score = 40.0 bits (92), Expect = 6e-04
Identities = 22/73 (30%), Positives = 35/73 (47%)
Query: 22 IGGMLSAGTTHLATTPLDVLKVNMQVHPIKYYSISSCFTTLLREQGPSVLWKGWTGKFFG 81
+ G L+ + +A PLDV+K +Q Y I+ CF ++++G +VLW+G
Sbjct: 205 VAGGLAGVASWVACYPLDVVKTRLQQGHGAYEGIADCFRKSVKQEGYTVLWRGLGTAVAR 264
Query: 82 YGAQGGCRFGLYE 94
G F YE
Sbjct: 265 AFVVNGAIFAAYE 277
Score = 37.4 bits (85), Expect = 0.004
Identities = 23/84 (27%), Positives = 39/84 (46%), Gaps = 1/84 (1%)
Query: 14 PRYYALCTIGGMLSAGTTHLATTPLDVLKVNMQVHPIKYYSISSCFTTLLREQGPSVLWK 73
P Y +GG+ + L TP++++K+ +Q+ K I+ ++LR QG L++
Sbjct: 103 PPSYRGVALGGVATGAVQSLLLTPVELIKIRLQLQQTKSGPITLA-KSILRRQGLQGLYR 161
Query: 74 GWTGKFFGYGAQGGCRFGLYEYFK 97
G T G F YEY +
Sbjct: 162 GLTITVLRDAPAHGLYFWTYEYVR 185
>At4g28390 ADP,ATP carrier-like protein
Length = 379
Score = 39.3 bits (90), Expect = 0.001
Identities = 24/87 (27%), Positives = 39/87 (44%), Gaps = 9/87 (10%)
Query: 24 GMLSAGTTHLATTPLDVLK--VNMQVHPIK-------YYSISSCFTTLLREQGPSVLWKG 74
G +SA + A P++ +K + Q IK Y IS CF ++++G LW+G
Sbjct: 86 GGVSAAVSKTAAAPIERVKLLIQNQDEMIKAGRLSEPYKGISDCFARTVKDEGMLALWRG 145
Query: 75 WTGKFFGYGAQGGCRFGLYEYFKGVYS 101
T Y F +YFK +++
Sbjct: 146 NTANVIRYFPTQALNFAFKDYFKRLFN 172
Score = 26.9 bits (58), Expect = 5.3
Identities = 13/45 (28%), Positives = 25/45 (54%), Gaps = 3/45 (6%)
Query: 33 LATTPLDVLKVNMQV---HPIKYYSISSCFTTLLREQGPSVLWKG 74
LA+ P+D ++ M + +KY S F+ +++ +G L+KG
Sbjct: 300 LASYPIDTVRRRMMMTSGEAVKYKSSLQAFSQIVKNEGAKSLFKG 344
>At4g03115 putative protein
Length = 300
Score = 38.5 bits (88), Expect = 0.002
Identities = 25/83 (30%), Positives = 39/83 (46%), Gaps = 8/83 (9%)
Query: 16 YYALCTIGGMLSAGTTHLATTPLDVLKVNMQVHPI----KYYSISSCFTTLLREQGPSVL 71
++ + I L+ G TH PLDV+KV +Q+ + ++ F L++ +G L
Sbjct: 66 HFGISGISVALATGVTH----PLDVVKVRLQMQHVGQRGPLIGMTGIFLQLMKNEGRRSL 121
Query: 72 WKGWTGKFFGYGAQGGCRFGLYE 94
+ G T GG R GLYE
Sbjct: 122 YLGLTPALTRSVLYGGLRLGLYE 144
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.352 0.156 0.557
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 3,293,400
Number of Sequences: 26719
Number of extensions: 123489
Number of successful extensions: 542
Number of sequences better than 10.0: 48
Number of HSP's better than 10.0 without gapping: 31
Number of HSP's successfully gapped in prelim test: 17
Number of HSP's that attempted gapping in prelim test: 458
Number of HSP's gapped (non-prelim): 86
length of query: 170
length of database: 11,318,596
effective HSP length: 92
effective length of query: 78
effective length of database: 8,860,448
effective search space: 691114944
effective search space used: 691114944
T: 11
A: 40
X1: 14 ( 7.1 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 38 (22.0 bits)
S2: 56 (26.2 bits)
Medicago: description of AC127019.17


