

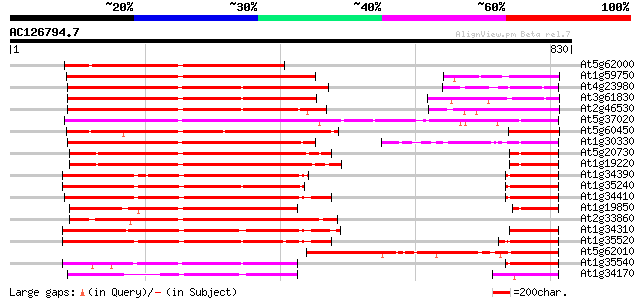
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC126794.7 - phase: 1 /pseudo
(830 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
At5g62000 auxin response factor - like protein 583 e-166
At1g59750 auxin response factor 1 513 e-145
At4g23980 auxin response factor 9 (ARF9) 457 e-129
At3g61830 auxin response factor-like protein 453 e-127
At2g46530 ARF1 family auxin responsive transcription factor like... 452 e-127
At5g37020 auxin response factor 8 (ARF8) 418 e-117
At5g60450 auxin response factor 4 402 e-112
At1g30330 putative protein 380 e-105
At5g20730 auxin response factor 7 (ARF7) 368 e-102
At1g19220 auxin response factor (ARF) like protein 367 e-101
At1g34390 auxin response factor, putative 363 e-100
At1g35240 hypothetical protein 359 4e-99
At1g34410 auxin response factor, putative 359 4e-99
At1g19850 transcription factor monopteros (MP) 359 4e-99
At2g33860 auxin response transcription factor 3 (ETTIN/ARF3) 355 8e-98
At1g34310 hypothetical protein 350 2e-96
At1g35520 auxin response factor, putative 348 7e-96
At5g62010 ARF1-binding protein 324 1e-88
At1g35540 auxin response factor, putative 318 6e-87
At1g34170 putative protein 282 6e-76
>At5g62000 auxin response factor - like protein
Length = 371
Score = 583 bits (1503), Expect = e-166
Identities = 284/326 (87%), Positives = 301/326 (92%), Gaps = 7/326 (2%)
Query: 81 EAEAALYRELWHACAGPLVTVPREGELVFYFPQGHIEQVEASTNQASEQHMPVYDLRPKI 140
+ EAALYRELWHACAGPLVTVPR+ + VFYFPQGHIEQ ASTNQA+EQ MP+YDL K+
Sbjct: 53 DPEAALYRELWHACAGPLVTVPRQDDRVFYFPQGHIEQ--ASTNQAAEQQMPLYDLPSKL 110
Query: 141 LCRVINVMLKAEPDTDEVFAQVTLVPEPNQDENAVEKEAPPAPPPRFHVHSFCKTLTASD 200
LCRVINV LKAE DTDEV+AQ+TL+PE NQDENA+EKEAP PPPRF VHSFCKTLTASD
Sbjct: 111 LCRVINVDLKAEADTDEVYAQITLLPEANQDENAIEKEAPLPPPPRFQVHSFCKTLTASD 170
Query: 201 TSTHGGFSVLRRHADECLPPLDMSKQPPTQELVAKDLHGNEWRFRHIFRGELISGQPRRH 260
TSTHGGFSVLRRHADECLPPLDMS+QPPTQELVAKDLH NEWRFRHIFRG QPRRH
Sbjct: 171 TSTHGGFSVLRRHADECLPPLDMSRQPPTQELVAKDLHANEWRFRHIFRG-----QPRRH 225
Query: 261 LLQSGWSVFVSSKRLVAGDAFIFLRGENGELRVGVRRAMRQQGNVPSSVISSHSMHLGVL 320
LLQSGWSVFVSSKRLVAGDAFIFLRGENGELRVGVRRAMRQQGNVPSSVISSHSMHLGVL
Sbjct: 226 LLQSGWSVFVSSKRLVAGDAFIFLRGENGELRVGVRRAMRQQGNVPSSVISSHSMHLGVL 285
Query: 321 ATAWHAVLTGTMFTVYYKPRTSPAEFIVPYDQYMESLKNNYTIGMRFKMRFEGEEAPEQR 380
ATAWHA+ TGTMFTVYYKPRTSP+EFIVP+DQYMES+KNNY+IGMRFKMRFEGEEAPEQR
Sbjct: 286 ATAWHAISTGTMFTVYYKPRTSPSEFIVPFDQYMESVKNNYSIGMRFKMRFEGEEAPEQR 345
Query: 381 FTGTIVGIEDSDSKRWPTSKWRCLKV 406
FTGTIVGIE+SD RWP SKWR LKV
Sbjct: 346 FTGTIVGIEESDPTRWPKSKWRSLKV 371
>At1g59750 auxin response factor 1
Length = 665
Score = 513 bits (1322), Expect = e-145
Identities = 251/370 (67%), Positives = 302/370 (80%), Gaps = 8/370 (2%)
Query: 85 ALYRELWHACAGPLVTVPREGELVFYFPQGHIEQVEASTNQASEQHMPVYDLRPKILCRV 144
AL RELWHACAGPLVT+PREGE V+YFP+GH+EQ+EAS +Q EQ MP ++L KILC+V
Sbjct: 18 ALCRELWHACAGPLVTLPREGERVYYFPEGHMEQLEASMHQGLEQQMPSFNLPSKILCKV 77
Query: 145 INVMLKAEPDTDEVFAQVTLVPEPNQDENAVEKEAPPAPPPRFHVHSFCKTLTASDTSTH 204
IN+ +AEP+TDEV+AQ+TL+PE +Q E +AP P + VHSFCKTLTASDTSTH
Sbjct: 78 INIQRRAEPETDEVYAQITLLPELDQSE-PTSPDAPVQEPEKCTVHSFCKTLTASDTSTH 136
Query: 205 GGFSVLRRHADECLPPLDMSKQPPTQELVAKDLHGNEWRFRHIFRGELISGQPRRHLLQS 264
GGFSVLRRHAD+CLPPLDMS+QPP QELVA DLH +EW FRHIFRG QPRRHLL +
Sbjct: 137 GGFSVLRRHADDCLPPLDMSQQPPWQELVATDLHNSEWHFRHIFRG-----QPRRHLLTT 191
Query: 265 GWSVFVSSKRLVAGDAFIFLRGENGELRVGVRRAMRQQGNVPSSVISSHSMHLGVLATAW 324
GWSVFVSSK+LVAGDAFIFLRGEN ELRVGVRR MRQQ N+PSSVISSHSMH+GVLATA
Sbjct: 192 GWSVFVSSKKLVAGDAFIFLRGENEELRVGVRRHMRQQTNIPSSVISSHSMHIGVLATAA 251
Query: 325 HAVLTGTMFTVYYKPRTSPAEFIVPYDQYMESLKNNYTIGMRFKMRFEGEEAPEQRFTGT 384
HA+ TGT+F+V+YKPRTS +EFIV ++Y+E+ ++GMRFKMRFEGEEAPE+RF+GT
Sbjct: 252 HAITTGTIFSVFYKPRTSRSEFIVSVNRYLEAKTQKLSVGMRFKMRFEGEEAPEKRFSGT 311
Query: 385 IVGIEDSDSKRWPTSKWRCLKVRWDETSNIPRPERVSPWKIEPALA--PPALNPLPMPRP 442
IVG++++ S W S+WR LKV+WDE S++ RPERVSPW++EP +A P+ P P R
Sbjct: 312 IVGVQENKSSVWHDSEWRSLKVQWDEPSSVFRPERVSPWELEPLVANSTPSSQPQPPQRN 371
Query: 443 KRPRANVVPS 452
KRPR +PS
Sbjct: 372 KRPRPPGLPS 381
Score = 100 bits (249), Expect = 3e-21
Identities = 59/174 (33%), Positives = 99/174 (55%), Gaps = 14/174 (8%)
Query: 643 SAKISEAAKPKDSD---CKLFGFSLLSSPTMLEPSLSQRNATSETSSHMQISSQHHTFEN 699
SA +E+ + K ++ C+LFGF L+ + + E S + + + + S F++
Sbjct: 448 SAFNNESTEKKQTNGNVCRLFGFELVENVNVDE-CFSAASVSGAVAVDQPVPSNE--FDS 504
Query: 700 DQKSEHSKSSKPADKLVIVDEHEKQLQTSQPHVKDVQLKPQSGSARSCTKVHKKGIALGR 759
Q+SE ++ D + L++ Q + QS RSCTKVH +G A+GR
Sbjct: 505 GQQSEPLNINQSDIPSGSGDPEKSSLRSPQ--------ESQSRQIRSCTKVHMQGSAVGR 556
Query: 760 SVDLTKFSDYDELTAELDQLFEFRGELISPQKDWLVVFTDNEGDMMLVGDDPWH 813
++DLT+ Y++L +L+++F+ +GEL+ K W VV+TD+E DMM+VGDDPW+
Sbjct: 557 AIDLTRSECYEDLFKKLEEMFDIKGELLESTKKWQVVYTDDEDDMMMVGDDPWN 610
>At4g23980 auxin response factor 9 (ARF9)
Length = 638
Score = 457 bits (1177), Expect = e-129
Identities = 229/387 (59%), Positives = 286/387 (73%), Gaps = 9/387 (2%)
Query: 86 LYRELWHACAGPLVTVPREGELVFYFPQGHIEQVEASTNQASEQHM-PVYDLRPKILCRV 144
LY ELW CAGPLV VP+ E V+YFPQGH+EQ+EAST Q M P++ L PKILC V
Sbjct: 9 LYDELWKLCAGPLVDVPQAQERVYYFPQGHMEQLEASTQQVDLNTMKPLFVLPPKILCNV 68
Query: 145 INVMLKAEPDTDEVFAQVTLVPEPNQDENAVEKEAPPAPPPRFHVHSFCKTLTASDTSTH 204
+NV L+AE DTDEV+AQ+TL+P + + + + P R VHSF K LTASDTSTH
Sbjct: 69 MNVSLQAEKDTDEVYAQITLIPVGTEVDEPMSPDPSPPELQRPKVHSFSKVLTASDTSTH 128
Query: 205 GGFSVLRRHADECLPPLDMSKQPPTQELVAKDLHGNEWRFRHIFRGELISGQPRRHLLQS 264
GGFSVLR+HA ECLPPLDM++Q PTQELVA+D+HG +W+F+HIFR GQPRRHLL +
Sbjct: 129 GGFSVLRKHATECLPPLDMTQQTPTQELVAEDVHGYQWKFKHIFR-----GQPRRHLLTT 183
Query: 265 GWSVFVSSKRLVAGDAFIFLRGENGELRVGVRRAMRQQGNVPSSVISSHSMHLGVLATAW 324
GWS FV+SKRLVAGD F+FLRGENGELRVGVRRA QQ ++PSSVISSHSMHLGVLATA
Sbjct: 184 GWSTFVTSKRLVAGDTFVFLRGENGELRVGVRRANLQQSSMPSSVISSHSMHLGVLATAR 243
Query: 325 HAVLTGTMFTVYYKPRTSPAEFIVPYDQYMESLKNNYTIGMRFKMRFEGEEAPEQRFTGT 384
HA T TMF VYYKPRTS +FI+ ++Y+E++ N +++GMRFKMRFEGE++PE+R++GT
Sbjct: 244 HATQTKTMFIVYYKPRTS--QFIISLNKYLEAMSNKFSVGMRFKMRFEGEDSPERRYSGT 301
Query: 385 IVGIEDSDSKRWPTSKWRCLKVRWDETSNIPRPERVSPWKIEPALAPPALNPLPMPRPKR 444
++G++D S W SKWRCL+V WDE ++I RP +VSPW+IEP + + M + KR
Sbjct: 302 VIGVKDC-SPHWKDSKWRCLEVHWDEPASISRPNKVSPWEIEPFVNSENVPKSVMLKNKR 360
Query: 445 PRANVVPSSPDSSVLTREASSKVSMDP 471
PR S+ D + S V P
Sbjct: 361 PRQVSEVSALDVGITASNLWSSVLTQP 387
Score = 102 bits (253), Expect = 1e-21
Identities = 65/172 (37%), Positives = 96/172 (55%), Gaps = 19/172 (11%)
Query: 641 QMSAKISEAAKPKDSDCKLFGFSLLSSPTMLEPSLSQRNATSETSSHMQISSQHHTFEND 700
QM + + + ++ +LFG L+SS SL+ + + IS +D
Sbjct: 437 QMVSPVEQKKPETTANYRLFGIDLMSS------SLAVPEEKTAPMRPINISKPTMDSHSD 490
Query: 701 QKSEHSKSSKPADKLVIVDEHEKQLQTSQPHVKDVQLKPQSGSARSCTKVHKKGIALGRS 760
KSE SK S+ EK+ + ++ K+VQ K QS S RS TKV +G+ +GR+
Sbjct: 491 PKSEISKVSE-----------EKKQEPAEGSPKEVQSK-QSSSTRSRTKVQMQGVPVGRA 538
Query: 761 VDLTKFSDYDELTAELDQLFEFRGELISPQKDWLVVFTDNEGDMMLVGDDPW 812
VDL Y+EL ++++LF+ +GEL S + W +VFTD+EGDMMLVGDDPW
Sbjct: 539 VDLNALKGYNELIDDIEKLFDIKGELRS-RNQWEIVFTDDEGDMMLVGDDPW 589
>At3g61830 auxin response factor-like protein
Length = 602
Score = 453 bits (1166), Expect = e-127
Identities = 231/370 (62%), Positives = 280/370 (75%), Gaps = 11/370 (2%)
Query: 86 LYRELWHACAGPLVTVPREGELVFYFPQGHIEQVEASTNQA-SEQHMPVYDLRPKILCRV 144
LY ELW CAGPLV VPR E VFYFPQGH+EQ+ ASTNQ + + +PV+DL PKILCRV
Sbjct: 22 LYTELWKVCAGPLVEVPRAQERVFYFPQGHMEQLVASTNQGINSEEIPVFDLPPKILCRV 81
Query: 145 INVMLKAEPDTDEVFAQVTLVPEPNQDENAVEKEAPPAPPPRFHVHSFCKTLTASDTSTH 204
++V LKAE +TDEV+AQ+TL PE +Q E + P P + HSF K LTASDTSTH
Sbjct: 82 LDVTLKAEHETDEVYAQITLQPEEDQSE-PTSLDPPIVGPTKQEFHSFVKILTASDTSTH 140
Query: 205 GGFSVLRRHADECLPPLDMSKQPPTQELVAKDLHGNEWRFRHIFRGELISGQPRRHLLQS 264
GGFSVLR+HA ECLP LDM++ PTQELV +DLHG EWRF+HIFRG QPRRHLL +
Sbjct: 141 GGFSVLRKHATECLPSLDMTQATPTQELVTRDLHGFEWRFKHIFRG-----QPRRHLLTT 195
Query: 265 GWSVFVSSKRLVAGDAFIFLRGENGELRVGVRRAMRQQGNVPSSVISSHSMHLGVLATAW 324
GWS FVSSKRLVAGDAF+FLRGENG+LRVGVRR R Q +P+SVISS SMHLGVLATA
Sbjct: 196 GWSTFVSSKRLVAGDAFVFLRGENGDLRVGVRRLARHQSTMPTSVISSQSMHLGVLATAS 255
Query: 325 HAVLTGTMFTVYYKPRTSPAEFIVPYDQYMESLKNNYTIGMRFKMRFEGEEAPEQRFTGT 384
HAV T T+F V+YKPR S +FIV ++YME++K+ +++G RF+MRFEGEE+PE+ FTGT
Sbjct: 256 HAVRTTTIFVVFYKPRIS--QFIVGVNKYMEAIKHGFSLGTRFRMRFEGEESPERIFTGT 313
Query: 385 IVGIEDSDSKRWPTSKWRCLKVRWDETSNIPRPERVSPWKIEPALA-PPALNPLPMPRPK 443
IVG D S +WP SKWR L+V+WDE + + RP++VSPW+IEP LA P P P+ K
Sbjct: 314 IVGSGDL-SSQWPASKWRSLQVQWDEPTTVQRPDKVSPWEIEPFLATSPISTPAQQPQSK 372
Query: 444 RPRANVVPSS 453
R+ + S
Sbjct: 373 CKRSRPIEPS 382
Score = 92.4 bits (228), Expect = 9e-19
Identities = 65/208 (31%), Positives = 95/208 (45%), Gaps = 20/208 (9%)
Query: 619 FLMPPPPPTQYESPHSRELSQKQMSAKISEAAKP-------KDSDCKLFGFSLLSSPTM- 670
FL P T + P S+ + + + A P + D L P++
Sbjct: 356 FLATSPISTPAQQPQSKCKRSRPIEPSVKTPAPPSFLYSLPQSQDSINASLKLFQDPSLE 415
Query: 671 -LEPSLSQRNATSETSSHMQISSQHHTFENDQKSEHS----KSSKPADKLVIVDEHEKQL 725
+ S N+ + + + F D S + +P D E
Sbjct: 416 RISGGYSSNNSFKPETPPPPTNCSYRLFGFDLTSNSPAPIPQDKQPMDTCGAAKCQEPIT 475
Query: 726 QTSQPHVKDVQLKPQSGSARSCTKVHKKGIALGRSVDLTKFSDYDELTAELDQLFEFRGE 785
TS K Q ++RS TKV +GIA+GR+VDLT YDEL EL+++FE +G+
Sbjct: 476 PTSMSEQKKQQ------TSRSRTKVQMQGIAVGRAVDLTLLKSYDELIDELEEMFEIQGQ 529
Query: 786 LISPQKDWLVVFTDNEGDMMLVGDDPWH 813
L++ K W+VVFTD+EGDMML GDDPW+
Sbjct: 530 LLARDK-WIVVFTDDEGDMMLAGDDPWN 556
>At2g46530 ARF1 family auxin responsive transcription factor like
protein
Length = 622
Score = 452 bits (1164), Expect = e-127
Identities = 235/389 (60%), Positives = 290/389 (74%), Gaps = 16/389 (4%)
Query: 86 LYRELWHACAGPLVTVPREGELVFYFPQGHIEQVEASTNQAS-EQHMPVYDLRPKILCRV 144
LY ELW ACAGPLV VPR GE VFYFPQGH+EQ+ ASTNQ +Q +PV++L PKILCRV
Sbjct: 39 LYTELWKACAGPLVEVPRYGERVFYFPQGHMEQLVASTNQGVVDQEIPVFNLPPKILCRV 98
Query: 145 INVMLKAEPDTDEVFAQVTLVPEPNQDENAVEKEAPPAPPPRFHVHSFCKTLTASDTSTH 204
++V LKAE +TDEV+AQ+TL PE +Q E + P P + V SF K LTASDTSTH
Sbjct: 99 LSVTLKAEHETDEVYAQITLQPEEDQSE-PTSLDPPLVEPAKPTVDSFVKILTASDTSTH 157
Query: 205 GGFSVLRRHADECLPPLDMSKQPPTQELVAKDLHGNEWRFRHIFRGELISGQPRRHLLQS 264
GGFSVLR+HA ECLP LDM++ PTQELVA+DLHG EWRF+HIFRG QPRRHLL +
Sbjct: 158 GGFSVLRKHATECLPSLDMTQPTPTQELVARDLHGYEWRFKHIFRG-----QPRRHLLTT 212
Query: 265 GWSVFVSSKRLVAGDAFIFLRGENGELRVGVRRAMRQQGNVPSSVISSHSMHLGVLATAW 324
GWS FV+SKRLVAGDAF+FLRGE G+LRVGVRR +QQ +P+SVISS SM LGVLATA
Sbjct: 213 GWSTFVTSKRLVAGDAFVFLRGETGDLRVGVRRLAKQQSTMPASVISSQSMRLGVLATAS 272
Query: 325 HAVLTGTMFTVYYKPRTSPAEFIVPYDQYMESLKNNYTIGMRFKMRFEGEEAPEQRFTGT 384
HAV T T+F V+YKPR S +FI+ ++YM ++KN +++GMR++MRFEGEE+PE+ FTGT
Sbjct: 273 HAVTTTTIFVVFYKPRIS--QFIISVNKYMMAMKNGFSLGMRYRMRFEGEESPERIFTGT 330
Query: 385 IVGIEDSDSKRWPTSKWRCLKVRWDETSNIPRPERVSPWKIEPALAPPALNPLP-----M 439
I+G D S +WP SKWR L+++WDE S+I RP +VSPW+IEP +P AL P P
Sbjct: 331 IIGSGDL-SSQWPASKWRSLQIQWDEPSSIQRPNKVSPWEIEP-FSPSALTPTPTQQQSK 388
Query: 440 PRPKRPRANVVPSSPDSSVLTREASSKVS 468
+ RP + + S SS L+ + S S
Sbjct: 389 SKRSRPISEITGSPVASSFLSSFSQSHES 417
Score = 97.4 bits (241), Expect = 3e-20
Identities = 74/209 (35%), Positives = 109/209 (51%), Gaps = 23/209 (11%)
Query: 620 LMPPPPPTQYESPHSRELSQKQMSAKISEAAKPKDSDCKLFGFSLLSSPTML----EPSL 675
L P P Q +S SR +S+ S S S F S S+P++ +P+
Sbjct: 378 LTPTPTQQQSKSKRSRPISEITGSPVAS-------SFLSSFSQSHESNPSVKLLFQDPAT 430
Query: 676 SQRNATSETSSHMQ-------ISSQHHTFENDQKSEHSKSSKPADK-LVIVDEH--EKQL 725
+ + S SS +Q ++S F D S+ + ++ P DK L+ VD + +
Sbjct: 431 ERNSNKSVFSSGLQCKITEAPVTSSCRLFGFDLTSKPASATIPHDKQLISVDSNISDSTT 490
Query: 726 QTSQPHVKDV-QLKPQSGSARSCTKVHKKGIALGRSVDLTKFSDYDELTAELDQLFEFRG 784
+ P+ + + + Q S RS KV +G A+GR+VDLT YDEL EL+++FE G
Sbjct: 491 KCQDPNSSNSPKEQKQQTSTRSRIKVQMQGTAVGRAVDLTLLRSYDELIKELEKMFEIEG 550
Query: 785 ELISPQKDWLVVFTDNEGDMMLVGDDPWH 813
EL SP+ W +VFTD+EGD MLVGDDPW+
Sbjct: 551 EL-SPKDKWAIVFTDDEGDRMLVGDDPWN 578
>At5g37020 auxin response factor 8 (ARF8)
Length = 811
Score = 418 bits (1074), Expect = e-117
Identities = 278/781 (35%), Positives = 392/781 (49%), Gaps = 71/781 (9%)
Query: 81 EAEAALYRELWHACAGPLVTVPREGELVFYFPQGHIEQVEASTNQASEQHMPVY-DLRPK 139
E E L ELWHACAGPLV++P G V YFPQGH EQV A+TN+ + H+P Y L P+
Sbjct: 14 EGEKCLNSELWHACAGPLVSLPSSGSRVVYFPQGHSEQVAATTNKEVDGHIPNYPSLPPQ 73
Query: 140 ILCRVINVMLKAEPDTDEVFAQVTLVPEPNQDENAVEKEAPPAPPPRFHVHSFCKTLTAS 199
++C++ NV + A+ +TDEV+AQ+TL P +++ P + + FCKTLTAS
Sbjct: 74 LICQLHNVTMHADVETDEVYAQMTLQPLTPEEQKETFVPIELGIPSKQPSNYFCKTLTAS 133
Query: 200 DTSTHGGFSVLRRHADECLPPLDMSKQPPTQELVAKDLHGNEWRFRHIFRGELISGQPRR 259
DTSTHGGFSV RR A++ PPLD + QPP QEL+A+DLH EW+FRHIFR GQP+R
Sbjct: 134 DTSTHGGFSVPRRAAEKVFPPLDYTLQPPAQELIARDLHDVEWKFRHIFR-----GQPKR 188
Query: 260 HLLQSGWSVFVSSKRLVAGDAFIFLRGENGELRVGVRRAMRQQGNVPSSVISSHSMHLGV 319
HLL +GWSVFVS+KRLVAGD+ IF+R E +L +G+R A R Q VPSSV+SS SMH+G+
Sbjct: 189 HLLTTGWSVFVSAKRLVAGDSVIFIRNEKNQLFLGIRHATRPQTIVPSSVLSSDSMHIGL 248
Query: 320 LATAWHAVLTGTMFTVYYKPRTSPAEFIVPYDQYMESL-KNNYTIGMRFKMRFEGEEAPE 378
LA A HA T + FTV++ PR S +EF++ +Y++++ ++GMRF+M FE EE+
Sbjct: 249 LAAAAHASATNSCFTVFFHPRASQSEFVIQLSKYIKAVFHTRISVGMRFRMLFETEESSV 308
Query: 379 QRFTGTIVGIEDSDSKRWPTSKWRCLKVRWDETSNIPRPERVSPWKIEPALAPPALNPLP 438
+R+ GTI GI D DS RWP S WR +KV WDE++ R RVS W+IEP P L
Sbjct: 309 RRYMGTITGISDLDSVRWPNSHWRSVKVGWDESTAGERQPRVSLWEIEPLTTFPMYPSLF 368
Query: 439 MPRPKRPRANVVPSSPDS--------SVLTREASSKVSMDPL--PTSGFQRVLQGQESST 488
R KRP S PD + L + + PL P+ G +Q + +
Sbjct: 369 PLRLKRPWHAGTSSLPDGRGDLGSGLTWLRGGGGEQQGLLPLNYPSVGLFPWMQQRLDLS 428
Query: 489 LRGNLAESNDSYTAEKSVAWTPATDEEKMDAVSTSRRYGSENWMPMSRQEPTYSDL-LSG 547
G ++N Y A + + + + ++ S SDL L
Sbjct: 429 QMG--TDNNQQYQAMLAAGLQNIGGGDPLRQQFVQLQEPHHQYLQQSASH--NSDLMLQQ 484
Query: 548 FGSTREGKHNMLTQWPVMPPGLSLNFLHSNMKGSAQGSDNATYQAQGNMRYSAFGDYSVL 607
+ +H M Q +M L + + G Q N +AF
Sbjct: 485 QQQQQASRHLMHAQTQIMSENLPQQNMRQEVSNQPAGQQQQLQQPDQNAYLNAF------ 538
Query: 608 HGHKVENPHGNFLMPPPPPTQYESPHSRELSQKQMSAKISEAAKPKDSDCKLFGFSLL-- 665
K++N H L ++ SP + S K + A P D L FS+
Sbjct: 539 ---KMQNGH---LQQWQQQSEMPSPSFMKSDFTDSSNKFATTASPASGDGNLLNFSITGQ 592
Query: 666 ---------------SSPTMLEP----------SLSQRNATSETSSHMQISSQHHTFEND 700
+S T EP SL+ + S + F
Sbjct: 593 SVLPEQLTTEGWSPKASNTFSEPLSLPQAYPGKSLALEPGNPQNPSLFGVDPDSGLFLPS 652
Query: 701 QKSEHSKSSKPAD--KLVIVDE------HEKQLQTSQPHVKDVQLKPQSGSARSCTKVHK 752
+ SS A+ + + D + T+ + S ++ KV+K
Sbjct: 653 TVPRFASSSGDAEASPMSLTDSGFQNSLYSCMQDTTHELLHGAGQINSSNQTKNFVKVYK 712
Query: 753 KGIALGRSVDLTKFSDYDELTAELDQLFEFRGELISP-QKDWLVVFTDNEGDMMLVGDDP 811
G ++GRS+D+++FS Y EL EL ++F G L P + W +VF D E D++L+GDDP
Sbjct: 713 SG-SVGRSLDISRFSSYHELREELGKMFAIEGLLEDPLRSGWQLVFVDKENDILLLGDDP 771
Query: 812 W 812
W
Sbjct: 772 W 772
>At5g60450 auxin response factor 4
Length = 788
Score = 402 bits (1033), Expect = e-112
Identities = 202/413 (48%), Positives = 279/413 (66%), Gaps = 20/413 (4%)
Query: 84 AALYRELWHACAGPLVTVPREGELVFYFPQGHIEQVEASTNQASEQHMPVYDLRPKILCR 143
+++Y ELWHACAGPL +P++G +V YFPQGH+EQ +A + +S +P +DL P+I+CR
Sbjct: 60 SSIYSELWHACAGPLTCLPKKGNVVVYFPQGHLEQ-DAMVSYSSPLEIPKFDLNPQIVCR 118
Query: 144 VINVMLKAEPDTDEVFAQVTLVP----------EPNQDENAVEKEAPPAPPPRFHVHSFC 193
V+NV L A DTDEV+ QVTL+P E E+E + + H FC
Sbjct: 119 VVNVQLLANKDTDEVYTQVTLLPLQEFSMLNGEGKEVKELGGEEERNGSSSVKRTPHMFC 178
Query: 194 KTLTASDTSTHGGFSVLRRHADECLPPLDMSKQPPTQELVAKDLHGNEWRFRHIFRGELI 253
KTLTASDTSTHGGFSV RR A++C PLD +Q P+QEL+AKDLHG EW+FRHI+RG
Sbjct: 179 KTLTASDTSTHGGFSVPRRAAEDCFAPLDYKQQRPSQELIAKDLHGVEWKFRHIYRG--- 235
Query: 254 SGQPRRHLLQSGWSVFVSSKRLVAGDAFIFLRGENGELRVGVRRAMRQQGNVPSSVISSH 313
QPRRHLL +GWS+FVS K LV+GDA +FLR E GELR+G+RRA R + +P S+I +
Sbjct: 236 --QPRRHLLTTGWSIFVSQKNLVSGDAVLFLRDEGGELRLGIRRAARPRNGLPDSIIEKN 293
Query: 314 SMHLGVLATAWHAVLTGTMFTVYYKPRTSPAEFIVPYDQYMESLKNNYTIGMRFKMRFEG 373
S +L+ +AV T +MF V+Y PR + AEF++PY++Y+ S+++ IG RF+MRFE
Sbjct: 294 SCS-NILSLVANAVSTKSMFHVFYSPRATHAEFVIPYEKYITSIRSPVCIGTRFRMRFEM 352
Query: 374 EEAPEQRFTGTIVGIEDSDSKRWPTSKWRCLKVRWDETSNIPRPERVSPWKIEPALAPPA 433
+++PE+R G + G+ D D RWP SKWRCL VRWDE+ ERVSPW+I+P+++ P
Sbjct: 353 DDSPERRCAGVVTGVCDLDPYRWPNSKWRCLLVRWDESFVSDHQERVSPWEIDPSVSLPH 412
Query: 434 LNPLPMPRPKRPRANVVPSSPDSSVLTREASSKVSMDPLPTSGFQRVLQGQES 486
L+ PRPKRP A ++ ++P + +T+ + + S +VLQGQE+
Sbjct: 413 LSIQSSPRPKRPWAGLLDTTPPGNPITKRGGFLDFEESVRPS---KVLQGQEN 462
Score = 100 bits (250), Expect = 3e-21
Identities = 42/76 (55%), Positives = 58/76 (76%)
Query: 738 KPQSGSARSCTKVHKKGIALGRSVDLTKFSDYDELTAELDQLFEFRGELISPQKDWLVVF 797
KPQS S R CTKVHK+G +GR++DL++ + YD+L EL++LF G L P+K W +++
Sbjct: 658 KPQSSSKRICTKVHKQGSQVGRAIDLSRLNGYDDLLMELERLFNMEGLLRDPEKGWRILY 717
Query: 798 TDNEGDMMLVGDDPWH 813
TD+E DMM+VGDDPWH
Sbjct: 718 TDSENDMMVVGDDPWH 733
>At1g30330 putative protein
Length = 933
Score = 380 bits (977), Expect = e-105
Identities = 197/370 (53%), Positives = 257/370 (69%), Gaps = 9/370 (2%)
Query: 86 LYRELWHACAGPLVTVPREGELVFYFPQGHIEQVEASTNQASEQHMPVY-DLRPKILCRV 144
L ELWHACAGPLV++P G V YFPQGH EQV ASTN+ + H+P Y L P+++C++
Sbjct: 20 LNSELWHACAGPLVSLPPVGSRVVYFPQGHSEQVAASTNKEVDAHIPNYPSLHPQLICQL 79
Query: 145 INVMLKAEPDTDEVFAQVTLVPEPNQDENAVEKEAPPAPPPRFHVHSFCKTLTASDTSTH 204
NV + A+ +TDEV+AQ+TL P Q++ A P R + FCKTLTASDTSTH
Sbjct: 80 HNVTMHADVETDEVYAQMTLQPLNAQEQKDPYLPAELGVPSRQPTNYFCKTLTASDTSTH 139
Query: 205 GGFSVLRRHADECLPPLDMSKQPPTQELVAKDLHGNEWRFRHIFRGELISGQPRRHLLQS 264
GGFSV RR A++ PPLD S+QPP QEL+A+DLH NEW+FRHIFR GQP+RHLL +
Sbjct: 140 GGFSVPRRAAEKVFPPLDYSQQPPAQELMARDLHDNEWKFRHIFR-----GQPKRHLLTT 194
Query: 265 GWSVFVSSKRLVAGDAFIFLRGENGELRVGVRRAMRQQGNVPSSVISSHSMHLGVLATAW 324
GWSVFVS+KRLVAGD+ +F+ + +L +G+RRA R Q +PSSV+SS SMHLG+LA A
Sbjct: 195 GWSVFVSAKRLVAGDSVLFIWNDKNQLLLGIRRANRPQTVMPSSVLSSDSMHLGLLAAAA 254
Query: 325 HAVLTGTMFTVYYKPRTSPAEFIVPYDQYMESL-KNNYTIGMRFKMRFEGEEAPEQRFTG 383
HA T + FT++Y PR SP+EF++P +Y++++ ++GMRF+M FE EE+ +R+ G
Sbjct: 255 HAAATNSRFTIFYNPRASPSEFVIPLAKYVKAVYHTRVSVGMRFRMLFETEESSVRRYMG 314
Query: 384 TIVGIEDSDSKRWPTSKWRCLKVRWDETSNIPRPERVSPWKIEPALAPPALNPLPMP-RP 442
TI GI D D RW S WR +KV WDE++ R RVS W+IEP P + P P P R
Sbjct: 315 TITGICDLDPTRWANSHWRSVKVGWDESTAGERQPRVSLWEIEPLTTFP-MYPSPFPLRL 373
Query: 443 KRPRANVVPS 452
KRP +PS
Sbjct: 374 KRPWPPGLPS 383
Score = 74.3 bits (181), Expect = 3e-13
Identities = 69/264 (26%), Positives = 111/264 (41%), Gaps = 43/264 (16%)
Query: 551 TREGKHNMLTQWPVMPPGLSLNFLHSNMKGSAQGSDNATYQAQGNMRYSAFGDYSVLHGH 610
TR + WP P + +F HS G+ + + + G H
Sbjct: 639 TRTNSAMTSSGWPSKRPAVDSSFQHS---GAGNNNTQSVLEQLGQ-------------SH 682
Query: 611 KVENPHGNFLMPPPPPTQYESPHSRELSQKQMSAKISEAAKPKDSDCKLFGFSLLSSPTM 670
P +PP P RE S +Q E + LFG ++ SS +
Sbjct: 683 TSNVPPNAVSLPP-------FPGGRECSIEQ------EGSASDPHSHLLFGVNIDSSSLL 729
Query: 671 LEPSLSQ-RNATSETSSHMQISSQHHTFENDQKSEHSKSSKPADKLVIVDEHEKQLQTSQ 729
+ +S R+ E + F ND + ++ + +DE LQ+S+
Sbjct: 730 MPNGMSNLRSIGIEGGDSTTLPFTSSNFNNDFSGNLAMTTPSS----CIDE-SGFLQSSE 784
Query: 730 PHVKDVQLKPQSGSARSCTKVHKKGIALGRSVDLTKFSDYDELTAELDQLFEFRGELISP 789
L ++ + + KV+K G + GRS+D++KFS Y EL +EL ++F G+L P
Sbjct: 785 ------NLGSENPQSNTFVKVYKSG-SFGRSLDISKFSSYHELRSELARMFGLEGQLEDP 837
Query: 790 -QKDWLVVFTDNEGDMMLVGDDPW 812
+ W +VF D E D++L+GDDPW
Sbjct: 838 VRSGWQLVFVDRENDVLLLGDDPW 861
>At5g20730 auxin response factor 7 (ARF7)
Length = 1164
Score = 368 bits (945), Expect = e-102
Identities = 197/388 (50%), Positives = 254/388 (64%), Gaps = 11/388 (2%)
Query: 89 ELWHACAGPLVTVPREGELVFYFPQGHIEQVEASTNQASEQHMPVY-DLRPKILCRVINV 147
ELWHACAGPL+++P G LV YFPQGH EQV AS + ++ +P Y +L K++C + NV
Sbjct: 24 ELWHACAGPLISLPPAGSLVVYFPQGHSEQVAASMQKQTD-FIPSYPNLPSKLICMLHNV 82
Query: 148 MLKAEPDTDEVFAQVTLVPEPNQDENAVEKEAPPAPPPRFHVHSFCKTLTASDTSTHGGF 207
L A+P+TDEV+AQ+TL P D +A+ R FCKTLTASDTSTHGGF
Sbjct: 83 TLNADPETDEVYAQMTLQPVNKYDRDALLASDMGLKLNRQPNEFFCKTLTASDTSTHGGF 142
Query: 208 SVLRRHADECLPPLDMSKQPPTQELVAKDLHGNEWRFRHIFRGELISGQPRRHLLQSGWS 267
SV RR A++ P LD S QPP QELVAKD+H N W FRHI+RG QP+RHLL +GWS
Sbjct: 143 SVPRRAAEKIFPALDFSMQPPCQELVAKDIHDNTWTFRHIYRG-----QPKRHLLTTGWS 197
Query: 268 VFVSSKRLVAGDAFIFLRGENGELRVGVRRAMRQQGNVPSSVISSHSMHLGVLATAWHAV 327
VFVS+KRL AGD+ +F+R +L +G+RRA RQQ + SSVISS SMH+GVLA A HA
Sbjct: 198 VFVSTKRLFAGDSVLFIRDGKAQLLLGIRRANRQQPALSSSVISSDSMHIGVLAAAAHAN 257
Query: 328 LTGTMFTVYYKPRTSPAEFIVPYDQYMESLKNNYTIGMRFKMRFEGEEAPEQRFTGTIVG 387
+ FT++Y PR +PAEF+VP +Y +++ ++GMRF+M FE EE +R+ GT+ G
Sbjct: 258 ANNSPFTIFYNPRAAPAEFVVPLAKYTKAMYAQVSLGMRFRMIFETEECGVRRYMGTVTG 317
Query: 388 IEDSDSKRWPTSKWRCLKVRWDETSNIPRPERVSPWKIEPALAPPALNPLPMPRPKRPRA 447
I D D RW S+WR L++ WDE++ RP RVS W IEP L P + P P RPR
Sbjct: 318 ISDLDPVRWKNSQWRNLQIGWDESAAGDRPSRVSVWDIEPVLTPFYICPPPF---FRPRF 374
Query: 448 NVVPSSPDSSVLTREASSKVSMDPLPTS 475
+ P PD E++ K +M L S
Sbjct: 375 SGQPGMPDDET-DMESALKRAMPWLDNS 401
Score = 73.2 bits (178), Expect = 6e-13
Identities = 34/74 (45%), Positives = 52/74 (69%), Gaps = 2/74 (2%)
Query: 740 QSGSARSCTKVHKKGIALGRSVDLTKFSDYDELTAELDQLFEFRGELISPQ-KDWLVVFT 798
Q+ R+ TKV K+G ++GRS+D+ ++ YDEL +L ++F G+L PQ DW +V+
Sbjct: 1032 QTQRMRTYTKVQKRG-SVGRSIDVNRYRGYDELRHDLARMFGIEGQLEDPQTSDWKLVYV 1090
Query: 799 DNEGDMMLVGDDPW 812
D+E D++LVGDDPW
Sbjct: 1091 DHENDILLVGDDPW 1104
>At1g19220 auxin response factor (ARF) like protein
Length = 1086
Score = 367 bits (942), Expect = e-101
Identities = 196/404 (48%), Positives = 258/404 (63%), Gaps = 14/404 (3%)
Query: 89 ELWHACAGPLVTVPREGELVFYFPQGHIEQVEASTNQASEQHMPVY-DLRPKILCRVINV 147
+LWHACAGPLV++P G LV YFPQGH EQV AS + ++ +P Y +L K++C + +V
Sbjct: 23 QLWHACAGPLVSLPPVGSLVVYFPQGHSEQVAASMQKQTD-FIPNYPNLPSKLICLLHSV 81
Query: 148 MLKAEPDTDEVFAQVTLVPEPNQDENAVEKEAPPAPPPRFHVHSFCKTLTASDTSTHGGF 207
L A+ +TDEV+AQ+TL P D A+ R FCKTLTASDTSTHGGF
Sbjct: 82 TLHADTETDEVYAQMTLQPVNKYDREALLASDMGLKLNRQPTEFFCKTLTASDTSTHGGF 141
Query: 208 SVLRRHADECLPPLDMSKQPPTQELVAKDLHGNEWRFRHIFRGELISGQPRRHLLQSGWS 267
SV RR A++ PPLD S QPP QE+VAKDLH W FRHI+RG QP+RHLL +GWS
Sbjct: 142 SVPRRAAEKIFPPLDFSMQPPAQEIVAKDLHDTTWTFRHIYRG-----QPKRHLLTTGWS 196
Query: 268 VFVSSKRLVAGDAFIFLRGENGELRVGVRRAMRQQGNVPSSVISSHSMHLGVLATAWHAV 327
VFVS+KRL AGD+ +F+R E +L +G+RRA RQ + SSVISS SMH+G+LA A HA
Sbjct: 197 VFVSTKRLFAGDSVLFVRDEKSQLMLGIRRANRQTPTLSSSVISSDSMHIGILAAAAHAN 256
Query: 328 LTGTMFTVYYKPRTSPAEFIVPYDQYMESLKNNYTIGMRFKMRFEGEEAPEQRFTGTIVG 387
+ FT+++ PR SP+EF+VP +Y ++L ++GMRF+M FE E+ +R+ GT+ G
Sbjct: 257 ANSSPFTIFFNPRASPSEFVVPLAKYNKALYAQVSLGMRFRMMFETEDCGVRRYMGTVTG 316
Query: 388 IEDSDSKRWPTSKWRCLKVRWDETSNIPRPERVSPWKIEPALAPPALNPLPMPRPKRPRA 447
I D D RW S+WR L+V WDE++ RP RVS W+IEP + P + P P RPK PR
Sbjct: 317 ISDLDPVRWKGSQWRNLQVGWDESTAGDRPSRVSIWEIEPVITPFYICPPPFFRPKYPRQ 376
Query: 448 NVVPSSPDSSVLTREASSKVSMDPLPTSGFQRVLQGQESSTLRG 491
P PD + A + +P G ++ +SS G
Sbjct: 377 ---PGMPDDELDMENAFKRA----MPWMGEDFGMKDAQSSMFPG 413
Score = 76.6 bits (187), Expect = 5e-14
Identities = 36/74 (48%), Positives = 54/74 (72%), Gaps = 2/74 (2%)
Query: 740 QSGSARSCTKVHKKGIALGRSVDLTKFSDYDELTAELDQLFEFRGELISP-QKDWLVVFT 798
Q+ R+ TKV K+G ++GRS+D+T++S YDEL +L ++F G+L P DW +V+T
Sbjct: 953 QTQRMRTYTKVQKRG-SVGRSIDVTRYSGYDELRHDLARMFGIEGQLEDPLTSDWKLVYT 1011
Query: 799 DNEGDMMLVGDDPW 812
D+E D++LVGDDPW
Sbjct: 1012 DHENDILLVGDDPW 1025
>At1g34390 auxin response factor, putative
Length = 600
Score = 363 bits (933), Expect = e-100
Identities = 184/365 (50%), Positives = 251/365 (68%), Gaps = 17/365 (4%)
Query: 78 IFPEAEAALYRELWHACAGPLVTVPREGELVFYFPQGHIEQVEASTNQASEQHMPVYDLR 137
I +++ +Y +LW CAGPL +P+ GE ++YFPQG+IE VEAST + + P+ DL
Sbjct: 16 IIDGSKSYMYEQLWKLCAGPLCDIPKLGEKIYYFPQGNIELVEASTREELNELKPICDLP 75
Query: 138 PKILCRVINVMLKAEPDTDEVFAQVTLVPEPNQDENAVEKEAPPAPPPRFHVHSFCKTLT 197
K+ CRVI + LK E ++DE +A++TL+P+ Q + E P V+SF K LT
Sbjct: 76 SKLQCRVIAIQLKVENNSDETYAEITLMPDTTQVVIPTQNENQFRPL----VNSFTKVLT 131
Query: 198 ASDTSTHGGFSVLRRHADECLPPLDMSKQPPTQELVAKDLHGNEWRFRHIFRGELISGQP 257
ASDTS GGF V ++HA ECLPPLDMS+ PTQEL+A DLHGN+WRF H +RG P
Sbjct: 132 ASDTS--GGFFVPKKHAIECLPPLDMSQPLPTQELLATDLHGNQWRFNHNYRGT-----P 184
Query: 258 RRHLLQSGWSVFVSSKRLVAGDAFIFLRGENGELRVGVRRAMRQQGNVPSSVISSHSMHL 317
+RHLL +GW+ F +SK+LVAGD +F+RGE GELRVG+RRA QQGN+PSS+IS SM
Sbjct: 185 QRHLLTTGWNAFTTSKKLVAGDVIVFVRGETGELRVGIRRAGHQQGNIPSSIISIESMRH 244
Query: 318 GVLATAWHAVLTGTMFTVYYKPRTSPAEFIVPYDQYMESLKNNYTIGMRFKMRFEGEEAP 377
GV+A+A HA MF V YKP ++FIV YD++++++ N + +G RF MRFEG++
Sbjct: 245 GVIASAKHAFDNQCMFIVVYKPSIRSSQFIVSYDKFLDAVNNKFNVGSRFTMRFEGDDFS 304
Query: 378 EQRFTGTIVGIEDSDSKRWPTSKWRCLKVRWDETSNIPRPERVSPWKIEPALAPPALNPL 437
E+R+ GTI+G+ D S W S+WR L+V+WDE ++ RP +VSPW+IE + PALN
Sbjct: 305 ERRYFGTIIGVSDF-SPHWKCSEWRNLEVQWDEFASFSRPNKVSPWEIEHLM--PALN-- 359
Query: 438 PMPRP 442
+PRP
Sbjct: 360 -VPRP 363
Score = 79.0 bits (193), Expect = 1e-14
Identities = 36/79 (45%), Positives = 55/79 (69%), Gaps = 2/79 (2%)
Query: 734 DVQLKPQSGSARSCTKVHKKGIALGRSVDLTKFSDYDELTAELDQLFEFRGELISPQKDW 793
++Q K Q S R+CTKV +G+ + R+VDL+ + YD+L EL++LF+ +G+L + W
Sbjct: 501 EIQSK-QFSSTRTCTKVQMQGVTIERAVDLSVLNGYDQLILELEELFDLKGQL-QTRNQW 558
Query: 794 LVVFTDNEGDMMLVGDDPW 812
+ FTD++ D MLVGDDPW
Sbjct: 559 EIAFTDSDDDKMLVGDDPW 577
>At1g35240 hypothetical protein
Length = 615
Score = 359 bits (921), Expect = 4e-99
Identities = 178/358 (49%), Positives = 249/358 (68%), Gaps = 14/358 (3%)
Query: 78 IFPEAEAALYRELWHACAGPLVTVPREGELVFYFPQGHIEQVEASTNQASEQHMPVYDLR 137
I +++ +Y +LW CAGPL +P+ GE V+YFPQG+IE V+AST + + P+ DL
Sbjct: 16 IIDGSKSYMYEQLWKLCAGPLCDIPKLGENVYYFPQGNIELVDASTREELNELQPICDLP 75
Query: 138 PKILCRVINVMLKAEPDTDEVFAQVTLVPEPNQDENAVEKEAPPAPPPRFHVHSFCKTLT 197
K+ CRVI + LK E ++DE +A++TL+P+ Q + E P V+SF K LT
Sbjct: 76 SKLQCRVIAIHLKVENNSDETYAEITLMPDTTQVVIPTQSENQFRPL----VNSFTKVLT 131
Query: 198 ASDTSTHGGFSVLRRHADECLPPLDMSKQPPTQELVAKDLHGNEWRFRHIFRGELISGQP 257
ASDTS +GGF V ++HA ECLPPLDMS+ P QEL+AKDLHGN+WRFRH +RG P
Sbjct: 132 ASDTSAYGGFFVPKKHAIECLPPLDMSQPLPAQELLAKDLHGNQWRFRHSYRGT-----P 186
Query: 258 RRHLLQSGWSVFVSSKRLVAGDAFIFLRGENGELRVGVRRAMRQQGNVPSSVISSHSMHL 317
+RH L +GW+ F +SK+LV GD +F+RGE GELRVG+RRA QQGN+PSS++S M
Sbjct: 187 QRHSLTTGWNEFTTSKKLVKGDVIVFVRGETGELRVGIRRARHQQGNIPSSIVSIDCMRH 246
Query: 318 GVLATAWHAVLTGTMFTVYYKPRTSPAEFIVPYDQYMESLKNNYTIGMRFKMRFEGEEAP 377
GV+A+A HA+ +F V YKPR+S +FIV YD++++++ N + +G RF MRFEG++
Sbjct: 247 GVIASAKHALDNQCIFIVVYKPRSS--QFIVSYDKFLDAMNNKFIVGSRFTMRFEGDDFS 304
Query: 378 EQRFTGTIVGIEDSDSKRWPTSKWRCLKVRWDETSNIPRPERVSPWKIEPALAPPALN 435
E+R+ GTI+G+ D S W S+WR L+V+WDE ++ RP +VSPW+IE ++ ALN
Sbjct: 305 ERRYFGTIIGVNDF-SPHWKCSEWRSLEVQWDEFASFSRPNKVSPWEIEHLMS--ALN 359
Score = 83.2 bits (204), Expect = 6e-16
Identities = 38/79 (48%), Positives = 57/79 (72%), Gaps = 2/79 (2%)
Query: 734 DVQLKPQSGSARSCTKVHKKGIALGRSVDLTKFSDYDELTAELDQLFEFRGELISPQKDW 793
++Q K Q GS R+CTKV +G+ +GR+VDL+ + YD+L EL++LF+ +G+L + W
Sbjct: 510 EIQSK-QFGSTRTCTKVQMQGVTIGRAVDLSVLNGYDQLILELEKLFDLKGQL-QTRNQW 567
Query: 794 LVVFTDNEGDMMLVGDDPW 812
+ FTD++G MLVGDDPW
Sbjct: 568 KIAFTDSDGYEMLVGDDPW 586
>At1g34410 auxin response factor, putative
Length = 620
Score = 359 bits (921), Expect = 4e-99
Identities = 182/395 (46%), Positives = 262/395 (66%), Gaps = 20/395 (5%)
Query: 82 AEAALYRELWHACAGPLVTVPREGELVFYFPQGHIEQVEASTNQASEQHMPVYDLRPKIL 141
+++ +Y +LW CAGPL +P+ GE V+YFPQG+IE V+AST + + P+ DL K+
Sbjct: 32 SKSYMYEQLWKLCAGPLCDIPKLGENVYYFPQGNIELVQASTREELNELQPICDLPSKLQ 91
Query: 142 CRVINVMLKAEPDTDEVFAQVTLVPEPNQDENAVEKEAPPAPPPRFHVHSFCKTLTASDT 201
CRVI + LK E ++DE++A++TL+P+ Q + E P V+SF K LTASDT
Sbjct: 92 CRVIAIHLKVENNSDEIYAEITLMPDTTQVVIPTQSENRFRPL----VNSFTKVLTASDT 147
Query: 202 STHGGFSVLRRHADECLPPLDMSKQPPTQELVAKDLHGNEWRFRHIFRGELISGQPRRHL 261
S +GGFSV ++HA ECLPPLDMS+ P QE++A DLH N+WRFRH +RG P+RH
Sbjct: 148 SAYGGFSVPKKHAIECLPPLDMSQPLPAQEILAIDLHDNQWRFRHNYRGT-----PQRHS 202
Query: 262 LQSGWSVFVSSKRLVAGDAFIFLRGENGELRVGVRRAMRQQGNVPSSVISSHSMHLGVLA 321
L +GW+ F++SK+LV GD +F+RGE GELRVG+RRA QQGN+PSS++S M GV+A
Sbjct: 203 LTTGWNEFITSKKLVKGDVIVFVRGETGELRVGIRRARHQQGNIPSSIVSIDCMRHGVIA 262
Query: 322 TAWHAVLTGTMFTVYYKPRTSPAEFIVPYDQYMESLKNNYTIGMRFKMRFEGEEAPEQRF 381
+A HA +F V YKP ++FIV YD++++++ N + +G RF MRFEG++ E+R+
Sbjct: 263 SAKHAFDNQCIFIVVYKPSIRSSQFIVSYDKFLDAVNNKFNVGSRFTMRFEGDDFSERRY 322
Query: 382 TGTIVGIEDSDSKRWPTSKWRCLKVRWDETSNIPRPERVSPWKIEPALAPPALNPLPMPR 441
GTI+G+ D S W S+WR L+V+WDE ++ RP +VSPW+IE + PALN
Sbjct: 323 FGTIIGVSDF-SPHWKCSEWRSLEVQWDEFASFSRPNKVSPWEIEHLV--PALNV----- 374
Query: 442 PKRPRANVVPSSPDSSVLTREASSKVSMDPLPTSG 476
PR++++ + V +SS + P+ T G
Sbjct: 375 ---PRSSLLKNKRLREVNEFGSSSSHLLPPILTQG 406
Score = 81.3 bits (199), Expect = 2e-15
Identities = 37/79 (46%), Positives = 57/79 (71%), Gaps = 2/79 (2%)
Query: 734 DVQLKPQSGSARSCTKVHKKGIALGRSVDLTKFSDYDELTAELDQLFEFRGELISPQKDW 793
++Q K Q S+R+CTKV +G+ +GR+VDL+ + YD+L EL++LF+ +G+L + W
Sbjct: 515 EIQSK-QFSSSRTCTKVQMQGVTIGRAVDLSVLNGYDQLILELEKLFDIKGQL-QTRNQW 572
Query: 794 LVVFTDNEGDMMLVGDDPW 812
+ FTD++G MLVGDDPW
Sbjct: 573 KIAFTDSDGYEMLVGDDPW 591
>At1g19850 transcription factor monopteros (MP)
Length = 902
Score = 359 bits (921), Expect = 4e-99
Identities = 185/348 (53%), Positives = 237/348 (67%), Gaps = 23/348 (6%)
Query: 89 ELWHACAGPLVTVPREGELVFYFPQGHIEQVEASTNQASEQHMPVY-DLRPKILCRVINV 147
ELWHACAGPLV +P+ G LV+YF QGH EQV ST +++ +P Y +L +++C+V NV
Sbjct: 54 ELWHACAGPLVCLPQVGSLVYYFSQGHSEQVAVSTRRSATTQVPNYPNLPSQLMCQVHNV 113
Query: 148 MLKAEPDTDEVFAQVTLVPEPNQDENAVEKEAPPAPPPRFHV--------HSFCKTLTAS 199
L A+ D+DE++AQ++L P V E P P F + FCKTLTAS
Sbjct: 114 TLHADKDSDEIYAQMSLQP--------VHSERDVFPVPDFGMLRGSKHPTEFFCKTLTAS 165
Query: 200 DTSTHGGFSVLRRHADECLPPLDMSKQPPTQELVAKDLHGNEWRFRHIFRGELISGQPRR 259
DTSTHGGFSV RR A++ PPLD S QPPTQELV +DLH N W FRHI+RG QP+R
Sbjct: 166 DTSTHGGFSVPRRAAEKLFPPLDYSAQPPTQELVVRDLHENTWTFRHIYRG-----QPKR 220
Query: 260 HLLQSGWSVFVSSKRLVAGDAFIFLRGENGELRVGVRRAMRQQGNVPSSVISSHSMHLGV 319
HLL +GWS+FV SKRL AGD+ +F+R E +L VGVRRA RQQ +PSSV+S+ SMH+GV
Sbjct: 221 HLLTTGWSLFVGSKRLRAGDSVLFIRDEKSQLMVGVRRANRQQTALPSSVLSADSMHIGV 280
Query: 320 LATAWHAVLTGTMFTVYYKPRTSPAEFIVPYDQYMESL-KNNYTIGMRFKMRFEGEEAPE 378
LA A HA T F ++Y PR PAEF++P +Y +++ + ++GMRF M FE E++ +
Sbjct: 281 LAAAAHATANRTPFLIFYNPRACPAEFVIPLAKYRKAICGSQLSVGMRFGMMFETEDSGK 340
Query: 379 QRFTGTIVGIEDSDSKRWPTSKWRCLKVRWDETSNIPRPERVSPWKIE 426
+R+ GTIVGI D D RWP SKWR L+V WDE +P RVSPW IE
Sbjct: 341 RRYMGTIVGISDLDPLRWPGSKWRNLQVEWDEPGCNDKPTRVSPWDIE 388
Score = 69.3 bits (168), Expect = 8e-12
Identities = 33/69 (47%), Positives = 47/69 (67%), Gaps = 2/69 (2%)
Query: 745 RSCTKVHKKGIALGRSVDLTKFSDYDELTAELDQLFEFRGELISPQKD-WLVVFTDNEGD 803
R+ TKV K G ++GRS+D+T F DY+EL + ++ +F G L PQ W +V+ D E D
Sbjct: 793 RTYTKVQKTG-SVGRSIDVTSFKDYEELKSAIECMFGLEGLLTHPQSSGWKLVYVDYESD 851
Query: 804 MMLVGDDPW 812
++LVGDDPW
Sbjct: 852 VLLVGDDPW 860
>At2g33860 auxin response transcription factor 3 (ETTIN/ARF3)
Length = 608
Score = 355 bits (910), Expect = 8e-98
Identities = 191/406 (47%), Positives = 253/406 (62%), Gaps = 25/406 (6%)
Query: 89 ELWHACAGPLVTVPREGELVFYFPQGHIEQVEASTNQASEQHMPVYDLRPKILCRVINVM 148
ELWHACAGPL+++P+ G LV YFPQGH+EQ A + +Y L P + CR+++V
Sbjct: 54 ELWHACAGPLISLPKRGSLVLYFPQGHLEQ-------APDFSAAIYGLPPHVFCRILDVK 106
Query: 149 LKAEPDTDEVFAQVTLVPEPNQDENAVEK--------EAPPAPPPRFHV-HSFCKTLTAS 199
L AE TDEV+AQV+L+PE E V + E R + H FCKTLTAS
Sbjct: 107 LHAETTTDEVYAQVSLLPESEDIERKVREGIIDVDGGEEDYEVLKRSNTPHMFCKTLTAS 166
Query: 200 DTSTHGGFSVLRRHADECLPPLDMSKQPPTQELVAKDLHGNEWRFRHIFRGELISGQPRR 259
DTSTHGGFSV RR A++C PPLD S+ P+QEL+A+DLHG EWRFRHI+RG QPRR
Sbjct: 167 DTSTHGGFSVPRRAAEDCFPPLDYSQPRPSQELLARDLHGLEWRFRHIYRG-----QPRR 221
Query: 260 HLLQSGWSVFVSSKRLVAGDAFIFLRGENGELRVGVRRAMRQQGNVPSSVISSHSMHLGV 319
HLL +GWS FV+ K+LV+GDA +FLRG++G+LR+GVRRA + +G S + +M+
Sbjct: 222 HLLTTGWSAFVNKKKLVSGDAVLFLRGDDGKLRLGVRRASQIEGTAALSAQYNQNMNHNN 281
Query: 320 LATAWHAVLTGTMFTVYYKPRTSPAEFIVPYDQYMESLKNNYTIGMRFKMRFEGEEAPEQ 379
+ HA+ T ++F++ Y P+ S + FI+P ++++ + + IGMRFK R E E+A E+
Sbjct: 282 FSEVAHAISTHSVFSISYNPKASWSNFIIPAPKFLKVVDYPFCIGMRFKARVESEDASER 341
Query: 380 RFTGTIVGIEDSDSKRWPTSKWRCLKVRWDETSNIPRPERVSPWKIEPALAPPALNPLPM 439
R G I GI D D RWP SKWRCL VRWD+ +RVSPW+IEP+ +
Sbjct: 342 RSPGIISGISDLDPIRWPGSKWRCLLVRWDDIVANGHQQRVSPWEIEPSGSISNSGSFVT 401
Query: 440 PRPKRPRANVVPSSPDSSVLTREASSKVSMDPLPTSGFQRVLQGQE 485
PKR R PD V + + D + FQRVLQGQE
Sbjct: 402 TGPKRSRIGFSSGKPDIPV----SEGIRATDFEESLRFQRVLQGQE 443
>At1g34310 hypothetical protein
Length = 591
Score = 350 bits (898), Expect = 2e-96
Identities = 188/414 (45%), Positives = 261/414 (62%), Gaps = 35/414 (8%)
Query: 78 IFPEAEAALYRELWHACAGPLVTVPREGELVFYFPQGHIEQVEASTNQASEQHMPVYDLR 137
I +++ +Y +LW CAGPL +P+ GE V+YFPQGHIE VE ST + + P+ DL
Sbjct: 16 IIDGSKSYVYEQLWKLCAGPLCDIPKLGEKVYYFPQGHIELVETSTREELNELQPICDLP 75
Query: 138 PKILCRVINVMLKAEPDTDEVFAQVTLVPEPNQDENAVEKEAPPAPPPRFH--VHSFCKT 195
K+ CRVI + LK E ++DE +A++TL+P+ EN ++ P +F V+SF K
Sbjct: 76 SKLQCRVIAIHLKVENNSDETYAEITLMPDTTVSEN-LQVVIPTQNENQFRPLVNSFTKV 134
Query: 196 LTASDTSTHGGFSVLRRHADECLPPLDMSKQPPTQELVAKDLHGNEWRFRHIFRGELISG 255
LTASDTS HGGF V ++HA ECLP LDMS+ P QEL+A DLHGN+WRF H +RG
Sbjct: 135 LTASDTSAHGGFFVPKKHAIECLPSLDMSQPLPAQELLAIDLHGNQWRFNHNYRGT---- 190
Query: 256 QPRRHLLQSGWSVFVSSKRLVAGDAFIFLRGENGELRVGVRRAMRQQGNVPSSVISSHSM 315
P+RHLL +GW+ F +SK+LVAGD +F+RGE GELRVG+RRA QQGN+PSS++S M
Sbjct: 191 -PQRHLLTTGWNAFTTSKKLVAGDVIVFVRGETGELRVGIRRARHQQGNIPSSIVSIDCM 249
Query: 316 HLGVLATAWHAVLTGTMFTVYYKPRTSPAEFIVPYDQYMESLKNNYTIGMRFKMRFEGEE 375
GV+A+A HA MFTV YKP YD++++++ N + +G RF MR EG++
Sbjct: 250 RHGVVASAKHAFDNQCMFTVVYKP---------SYDKFLDAVNNKFNVGSRFTMRLEGDD 300
Query: 376 APEQRFTGTIVGIEDSDSKRWPTSKWRCLKVRWDETSNIPRPERVSPWKIEPALAPPALN 435
E+R GTI+G+ D S W S+WR L+V+WDE ++ P P++VSPW IE + PA+N
Sbjct: 301 FSERRCFGTIIGVSDF-SPHWKCSEWRSLEVQWDEFTSFPGPKKVSPWDIEHLM--PAIN 357
Query: 436 PLPMPRPKRPRANVVPSSPDSSVLTREASSKVSMDPLPTSGFQRVLQGQESSTL 489
PR+ ++ + V +SS + P+ T QGQE+ L
Sbjct: 358 V--------PRSFLLKNKRLREVNEIGSSSSHLLPPILT-------QGQENEQL 396
Score = 81.3 bits (199), Expect = 2e-15
Identities = 35/73 (47%), Positives = 53/73 (71%), Gaps = 1/73 (1%)
Query: 740 QSGSARSCTKVHKKGIALGRSVDLTKFSDYDELTAELDQLFEFRGELISPQKDWLVVFTD 799
Q S+R+CTKV +G+ +GR+VDL+ + YD+L EL++LF+ +G+L + W + FTD
Sbjct: 504 QFSSSRTCTKVQMQGVTIGRAVDLSVLNGYDQLILELEKLFDIKGQL-QTRNQWEIAFTD 562
Query: 800 NEGDMMLVGDDPW 812
++ D MLVGDDPW
Sbjct: 563 SDEDKMLVGDDPW 575
>At1g35520 auxin response factor, putative
Length = 576
Score = 348 bits (893), Expect = 7e-96
Identities = 185/399 (46%), Positives = 258/399 (64%), Gaps = 24/399 (6%)
Query: 78 IFPEAEAALYRELWHACAGPLVTVPREGELVFYFPQGHIEQVEASTNQASEQHMPVYDLR 137
I +++ +Y +LW CAGPL +P+ GE V+YFPQG+IE VEAST + + P+ DL
Sbjct: 16 IIDRSKSYMYEQLWKLCAGPLCDIPKLGEKVYYFPQGNIELVEASTREELNELQPICDLP 75
Query: 138 PKILCRVINVMLKAEPDTDEVFAQVTLVPEPNQDENAVEKEAPPAPPPRFHVHSFCKTLT 197
K+ CRVI + LK E ++DE +A++TL+P+ Q + E P V+SF K LT
Sbjct: 76 SKLQCRVIAIHLKVENNSDETYAKITLMPDTTQVVIPTQNENQFRPL----VNSFTKVLT 131
Query: 198 ASDTSTHGGFSVLRRHADECLPPLDMSKQPPTQELVAKDLHGNEWRFRHIFRGELISGQP 257
ASD S +G FSV ++HA ECLPPLDMS+ P QEL+A DLHGN+W FRH +RG P
Sbjct: 132 ASDISANGVFSVPKKHAIECLPPLDMSQPLPAQELLAIDLHGNQWSFRHSYRGT-----P 186
Query: 258 RRHLLQSGWSVFVSSKRLVAGDAFIFLRGENGELRVGVRRAMRQQGNVPSSVISSHSMHL 317
+RHLL +GW+ F +SK+LV GD +F+RGE GELRVG+RRA QQGN+PSS++S M
Sbjct: 187 QRHLLTTGWNEFTTSKKLVKGDVIVFVRGETGELRVGIRRARHQQGNIPSSIVSIDCMRH 246
Query: 318 GVLATAWHAVLTGTMFTVYYKPRTSPAEFIVPYDQYMESLKNNYTIGMRFKMRFEGEEAP 377
GV+A+A HA MF V YKPR+S +FIV YD++++++ N + +G RF MRFEG++
Sbjct: 247 GVIASAKHAFDNQCMFIVVYKPRSS--QFIVSYDKFLDAVNNKFNVGSRFTMRFEGDDLS 304
Query: 378 EQRFTGTIVGIEDSDSKRWPTSKWRCLKVRWDETSNIPRPERVSPWKIEPALAPPALNPL 437
E+R+ GTI+G+ + S W S WR L+ WDE ++ RP +VSPW+IE + PALN
Sbjct: 305 ERRYFGTIIGVSNF-SPHWKCSDWRSLE--WDEFASFLRPNKVSPWEIEHLM--PALNV- 358
Query: 438 PMPRPKRPRANVVPSSPDSSVLTREASSKVSMDPLPTSG 476
PR++ + + V +SS + P+ T G
Sbjct: 359 -------PRSSFLKNKRLREVNEFGSSSSHLLPPILTQG 390
Score = 79.7 bits (195), Expect = 6e-15
Identities = 40/89 (44%), Positives = 60/89 (66%), Gaps = 3/89 (3%)
Query: 724 QLQTSQPHVKDVQLKPQSGSARSCTKVHKKGIALGRSVDLTKFSDYDELTAELDQLFEFR 783
Q QT + K +Q K Q S R+CTKV +G+ +GR+VDL+ + YD+L EL++LF+ +
Sbjct: 490 QSQTLRSPTK-IQSK-QFSSTRTCTKVQMQGVTIGRAVDLSVLNGYDQLILELEKLFDLK 547
Query: 784 GELISPQKDWLVVFTDNEGDMMLVGDDPW 812
G+L + W ++FT ++ D MLVGDDPW
Sbjct: 548 GQL-QTRNQWKIIFTGSDEDEMLVGDDPW 575
>At5g62010 ARF1-binding protein
Length = 454
Score = 324 bits (830), Expect = 1e-88
Identities = 191/413 (46%), Positives = 250/413 (60%), Gaps = 57/413 (13%)
Query: 439 MPRPKRPRANVVPSSPDSSVLTREASSKVSMDPLPTSGFQRVLQGQESSTLRGNLAESND 498
MPRPKRPR+N+ PSSPDSS+LTRE ++K +MDPLP SG RVLQGQE STLR ES +
Sbjct: 1 MPRPKRPRSNIAPSSPDSSMLTREGTTKANMDPLPASGLSRVLQGQEYSTLRTKHTESVE 60
Query: 499 SYTAEKSVAWTPATDEEKMDAVSTSRRYGSENWMPMSRQEPTYSDLLSGFG--------- 549
E SV W + D++K+D VS SRRYGSENWM +R EPTY+DLLSGFG
Sbjct: 61 CDAPENSVVWQSSADDDKVDVVSGSRRYGSENWMSSARHEPTYTDLLSGFGTNIDPSHGQ 120
Query: 550 -------------------STREGKHNML-TQWPVMPPGLSLNFLHSNMKGSAQGSDNAT 589
S EGK + L QW ++ GLSL LH + K A + +A+
Sbjct: 121 RIPFYDHSSSPSMPAKRILSDSEGKFDYLANQWQMIHSGLSLK-LHESPKVPA--ATDAS 177
Query: 590 YQAQGNMRYSAFGDYSVLHGHKVENPHGNFLMPPPPPTQYE-------SPHSRELSQKQM 642
Q + N++YS +Y VL+G EN GN+ + P YE +RE KQ
Sbjct: 178 LQGRCNVKYS---EYPVLNGLSTENAGGNWPIRPRALNYYEEVVNAQAQAQAREQVTKQP 234
Query: 643 SAKISEAAKPKDSDCKLFGFSLLSSPTMLEPSLSQRNATSETSSHMQISSQHHTFENDQK 702
E AK ++ +C+LFG L ++ + ++SQRN ++ + QI+S +
Sbjct: 235 FTIQEETAKSREGNCRLFGIPLTNNMNGTDSTMSQRNNLNDAAGLTQIASP----KVQDL 290
Query: 703 SEHSKSSKPADKLVIVDEHEKQ---LQTSQPHVKDVQLKPQSGSARSCTKVHKKGIALGR 759
S+ SK SK ++H +Q QT+ PH KD Q K + S+RSCTKVHK+GIALGR
Sbjct: 291 SDQSKGSKS------TNDHREQGRPFQTNNPHPKDAQTK--TNSSRSCTKVHKQGIALGR 342
Query: 760 SVDLTKFSDYDELTAELDQLFEFRGELISPQKDWLVVFTDNEGDMMLVGDDPW 812
SVDL+KF +Y+EL AELD+LFEF GEL++P+KDWL+V+TD E DMMLVGDDPW
Sbjct: 343 SVDLSKFQNYEELVAELDRLFEFNGELMAPKKDWLIVYTDEENDMMLVGDDPW 395
>At1g35540 auxin response factor, putative
Length = 661
Score = 318 bits (816), Expect = 6e-87
Identities = 172/405 (42%), Positives = 239/405 (58%), Gaps = 68/405 (16%)
Query: 78 IFPEAEAALYRELWHACAGPLVTVPREGELVFYFPQGHIEQVEA---------------- 121
I +++ +Y +LW CAGPL +P+ GE V+YFPQGHIE V +
Sbjct: 16 IIDGSKSYMYEQLWKLCAGPLCDIPKLGEKVYYFPQGHIELVSSLSLYSFSLLSLSLSLS 75
Query: 122 -------STNQASEQHMPVYDLRPKILCRVINVML------------------------- 149
S + + ++ L P L ++++L
Sbjct: 76 LSLSLSLSLSLSPFSLFSLFSLSPLSLSLSLSLLLSHVEASTREELNELQPICDFPSKLQ 135
Query: 150 --------KAEPDTDEVFAQVTLVPEPNQDENAVEKEAPPAPPPRFHVHSFCKTLTASDT 201
K E ++DE +A++TL+P+ Q + + P V+SF K LTASDT
Sbjct: 136 CRVIAIQLKVENNSDETYAEITLMPDTTQVVIPTQNQNQFRP----LVNSFTKVLTASDT 191
Query: 202 STHGGFSVLRRHADECLPPLDMSKQPPTQELVAKDLHGNEWRFRHIFRGELISGQPRRHL 261
S HGGFSV ++HA ECLPPLDMS+ PTQE++A DLHGN+WRFRHI+R G +RHL
Sbjct: 192 SVHGGFSVPKKHAIECLPPLDMSQPLPTQEILAIDLHGNQWRFRHIYR-----GTAQRHL 246
Query: 262 LQSGWSVFVSSKRLVAGDAFIFLRGENGELRVGVRRAMRQQGNVPSSVISSHSMHLGVLA 321
L GW+ F +SK+LV GD +F+RGE GELRVG+RRA QQGN+PSS++S SM G++A
Sbjct: 247 LTIGWNAFTTSKKLVEGDVIVFVRGETGELRVGIRRAGHQQGNIPSSIVSIESMRHGIIA 306
Query: 322 TAWHAVLTGTMFTVYYKPRTSPAEFIVPYDQYMESLKNNYTIGMRFKMRFEGEEAPEQRF 381
+A HA MF V YKPR+S +FIV YD++++ + N + +G RF MRFEG++ E+R
Sbjct: 307 SAKHAFDNQCMFIVVYKPRSS--QFIVSYDKFLDVVNNKFNVGSRFTMRFEGDDFSERRS 364
Query: 382 TGTIVGIEDSDSKRWPTSKWRCLKVRWDETSNIPRPERVSPWKIE 426
GTI+G+ D S W S+WR L+V+WDE ++ PRP +VSPW IE
Sbjct: 365 FGTIIGVSDF-SPHWKCSEWRSLEVQWDEFASFPRPNQVSPWDIE 408
Score = 80.9 bits (198), Expect = 3e-15
Identities = 37/79 (46%), Positives = 56/79 (70%), Gaps = 2/79 (2%)
Query: 734 DVQLKPQSGSARSCTKVHKKGIALGRSVDLTKFSDYDELTAELDQLFEFRGELISPQKDW 793
++Q K Q S R+CTKV +G+ +GR+VDL+ + YD+L EL++LF+ +G+L + W
Sbjct: 556 EIQSK-QFSSTRTCTKVQMQGVTIGRAVDLSVLNGYDQLILELEKLFDLKGQL-QARNQW 613
Query: 794 LVVFTDNEGDMMLVGDDPW 812
+ FT+NE D MLVG+DPW
Sbjct: 614 EIAFTNNEEDKMLVGEDPW 632
>At1g34170 putative protein
Length = 505
Score = 282 bits (721), Expect = 6e-76
Identities = 146/343 (42%), Positives = 202/343 (58%), Gaps = 69/343 (20%)
Query: 86 LYRELWHACAGPLVTVPREGELVFYFPQGHIEQVEASTNQASEQHMPVYDLRPKILCRVI 145
+Y +LW+ CAGPL +P+ GE V+YFPQGHIE +E ST + P++DL K+ CRV+
Sbjct: 24 MYEKLWNICAGPLCVLPKPGEKVYYFPQGHIELIENSTRDELDHIRPIFDLPSKLRCRVV 83
Query: 146 NVMLKAEPDTDEVFAQVTLVPEPNQDENAVEKEAPPAPPPRFHVHSFCKTLTASDTSTHG 205
+ K + +TDEV+AQ++L+P+
Sbjct: 84 AIDRKVDKNTDEVYAQISLMPDTT------------------------------------ 107
Query: 206 GFSVLRRHADECLPPLDMSKQPPTQELVAKDLHGNEWRFRHIFRGELISGQPRRHLLQSG 265
DMS+ TQ LVAKDL+G EW F+H+FRG P+RH+ SG
Sbjct: 108 ----------------DMSQPISTQNLVAKDLYGQEWSFKHVFRGT-----PQRHMFTSG 146
Query: 266 --WSVFVSSKRLVAGDAFIFLRGENGELRVGVRRAMRQQGNVPSSVISSHSMHLGVLATA 323
WSVF ++KRL+ GD F+ LRGENGELR G+RRA QQG++PSSVIS++ M GV+A+
Sbjct: 147 GGWSVFATTKRLIVGDIFVLLRGENGELRFGIRRAKHQQGHIPSSVISANCMQHGVIASV 206
Query: 324 WHAVLTGTMFTVYYKPRTSPAEFIVPYDQYMESLKNNYTIGMRFKMRFEGEEAPEQRFTG 383
+A T MF V YKP YD++++++ NNY +G RF+M+FEG++ E+R+ G
Sbjct: 207 VNAFKTKCMFNVVYKP---------SYDKFVDAMNNNYIVGSRFRMQFEGKDFSEKRYDG 257
Query: 384 TIVGIEDSDSKRWPTSKWRCLKVRWDETSNIPRPERVSPWKIE 426
TI+G+ D S W S+WR LKV+WDE S RP +VSPW IE
Sbjct: 258 TIIGVNDM-SPHWKDSEWRSLKVQWDELSPFLRPNQVSPWDIE 299
Score = 76.3 bits (186), Expect = 7e-14
Identities = 40/101 (39%), Positives = 60/101 (58%), Gaps = 4/101 (3%)
Query: 715 LVIVDEHEKQLQTSQPHVKDVQLKPQSGSAR---SCTKVHKKGIALGRSVDLTKFSDYDE 771
L I +E+ Q QP +D+ + + TKVH +G+A+ R+VDLT Y++
Sbjct: 359 LAIPNENYNSDQMIQPRKEDITTEATTSCLLFGVDLTKVHMQGVAISRAVDLTAMHGYNQ 418
Query: 772 LTAELDQLFEFRGELISPQKDWLVVFTDNEGDMMLVGDDPW 812
L +L++LF+ + EL + W +VFT+NEG MLVGDDPW
Sbjct: 419 LIQKLEELFDLKDEL-RTRNQWEIVFTNNEGAEMLVGDDPW 458
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.322 0.136 0.426
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 19,841,006
Number of Sequences: 26719
Number of extensions: 916601
Number of successful extensions: 4490
Number of sequences better than 10.0: 104
Number of HSP's better than 10.0 without gapping: 56
Number of HSP's successfully gapped in prelim test: 50
Number of HSP's that attempted gapping in prelim test: 4192
Number of HSP's gapped (non-prelim): 189
length of query: 830
length of database: 11,318,596
effective HSP length: 108
effective length of query: 722
effective length of database: 8,432,944
effective search space: 6088585568
effective search space used: 6088585568
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.9 bits)
S2: 64 (29.3 bits)
Medicago: description of AC126794.7


