

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC126792.3 + phase: 0
(987 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
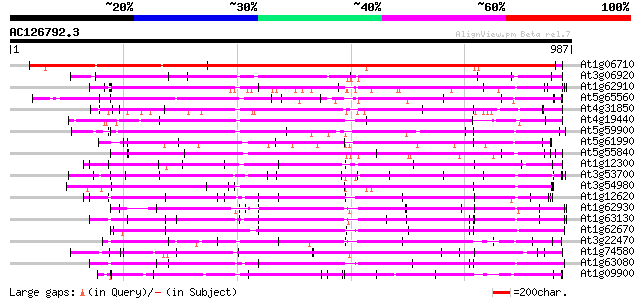
Score E
Sequences producing significant alignments: (bits) Value
At1g06710 hypothetical protein 1157 0.0
At3g06920 hypothetical protein 298 1e-80
At1g62910 unknown protein 298 1e-80
At5g65560 putative protein 293 3e-79
At4g31850 putative protein 286 4e-77
At4g19440 unknown protein 285 7e-77
At5g59900 putative protein 274 2e-73
At5g61990 unknown protein 273 3e-73
At5g55840 putative protein 273 3e-73
At1g12300 hypothetical protein 273 3e-73
At3g53700 putative protein 273 3e-73
At3g54980 unknown protein (At3g54980) 271 2e-72
At1g12620 putative protein 268 1e-71
At1g62930 unknown protein 267 2e-71
At1g63130 unknown protein 266 3e-71
At1g62670 PPR-repeat protein 266 6e-71
At3g22470 hypothetical protein 264 2e-70
At1g74580 hypothetical protein 263 3e-70
At1g63080 unknown protein 263 4e-70
At1g09900 hypothetical protein 259 4e-69
>At1g06710 hypothetical protein
Length = 946
Score = 1157 bits (2993), Expect = 0.0
Identities = 575/935 (61%), Positives = 716/935 (76%), Gaps = 12/935 (1%)
Query: 35 DSTFSPSTPTPR-DLSQDYAFLRNTLIN-------STSPQSTPSSGDD-AISTISKALKT 85
D FSPS DL+++Y+FL ++L++ P T SS D AI+ +
Sbjct: 13 DDPFSPSDSREVVDLTKEYSFLHDSLVDYGNVNVHQVVPIITQSSIDARAIADAVSGVDD 72
Query: 86 GFNIETHQFFRQFRNQLNDSLVVEVMNNVKNPELCVKFFLWAGRQIGYSHTPQVFDKLLD 145
F ++ +F RQFR +L++SLV+EV+ + P + FF+WAGRQIGY HT V++ L+D
Sbjct: 73 VFGRKSQKFLRQFREKLSESLVIEVLRLIARPSAVISFFVWAGRQIGYKHTAPVYNALVD 132
Query: 146 LLGCNVNADDRVPLKFLMEIKDDDHELLRRLLNFLVRKCCRNGWWNMALEELGRLKDFGY 205
L+ + D++VP +FL +I+DDD E+ LN LVRK CRNG +++ALEELGRLKDF +
Sbjct: 133 LIVRD--DDEKVPEEFLQQIRDDDKEVFGEFLNVLVRKHCRNGSFSIALEELGRLKDFRF 190
Query: 206 KPSQTTYNALIQVFLRADKLDTAYLVKREMLSYAFVMDRYTLSCFAYSLCKGGKCREAFD 265
+PS++TYN LIQ FL+AD+LD+A L+ REM MD +TL CFAYSLCK GK REA
Sbjct: 191 RPSRSTYNCLIQAFLKADRLDSASLIHREMSLANLRMDGFTLRCFAYSLCKVGKWREALT 250
Query: 266 LIDEAEDFVPDTVFYNRMVSGLCEASLFEEAMDILHRMRSSSCIPNVVTYRILLSGCLRK 325
L+ E E+FVPDTVFY +++SGLCEASLFEEAMD L+RMR++SC+PNVVTY LL GCL K
Sbjct: 251 LV-ETENFVPDTVFYTKLISGLCEASLFEEAMDFLNRMRATSCLPNVVTYSTLLCGCLNK 309
Query: 326 GQLGRCKRILSMMITEGCYPNREIFNSLIHAYCKSRDYSYAYKLFKKMIKCGCQPGYLVY 385
QLGRCKR+L+MM+ EGCYP+ +IFNSL+HAYC S D+SYAYKL KKM+KCG PGY+VY
Sbjct: 310 KQLGRCKRVLNMMMMEGCYPSPKIFNSLVHAYCTSGDHSYAYKLLKKMVKCGHMPGYVVY 369
Query: 386 NIFIGSVCSNEEQPSSDILDLVEKAYSEMLDLGVVLNKVNVSNFARCLCGAGKFDQAFKI 445
NI IGS+C +++ + D+LDL EKAYSEML GVVLNK+NVS+F RCLC AGK+++AF +
Sbjct: 370 NILIGSICGDKDSLNCDLLDLAEKAYSEMLAAGVVLNKINVSSFTRCLCSAGKYEKAFSV 429
Query: 446 ICEMMGKGFVPDDSTYSKVIGFLCDASKVEKAFSLFEEMKRNGIVPSVYTYTILIDSFCK 505
I EM+G+GF+PD STYSKV+ +LC+ASK+E AF LFEEMKR G+V VYTYTI++DSFCK
Sbjct: 430 IREMIGQGFIPDTSTYSKVLNYLCNASKMELAFLLFEEMKRGGLVADVYTYTIMVDSFCK 489
Query: 506 AGLIQQARKWFDEMLHKGCTPNVVTYTALIHAYLKAKQMPVADELFEMMLLEGCKPNVVT 565
AGLI+QARKWF+EM GCTPNVVTYTALIHAYLKAK++ A+ELFE ML EGC PN+VT
Sbjct: 490 AGLIEQARKWFNEMREVGCTPNVVTYTALIHAYLKAKKVSYANELFETMLSEGCLPNIVT 549
Query: 566 YTALIDGHCKAGQIEKACQIYARMRGDIESSDMDKYFKLDHNNCEGPNVITYGALVDGLC 625
Y+ALIDGHCKAGQ+EKACQI+ RM G + D+D YFK +N E PNV+TYGAL+DG C
Sbjct: 550 YSALIDGHCKAGQVEKACQIFERMCGSKDVPDVDMYFKQYDDNSERPNVVTYGALLDGFC 609
Query: 626 KANRVKEAHELLDTMLAHGCEPNQIVYDAVIDGFCKIGKLQDAQEVFTKMSERGYSPNLY 685
K++RV+EA +LLD M GCEPNQIVYDA+IDG CK+GKL +AQEV T+MSE G+ LY
Sbjct: 610 KSHRVEEARKLLDAMSMEGCEPNQIVYDALIDGLCKVGKLDEAQEVKTEMSEHGFPATLY 669
Query: 686 TYSSFIDCLFKDNRLDLVLKVLSKMLENSCTPNVVIYTEMVDGLCKIGKTDEAYKLMLKM 745
TYSS ID FK R DL KVLSKMLENSC PNVVIYTEM+DGLCK+GKTDEAYKLM M
Sbjct: 670 TYSSLIDRYFKVKRQDLASKVLSKMLENSCAPNVVIYTEMIDGLCKVGKTDEAYKLMQMM 729
Query: 746 EEKGCNPNVVTYTAMIDGFGKSGKIEQCLELFRDMCSKGCAPNFITYRVLINHCCSNGLL 805
EEKGC PNVVTYTAMIDGFG GKIE CLEL M SKG APN++TYRVLI+HCC NG L
Sbjct: 730 EEKGCQPNVVTYTAMIDGFGMIGKIETCLELLERMGSKGVAPNYVTYRVLIDHCCKNGAL 789
Query: 806 DEAYKLLDEMKQTYWPKHILSHRKIIEGFSQEFITSIGLLDELSENESVPVDSLYRILID 865
D A+ LL+EMKQT+WP H +RK+IEGF++EFI S+GLLDE+ ++++ P S+YR+LID
Sbjct: 790 DVAHNLLEEMKQTHWPTHTAGYRKVIEGFNKEFIESLGLLDEIGQDDTAPFLSVYRLLID 849
Query: 866 NYIKAGRLEVALDLLEEISSSPSHAVSNKYLYASLIENLSHASKVDKALELYASMISKNV 925
N IKA RLE+AL LLEE+++ + V Y SLIE+L A+KV+ A +L++ M K V
Sbjct: 850 NLIKAQRLEMALRLLEEVATFSATLVDYSSTYNSLIESLCLANKVETAFQLFSEMTKKGV 909
Query: 926 VPELSILVHLIKGLIKVDKWQEALQLSDSICQMVC 960
+PE+ LIKGL + K EAL L D I MVC
Sbjct: 910 IPEMQSFCSLIKGLFRNSKISEALLLLDFISHMVC 944
Score = 228 bits (582), Expect = 1e-59
Identities = 166/649 (25%), Positives = 280/649 (42%), Gaps = 86/649 (13%)
Query: 348 EIFNSLIHAYCKSRDYSYAYKLFKKMIKCGCQPGYLVYNIFIGSVCSNEEQPSSDILDLV 407
E N L+ +C++ +S A + ++ +P YN I + +D LD
Sbjct: 160 EFLNVLVRKHCRNGSFSIALEELGRLKDFRFRPSRSTYNCLIQAFLK------ADRLDSA 213
Query: 408 EKAYSEMLDLGVVLNKVNVSNFARCLCGAGKFDQAFKIICEMMGKGFVPDDSTYSKVIGF 467
+ EM + ++ + FA LC GK+ +A ++
Sbjct: 214 SLIHREMSLANLRMDGFTLRCFAYSLCKVGKWREALTLV--------------------- 252
Query: 468 LCDASKVEKAFSLFEEMKRNGIVPSVYTYTILIDSFCKAGLIQQARKWFDEMLHKGCTPN 527
+ VP YT LI C+A L ++A + + M C PN
Sbjct: 253 -----------------ETENFVPDTVFYTKLISGLCEASLFEEAMDFLNRMRATSCLPN 295
Query: 528 VVTYTALIHAYLKAKQMPVADELFEMMLLEGCKPNVVTYTALIDGHCKAGQIEKACQIYA 587
VVTY+ L+ L KQ+ + MM++EGC P+ + +L+ +C +G A ++
Sbjct: 296 VVTYSTLLCGCLNKKQLGRCKRVLNMMMMEGCYPSPKIFNSLVHAYCTSGDHSYAYKLLK 355
Query: 588 RMRGDIESSDMDKYFKLDHNNCEGPNVITYGALVDGLC------KANRVKEAHELLDTML 641
+M K H P + Y L+ +C + + A + ML
Sbjct: 356 KM------------VKCGHM----PGYVVYNILIGSICGDKDSLNCDLLDLAEKAYSEML 399
Query: 642 AHGCEPNQIVYDAVIDGFCKIGKLQDAQEVFTKMSERGYSPNLYTYSSFIDCLFKDNRLD 701
A G N+I + C GK + A V +M +G+ P+ TYS ++ L ++++
Sbjct: 400 AAGVVLNKINVSSFTRCLCSAGKYEKAFSVIREMIGQGFIPDTSTYSKVLNYLCNASKME 459
Query: 702 LVLKVLSKMLENSCTPNVVIYTEMVDGLCKIGKTDEAYKLMLKMEEKGCNPNVVTYTAMI 761
L + +M +V YT MVD CK G ++A K +M E GC PNVVTYTA+I
Sbjct: 460 LAFLLFEEMKRGGLVADVYTYTIMVDSFCKAGLIEQARKWFNEMREVGCTPNVVTYTALI 519
Query: 762 DGFGKSGKIEQCLELFRDMCSKGCAPNFITYRVLINHCCSNGLLDEAYKLLDEM------ 815
+ K+ K+ ELF M S+GC PN +TY LI+ C G +++A ++ + M
Sbjct: 520 HAYLKAKKVSYANELFETMLSEGCLPNIVTYSALIDGHCKAGQVEKACQIFERMCGSKDV 579
Query: 816 --KQTYWPKH--------ILSHRKIIEGF--SQEFITSIGLLDELSENESVPVDSLYRIL 863
Y+ ++ ++++ +++GF S + LLD +S P +Y L
Sbjct: 580 PDVDMYFKQYDDNSERPNVVTYGALLDGFCKSHRVEEARKLLDAMSMEGCEPNQIVYDAL 639
Query: 864 IDNYIKAGRLEVALDLLEEISSSPSHAVSNKYLYASLIENLSHASKVDKALELYASMISK 923
ID K G+L+ A ++ E+S A Y Y+SLI+ + D A ++ + M+
Sbjct: 640 IDGLCKVGKLDEAQEVKTEMSEHGFPATL--YTYSSLIDRYFKVKRQDLASKVLSKMLEN 697
Query: 924 NVVPELSILVHLIKGLIKVDKWQEALQLSDSICQMVCLTLSQTFQPLVN 972
+ P + I +I GL KV K EA +L + + C T+ +++
Sbjct: 698 SCAPNVVIYTEMIDGLCKVGKTDEAYKLMQMMEEKGCQPNVVTYTAMID 746
>At3g06920 hypothetical protein
Length = 871
Score = 298 bits (763), Expect = 1e-80
Identities = 216/803 (26%), Positives = 362/803 (44%), Gaps = 75/803 (9%)
Query: 107 VVEVMNNVKNPELCVKFFLWAGRQIGYSHTPQVFDKLLDLLGCNVNADDRVPLKFLMEIK 166
V+ V+ +K+ +++F W R+ H P+ ++ LL
Sbjct: 68 VIGVLRRLKDVNRAIEYFRWYERRTELPHCPESYNSLL---------------------- 105
Query: 167 DDDHELLRRLLNFLVRKCCRNGWWNMALEELGRLKDFGYKPSQTTYNALIQVFLRADKLD 226
LV CRN ++ + LG + G+ PS T ++ ++A+KL
Sbjct: 106 -------------LVMARCRN--FDALDQILGEMSVAGFGPSVNTCIEMVLGCVKANKLR 150
Query: 227 TAYLVKREMLSYAF--VMDRYTLSCFAYSLCKGGKCREAFDLIDEAEDFVPDTVFYNRMV 284
Y V + M + F YT A+S + + P + ++
Sbjct: 151 EGYDVVQMMRKFKFRPAFSAYTTLIGAFSAVNHSDMMLTLFQQMQELGYEPTVHLFTTLI 210
Query: 285 SGLCEASLFEEAMDILHRMRSSSCIPNVVTYRILLSGCLRKGQLGRCKRILSMMITEGCY 344
G + + A+ +L M+SSS ++V Y + + + G++ + + G
Sbjct: 211 RGFAKEGRVDSALSLLDEMKSSSLDADIVLYNVCIDSFGKVGKVDMAWKFFHEIEANGLK 270
Query: 345 PNREIFNSLIHAYCKSRDYSYAYKLFKKMIKCGCQPGYLVYNIFI---GSVCSNEEQPSS 401
P+ + S+I CK+ A ++F+ + K P YN I GS +E S
Sbjct: 271 PDEVTYTSMIGVLCKANRLDEAVEMFEHLEKNRRVPCTYAYNTMIMGYGSAGKFDEAYS- 329
Query: 402 DILDLVEKAYSEMLDLGVVLNKVNVSNFARCLCGAGKFDQAFKIICEMMGKGFVPDDSTY 461
L+E+ ++ G + + + + CL GK D+A K+ E M K P+ STY
Sbjct: 330 ----LLERQRAK----GSIPSVIAYNCILTCLRKMGKVDEALKVF-EEMKKDAAPNLSTY 380
Query: 462 SKVIGFLCDASKVEKAFSLFEEMKRNGIVPSVYTYTILIDSFCKAGLIQQARKWFDEMLH 521
+ +I LC A K++ AF L + M++ G+ P+V T I++D CK+ + +A F+EM +
Sbjct: 381 NILIDMLCRAGKLDTAFELRDSMQKAGLFPNVRTVNIMVDRLCKSQKLDEACAMFEEMDY 440
Query: 522 KGCTPNVVTYTALIHAYLKAKQMPVADELFEMMLLEGCKPNVVTYTALIDGHCKAGQIEK 581
K CTP+ +T+ +LI K ++ A +++E ML C+ N + YT+LI G+ E
Sbjct: 441 KVCTPDEITFCSLIDGLGKVGRVDDAYKVYEKMLDSDCRTNSIVYTSLIKNFFNHGRKED 500
Query: 582 ACQIYARMRGDIESSD-------MDKYFKLDHNNCEG-------------PNVITYGALV 621
+IY M S D MD FK +G P+ +Y L+
Sbjct: 501 GHKIYKDMINQNCSPDLQLLNTYMDCMFKAGEPE-KGRAMFEEIKARRFVPDARSYSILI 559
Query: 622 DGLCKANRVKEAHELLDTMLAHGCEPNQIVYDAVIDGFCKIGKLQDAQEVFTKMSERGYS 681
GL KA E +EL +M GC + Y+ VIDGFCK GK+ A ++ +M +G+
Sbjct: 560 HGLIKAGFANETYELFYSMKEQGCVLDTRAYNIVIDGFCKCGKVNKAYQLLEEMKTKGFE 619
Query: 682 PNLYTYSSFIDCLFKDNRLDLVLKVLSKMLENSCTPNVVIYTEMVDGLCKIGKTDEAYKL 741
P + TY S ID L K +RLD + + NVVIY+ ++DG K+G+ DEAY +
Sbjct: 620 PTVVTYGSVIDGLAKIDRLDEAYMLFEEAKSKRIELNVVIYSSLIDGFGKVGRIDEAYLI 679
Query: 742 MLKMEEKGCNPNVVTYTAMIDGFGKSGKIEQCLELFRDMCSKGCAPNFITYRVLINHCCS 801
+ ++ +KG PN+ T+ +++D K+ +I + L F+ M C PN +TY +LIN C
Sbjct: 680 LEELMQKGLTPNLYTWNSLLDALVKAEEINEALVCFQSMKELKCTPNQVTYGILINGLCK 739
Query: 802 NGLLDEAYKLLDEMKQTYWPKHILSHRKIIEGFSQ--EFITSIGLLDELSENESVPVDSL 859
++A+ EM++ +S+ +I G ++ + L D N VP +
Sbjct: 740 VRKFNKAFVFWQEMQKQGMKPSTISYTTMISGLAKAGNIAEAGALFDRFKANGGVPDSAC 799
Query: 860 YRILIDNYIKAGRLEVALDLLEE 882
Y +I+ R A L EE
Sbjct: 800 YNAMIEGLSNGNRAMDAFSLFEE 822
Score = 291 bits (745), Expect = 1e-78
Identities = 187/676 (27%), Positives = 321/676 (46%), Gaps = 28/676 (4%)
Query: 151 VNADDRVPLKFLMEIKDDDHELLRRLLNFLVRKCCRNGWWNMALEELGRLKDFGYKPSQT 210
VN D + L ++++ +E L L+R + G + AL L +K
Sbjct: 181 VNHSDMM-LTLFQQMQELGYEPTVHLFTTLIRGFAKEGRVDSALSLLDEMKSSSLDADIV 239
Query: 211 TYNALIQVFLRADKLDTAYLVKREMLSYAFVMDRYTLSCFAYSLCKGGKCREAFDLIDEA 270
YN I F + K+D A+ E+ + D T + LCK + EA ++ +
Sbjct: 240 LYNVCIDSFGKVGKVDMAWKFFHEIEANGLKPDEVTYTSMIGVLCKANRLDEAVEMFEHL 299
Query: 271 EDF--VPDTVFYNRMVSGLCEASLFEEAMDILHRMRSSSCIPNVVTYRILLSGCLRK-GQ 327
E VP T YN M+ G A F+EA +L R R+ IP+V+ Y +L+ CLRK G+
Sbjct: 300 EKNRRVPCTYAYNTMIMGYGSAGKFDEAYSLLERQRAKGSIPSVIAYNCILT-CLRKMGK 358
Query: 328 LGRCKRILSMMITEGCYPNREIFNSLIHAYCKSRDYSYAYKLFKKMIKCGCQPGYLVYNI 387
+ ++ M + PN +N LI C++ A++L M K G P NI
Sbjct: 359 VDEALKVFEEMKKDAA-PNLSTYNILIDMLCRAGKLDTAFELRDSMQKAGLFPNVRTVNI 417
Query: 388 FIGSVCSNEEQPSSDILDLVEKAYSEMLDLGVVLNKVNVSNFARCLCGAGKFDQAFKIIC 447
+ +C +++ LD + EM +++ + L G+ D A+K+
Sbjct: 418 MVDRLCKSQK------LDEACAMFEEMDYKVCTPDEITFCSLIDGLGKVGRVDDAYKVYE 471
Query: 448 EMMGKGFVPDDSTYSKVIGFLCDASKVEKAFSLFEEMKRNGIVPSVYTYTILIDSFCKAG 507
+M+ + Y+ +I + + E ++++M P + +D KAG
Sbjct: 472 KMLDSDCRTNSIVYTSLIKNFFNHGRKEDGHKIYKDMINQNCSPDLQLLNTYMDCMFKAG 531
Query: 508 LIQQARKWFDEMLHKGCTPNVVTYTALIHAYLKAKQMPVADELFEMMLLEGCKPNVVTYT 567
++ R F+E+ + P+ +Y+ LIH +KA ELF M +GC + Y
Sbjct: 532 EPEKGRAMFEEIKARRFVPDARSYSILIHGLIKAGFANETYELFYSMKEQGCVLDTRAYN 591
Query: 568 ALIDGHCKAGQIEKACQIYARMRGDIESSDMDKYFKLDHNNCEGPNVITYGALVDGLCKA 627
+IDG CK G++ KA Q+ M+ K F+ P V+TYG+++DGL K
Sbjct: 592 IVIDGFCKCGKVNKAYQLLEEMK--------TKGFE--------PTVVTYGSVIDGLAKI 635
Query: 628 NRVKEAHELLDTMLAHGCEPNQIVYDAVIDGFCKIGKLQDAQEVFTKMSERGYSPNLYTY 687
+R+ EA+ L + + E N ++Y ++IDGF K+G++ +A + ++ ++G +PNLYT+
Sbjct: 636 DRLDEAYMLFEEAKSKRIELNVVIYSSLIDGFGKVGRIDEAYLILEELMQKGLTPNLYTW 695
Query: 688 SSFIDCLFKDNRLDLVLKVLSKMLENSCTPNVVIYTEMVDGLCKIGKTDEAYKLMLKMEE 747
+S +D L K ++ L M E CTPN V Y +++GLCK+ K ++A+ +M++
Sbjct: 696 NSLLDALVKAEEINEALVCFQSMKELKCTPNQVTYGILINGLCKVRKFNKAFVFWQEMQK 755
Query: 748 KGCNPNVVTYTAMIDGFGKSGKIEQCLELFRDMCSKGCAPNFITYRVLINHCCSNGLLDE 807
+G P+ ++YT MI G K+G I + LF + G P+ Y +I + +
Sbjct: 756 QGMKPSTISYTTMISGLAKAGNIAEAGALFDRFKANGGVPDSACYNAMIEGLSNGNRAMD 815
Query: 808 AYKLLDEMKQTYWPKH 823
A+ L +E ++ P H
Sbjct: 816 AFSLFEETRRRGLPIH 831
Score = 275 bits (704), Expect = 7e-74
Identities = 188/699 (26%), Positives = 331/699 (46%), Gaps = 38/699 (5%)
Query: 280 YNRMVSGLCEASLFEEAMDILHRMRSSSCIPNVVTYRILLSGCLRKGQLGRCKRILSMMI 339
YN ++ + F+ IL M + P+V T ++ GC++ +L ++ MM
Sbjct: 101 YNSLLLVMARCRNFDALDQILGEMSVAGFGPSVNTCIEMVLGCVKANKLREGYDVVQMMR 160
Query: 340 TEGCYPNREIFNSLIHAYCKSRDYSYAYKLFKKMIKCGCQPGYLVYNIFIGSVCSNEEQP 399
P + +LI A+ LF++M + G +P ++ I
Sbjct: 161 KFKFRPAFSAYTTLIGAFSAVNHSDMMLTLFQQMQELGYEPTVHLFTTLIRGFAKEGRVD 220
Query: 400 SSDILDLVEKAYSEMLDLGVVLNKVNVSNFARCLCGAGKFDQAFKIICEMMGKGFVPDDS 459
S+ L L+++ S LD +VL V + +F + GK D A+K E+ G PD+
Sbjct: 221 SA--LSLLDEMKSSSLDADIVLYNVCIDSFGK----VGKVDMAWKFFHEIEANGLKPDEV 274
Query: 460 TYSKVIGFLCDASKVEKAFSLFEEMKRNGIVPSVYTYTILIDSFCKAGLIQQARKWFDEM 519
TY+ +IG LC A+++++A +FE +++N VP Y Y +I + AG +A +
Sbjct: 275 TYTSMIGVLCKANRLDEAVEMFEHLEKNRRVPCTYAYNTMIMGYGSAGKFDEAYSLLERQ 334
Query: 520 LHKGCTPNVVTYTALIHAYLKAKQMPVADELFEMMLLEGCKPNVVTYTALIDGHCKAGQI 579
KG P+V+ Y ++ K ++ A ++FE M + PN+ TY LID C+AG++
Sbjct: 335 RAKGSIPSVIAYNCILTCLRKMGKVDEALKVFEEMKKDAA-PNLSTYNILIDMLCRAGKL 393
Query: 580 EKACQIYARMRGD----------------IESSDMDK----YFKLDHNNCEGPNVITYGA 619
+ A ++ M+ +S +D+ + ++D+ C P+ IT+ +
Sbjct: 394 DTAFELRDSMQKAGLFPNVRTVNIMVDRLCKSQKLDEACAMFEEMDYKVCT-PDEITFCS 452
Query: 620 LVDGLCKANRVKEAHELLDTMLAHGCEPNQIVYDAVIDGFCKIGKLQDAQEVFTKMSERG 679
L+DGL K RV +A+++ + ML C N IVY ++I F G+ +D +++ M +
Sbjct: 453 LIDGLGKVGRVDDAYKVYEKMLDSDCRTNSIVYTSLIKNFFNHGRKEDGHKIYKDMINQN 512
Query: 680 YSPNLYTYSSFIDCLFKDNRLDLVLKVLSKMLENSCTPNVVIYTEMVDGLCKIGKTDEAY 739
SP+L ++++DC+FK + + ++ P+ Y+ ++ GL K G +E Y
Sbjct: 513 CSPDLQLLNTYMDCMFKAGEPEKGRAMFEEIKARRFVPDARSYSILIHGLIKAGFANETY 572
Query: 740 KLMLKMEEKGCNPNVVTYTAMIDGFGKSGKIEQCLELFRDMCSKGCAPNFITYRVLINHC 799
+L M+E+GC + Y +IDGF K GK+ + +L +M +KG P +TY +I+
Sbjct: 573 ELFYSMKEQGCVLDTRAYNIVIDGFCKCGKVNKAYQLLEEMKTKGFEPTVVTYGSVIDGL 632
Query: 800 CSNGLLDEAYKLLDEMKQTYWPKHILSHRKIIEGFSQ--EFITSIGLLDELSENESVPVD 857
LDEAY L +E K +++ + +I+GF + + +L+EL + P
Sbjct: 633 AKIDRLDEAYMLFEEAKSKRIELNVVIYSSLIDGFGKVGRIDEAYLILEELMQKGLTPNL 692
Query: 858 SLYRILIDNYIKAGRLEVAL---DLLEEISSSPSHAVSNKYLYASLIENLSHASKVDKAL 914
+ L+D +KA + AL ++E+ +P N+ Y LI L K +KA
Sbjct: 693 YTWNSLLDALVKAEEINEALVCFQSMKELKCTP-----NQVTYGILINGLCKVRKFNKAF 747
Query: 915 ELYASMISKNVVPELSILVHLIKGLIKVDKWQEALQLSD 953
+ M + + P +I GL K EA L D
Sbjct: 748 VFWQEMQKQGMKPSTISYTTMISGLAKAGNIAEAGALFD 786
Score = 238 bits (608), Expect = 1e-62
Identities = 166/661 (25%), Positives = 292/661 (44%), Gaps = 27/661 (4%)
Query: 314 TYRILLSGCLRKGQLGRCKRILSMMITEGCYPNREIFNSLIHAYCKSRDYSYAYKLFKKM 373
+Y LL R +IL M G P+ ++ K+ Y + + M
Sbjct: 100 SYNSLLLVMARCRNFDALDQILGEMSVAGFGPSVNTCIEMVLGCVKANKLREGYDVVQMM 159
Query: 374 IKCGCQPGYLVYNIFIGSVCSNEEQPSSDILDLVEKAYSEMLDLGVVLNKVNVSNFARCL 433
K +P + Y IG+ + D++ + +M +LG + R
Sbjct: 160 RKFKFRPAFSAYTTLIGAFSAVNHS------DMMLTLFQQMQELGYEPTVHLFTTLIRGF 213
Query: 434 CGAGKFDQAFKIICEMMGKGFVPDDSTYSKVIGFLCDASKVEKAFSLFEEMKRNGIVPSV 493
G+ D A ++ EM D Y+ I KV+ A+ F E++ NG+ P
Sbjct: 214 AKEGRVDSALSLLDEMKSSSLDADIVLYNVCIDSFGKVGKVDMAWKFFHEIEANGLKPDE 273
Query: 494 YTYTILIDSFCKAGLIQQARKWFDEMLHKGCTPNVVTYTALIHAYLKAKQMPVADELFEM 553
TYT +I CKA + +A + F+ + P Y +I Y A + A L E
Sbjct: 274 VTYTSMIGVLCKANRLDEAVEMFEHLEKNRRVPCTYAYNTMIMGYGSAGKFDEAYSLLER 333
Query: 554 MLLEGCKPNVVTYTALIDGHCKAGQIEKACQIYARMRGDIESSDMDKYFKLDHNNCEGPN 613
+G P+V+ Y ++ K G++++A +++ M+ D PN
Sbjct: 334 QRAKGSIPSVIAYNCILTCLRKMGKVDEALKVFEEMKKDA-----------------APN 376
Query: 614 VITYGALVDGLCKANRVKEAHELLDTMLAHGCEPNQIVYDAVIDGFCKIGKLQDAQEVFT 673
+ TY L+D LC+A ++ A EL D+M G PN + ++D CK KL +A +F
Sbjct: 377 LSTYNILIDMLCRAGKLDTAFELRDSMQKAGLFPNVRTVNIMVDRLCKSQKLDEACAMFE 436
Query: 674 KMSERGYSPNLYTYSSFIDCLFKDNRLDLVLKVLSKMLENSCTPNVVIYTEMVDGLCKIG 733
+M + +P+ T+ S ID L K R+D KV KML++ C N ++YT ++ G
Sbjct: 437 EMDYKVCTPDEITFCSLIDGLGKVGRVDDAYKVYEKMLDSDCRTNSIVYTSLIKNFFNHG 496
Query: 734 KTDEAYKLMLKMEEKGCNPNVVTYTAMIDGFGKSGKIEQCLELFRDMCSKGCAPNFITYR 793
+ ++ +K+ M + C+P++ +D K+G+ E+ +F ++ ++ P+ +Y
Sbjct: 497 RKEDGHKIYKDMINQNCSPDLQLLNTYMDCMFKAGEPEKGRAMFEEIKARRFVPDARSYS 556
Query: 794 VLINHCCSNGLLDEAYKLLDEMKQTYWPKHILSHRKIIEGFSQ--EFITSIGLLDELSEN 851
+LI+ G +E Y+L MK+ ++ +I+GF + + + LL+E+
Sbjct: 557 ILIHGLIKAGFANETYELFYSMKEQGCVLDTRAYNIVIDGFCKCGKVNKAYQLLEEMKTK 616
Query: 852 ESVPVDSLYRILIDNYIKAGRLEVALDLLEEISSSPSHAVSNKYLYASLIENLSHASKVD 911
P Y +ID K RL+ A L EE S N +Y+SLI+ ++D
Sbjct: 617 GFEPTVVTYGSVIDGLAKIDRLDEAYMLFEEAKSKRIEL--NVVIYSSLIDGFGKVGRID 674
Query: 912 KALELYASMISKNVVPELSILVHLIKGLIKVDKWQEALQLSDSICQMVCLTLSQTFQPLV 971
+A + ++ K + P L L+ L+K ++ EAL S+ ++ C T+ L+
Sbjct: 675 EAYLILEELMQKGLTPNLYTWNSLLDALVKAEEINEALVCFQSMKELKCTPNQVTYGILI 734
Query: 972 N 972
N
Sbjct: 735 N 735
Score = 134 bits (338), Expect = 2e-31
Identities = 93/376 (24%), Positives = 170/376 (44%), Gaps = 6/376 (1%)
Query: 600 KYFKLDHNNCEGPNVI-TYGALVDGLCKANRVKEAHELLDTMLAHGCEPNQIVYDAVIDG 658
+YF+ E P+ +Y +L+ + + ++L M G P+ ++ G
Sbjct: 83 EYFRWYERRTELPHCPESYNSLLLVMARCRNFDALDQILGEMSVAGFGPSVNTCIEMVLG 142
Query: 659 FCKIGKLQDAQEVFTKMSERGYSPNLYTYSSFIDCLFKDNRLDLVLKVLSKMLENSCTPN 718
K KL++ +V M + + P Y++ I N D++L + +M E P
Sbjct: 143 CVKANKLREGYDVVQMMRKFKFRPAFSAYTTLIGAFSAVNHSDMMLTLFQQMQELGYEPT 202
Query: 719 VVIYTEMVDGLCKIGKTDEAYKLMLKMEEKGCNPNVVTYTAMIDGFGKSGKIEQCLELFR 778
V ++T ++ G K G+ D A L+ +M+ + ++V Y ID FGK GK++ + F
Sbjct: 203 VHLFTTLIRGFAKEGRVDSALSLLDEMKSSSLDADIVLYNVCIDSFGKVGKVDMAWKFFH 262
Query: 779 DMCSKGCAPNFITYRVLINHCCSNGLLDEAYKLLDEMKQTYWPKHILSHRKIIEGFSQ-- 836
++ + G P+ +TY +I C LDEA ++ + +++ ++ +I G+
Sbjct: 263 EIEANGLKPDEVTYTSMIGVLCKANRLDEAVEMFEHLEKNRRVPCTYAYNTMIMGYGSAG 322
Query: 837 EFITSIGLLDELSENESVPVDSLYRILIDNYIKAGRLEVALDLLEEISSSPSHAVSNKYL 896
+F + LL+ S+P Y ++ K G+++ AL + EE+ A N
Sbjct: 323 KFDEAYSLLERQRAKGSIPSVIAYNCILTCLRKMGKVDEALKVFEEMKKD---AAPNLST 379
Query: 897 YASLIENLSHASKVDKALELYASMISKNVVPELSILVHLIKGLIKVDKWQEALQLSDSIC 956
Y LI+ L A K+D A EL SM + P + + ++ L K K EA + + +
Sbjct: 380 YNILIDMLCRAGKLDTAFELRDSMQKAGLFPNVRTVNIMVDRLCKSQKLDEACAMFEEMD 439
Query: 957 QMVCLTLSQTFQPLVN 972
VC TF L++
Sbjct: 440 YKVCTPDEITFCSLID 455
>At1g62910 unknown protein
Length = 1133
Score = 298 bits (763), Expect = 1e-80
Identities = 198/709 (27%), Positives = 322/709 (44%), Gaps = 55/709 (7%)
Query: 180 LVRKCCRNGWWNMALEELGRLKDFGYKPSQTTYNALIQVFLRADKLDTAYLVKREMLSYA 239
L++ C+ +E + G + TY LI F +A D A +V ++M+S
Sbjct: 404 LIKGFCKAKRVEEGMELFREMSQRGLVGNTVTYTTLIHGFFQARDCDNAQMVFKQMVSVG 463
Query: 240 FVMDRYTLSCFAYSLCKGGKCREAFDLID--EAEDFVPDTVFYNRMVSGLCEASLFEEAM 297
+ T + LCK GK +A + + + PD YN M+ G+C+A E+
Sbjct: 464 VHPNILTYNILLDGLCKNGKLAKAMVVFEYLQRSTMEPDIYTYNIMIEGMCKAGKVEDGW 523
Query: 298 DILHRMRSSSCIPNVVTYRILLSGCLRKGQLGRCKRILSMMITEGCYPNREIFNSLIHAY 357
++ + PNV+ Y ++SG RKG +L M +G PN +N+LI A
Sbjct: 524 ELFCNLSLKGVSPNVIAYNTMISGFCRKGSKEEADSLLKKMKEDGPLPNSGTYNTLIRAR 583
Query: 358 CKSRDYSYAYKLFKKMIKCGCQPGYLVYN---------------------IFIGSVCSNE 396
+ D + +L K+M CG IF +CS+E
Sbjct: 584 LRDGDREASAELIKEMRSCGFAGDASTIGLSSKFKREIEKPNRENACKNIIFSEEICSSE 643
Query: 397 EQPSS-DILDLVEKAYSEM-LD-----LGVVLNK------VNVSNFARCLCGAGKFDQAF 443
S D +++ S++ LD GV+ + S + KFD
Sbjct: 644 SCGESYDYREVLRTGLSDIELDDAIGLFGVMAQSRPFPSIIEFSKLLSAIAKMNKFDLVI 703
Query: 444 KIICEMMGKGFVPDDSTYSKVIGFLCDASKVEKAFSLFEEMKRNGIVPSVYTYTILIDSF 503
+M G + TY+ +I C S++ A +L +M + G P + T L++ F
Sbjct: 704 SFGEKMEILGISHNLYTYNILINCFCRCSRLSLALALLGKMMKLGYEPDIVTLNSLLNGF 763
Query: 504 CKAGLIQQARKWFDEMLHKGCTPNVVTYTALIHAYLKAKQMPVADELFEMMLLEGCKPNV 563
C I A D+M+ G P+ VT+T LIH + A L + M+ GC+P++
Sbjct: 764 CHGNRISDAVALVDQMVEMGYKPDTVTFTTLIHGLFLHNKASEAVALIDRMVQRGCQPDL 823
Query: 564 VTYTALIDGHCKAGQIEKACQIYARMRGD-------IESSDMDKYFKLDH---------- 606
VTY A+++G CK G + A + +M I S+ +D K H
Sbjct: 824 VTYGAVVNGLCKRGDTDLALNLLNKMEAAKIEANVVIYSTVIDSLCKYRHEDDALNLFTE 883
Query: 607 --NNCEGPNVITYGALVDGLCKANRVKEAHELLDTMLAHGCEPNQIVYDAVIDGFCKIGK 664
N PNVITY +L+ LC R +A LL M+ PN + + A+ID F K GK
Sbjct: 884 MENKGVRPNVITYSSLISCLCNYGRWSDASRLLSDMIERKINPNLVTFSALIDAFVKKGK 943
Query: 665 LQDAQEVFTKMSERGYSPNLYTYSSFIDCLFKDNRLDLVLKVLSKMLENSCTPNVVIYTE 724
L A++++ +M +R PN++TYSS I+ +RL ++L M+ C PNVV Y
Sbjct: 944 LVKAEKLYEEMIKRSIDPNIFTYSSLINGFCMLDRLGEAKQMLELMIRKDCLPNVVTYNT 1003
Query: 725 MVDGLCKIGKTDEAYKLMLKMEEKGCNPNVVTYTAMIDGFGKSGKIEQCLELFRDMCSKG 784
+++G CK + D+ +L +M ++G N VTYT +I GF ++ + +F+ M S G
Sbjct: 1004 LINGFCKAKRVDKGMELFREMSQRGLVGNTVTYTTLIHGFFQARDCDNAQMVFKQMVSVG 1063
Query: 785 CAPNFITYRVLINHCCSNGLLDEAYKLLDEMKQTYWPKHILSHRKIIEG 833
PN +TY +L++ C NG L +A + + ++++ I ++ +IEG
Sbjct: 1064 VHPNILTYNILLDGLCKNGKLAKAMVVFEYLQRSTMEPDIYTYNIMIEG 1112
Score = 290 bits (741), Expect = 4e-78
Identities = 217/859 (25%), Positives = 364/859 (42%), Gaps = 71/859 (8%)
Query: 177 LNFLVRKCCRNGWWNMALEELGRLKDFGYKPSQTTYNALIQVFLRADKLDTAYLVKREML 236
L+ L+ C + + A+ + ++ + GYKP T+ LI +K A + +M+
Sbjct: 156 LSSLLNGYCHSKRISDAVALVDQMVEMGYKPDTFTFTTLIHGLFLHNKASEAVALVDQMV 215
Query: 237 SYAFVMDRYTLSCFAYSLCKGGKCREAFDLIDEAED--FVPDTVFYNRMVSGLCEASLFE 294
D T LCK G A L+ + E D V YN ++ GLC+ +
Sbjct: 216 QRGCQPDLVTYGTVVNGLCKRGDIDLALSLLKKMEKGKIEADVVIYNTIIDGLCKYKHMD 275
Query: 295 EAMDILHRMRSSSCIPNVVTYRILLSGCLRKGQLGRCKRILSMMITEGCYPNREIFNSLI 354
+A+++ M + P+V TY L+S G+ R+LS MI PN F++LI
Sbjct: 276 DALNLFTEMDNKGIRPDVFTYSSLISCLCNYGRWSDASRLLSDMIERKINPNVVTFSALI 335
Query: 355 HAYCKSRDYSYAYKLFKKMIKCGCQPGYLVYNIFIGSVCSNEEQPSSDILDLVEKAYSEM 414
A+ K A KL+ +MIK P Y+ I C + D LD + + M
Sbjct: 336 DAFVKEGKLVEAEKLYDEMIKRSIDPDIFTYSSLINGFCMH------DRLDEAKHMFELM 389
Query: 415 LDLGVVLNKVNVSNFARCLCGAGKFDQAFKIICEMMGKGFVPDDSTYSKVI-GF------ 467
+ N V S + C A + ++ ++ EM +G V + TY+ +I GF
Sbjct: 390 ISKDCFPNVVTYSTLIKGFCKAKRVEEGMELFREMSQRGLVGNTVTYTTLIHGFFQARDC 449
Query: 468 ----------------------------LCDASKVEKAFSLFEEMKRNGIVPSVYTYTIL 499
LC K+ KA +FE ++R+ + P +YTY I+
Sbjct: 450 DNAQMVFKQMVSVGVHPNILTYNILLDGLCKNGKLAKAMVVFEYLQRSTMEPDIYTYNIM 509
Query: 500 IDSFCKAGLIQQARKWFDEMLHKGCTPNVVTYTALIHAYLKAKQMPVADELFEMMLLEGC 559
I+ CKAG ++ + F + KG +PNV+ Y +I + + AD L + M +G
Sbjct: 510 IEGMCKAGKVEDGWELFCNLSLKGVSPNVIAYNTMISGFCRKGSKEEADSLLKKMKEDGP 569
Query: 560 KPNVVTYTALIDGHCKAGQIEKACQIYARMR-----GDIESSDMDKYFKLDHNNCEGPNV 614
PN TY LI + G E + ++ MR GD + + FK + N
Sbjct: 570 LPNSGTYNTLIRARLRDGDREASAELIKEMRSCGFAGDASTIGLSSKFKREIEKPNRENA 629
Query: 615 ITYGALVDGLCKANRVKEAHE-------------------LLDTMLAHGCEPNQIVYDAV 655
+ +C + E+++ L M P+ I + +
Sbjct: 630 CKNIIFSEEICSSESCGESYDYREVLRTGLSDIELDDAIGLFGVMAQSRPFPSIIEFSKL 689
Query: 656 IDGFCKIGKLQDAQEVFTKMSERGYSPNLYTYSSFIDCLFKDNRLDLVLKVLSKMLENSC 715
+ K+ K KM G S NLYTY+ I+C + +RL L L +L KM++
Sbjct: 690 LSAIAKMNKFDLVISFGEKMEILGISHNLYTYNILINCFCRCSRLSLALALLGKMMKLGY 749
Query: 716 TPNVVIYTEMVDGLCKIGKTDEAYKLMLKMEEKGCNPNVVTYTAMIDGFGKSGKIEQCLE 775
P++V +++G C + +A L+ +M E G P+ VT+T +I G K + +
Sbjct: 750 EPDIVTLNSLLNGFCHGNRISDAVALVDQMVEMGYKPDTVTFTTLIHGLFLHNKASEAVA 809
Query: 776 LFRDMCSKGCAPNFITYRVLINHCCSNGLLDEAYKLLDEMKQTYWPKHILSHRKIIEGFS 835
L M +GC P+ +TY ++N C G D A LL++M+ +++ + +I+
Sbjct: 810 LIDRMVQRGCQPDLVTYGAVVNGLCKRGDTDLALNLLNKMEAAKIEANVVIYSTVIDSLC 869
Query: 836 Q--EFITSIGLLDELSENESVPVDSLYRILIDNYIKAGRLEVALDLLEEISSSPSHAVSN 893
+ ++ L E+ P Y LI GR A LL ++ + N
Sbjct: 870 KYRHEDDALNLFTEMENKGVRPNVITYSSLISCLCNYGRWSDASRLLSDMIERKINP--N 927
Query: 894 KYLYASLIENLSHASKVDKALELYASMISKNVVPELSILVHLIKGLIKVDKWQEALQLSD 953
+++LI+ K+ KA +LY MI +++ P + LI G +D+ EA Q+ +
Sbjct: 928 LVTFSALIDAFVKKGKLVKAEKLYEEMIKRSIDPNIFTYSSLINGFCMLDRLGEAKQMLE 987
Query: 954 SICQMVCLTLSQTFQPLVN 972
+ + CL T+ L+N
Sbjct: 988 LMIRKDCLPNVVTYNTLIN 1006
Score = 276 bits (705), Expect = 5e-74
Identities = 205/826 (24%), Positives = 368/826 (43%), Gaps = 64/826 (7%)
Query: 168 DDHELLRRLLNFLVRKCCRNGWWNMALEELGRLKDFGYKPSQTTYNALIQVFLRADKLDT 227
D E+LR L+ +++ + A++ G + PS +N L+ + +K +
Sbjct: 49 DYREILRNRLSDIIKV-------DDAVDLFGDMVKSRPFPSIVEFNKLLSAVAKMNKFEL 101
Query: 228 AYLVKREMLSYAFVMDRYTLSCFAYSLCKGGKCREAFDLIDEAE--DFVPDTVFYNRMVS 285
+ +M + D YT S F C+ + A ++ + + PD V + +++
Sbjct: 102 VISLGEQMQTLGISHDLYTYSIFINCFCRRSQLSLALAVLAKMMKLGYEPDIVTLSSLLN 161
Query: 286 GLCEASLFEEAMDILHRMRSSSCIPNVVTYRILLSGCLRKGQLGRCKRILSMMITEGCYP 345
G C + +A+ ++ +M P+ T+ L+ G + ++ M+ GC P
Sbjct: 162 GYCHSKRISDAVALVDQMVEMGYKPDTFTFTTLIHGLFLHNKASEAVALVDQMVQRGCQP 221
Query: 346 NREIFNSLIHAYCKSRDYSYAYKLFKKMIKCGCQPGYLVYNIFIGSVCSNEEQPSSDILD 405
+ + ++++ CK D A L KKM K + ++YN I +C + D L+
Sbjct: 222 DLVTYGTVVNGLCKRGDIDLALSLLKKMEKGKIEADVVIYNTIIDGLCKYKHM--DDALN 279
Query: 406 LVEKAYSEMLDLGVVLNKVNVSNFARCLCGAGKFDQAFKIICEMMGKGFVPDDSTYSKVI 465
L ++EM + G+ + S+ CLC G++ A +++ +M+ + P+ T+S +I
Sbjct: 280 L----FTEMDNKGIRPDVFTYSSLISCLCNYGRWSDASRLLSDMIERKINPNVVTFSALI 335
Query: 466 GFLCDASKVEKAFSLFEEMKRNGIVPSVYTYTILIDSFCKAGLIQQARKWFDEMLHKGCT 525
K+ +A L++EM + I P ++TY+ LI+ FC + +A+ F+ M+ K C
Sbjct: 336 DAFVKEGKLVEAEKLYDEMIKRSIDPDIFTYSSLINGFCMHDRLDEAKHMFELMISKDCF 395
Query: 526 PNVVTYTALIHAYLKAKQMPVADELFEMMLLEGCKPNVVTYTALIDGHCKAGQIEKACQI 585
PNVVTY+ LI + KAK++ ELF M G N VTYT LI G +A + A +
Sbjct: 396 PNVVTYSTLIKGFCKAKRVEEGMELFREMSQRGLVGNTVTYTTLIHGFFQARDCDNAQMV 455
Query: 586 YARMRGDIESSDMDKYFKLDHNNCEGPNVITYGALVDGLCKANRVKEAHELLDTMLAHGC 645
+ +M + PN++TY L+DGLCK ++ +A + + +
Sbjct: 456 FKQM----------------VSVGVHPNILTYNILLDGLCKNGKLAKAMVVFEYLQRSTM 499
Query: 646 EPNQIVYDAVIDGFCKIGKLQDAQEVFTKMSERGYSPNLYTYSSFIDCLFKDNRLDLVLK 705
EP+ Y+ +I+G CK GK++D E+F +S +G SPN+ Y++ I + +
Sbjct: 500 EPDIYTYNIMIEGMCKAGKVEDGWELFCNLSLKGVSPNVIAYNTMISGFCRKGSKEEADS 559
Query: 706 VLSKMLENSCTPNVVIYTEMVDGLCKIGKTDEAYKLMLKMEEKGCNPNVVTYTAMIDGFG 765
+L KM E+ PN Y ++ + G + + +L+ +M G + T G
Sbjct: 560 LLKKMKEDGPLPNSGTYNTLIRARLRDGDREASAELIKEMRSCGFAGDAST-------IG 612
Query: 766 KSGKIEQCLE------------LFRDMCSKGCAPNFITYRVLINHCCSNGLLDEAYKLLD 813
S K ++ +E ++CS YR ++ S+ LD+A L
Sbjct: 613 LSSKFKREIEKPNRENACKNIIFSEEICSSESCGESYDYREVLRTGLSDIELDDAIGLFG 672
Query: 814 EMKQTYWPKHILSHRKIIEGFSQEFITSIGLLDELSENESVPVDS----LYRILIDNYIK 869
M Q+ I+ K++ ++ + L+ E + S Y ILI+ + +
Sbjct: 673 VMAQSRPFPSIIEFSKLLSAIAK--MNKFDLVISFGEKMEILGISHNLYTYNILINCFCR 730
Query: 870 AGRLEVALDLL---EEISSSPSHAVSNKYLYASLIENLSHASKVDKALELYASMISKNVV 926
RL +AL LL ++ P N SL+ H +++ A+ L M+
Sbjct: 731 CSRLSLALALLGKMMKLGYEPDIVTLN-----SLLNGFCHGNRISDAVALVDQMVEMGYK 785
Query: 927 PELSILVHLIKGLIKVDKWQEALQLSDSICQMVCLTLSQTFQPLVN 972
P+ LI GL +K EA+ L D + Q C T+ +VN
Sbjct: 786 PDTVTFTTLIHGLFLHNKASEAVALIDRMVQRGCQPDLVTYGAVVN 831
Score = 274 bits (700), Expect = 2e-73
Identities = 201/810 (24%), Positives = 356/810 (43%), Gaps = 68/810 (8%)
Query: 176 LLNFLVRKCCRNGWWNMALEELGRLKDFGYKPSQTTYNALIQVFLRADKLDTAYLVKREM 235
+ N ++ C+ + AL + + G +P TY++LI + A + +M
Sbjct: 260 IYNTIIDGLCKYKHMDDALNLFTEMDNKGIRPDVFTYSSLISCLCNYGRWSDASRLLSDM 319
Query: 236 LSYAFVMDRYTLSCFAYSLCKGGKCREAFDLIDEA--EDFVPDTVFYNRMVSGLCEASLF 293
+ + T S + K GK EA L DE PD Y+ +++G C
Sbjct: 320 IERKINPNVVTFSALIDAFVKEGKLVEAEKLYDEMIKRSIDPDIFTYSSLINGFCMHDRL 379
Query: 294 EEAMDILHRMRSSSCIPNVVTYRILLSGCLRKGQLGRCKRILSMMITEGCYPNREIFNSL 353
+EA + M S C PNVVTY L+ G + ++ + M G N + +L
Sbjct: 380 DEAKHMFELMISKDCFPNVVTYSTLIKGFCKAKRVEEGMELFREMSQRGLVGNTVTYTTL 439
Query: 354 IHAYCKSRDYSYAYKLFKKMIKCGCQPGYLVYNIFIGSVCSNEEQPSSDILDLVEKAYSE 413
IH + ++RD A +FK+M+ G P L YNI + +C N + + ++ E
Sbjct: 440 IHGFFQARDCDNAQMVFKQMVSVGVHPNILTYNILLDGLCKNGKLAKAMVV--FEYLQRS 497
Query: 414 MLDLGVVLNKVNVSNFARCLCGAGKFDQAFKIICEMMGKGFVPDDSTYSKVIGFLCDASK 473
++ + + + +C AGK + +++ C + KG P+ Y+ +I C
Sbjct: 498 TMEPDIYTYNIMIEG----MCKAGKVEDGWELFCNLSLKGVSPNVIAYNTMISGFCRKGS 553
Query: 474 VEKAFSLFEEMKRNGIVPSVYTYTILI-------DSFCKAGLIQQAR------------- 513
E+A SL ++MK +G +P+ TY LI D A LI++ R
Sbjct: 554 KEEADSLLKKMKEDGPLPNSGTYNTLIRARLRDGDREASAELIKEMRSCGFAGDASTIGL 613
Query: 514 -----KWFDEMLHKGCTPNVV---------------TYTALIHAYLKAKQMPVADELFEM 553
+ ++ + N++ Y ++ L ++ A LF +
Sbjct: 614 SSKFKREIEKPNRENACKNIIFSEEICSSESCGESYDYREVLRTGLSDIELDDAIGLFGV 673
Query: 554 MLLEGCKPNVVTYTALIDGHCKAGQIEKACQIYARMRGDIESSDMDKYFKLDHNNCEGPN 613
M P+++ ++ L+ K + + +M + + HN
Sbjct: 674 MAQSRPFPSIIEFSKLLSAIAKMNKFDLVISFGEKM----------EILGISHN------ 717
Query: 614 VITYGALVDGLCKANRVKEAHELLDTMLAHGCEPNQIVYDAVIDGFCKIGKLQDAQEVFT 673
+ TY L++ C+ +R+ A LL M+ G EP+ + +++++GFC ++ DA +
Sbjct: 718 LYTYNILINCFCRCSRLSLALALLGKMMKLGYEPDIVTLNSLLNGFCHGNRISDAVALVD 777
Query: 674 KMSERGYSPNLYTYSSFIDCLFKDNRLDLVLKVLSKMLENSCTPNVVIYTEMVDGLCKIG 733
+M E GY P+ T+++ I LF N+ + ++ +M++ C P++V Y +V+GLCK G
Sbjct: 778 QMVEMGYKPDTVTFTTLIHGLFLHNKASEAVALIDRMVQRGCQPDLVTYGAVVNGLCKRG 837
Query: 734 KTDEAYKLMLKMEEKGCNPNVVTYTAMIDGFGKSGKIEQCLELFRDMCSKGCAPNFITYR 793
TD A L+ KME NVV Y+ +ID K + L LF +M +KG PN ITY
Sbjct: 838 DTDLALNLLNKMEAAKIEANVVIYSTVIDSLCKYRHEDDALNLFTEMENKGVRPNVITYS 897
Query: 794 VLINHCCSNGLLDEAYKLLDEMKQTYWPKHILSHRKIIEGFSQE--FITSIGLLDELSEN 851
LI+ C+ G +A +LL +M + ++++ +I+ F ++ + + L +E+ +
Sbjct: 898 SLISCLCNYGRWSDASRLLSDMIERKINPNLVTFSALIDAFVKKGKLVKAEKLYEEMIKR 957
Query: 852 ESVPVDSLYRILIDNYIKAGRLEVALDLLEEISSSPSHAVSNKYLYASLIENLSHASKVD 911
P Y LI+ + RL A +LE + + N Y +LI A +VD
Sbjct: 958 SIDPNIFTYSSLINGFCMLDRLGEAKQMLELMIRKD--CLPNVVTYNTLINGFCKAKRVD 1015
Query: 912 KALELYASMISKNVVPELSILVHLIKGLIK 941
K +EL+ M + +V LI G +
Sbjct: 1016 KGMELFREMSQRGLVGNTVTYTTLIHGFFQ 1045
Score = 273 bits (697), Expect = 5e-73
Identities = 208/884 (23%), Positives = 378/884 (42%), Gaps = 81/884 (9%)
Query: 140 FDKLLDLLGCNVNADDRVPLKFLMEIKDDDHELLRRLLNFLVRKCCRNGWWNMALEELGR 199
F+KLL + + + L M+ H+L + + CR ++AL L +
Sbjct: 86 FNKLLSAVAKMNKFELVISLGEQMQTLGISHDLYT--YSIFINCFCRRSQLSLALAVLAK 143
Query: 200 LKDFGYKPSQTTYNALIQVFLRADKLDTAYLVKREMLSYAFVMDRYTLSCFAYSLCKGGK 259
+ GY+P T ++L+ + + ++ A + +M+ + D +T + + L K
Sbjct: 144 MMKLGYEPDIVTLSSLLNGYCHSKRISDAVALVDQMVEMGYKPDTFTFTTLIHGLFLHNK 203
Query: 260 CREAFDLIDE--AEDFVPDTVFYNRMVSGLCEASLFEEAMDILHRMRSSSCIPNVVTYRI 317
EA L+D+ PD V Y +V+GLC+ + A+ +L +M +VV Y
Sbjct: 204 ASEAVALVDQMVQRGCQPDLVTYGTVVNGLCKRGDIDLALSLLKKMEKGKIEADVVIYNT 263
Query: 318 LLSGCLRKGQLGRCKRILSMMITEGCYPNREIFNSLIHAYCKSRDYSYAYKLFKKMIKCG 377
++ G + + + + M +G P+ ++SLI C +S A +L MI+
Sbjct: 264 IIDGLCKYKHMDDALNLFTEMDNKGIRPDVFTYSSLISCLCNYGRWSDASRLLSDMIERK 323
Query: 378 CQPGYLVYNIFIGSVCSNEEQPSSDILDLVEKAYSEMLDLGVVLNKVNVSNFARCLCGAG 437
P + ++ I + + L EK Y EM+ + + S+ C
Sbjct: 324 INPNVVTFSALIDAFVKEGK------LVEAEKLYDEMIKRSIDPDIFTYSSLINGFCMHD 377
Query: 438 KFDQAFKIICEMMGKGFVPDDSTYSKVIGFLCDASKVEKAFSLFEEMKRNGIVPSVYTYT 497
+ D+A + M+ K P+ TYS +I C A +VE+ LF EM + G+V + TYT
Sbjct: 378 RLDEAKHMFELMISKDCFPNVVTYSTLIKGFCKAKRVEEGMELFREMSQRGLVGNTVTYT 437
Query: 498 ILIDSFCKAGLIQQARKWFDEMLHKGCTPNVVTYTALIHAYLKAKQMPVADELFEMMLLE 557
LI F +A A+ F +M+ G PN++TY L+ K ++ A +FE +
Sbjct: 438 TLIHGFFQARDCDNAQMVFKQMVSVGVHPNILTYNILLDGLCKNGKLAKAMVVFEYLQRS 497
Query: 558 GCKPNVVTYTALIDGHCKAGQIEKACQIYARMRGDIESSDMDKYFKLDHNNCEGPNVITY 617
+P++ TY +I+G CKAG++E +++ + S PNVI Y
Sbjct: 498 TMEPDIYTYNIMIEGMCKAGKVEDGWELFCNLSLKGVS----------------PNVIAY 541
Query: 618 GALVDGLCKANRVKEAHELLDTMLAHGCEPNQIVYDAVIDGFCKIGKLQDAQEVFTKMSE 677
++ G C+ +EA LL M G PN Y+ +I + G + + E+ +M
Sbjct: 542 NTMISGFCRKGSKEEADSLLKKMKEDGPLPNSGTYNTLIRARLRDGDREASAELIKEMRS 601
Query: 678 RGYS------------------PN----------------------LYTYSSFIDCLFKD 697
G++ PN Y Y + D
Sbjct: 602 CGFAGDASTIGLSSKFKREIEKPNRENACKNIIFSEEICSSESCGESYDYREVLRTGLSD 661
Query: 698 NRLDLVLKVLSKMLENSCTPNVVIYTEMVDGLCKIGKTDEAYKLMLKMEEKGCNPNVVTY 757
LD + + M ++ P+++ +++++ + K+ K D KME G + N+ TY
Sbjct: 662 IELDDAIGLFGVMAQSRPFPSIIEFSKLLSAIAKMNKFDLVISFGEKMEILGISHNLYTY 721
Query: 758 TAMIDGFGKSGKIEQCLELFRDMCSKGCAPNFITYRVLINHCCSNGLLDEAYKLLDEMKQ 817
+I+ F + ++ L L M G P+ +T L+N C + +A L+D+M +
Sbjct: 722 NILINCFCRCSRLSLALALLGKMMKLGYEPDIVTLNSLLNGFCHGNRISDAVALVDQMVE 781
Query: 818 TYWPKHILSHRKIIEG--FSQEFITSIGLLDELSENESVPVDSLYRILIDNYIKAGRLEV 875
+ ++ +I G + ++ L+D + + P Y +++ K G ++
Sbjct: 782 MGYKPDTVTFTTLIHGLFLHNKASEAVALIDRMVQRGCQPDLVTYGAVVNGLCKRGDTDL 841
Query: 876 ALDLLEEISSSPSHAVSNKYLYASLIENLSHASKVDKALELYASMISKNVVPELSILVHL 935
AL+LL ++ ++ A N +Y+++I++L D AL L+ M +K V P + L
Sbjct: 842 ALNLLNKMEAAKIEA--NVVIYSTVIDSLCKYRHEDDALNLFTEMENKGVRPNVITYSSL 899
Query: 936 IKGLIKVDKWQEALQLSDSICQMVCLTLSQTFQPLVNLNLLTLS 979
I L +W +A +L LS + +N NL+T S
Sbjct: 900 ISCLCNYGRWSDASRL-----------LSDMIERKINPNLVTFS 932
Score = 272 bits (696), Expect = 6e-73
Identities = 204/840 (24%), Positives = 362/840 (42%), Gaps = 91/840 (10%)
Query: 180 LVRKCCRNGWWNMALEELGRLKDFGYKPSQTTYNALIQVFLRADKLDTAYLVKREMLSYA 239
L+ C G W+ A L + + P+ T++ALI F++ KL A + EM+ +
Sbjct: 299 LISCLCNYGRWSDASRLLSDMIERKINPNVVTFSALIDAFVKEGKLVEAEKLYDEMIKRS 358
Query: 240 FVMDRYTLSCFAYSLCKGGKCREAFDLIDE--AEDFVPDTVFYNRMVSGLCEASLFEEAM 297
D +T S C + EA + + ++D P+ V Y+ ++ G C+A EE M
Sbjct: 359 IDPDIFTYSSLINGFCMHDRLDEAKHMFELMISKDCFPNVVTYSTLIKGFCKAKRVEEGM 418
Query: 298 DILHRMRSSSCIPNVVTYRILLSGCLRKGQLGRCKRILSMMITEGCYPNREIFNSLIHAY 357
++ M + N VTY L+ G + + + M++ G +PN +N L+
Sbjct: 419 ELFREMSQRGLVGNTVTYTTLIHGFFQARDCDNAQMVFKQMVSVGVHPNILTYNILLDGL 478
Query: 358 CKSRDYSYAYKLFKKMIKCGCQPGYLVYNIFIGSVCS---------------------NE 396
CK+ + A +F+ + + +P YNI I +C N
Sbjct: 479 CKNGKLAKAMVVFEYLQRSTMEPDIYTYNIMIEGMCKAGKVEDGWELFCNLSLKGVSPNV 538
Query: 397 EQPSSDILDLVEKAYSEMLDL--------GVVLNKVNVSNFARCLCGAGKFDQAFKIICE 448
++ I K E D G + N + R G + + ++I E
Sbjct: 539 IAYNTMISGFCRKGSKEEADSLLKKMKEDGPLPNSGTYNTLIRARLRDGDREASAELIKE 598
Query: 449 MMGKGFVPDDST----------------------------------------YSKVIGFL 468
M GF D ST Y +V+
Sbjct: 599 MRSCGFAGDASTIGLSSKFKREIEKPNRENACKNIIFSEEICSSESCGESYDYREVLRTG 658
Query: 469 CDASKVEKAFSLFEEMKRNGIVPSVYTYTILIDSFCKAGLIQQARKWFDEMLHKGCTPNV 528
+++ A LF M ++ PS+ ++ L+ + K + ++M G + N+
Sbjct: 659 LSDIELDDAIGLFGVMAQSRPFPSIIEFSKLLSAIAKMNKFDLVISFGEKMEILGISHNL 718
Query: 529 VTYTALIHAYLKAKQMPVADELFEMMLLEGCKPNVVTYTALIDGHCKAGQIEKACQIYAR 588
TY LI+ + + ++ +A L M+ G +P++VT +L++G C +I A + +
Sbjct: 719 YTYNILINCFCRCSRLSLALALLGKMMKLGYEPDIVTLNSLLNGFCHGNRISDAVALVDQ 778
Query: 589 MRGDIESSDMDKYFKLDHNNCEGPNVITYGALVDGLCKANRVKEAHELLDTMLAHGCEPN 648
M ++ +K P+ +T+ L+ GL N+ EA L+D M+ GC+P+
Sbjct: 779 M--------VEMGYK--------PDTVTFTTLIHGLFLHNKASEAVALIDRMVQRGCQPD 822
Query: 649 QIVYDAVIDGFCKIGKLQDAQEVFTKMSERGYSPNLYTYSSFIDCLFKDNRLDLVLKVLS 708
+ Y AV++G CK G A + KM N+ YS+ ID L K D L + +
Sbjct: 823 LVTYGAVVNGLCKRGDTDLALNLLNKMEAAKIEANVVIYSTVIDSLCKYRHEDDALNLFT 882
Query: 709 KMLENSCTPNVVIYTEMVDGLCKIGKTDEAYKLMLKMEEKGCNPNVVTYTAMIDGFGKSG 768
+M PNV+ Y+ ++ LC G+ +A +L+ M E+ NPN+VT++A+ID F K G
Sbjct: 883 EMENKGVRPNVITYSSLISCLCNYGRWSDASRLLSDMIERKINPNLVTFSALIDAFVKKG 942
Query: 769 KIEQCLELFRDMCSKGCAPNFITYRVLINHCCSNGLLDEAYKLLDEMKQTYWPKHILSHR 828
K+ + +L+ +M + PN TY LIN C L EA ++L+ M + +++++
Sbjct: 943 KLVKAEKLYEEMIKRSIDPNIFTYSSLINGFCMLDRLGEAKQMLELMIRKDCLPNVVTYN 1002
Query: 829 KIIEGFSQEFITSIG--LLDELSENESVPVDSLYRILIDNYIKAGRLEVALDLLEEISSS 886
+I GF + G L E+S+ V Y LI + +A + A + +++ S
Sbjct: 1003 TLINGFCKAKRVDKGMELFREMSQRGLVGNTVTYTTLIHGFFQARDCDNAQMVFKQMVSV 1062
Query: 887 PSHAVSNKYLYASLIENLSHASKVDKALELYASMISKNVVPELSILVHLIKGLIKVDKWQ 946
H N Y L++ L K+ KA+ ++ + + P++ +I+G+ K KW+
Sbjct: 1063 GVHP--NILTYNILLDGLCKNGKLAKAMVVFEYLQRSTMEPDIYTYNIMIEGMCKAGKWK 1120
Score = 241 bits (616), Expect = 1e-63
Identities = 156/540 (28%), Positives = 266/540 (48%), Gaps = 20/540 (3%)
Query: 438 KFDQAFKIICEMMGKGFVPDDSTYSKVIGFLCDASKVEKAFSLFEEMKRNGIVPSVYTYT 497
K D A + +M+ P ++K++ + +K E SL E+M+ GI +YTY+
Sbjct: 63 KVDDAVDLFGDMVKSRPFPSIVEFNKLLSAVAKMNKFELVISLGEQMQTLGISHDLYTYS 122
Query: 498 ILIDSFCKAGLIQQARKWFDEMLHKGCTPNVVTYTALIHAYLKAKQMPVADELFEMMLLE 557
I I+ FC+ + A +M+ G P++VT ++L++ Y +K++ A L + M+
Sbjct: 123 IFINCFCRRSQLSLALAVLAKMMKLGYEPDIVTLSSLLNGYCHSKRISDAVALVDQMVEM 182
Query: 558 GCKPNVVTYTALIDGHCKAGQIEKACQIYARMRGDIESSDMDKYFKLDHNNCEGPNVITY 617
G KP+ T+T LI G + +A + +M C+ P+++TY
Sbjct: 183 GYKPDTFTFTTLIHGLFLHNKASEAVALVDQMV---------------QRGCQ-PDLVTY 226
Query: 618 GALVDGLCKANRVKEAHELLDTMLAHGCEPNQIVYDAVIDGFCKIGKLQDAQEVFTKMSE 677
G +V+GLCK + A LL M E + ++Y+ +IDG CK + DA +FT+M
Sbjct: 227 GTVVNGLCKRGDIDLALSLLKKMEKGKIEADVVIYNTIIDGLCKYKHMDDALNLFTEMDN 286
Query: 678 RGYSPNLYTYSSFIDCLFKDNRLDLVLKVLSKMLENSCTPNVVIYTEMVDGLCKIGKTDE 737
+G P+++TYSS I CL R ++LS M+E PNVV ++ ++D K GK E
Sbjct: 287 KGIRPDVFTYSSLISCLCNYGRWSDASRLLSDMIERKINPNVVTFSALIDAFVKEGKLVE 346
Query: 738 AYKLMLKMEEKGCNPNVVTYTAMIDGFGKSGKIEQCLELFRDMCSKGCAPNFITYRVLIN 797
A KL +M ++ +P++ TY+++I+GF ++++ +F M SK C PN +TY LI
Sbjct: 347 AEKLYDEMIKRSIDPDIFTYSSLINGFCMHDRLDEAKHMFELMISKDCFPNVVTYSTLIK 406
Query: 798 HCCSNGLLDEAYKLLDEMKQTYWPKHILSHRKIIEGFSQ--EFITSIGLLDELSENESVP 855
C ++E +L EM Q + +++ +I GF Q + + + ++ P
Sbjct: 407 GFCKAKRVEEGMELFREMSQRGLVGNTVTYTTLIHGFFQARDCDNAQMVFKQMVSVGVHP 466
Query: 856 VDSLYRILIDNYIKAGRLEVALDLLEEISSSPSHAVSNKYLYASLIENLSHASKVDKALE 915
Y IL+D K G+L A+ + E + S + Y Y +IE + A KV+ E
Sbjct: 467 NILTYNILLDGLCKNGKLAKAMVVFEYLQRSTME--PDIYTYNIMIEGMCKAGKVEDGWE 524
Query: 916 LYASMISKNVVPELSILVHLIKGLIKVDKWQEALQLSDSICQMVCLTLSQTFQPLVNLNL 975
L+ ++ K V P + +I G + +EA L + + L S T+ L+ L
Sbjct: 525 LFCNLSLKGVSPNVIAYNTMISGFCRKGSKEEADSLLKKMKEDGPLPNSGTYNTLIRARL 584
Score = 176 bits (446), Expect = 6e-44
Identities = 121/441 (27%), Positives = 202/441 (45%), Gaps = 10/441 (2%)
Query: 140 FDKLLDLLGCNVNADDRVPLKFLMEIKDDDHELLRRLLNFLVRKCCRNGWWNMALEELGR 199
F KLL + D + MEI H L N L+ CR ++AL LG+
Sbjct: 686 FSKLLSAIAKMNKFDLVISFGEKMEILGISHNLYT--YNILINCFCRCSRLSLALALLGK 743
Query: 200 LKDFGYKPSQTTYNALIQVFLRADKLDTAYLVKREMLSYAFVMDRYTLSCFAYSLCKGGK 259
+ GY+P T N+L+ F +++ A + +M+ + D T + + L K
Sbjct: 744 MMKLGYEPDIVTLNSLLNGFCHGNRISDAVALVDQMVEMGYKPDTVTFTTLIHGLFLHNK 803
Query: 260 CREAFDLIDE--AEDFVPDTVFYNRMVSGLCEASLFEEAMDILHRMRSSSCIPNVVTYRI 317
EA LID PD V Y +V+GLC+ + A+++L++M ++ NVV Y
Sbjct: 804 ASEAVALIDRMVQRGCQPDLVTYGAVVNGLCKRGDTDLALNLLNKMEAAKIEANVVIYST 863
Query: 318 LLSGCLRKGQLGRCKRILSMMITEGCYPNREIFNSLIHAYCKSRDYSYAYKLFKKMIKCG 377
++ + + + M +G PN ++SLI C +S A +L MI+
Sbjct: 864 VIDSLCKYRHEDDALNLFTEMENKGVRPNVITYSSLISCLCNYGRWSDASRLLSDMIERK 923
Query: 378 CQPGYLVYNIFIGSVCSNEEQPSSDILDLVEKAYSEMLDLGVVLNKVNVSNFARCLCGAG 437
P + ++ I + + L EK Y EM+ + N S+ C
Sbjct: 924 INPNLVTFSALIDAFVKKGK------LVKAEKLYEEMIKRSIDPNIFTYSSLINGFCMLD 977
Query: 438 KFDQAFKIICEMMGKGFVPDDSTYSKVIGFLCDASKVEKAFSLFEEMKRNGIVPSVYTYT 497
+ +A +++ M+ K +P+ TY+ +I C A +V+K LF EM + G+V + TYT
Sbjct: 978 RLGEAKQMLELMIRKDCLPNVVTYNTLINGFCKAKRVDKGMELFREMSQRGLVGNTVTYT 1037
Query: 498 ILIDSFCKAGLIQQARKWFDEMLHKGCTPNVVTYTALIHAYLKAKQMPVADELFEMMLLE 557
LI F +A A+ F +M+ G PN++TY L+ K ++ A +FE +
Sbjct: 1038 TLIHGFFQARDCDNAQMVFKQMVSVGVHPNILTYNILLDGLCKNGKLAKAMVVFEYLQRS 1097
Query: 558 GCKPNVVTYTALIDGHCKAGQ 578
+P++ TY +I+G CKAG+
Sbjct: 1098 TMEPDIYTYNIMIEGMCKAGK 1118
>At5g65560 putative protein
Length = 915
Score = 293 bits (750), Expect = 3e-79
Identities = 247/922 (26%), Positives = 401/922 (42%), Gaps = 100/922 (10%)
Query: 40 PSTPTPRDLSQDYAFLRNTLINSTSPQSTPSSGDDAISTISKALKTGFNIETHQFFRQFR 99
PS T R LRN + S P +S +SK N +
Sbjct: 34 PSPVTRRQFCSVSPLLRNLPEEESDSMSVPHR---LLSILSKP-----NWHKSPSLKSMV 85
Query: 100 NQLNDSLVVEVMNNVKNPELCVKFFLWAGRQIGYSHTPQVFDKLLDLLGCNVNADDRVPL 159
+ ++ S V + + +P+ + F W + Y H+ + LL LL N +
Sbjct: 86 SAISPSHVSSLFSLDLDPKTALNFSHWISQNPRYKHSVYSYASLLTLLINNGYVGVVFKI 145
Query: 160 KFLMEIKDDDHELLRRLLNFLVRKCCRNGWWNMALEELGRLKDFGYKPSQTTYNALIQVF 219
+ LM IK D + L RK M +E LK YK YN L+
Sbjct: 146 RLLM-IKSCDSVGDALYVLDLCRK--------MNKDERFELK---YKLIIGCYNTLLNSL 193
Query: 220 LRADKLDTAYLVKREMLSYAFVMDRYTLSCFAYSLCKGGKCREAFDLIDEAEDFVPDTVF 279
R +D V EML + P+
Sbjct: 194 ARFGLVDEMKQVYMEMLE---------------------------------DKVCPNIYT 220
Query: 280 YNRMVSGLCEASLFEEAMDILHRMRSSSCIPNVVTYRILLSGCLRKGQLGRCKRILSMMI 339
YN+MV+G C+ EEA + ++ + P+ TY L+ G ++ L ++ + M
Sbjct: 221 YNKMVNGYCKLGNVEEANQYVSKIVEAGLDPDFFTYTSLIMGYCQRKDLDSAFKVFNEMP 280
Query: 340 TEGCYPNREIFNSLIHAYCKSRDYSYAYKLFKKMIKCGCQPGYLVYNIFIGSVCSNEEQP 399
+GC N + LIH C +R A LF KM C P Y + I S+C +E +
Sbjct: 281 LKGCRRNEVAYTHLIHGLCVARRIDEAMDLFVKMKDDECFPTVRTYTVLIKSLCGSERK- 339
Query: 400 SSDILDLVEKAYSEMLDLGVVLNKVNVSNFARCLCGAGKFDQAFKIICEMMGKGFVPDDS 459
S+ L+LV+ EM + G+ N + LC KF++A +++ +M+ KG +P+
Sbjct: 340 -SEALNLVK----EMEETGIKPNIHTYTVLIDSLCSQCKFEKARELLGQMLEKGLMPNVI 394
Query: 460 TYSKVIGFLCDASKVEKAFSLFEEMKRNGIVPSVYTYTILIDSFCKAGLIQQARKWFDEM 519
TY+ +I C +E A + E M+ + P+ TY LI +CK+ + +A ++M
Sbjct: 395 TYNALINGYCKRGMIEDAVDVVELMESRKLSPNTRTYNELIKGYCKSN-VHKAMGVLNKM 453
Query: 520 LHKGCTPNVVTYTALIHAYLKAKQMPVADELFEMMLLEGCKPNVVTYTALIDGHCKAGQI 579
L + P+VVTY +LI ++ A L +M G P+ TYT++ID CK+ ++
Sbjct: 454 LERKVLPDVVTYNSLIDGQCRSGNFDSAYRLLSLMNDRGLVPDQWTYTSMIDSLCKSKRV 513
Query: 580 EKACQIYARMRGDIESSDMDKYFKLDHNNCEGPNVITYGALVDGLCKANRVKEAHELLDT 639
E+AC ++ +E ++ PNV+ Y AL+DG CKA +V EAH +L+
Sbjct: 514 EEACDLF----DSLEQKGVN------------PNVVMYTALIDGYCKAGKVDEAHLMLEK 557
Query: 640 MLAHGCEPNQIVYDAVIDGFCKIGKLQDAQEVFTKMSERGYSPNLYTYSSFIDCLFKDNR 699
ML+ C PN + ++A+I G C GKL++A + KM + G P + T + I L KD
Sbjct: 558 MLSKNCLPNSLTFNALIHGLCADGKLKEATLLEEKMVKIGLQPTVSTDTILIHRLLKDGD 617
Query: 700 LDLVLKVLSKMLENSCTPNVVIYTEMVDGLCKIGKTDEAYKLMLKMEEKGCNPNVVTYTA 759
D +ML + P+ YT + C+ G+ +A +M KM E G +P++ TY++
Sbjct: 618 FDHAYSRFQQMLSSGTKPDAHTYTTFIQTYCREGRLLDAEDMMAKMRENGVSPDLFTYSS 677
Query: 760 MIDGFGKSGKIEQCLELFRDMCSKGCAPNFITYRVLINHCCSNGLLDEAYKLLDEMKQTY 819
+I G+G G+ ++ + M GC P+ T+ LI H LL+ Y KQ
Sbjct: 678 LIKGYGDLGQTNFAFDVLKRMRDTGCEPSQHTFLSLIKH-----LLEMKYG-----KQKG 727
Query: 820 WPKHILSHRKIIEGFSQEFITSIGLLDELSENESVPVDSLYRILIDNYIKAGRLEVALDL 879
+ + ++ EF T + LL+++ E+ P Y LI + G L VA +
Sbjct: 728 SEPELCAMSNMM-----EFDTVVELLEKMVEHSVTPNAKSYEKLILGICEVGNLRVAEKV 782
Query: 880 LEEISS----SPSHAVSNKYLYASLIENLSHASKVDKALELYASMISKNVVPELSILVHL 935
+ + SPS V N +L+ K ++A ++ MI +P+L L
Sbjct: 783 FDHMQRNEGISPSELVFN-----ALLSCCCKLKKHNEAAKVVDDMICVGHLPQLESCKVL 837
Query: 936 IKGLIKVDKWQEALQLSDSICQ 957
I GL K + + + ++ Q
Sbjct: 838 ICGLYKKGEKERGTSVFQNLLQ 859
Score = 265 bits (676), Expect = 1e-70
Identities = 182/711 (25%), Positives = 328/711 (45%), Gaps = 46/711 (6%)
Query: 178 NFLVRKCCRNGWWNMALEELGRLKDFGYKPSQTTYNALIQVFLRADKLDTAYLVKREMLS 237
N +V C+ G A + + ++ + G P TY +LI + + LD+A+ V EM
Sbjct: 222 NKMVNGYCKLGNVEEANQYVSKIVEAGLDPDFFTYTSLIMGYCQRKDLDSAFKVFNEMPL 281
Query: 238 YAFVMDRYTLSCFAYSLCKGGKCREAFDLIDEAED--FVPDTVFYNRMVSGLCEASLFEE 295
+ + + LC + EA DL + +D P Y ++ LC + E
Sbjct: 282 KGCRRNEVAYTHLIHGLCVARRIDEAMDLFVKMKDDECFPTVRTYTVLIKSLCGSERKSE 341
Query: 296 AMDILHRMRSSSCIPNVVTYRILLSGCLRKGQLGRCKRILSMMITEGCYPNREIFNSLIH 355
A++++ M + PN+ TY +L+ + + + + +L M+ +G PN +N+LI+
Sbjct: 342 ALNLVKEMEETGIKPNIHTYTVLIDSLCSQCKFEKARELLGQMLEKGLMPNVITYNALIN 401
Query: 356 AYCKSRDYSYAYKLFKKMIKCGCQPGYLVYNIFIGSVCSNEEQPSSDILDLVEKAYSEML 415
YCK A + + M P YN I C + + +L ++ML
Sbjct: 402 GYCKRGMIEDAVDVVELMESRKLSPNTRTYNELIKGYCKSNVHKAMGVL-------NKML 454
Query: 416 DLGVVLNKVNVSNFARCLCGAGKFDQAFKIICEMMGKGFVPDDSTYSKVIGFLCDASKVE 475
+ V+ + V ++ C +G FD A++++ M +G VPD TY+ +I LC + +VE
Sbjct: 455 ERKVLPDVVTYNSLIDGQCRSGNFDSAYRLLSLMNDRGLVPDQWTYTSMIDSLCKSKRVE 514
Query: 476 KAFSLFEEMKRNGIVPSVYTYTILIDSFCKAGLIQQARKWFDEMLHKGCTPNVVTYTALI 535
+A LF+ +++ G+ P+V YT LID +CKAG + +A ++ML K C PN +T+ ALI
Sbjct: 515 EACDLFDSLEQKGVNPNVVMYTALIDGYCKAGKVDEAHLMLEKMLSKNCLPNSLTFNALI 574
Query: 536 HAYLKAKQMPVADELFEMMLLEGCKPNVVTYTALIDGHCKAGQIEKACQIYARMRGDIES 595
H ++ A L E M+ G +P V T T LI K G + A Y+R + + S
Sbjct: 575 HGLCADGKLKEATLLEEKMVKIGLQPTVSTDTILIHRLLKDGDFDHA---YSRFQQMLSS 631
Query: 596 SDMDKYFKLDHNNCEGPNVITYGALVDGLCKANRVKEAHELLDTMLAHGCEPNQIVYDAV 655
P+ TY + C+ R+ +A +++ M +G P+ Y ++
Sbjct: 632 GTK-------------PDAHTYTTFIQTYCREGRLLDAEDMMAKMRENGVSPDLFTYSSL 678
Query: 656 IDGFCKIGKLQDAQEVFTKMSERGYSPNLYTYSSFIDCLFKDN----------------- 698
I G+ +G+ A +V +M + G P+ +T+ S I L +
Sbjct: 679 IKGYGDLGQTNFAFDVLKRMRDTGCEPSQHTFLSLIKHLLEMKYGKQKGSEPELCAMSNM 738
Query: 699 -RLDLVLKVLSKMLENSCTPNVVIYTEMVDGLCKIGKTDEAYKLMLKME-EKGCNPNVVT 756
D V+++L KM+E+S TPN Y +++ G+C++G A K+ M+ +G +P+ +
Sbjct: 739 MEFDTVVELLEKMVEHSVTPNAKSYEKLILGICEVGNLRVAEKVFDHMQRNEGISPSELV 798
Query: 757 YTAMIDGFGKSGKIEQCLELFRDMCSKGCAPNFITYRVLINHCCSNGLLDEAYKLLDEMK 816
+ A++ K K + ++ DM G P + +VLI G + + +
Sbjct: 799 FNALLSCCCKLKKHNEAAKVVDDMICVGHLPQLESCKVLICGLYKKGEKERGTSVFQNLL 858
Query: 817 QTYWPKHILSHRKIIEGFSQEFITS--IGLLDELSENESVPVDSLYRILID 865
Q + + L+ + II+G ++ + L + + +N Y +LI+
Sbjct: 859 QCGYYEDELAWKIIIDGVGKQGLVEAFYELFNVMEKNGCKFSSQTYSLLIE 909
Score = 256 bits (655), Expect = 3e-68
Identities = 161/532 (30%), Positives = 266/532 (49%), Gaps = 24/532 (4%)
Query: 461 YSKVIGFLCDASKVEKAFSLFEEMKRNGIVPSVYTYTILIDSFCKAGLIQQARKWFDEML 520
Y+ ++ L V++ ++ EM + + P++YTY +++ +CK G +++A ++ +++
Sbjct: 186 YNTLLNSLARFGLVDEMKQVYMEMLEDKVCPNIYTYNKMVNGYCKLGNVEEANQYVSKIV 245
Query: 521 HKGCTPNVVTYTALIHAYLKAKQMPVADELFEMMLLEGCKPNVVTYTALIDGHCKAGQIE 580
G P+ TYT+LI Y + K + A ++F M L+GC+ N V YT LI G C A +I+
Sbjct: 246 EAGLDPDFFTYTSLIMGYCQRKDLDSAFKVFNEMPLKGCRRNEVAYTHLIHGLCVARRID 305
Query: 581 KACQIYARMRGDIESSDMDKYFKLDHNNCEG-------------------PNVITYGALV 621
+A ++ +M+ D + Y L + C PN+ TY L+
Sbjct: 306 EAMDLFVKMKDDECFPTVRTYTVLIKSLCGSERKSEALNLVKEMEETGIKPNIHTYTVLI 365
Query: 622 DGLCKANRVKEAHELLDTMLAHGCEPNQIVYDAVIDGFCKIGKLQDAQEVFTKMSERGYS 681
D LC + ++A ELL ML G PN I Y+A+I+G+CK G ++DA +V M R S
Sbjct: 366 DSLCSQCKFEKARELLGQMLEKGLMPNVITYNALINGYCKRGMIEDAVDVVELMESRKLS 425
Query: 682 PNLYTYSSFIDCLFKDNRLDLVLKVLSKMLENSCTPNVVIYTEMVDGLCKIGKTDEAYKL 741
PN TY+ I K N + + VL+KMLE P+VV Y ++DG C+ G D AY+L
Sbjct: 426 PNTRTYNELIKGYCKSN-VHKAMGVLNKMLERKVLPDVVTYNSLIDGQCRSGNFDSAYRL 484
Query: 742 MLKMEEKGCNPNVVTYTAMIDGFGKSGKIEQCLELFRDMCSKGCAPNFITYRVLINHCCS 801
+ M ++G P+ TYT+MID KS ++E+ +LF + KG PN + Y LI+ C
Sbjct: 485 LSLMNDRGLVPDQWTYTSMIDSLCKSKRVEEACDLFDSLEQKGVNPNVVMYTALIDGYCK 544
Query: 802 NGLLDEAYKLLDEMKQTYWPKHILSHRKIIEGFSQEF-ITSIGLLDELSENESV-PVDSL 859
G +DEA+ +L++M + L+ +I G + + LL+E + P S
Sbjct: 545 AGKVDEAHLMLEKMLSKNCLPNSLTFNALIHGLCADGKLKEATLLEEKMVKIGLQPTVST 604
Query: 860 YRILIDNYIKAGRLEVALDLLEEISSSPSHAVSNKYLYASLIENLSHASKVDKALELYAS 919
ILI +K G + A +++ SS + + + Y + I+ ++ A ++ A
Sbjct: 605 DTILIHRLLKDGDFDHAYSRFQQMLSSGTKP--DAHTYTTFIQTYCREGRLLDAEDMMAK 662
Query: 920 MISKNVVPELSILVHLIKGLIKVDKWQEALQLSDSICQMVCLTLSQTFQPLV 971
M V P+L LIKG + + A + + C TF L+
Sbjct: 663 MRENGVSPDLFTYSSLIKGYGDLGQTNFAFDVLKRMRDTGCEPSQHTFLSLI 714
Score = 226 bits (575), Expect = 6e-59
Identities = 158/615 (25%), Positives = 291/615 (46%), Gaps = 22/615 (3%)
Query: 159 LKFLMEIKDDDHELLRRLLNFLVRKCCRNGWWNMALEELGRLKDFGYKPSQTTYNALIQV 218
+ +++KDD+ R L++ C + + AL + +++ G KP+ TY LI
Sbjct: 308 MDLFVKMKDDECFPTVRTYTVLIKSLCGSERKSEALNLVKEMEETGIKPNIHTYTVLIDS 367
Query: 219 FLRADKLDTAYLVKREMLSYAFVMDRYTLSCFAYSLCKGGKCREAFDLID--EAEDFVPD 276
K + A + +ML + + T + CK G +A D+++ E+ P+
Sbjct: 368 LCSQCKFEKARELLGQMLEKGLMPNVITYNALINGYCKRGMIEDAVDVVELMESRKLSPN 427
Query: 277 TVFYNRMVSGLCEASLFEEAMDILHRMRSSSCIPNVVTYRILLSGCLRKGQLGRCKRILS 336
T YN ++ G C++++ +AM +L++M +P+VVTY L+ G R G R+LS
Sbjct: 428 TRTYNELIKGYCKSNV-HKAMGVLNKMLERKVLPDVVTYNSLIDGQCRSGNFDSAYRLLS 486
Query: 337 MMITEGCYPNREIFNSLIHAYCKSRDYSYAYKLFKKMIKCGCQPGYLVYNIFIGSVCSNE 396
+M G P++ + S+I + CKS+ A LF + + G P ++Y I C
Sbjct: 487 LMNDRGLVPDQWTYTSMIDSLCKSKRVEEACDLFDSLEQKGVNPNVVMYTALIDGYCKAG 546
Query: 397 EQPSSDILDLVEKAYSEMLDLGVVLNKVNVSNFARCLCGAGKFDQAFKIICEMMGKGFVP 456
+ + ++ +ML + N + + LC GK +A + +M+ G P
Sbjct: 547 KVDEAHLM------LEKMLSKNCLPNSLTFNALIHGLCADGKLKEATLLEEKMVKIGLQP 600
Query: 457 DDSTYSKVIGFLCDASKVEKAFSLFEEMKRNGIVPSVYTYTILIDSFCKAGLIQQARKWF 516
ST + +I L + A+S F++M +G P +TYT I ++C+ G + A
Sbjct: 601 TVSTDTILIHRLLKDGDFDHAYSRFQQMLSSGTKPDAHTYTTFIQTYCREGRLLDAEDMM 660
Query: 517 DEMLHKGCTPNVVTYTALIHAYLKAKQMPVADELFEMMLLEGCKPNVVTYTALID----- 571
+M G +P++ TY++LI Y Q A ++ + M GC+P+ T+ +LI
Sbjct: 661 AKMRENGVSPDLFTYSSLIKGYGDLGQTNFAFDVLKRMRDTGCEPSQHTFLSLIKHLLEM 720
Query: 572 --GHCKAGQIEKACQIYARMRGDIESSDMDKYFKLDHNNCEGPNVITYGALVDGLCKANR 629
G K + E C + M D ++K ++H+ PN +Y L+ G+C+
Sbjct: 721 KYGKQKGSEPE-LCAMSNMMEFDTVVELLEKM--VEHSVT--PNAKSYEKLILGICEVGN 775
Query: 630 VKEAHELLDTMLAH-GCEPNQIVYDAVIDGFCKIGKLQDAQEVFTKMSERGYSPNLYTYS 688
++ A ++ D M + G P+++V++A++ CK+ K +A +V M G+ P L +
Sbjct: 776 LRVAEKVFDHMQRNEGISPSELVFNALLSCCCKLKKHNEAAKVVDDMICVGHLPQLESCK 835
Query: 689 SFIDCLFKDNRLDLVLKVLSKMLENSCTPNVVIYTEMVDGLCKIGKTDEAYKLMLKMEEK 748
I L+K + V +L+ + + + ++DG+ K G + Y+L ME+
Sbjct: 836 VLICGLYKKGEKERGTSVFQNLLQCGYYEDELAWKIIIDGVGKQGLVEAFYELFNVMEKN 895
Query: 749 GCNPNVVTYTAMIDG 763
GC + TY+ +I+G
Sbjct: 896 GCKFSSQTYSLLIEG 910
Score = 201 bits (511), Expect = 2e-51
Identities = 125/428 (29%), Positives = 217/428 (50%), Gaps = 27/428 (6%)
Query: 548 DELFEMM--LLEGCKPNVVTYTALIDGHCKAGQIEKACQIYARMRGDIESSDMDKYFKLD 605
DE FE+ L+ GC Y L++ + G +++ Q+Y M L+
Sbjct: 172 DERFELKYKLIIGC------YNTLLNSLARFGLVDEMKQVYMEM--------------LE 211
Query: 606 HNNCEGPNVITYGALVDGLCKANRVKEAHELLDTMLAHGCEPNQIVYDAVIDGFCKIGKL 665
C PN+ TY +V+G CK V+EA++ + ++ G +P+ Y ++I G+C+ L
Sbjct: 212 DKVC--PNIYTYNKMVNGYCKLGNVEEANQYVSKIVEAGLDPDFFTYTSLIMGYCQRKDL 269
Query: 666 QDAQEVFTKMSERGYSPNLYTYSSFIDCLFKDNRLDLVLKVLSKMLENSCTPNVVIYTEM 725
A +VF +M +G N Y+ I L R+D + + KM ++ C P V YT +
Sbjct: 270 DSAFKVFNEMPLKGCRRNEVAYTHLIHGLCVARRIDEAMDLFVKMKDDECFPTVRTYTVL 329
Query: 726 VDGLCKIGKTDEAYKLMLKMEEKGCNPNVVTYTAMIDGFGKSGKIEQCLELFRDMCSKGC 785
+ LC + EA L+ +MEE G PN+ TYT +ID K E+ EL M KG
Sbjct: 330 IKSLCGSERKSEALNLVKEMEETGIKPNIHTYTVLIDSLCSQCKFEKARELLGQMLEKGL 389
Query: 786 APNFITYRVLINHCCSNGLLDEAYKLLDEMKQTYWPKHILSHRKIIEGFSQEFI-TSIGL 844
PN ITY LIN C G++++A +++ M+ + ++ ++I+G+ + + ++G+
Sbjct: 390 MPNVITYNALINGYCKRGMIEDAVDVVELMESRKLSPNTRTYNELIKGYCKSNVHKAMGV 449
Query: 845 LDELSENESVPVDSLYRILIDNYIKAGRLEVALDLLEEISSSPSHAVSNKYLYASLIENL 904
L+++ E + +P Y LID ++G + A LL ++ V +++ Y S+I++L
Sbjct: 450 LNKMLERKVLPDVVTYNSLIDGQCRSGNFDSAYRLLSLMND--RGLVPDQWTYTSMIDSL 507
Query: 905 SHASKVDKALELYASMISKNVVPELSILVHLIKGLIKVDKWQEALQLSDSICQMVCLTLS 964
+ +V++A +L+ S+ K V P + + LI G K K EA + + + CL S
Sbjct: 508 CKSKRVEEACDLFDSLEQKGVNPNVVMYTALIDGYCKAGKVDEAHLMLEKMLSKNCLPNS 567
Query: 965 QTFQPLVN 972
TF L++
Sbjct: 568 LTFNALIH 575
Score = 191 bits (486), Expect = 1e-48
Identities = 134/488 (27%), Positives = 238/488 (48%), Gaps = 27/488 (5%)
Query: 479 SLFEEMKRNGIVPSVYTYTILIDSFCKAG-----LIQQARKWF-DEMLHKGCTPNVVTYT 532
SL + NG V V+ +L+ C + ++ RK DE + Y
Sbjct: 128 SLLTLLINNGYVGVVFKIRLLMIKSCDSVGDALYVLDLCRKMNKDERFELKYKLIIGCYN 187
Query: 533 ALIHAYLKAKQMPVADELFEMMLLEGCKPNVVTYTALIDGHCKAGQIEKACQIYARMRGD 592
L+++ + + +++ ML + PN+ TY +++G+CK G +E+A Q ++
Sbjct: 188 TLLNSLARFGLVDEMKQVYMEMLEDKVCPNIYTYNKMVNGYCKLGNVEEANQYVSK---- 243
Query: 593 IESSDMDKYFKLDHNNCEGPNVITYGALVDGLCKANRVKEAHELLDTMLAHGCEPNQIVY 652
I + +D P+ TY +L+ G C+ + A ++ + M GC N++ Y
Sbjct: 244 IVEAGLD------------PDFFTYTSLIMGYCQRKDLDSAFKVFNEMPLKGCRRNEVAY 291
Query: 653 DAVIDGFCKIGKLQDAQEVFTKMSERGYSPNLYTYSSFIDCLFKDNRLDLVLKVLSKMLE 712
+I G C ++ +A ++F KM + P + TY+ I L R L ++ +M E
Sbjct: 292 THLIHGLCVARRIDEAMDLFVKMKDDECFPTVRTYTVLIKSLCGSERKSEALNLVKEMEE 351
Query: 713 NSCTPNVVIYTEMVDGLCKIGKTDEAYKLMLKMEEKGCNPNVVTYTAMIDGFGKSGKIEQ 772
PN+ YT ++D LC K ++A +L+ +M EKG PNV+TY A+I+G+ K G IE
Sbjct: 352 TGIKPNIHTYTVLIDSLCSQCKFEKARELLGQMLEKGLMPNVITYNALINGYCKRGMIED 411
Query: 773 CLELFRDMCSKGCAPNFITYRVLINHCCSNGLLDEAYKLLDEMKQTYWPKHILSHRKIIE 832
+++ M S+ +PN TY LI C + + +A +L++M + ++++ +I+
Sbjct: 412 AVDVVELMESRKLSPNTRTYNELIKGYCKSNV-HKAMGVLNKMLERKVLPDVVTYNSLID 470
Query: 833 GF--SQEFITSIGLLDELSENESVPVDSLYRILIDNYIKAGRLEVALDLLEEISSSPSHA 890
G S F ++ LL +++ VP Y +ID+ K+ R+E A DL + + +
Sbjct: 471 GQCRSGNFDSAYRLLSLMNDRGLVPDQWTYTSMIDSLCKSKRVEEACDLFDSLEQKGVNP 530
Query: 891 VSNKYLYASLIENLSHASKVDKALELYASMISKNVVPELSILVHLIKGLIKVDKWQEALQ 950
N +Y +LI+ A KVD+A + M+SKN +P LI GL K +EA
Sbjct: 531 --NVVMYTALIDGYCKAGKVDEAHLMLEKMLSKNCLPNSLTFNALIHGLCADGKLKEATL 588
Query: 951 LSDSICQM 958
L + + ++
Sbjct: 589 LEEKMVKI 596
>At4g31850 putative protein
Length = 1112
Score = 286 bits (732), Expect = 4e-77
Identities = 225/819 (27%), Positives = 359/819 (43%), Gaps = 68/819 (8%)
Query: 181 VRKCCRNGWWNMALEELGRLKDFGYKPSQTTYNALIQVFLRADKLDTAYLVKREMLSYAF 240
+R R G N A E L R+ D G P TY LI A KLD A V +M +
Sbjct: 265 IRVLGRAGKINEAYEILKRMDDEGCGPDVVTYTVLIDALCTARKLDCAKEVFEKMKTGRH 324
Query: 241 VMDRYTLSCFA--YSLCKGGKCREAFDLIDEAEDFVPDTVFYNRMVSGLCEASLFEEAMD 298
DR T +S + + F E + VPD V + +V LC+A F EA D
Sbjct: 325 KPDRVTYITLLDRFSDNRDLDSVKQFWSEMEKDGHVPDVVTFTILVDALCKAGNFGEAFD 384
Query: 299 ILHRMRSSSCIPNVVTYRILLSGCLRKGQLGRCKRILSMMITEGCYPNREIFNSLIHAYC 358
L MR +PN+ TY L+ G LR +L + M + G P + I Y
Sbjct: 385 TLDVMRDQGILPNLHTYNTLICGLLRVHRLDDALELFGNMESLGVKPTAYTYIVFIDYYG 444
Query: 359 KSRDYSYAYKLFKKMIKCGCQPGYLVYNIFIGSVCSNEEQPSSDILDLVEKAYSEMLDLG 418
KS D A + F+KM G P + N + S+ + ++ + + D+G
Sbjct: 445 KSGDSVSALETFEKMKTKGIAPNIVACNASLYSLAKAGRDREA------KQIFYGLKDIG 498
Query: 419 VVLNKVNVSNFARCLCGAGKFDQAFKIICEMMGKGFVPDDSTYSKVIGFLCDASKVEKAF 478
+V + V + +C G+ D+A K++ EMM G PD + +I L A +V++A+
Sbjct: 499 LVPDSVTYNMMMKCYSKVGEIDEAIKLLSEMMENGCEPDVIVVNSLINTLYKADRVDEAW 558
Query: 479 SLFEEMKRNGIVPSVYTYTILIDSFCKAGLIQQARKWFDEMLHKGCTPNVVTYTALIHAY 538
+F MK + P+V TY L+ K G IQ+A + F+ M+ KGC PN +T+ L
Sbjct: 559 KMFMRMKEMKLKPTVVTYNTLLAGLGKNGKIQEAIELFEGMVQKGCPPNTITFNTLFDCL 618
Query: 539 LKAKQMPVADELFEMMLLEGCKPNVVTYTALIDGHCKAGQIEKACQIYARMR-------- 590
K ++ +A ++ M+ GC P+V TY +I G K GQ+++A + +M+
Sbjct: 619 CKNDEVTLALKMLFKMMDMGCVPDVFTYNTIIFGLVKNGQVKEAMCFFHQMKKLVYPDFV 678
Query: 591 -------GDIESSDMDKYFKLDHN---NCE---------------------------GPN 613
G +++S ++ +K+ N NC
Sbjct: 679 TLCTLLPGVVKASLIEDAYKIITNFLYNCADQPANLFWEDLIGSILAEAGIDNAVSFSER 738
Query: 614 VITYGALVDG----------LCKANRVKEAHELLDTMLAH-GCEPNQIVYDAVIDGFCKI 662
++ G DG CK N V A L + G +P Y+ +I G +
Sbjct: 739 LVANGICRDGDSILVPIIRYSCKHNNVSGARTLFEKFTKDLGVQPKLPTYNLLIGGLLEA 798
Query: 663 GKLQDAQEVFTKMSERGYSPNLYTYSSFIDCLFKDNRLDLVLKVLSKMLENSCTPNVVIY 722
++ AQ+VF ++ G P++ TY+ +D K ++D + ++ +M + C N + +
Sbjct: 799 DMIEIAQDVFLQVKSTGCIPDVATYNFLLDAYGKSGKIDELFELYKEMSTHECEANTITH 858
Query: 723 TEMVDGLCKIGKTDEAYKLMLK-MEEKGCNPNVVTYTAMIDGFGKSGKIEQCLELFRDMC 781
++ GL K G D+A L M ++ +P TY +IDG KSG++ + +LF M
Sbjct: 859 NIVISGLVKAGNVDDALDLYYDLMSDRDFSPTACTYGPLIDGLSKSGRLYEAKQLFEGML 918
Query: 782 SKGCAPNFITYRVLINHCCSNGLLDEAYKLLDEMKQTYWPKHILSHRKIIEGFSQEFITS 841
GC PN Y +LIN G D A L M + + ++ +++
Sbjct: 919 DYGCRPNCAIYNILINGFGKAGEADAACALFKRMVKEGVRPDLKTYSVLVDCLCMVGRVD 978
Query: 842 IGL--LDELSENESVPVDSLYRILIDNYIKAGRLEVALDLLEEISSSPSHAVSNKYLYAS 899
GL EL E+ P Y ++I+ K+ RLE AL L E+ +S + Y Y S
Sbjct: 979 EGLHYFKELKESGLNPDVVCYNLIINGLGKSHRLEEALVLFNEMKTSRG-ITPDLYTYNS 1037
Query: 900 LIENLSHASKVDKALELYASMISKNVVPELSILVHLIKG 938
LI NL A V++A ++Y + + P + LI+G
Sbjct: 1038 LILNLGIAGMVEEAGKIYNEIQRAGLEPNVFTFNALIRG 1076
Score = 278 bits (710), Expect = 1e-74
Identities = 224/834 (26%), Positives = 378/834 (44%), Gaps = 80/834 (9%)
Query: 193 ALEELGRLKDFGY----------KPSQTTYNALIQVFLRADKLDTAYLVKREMLSYAFVM 242
AL G+L++ Y K TY + + L A R+M + FV+
Sbjct: 127 ALRVDGKLEEMAYVFDLMQKRIIKRDTNTYLTIFKSLSVKGGLKQAPYALRKMREFGFVL 186
Query: 243 DRYTLSCFAYSLCKGGKCREAFDLIDEA--EDFVPDTVFYNRMVSGLCEASLFEEAMDIL 300
+ Y+ + + L K C EA ++ E F P Y+ ++ GL + + M +L
Sbjct: 187 NAYSYNGLIHLLLKSRFCTEAMEVYRRMILEGFRPSLQTYSSLMVGLGKRRDIDSVMGLL 246
Query: 301 HRMRSSSCIPNVVTYRILLSGCLRKGQLGRCKRILSMMITEGCYPNREIFNSLIHAYCKS 360
M + PNV T+ I + R G++ IL M EGC P+ + LI A C +
Sbjct: 247 KEMETLGLKPNVYTFTICIRVLGRAGKINEAYEILKRMDDEGCGPDVVTYTVLIDALCTA 306
Query: 361 RDYSYAYKLFKKMIKCGCQPGYLVYNIFIGSVCSNEEQPSSDILDLVEKAYSEMLDLGVV 420
R A ++F+KM +P + Y + N + LD V++ +SEM G V
Sbjct: 307 RKLDCAKEVFEKMKTGRHKPDRVTYITLLDRFSDNRD------LDSVKQFWSEMEKDGHV 360
Query: 421 LNKVNVSNFARCLCGAGKFDQAFKIICEMMGKGFVPDDSTYSKVIGFLCDASKVEKAFSL 480
+ V + LC AG F +AF + M +G +P+ TY+ +I L +++ A L
Sbjct: 361 PDVVTFTILVDALCKAGNFGEAFDTLDVMRDQGILPNLHTYNTLICGLLRVHRLDDALEL 420
Query: 481 FEEMKRNGIVPSVYTYTILIDSFCKAGLIQQARKWFDEMLHKGCTPNVVTYTALIHAYLK 540
F M+ G+ P+ YTY + ID + K+G A + F++M KG PN+V A +++ K
Sbjct: 421 FGNMESLGVKPTAYTYIVFIDYYGKSGDSVSALETFEKMKTKGIAPNIVACNASLYSLAK 480
Query: 541 AKQMPVADELFEMMLLEGCKPNVVTYTALIDGHCKAGQIEKACQIYARMRGDIESSDMDK 600
A + A ++F + G P+ VTY ++ + K G+I++A ++ + M
Sbjct: 481 AGRDREAKQIFYGLKDIGLVPDSVTYNMMMKCYSKVGEIDEAIKLLSEMM---------- 530
Query: 601 YFKLDHNNCEGPNVITYGALVDGLCKANRVKEAHELLDTMLAHGCEPNQIVYDAVIDGFC 660
N CE P+VI +L++ L KA+RV EA ++ M +P + Y+ ++ G
Sbjct: 531 -----ENGCE-PDVIVVNSLINTLYKADRVDEAWKMFMRMKEMKLKPTVVTYNTLLAGLG 584
Query: 661 KIGKLQDAQEVFTKMSERGYSPNLYTYSSFIDCLFKDNRLDLVLKVLSKMLENSCTPNVV 720
K GK+Q+A E+F M ++G PN T+++ DCL K++ + L LK+L KM++ C P+V
Sbjct: 585 KNGKIQEAIELFEGMVQKGCPPNTITFNTLFDCLCKNDEVTLALKMLFKMMDMGCVPDVF 644
Query: 721 IYTEMVDGLCKIGKTDEAYKLMLKMEEKGCNPNVVTYTAMIDGFGKSGKIEQCLELFRDM 780
Y ++ GL K G+ EA +M +K P+ VT ++ G K+ IE ++ +
Sbjct: 645 TYNTIIFGLVKNGQVKEAMCFFHQM-KKLVYPDFVTLCTLLPGVVKASLIEDAYKIITNF 703
Query: 781 CSKGCA--PNFITYRVLINHCCSNGLLDEAYKLLDEMKQT---------------YWPKH 823
CA P + + LI + +D A + + Y KH
Sbjct: 704 L-YNCADQPANLFWEDLIGSILAEAGIDNAVSFSERLVANGICRDGDSILVPIIRYSCKH 762
Query: 824 --ILSHRKIIEGFSQEF----------ITSIGLLD------------ELSENESVPVDSL 859
+ R + E F+++ + GLL+ ++ +P +
Sbjct: 763 NNVSGARTLFEKFTKDLGVQPKLPTYNLLIGGLLEADMIEIAQDVFLQVKSTGCIPDVAT 822
Query: 860 YRILIDNYIKAGRLEVALDLLEEISSSPSHAVSNKYLYASLIENLSHASKVDKALELYAS 919
Y L+D Y K+G+++ +L +E+S+ A N + +I L A VD AL+LY
Sbjct: 823 YNFLLDAYGKSGKIDELFELYKEMSTHECEA--NTITHNIVISGLVKAGNVDDALDLYYD 880
Query: 920 MIS-KNVVPELSILVHLIKGLIKVDKWQEALQLSDSICQMVCLTLSQTFQPLVN 972
++S ++ P LI GL K + EA QL + + C + L+N
Sbjct: 881 LMSDRDFSPTACTYGPLIDGLSKSGRLYEAKQLFEGMLDYGCRPNCAIYNILIN 934
Score = 272 bits (696), Expect = 6e-73
Identities = 215/778 (27%), Positives = 344/778 (43%), Gaps = 123/778 (15%)
Query: 143 LLDLLGCNVNADDRVPLKFLMEIKDDDHELLRRLLNFLVRKCCRNGWWNMALEELGRLKD 202
LLD N + D +F E++ D H LV C+ G + A + L ++D
Sbjct: 334 LLDRFSDNRDLDS--VKQFWSEMEKDGHVPDVVTFTILVDALCKAGNFGEAFDTLDVMRD 391
Query: 203 FGYKPSQTTYNALIQVFLRADKLDTAYL---------VKREMLSYAFVMDRY-------- 245
G P+ TYN LI LR +LD A VK +Y +D Y
Sbjct: 392 QGILPNLHTYNTLICGLLRVHRLDDALELFGNMESLGVKPTAYTYIVFIDYYGKSGDSVS 451
Query: 246 ----------------TLSCFA--YSLCKGGKCREAFDLIDEAEDF--VPDTVFYNRMVS 285
++C A YSL K G+ REA + +D VPD+V YN M+
Sbjct: 452 ALETFEKMKTKGIAPNIVACNASLYSLAKAGRDREAKQIFYGLKDIGLVPDSVTYNMMMK 511
Query: 286 GLCEASLFEEAMDILHRMRSSSCIPNV--------------------------------- 312
+ +EA+ +L M + C P+V
Sbjct: 512 CYSKVGEIDEAIKLLSEMMENGCEPDVIVVNSLINTLYKADRVDEAWKMFMRMKEMKLKP 571
Query: 313 --VTYRILLSGCLRKGQLGRCKRILSMMITEGCYPNREIFNSLIHAYCKSRDYSYAYKLF 370
VTY LL+G + G++ + M+ +GC PN FN+L CK+ + + A K+
Sbjct: 572 TVVTYNTLLAGLGKNGKIQEAIELFEGMVQKGCPPNTITFNTLFDCLCKNDEVTLALKML 631
Query: 371 KKMIKCGCQPGYLVYN-IFIGSVCSNEEQPSSDILDLVEK-AYSEMLDLGVVLNKVN--- 425
KM+ GC P YN I G V + + + + ++K Y + + L +L V
Sbjct: 632 FKMMDMGCVPDVFTYNTIIFGLVKNGQVKEAMCFFHQMKKLVYPDFVTLCTLLPGVVKAS 691
Query: 426 --------VSNF-----------------ARCLCGAGKFDQAFKIICEMMGKGFVPD-DS 459
++NF L AG D A ++ G D DS
Sbjct: 692 LIEDAYKIITNFLYNCADQPANLFWEDLIGSILAEAG-IDNAVSFSERLVANGICRDGDS 750
Query: 460 TYSKVIGFLCDASKVEKAFSLFEEMKRN-GIVPSVYTYTILIDSFCKAGLIQQARKWFDE 518
+I + C + V A +LFE+ ++ G+ P + TY +LI +A +I+ A+ F +
Sbjct: 751 ILVPIIRYSCKHNNVSGARTLFEKFTKDLGVQPKLPTYNLLIGGLLEADMIEIAQDVFLQ 810
Query: 519 MLHKGCTPNVVTYTALIHAYLKAKQMPVADELFEMMLLEGCKPNVVTYTALIDGHCKAGQ 578
+ GC P+V TY L+ AY K+ ++ EL++ M C+ N +T+ +I G KAG
Sbjct: 811 VKSTGCIPDVATYNFLLDAYGKSGKIDELFELYKEMSTHECEANTITHNIVISGLVKAGN 870
Query: 579 IEKACQIYARMRGDIESSDMDKYFKLDHNNCEGPNVITYGALVDGLCKANRVKEAHELLD 638
++ A +Y + D + S P TYG L+DGL K+ R+ EA +L +
Sbjct: 871 VDDALDLYYDLMSDRDFS---------------PTACTYGPLIDGLSKSGRLYEAKQLFE 915
Query: 639 TMLAHGCEPNQIVYDAVIDGFCKIGKLQDAQEVFTKMSERGYSPNLYTYSSFIDCLFKDN 698
ML +GC PN +Y+ +I+GF K G+ A +F +M + G P+L TYS +DCL
Sbjct: 916 GMLDYGCRPNCAIYNILINGFGKAGEADAACALFKRMVKEGVRPDLKTYSVLVDCLCMVG 975
Query: 699 RLDLVLKVLSKMLENSCTPNVVIYTEMVDGLCKIGKTDEAYKLMLKME-EKGCNPNVVTY 757
R+D L ++ E+ P+VV Y +++GL K + +EA L +M+ +G P++ TY
Sbjct: 976 RVDEGLHYFKELKESGLNPDVVCYNLIINGLGKSHRLEEALVLFNEMKTSRGITPDLYTY 1035
Query: 758 TAMIDGFGKSGKIEQCLELFRDMCSKGCAPNFITYRVLINHCCSNGLLDEAYKLLDEM 815
++I G +G +E+ +++ ++ G PN T+ LI +G + AY + M
Sbjct: 1036 NSLILNLGIAGMVEEAGKIYNEIQRAGLEPNVFTFNALIRGYSLSGKPEHAYAVYQTM 1093
Score = 249 bits (637), Expect = 4e-66
Identities = 182/672 (27%), Positives = 317/672 (47%), Gaps = 33/672 (4%)
Query: 272 DFVPDTVFYNRMVSGLCEASLFEEAMDILHRMRSSSCIPNVVTYRILLSGCLRKGQLGRC 331
+ V T N M+ L EE + M+ + TY + KG L +
Sbjct: 113 NLVHTTETCNYMLEALRVDGKLEEMAYVFDLMQKRIIKRDTNTYLTIFKSLSVKGGLKQA 172
Query: 332 KRILSMMITEGCYPNREIFNSLIHAYCKSRDYSYAYKLFKKMIKCGCQPGYLVYNIFIGS 391
L M G N +N LIH KSR + A +++++MI G +P Y+ +
Sbjct: 173 PYALRKMREFGFVLNAYSYNGLIHLLLKSRFCTEAMEVYRRMILEGFRPSLQTYSSLMVG 232
Query: 392 VCSNEEQPSSDILDLVEKAYSEMLDLGVVLNKVNVSNFARCLCGAGKFDQAFKIICEMMG 451
+ + +D V EM LG+ N + R L AGK ++A++I+ M
Sbjct: 233 LGKRRD------IDSVMGLLKEMETLGLKPNVYTFTICIRVLGRAGKINEAYEILKRMDD 286
Query: 452 KGFVPDDSTYSKVIGFLCDASKVEKAFSLFEEMKRNGIVPSVYTYTILIDSFCKAGLIQQ 511
+G PD TY+ +I LC A K++ A +FE+MK P TY L+D F +
Sbjct: 287 EGCGPDVVTYTVLIDALCTARKLDCAKEVFEKMKTGRHKPDRVTYITLLDRFSDNRDLDS 346
Query: 512 ARKWFDEMLHKGCTPNVVTYTALIHAYLKAKQMPVADELFEMMLLEGCKPNVVTYTALID 571
++++ EM G P+VVT+T L+ A KA A + ++M +G PN+ TY LI
Sbjct: 347 VKQFWSEMEKDGHVPDVVTFTILVDALCKAGNFGEAFDTLDVMRDQGILPNLHTYNTLIC 406
Query: 572 GHCKAGQIEKACQIYARMRGDIESSDMDKYFKLDHNNCEGPNVITYGALVDGLCKANRVK 631
G + +++ A +++ G++ES + P TY +D K+
Sbjct: 407 GLLRVHRLDDALELF----GNMESLGVK------------PTAYTYIVFIDYYGKSGDSV 450
Query: 632 EAHELLDTMLAHGCEPNQIVYDAVIDGFCKIGKLQDAQEVFTKMSERGYSPNLYTYSSFI 691
A E + M G PN + +A + K G+ ++A+++F + + G P+ TY+ +
Sbjct: 451 SALETFEKMKTKGIAPNIVACNASLYSLAKAGRDREAKQIFYGLKDIGLVPDSVTYNMMM 510
Query: 692 DCLFKDNRLDLVLKVLSKMLENSCTPNVVIYTEMVDGLCKIGKTDEAYKLMLKMEEKGCN 751
C K +D +K+LS+M+EN C P+V++ +++ L K + DEA+K+ ++M+E
Sbjct: 511 KCYSKVGEIDEAIKLLSEMMENGCEPDVIVVNSLINTLYKADRVDEAWKMFMRMKEMKLK 570
Query: 752 PNVVTYTAMIDGFGKSGKIEQCLELFRDMCSKGCAPNFITYRVLINHCCSNGLLDEAYKL 811
P VVTY ++ G GK+GKI++ +ELF M KGC PN IT+ L + C N + A K+
Sbjct: 571 PTVVTYNTLLAGLGKNGKIQEAIELFEGMVQKGCPPNTITFNTLFDCLCKNDEVTLALKM 630
Query: 812 LDEMKQTYWPKHILSHRKII-----EGFSQEFITSIGLLDELSENESVPVDSLYRILIDN 866
L +M + ++ II G +E + + +L + V + + L+
Sbjct: 631 LFKMMDMGCVPDVFTYNTIIFGLVKNGQVKEAMCFFHQMKKLVYPDFVTLCT----LLPG 686
Query: 867 YIKAGRLEVALDLLEEISSSPSHAVSNKYLYASLIENLSHASKVDKALELYASMISKNVV 926
+KA +E A ++ + + +N + + LI ++ + +D A+ +++ +
Sbjct: 687 VVKASLIEDAYKIITNFLYNCADQPANLF-WEDLIGSILAEAGIDNAVSFSERLVANGIC 745
Query: 927 PE-LSILVHLIK 937
+ SILV +I+
Sbjct: 746 RDGDSILVPIIR 757
Score = 177 bits (450), Expect = 2e-44
Identities = 145/613 (23%), Positives = 267/613 (42%), Gaps = 73/613 (11%)
Query: 159 LKFLMEIKDDDHE----LLRRLLNFLVRKCCRNGWWNMALEELGRLKDFGYKPSQTTYNA 214
+K L E+ ++ E ++ L+N L + + W M + R+K+ KP+ TYN
Sbjct: 523 IKLLSEMMENGCEPDVIVVNSLINTLYKADRVDEAWKMFM----RMKEMKLKPTVVTYNT 578
Query: 215 LIQVFLRADKLDTAYLVKREMLSYAFVMDRYTLSCFAYSLCKGGKCREAFDLIDEAEDF- 273
L+ + K+ A + M+ + T + LCK + A ++ + D
Sbjct: 579 LLAGLGKNGKIQEAIELFEGMVQKGCPPNTITFNTLFDCLCKNDEVTLALKMLFKMMDMG 638
Query: 274 -VPDTVFYNRMVSGLCEASLFEEAMDILHRMRSSSCIPNVVTYRILLSGCLRKGQLGRCK 332
VPD YN ++ GL + +EAM H+M+ P+ VT LL G ++ +
Sbjct: 639 CVPDVFTYNTIIFGLVKNGQVKEAMCFFHQMK-KLVYPDFVTLCTLLPGVVKASLIEDAY 697
Query: 333 RILS------------------------------------MMITEG-CYPNREIFNSLIH 355
+I++ ++ G C I +I
Sbjct: 698 KIITNFLYNCADQPANLFWEDLIGSILAEAGIDNAVSFSERLVANGICRDGDSILVPIIR 757
Query: 356 AYCKSRDYSYAYKLFKKMIK-CGCQPGYLVYNIFIGSVCSNEEQPSSDILDLVEKAYSEM 414
CK + S A LF+K K G QP YN+ IG + +D++++ + + ++
Sbjct: 758 YSCKHNNVSGARTLFEKFTKDLGVQPKLPTYNLLIGGLL------EADMIEIAQDVFLQV 811
Query: 415 LDLGVVLNKVNVSNFARCLCGAGKFDQAFKIICEMMGKGFVPDDSTYSKVIGFLCDASKV 474
G + + + +GK D+ F++ EM + T++ VI L A V
Sbjct: 812 KSTGCIPDVATYNFLLDAYGKSGKIDELFELYKEMSTHECEANTITHNIVISGLVKAGNV 871
Query: 475 EKAFSLFEE-MKRNGIVPSVYTYTILIDSFCKAGLIQQARKWFDEMLHKGCTPNVVTYTA 533
+ A L+ + M P+ TY LID K+G + +A++ F+ ML GC PN Y
Sbjct: 872 DDALDLYYDLMSDRDFSPTACTYGPLIDGLSKSGRLYEAKQLFEGMLDYGCRPNCAIYNI 931
Query: 534 LIHAYLKAKQMPVADELFEMMLLEGCKPNVVTYTALIDGHCKAGQIEKACQIYARMRGDI 593
LI+ + KA + A LF+ M+ EG +P++ TY+ L+D C G++++
Sbjct: 932 LINGFGKAGEADAACALFKRMVKEGVRPDLKTYSVLVDCLCMVGRVDEGLH--------- 982
Query: 594 ESSDMDKYFKLDHNNCEGPNVITYGALVDGLCKANRVKEAHELLDTM-LAHGCEPNQIVY 652
YFK + P+V+ Y +++GL K++R++EA L + M + G P+ Y
Sbjct: 983 -------YFKELKESGLNPDVVCYNLIINGLGKSHRLEEALVLFNEMKTSRGITPDLYTY 1035
Query: 653 DAVIDGFCKIGKLQDAQEVFTKMSERGYSPNLYTYSSFIDCLFKDNRLDLVLKVLSKMLE 712
+++I G +++A +++ ++ G PN++T+++ I + + V M+
Sbjct: 1036 NSLILNLGIAGMVEEAGKIYNEIQRAGLEPNVFTFNALIRGYSLSGKPEHAYAVYQTMVT 1095
Query: 713 NSCTPNVVIYTEM 725
+PN Y ++
Sbjct: 1096 GGFSPNTGTYEQL 1108
>At4g19440 unknown protein
Length = 825
Score = 285 bits (730), Expect = 7e-77
Identities = 205/742 (27%), Positives = 341/742 (45%), Gaps = 56/742 (7%)
Query: 104 DSLVVEVMNNVKNPELCVKFFLWAGRQIGYSHTPQVFDKLLDLL-GCNVNADDRVPLKFL 162
D L E + V NP+ + FF A +S + + + L+ LL N+ + RV L L
Sbjct: 94 DRLFPEFRSKV-NPKTALDFFRLASDSFSFSFSLRSYCLLIGLLLDANLLSAARVVLIRL 152
Query: 163 ME---------IKDD---------------DHELLRRLLNFLVRKCC----RNGWWNMAL 194
+ ++D D E+ R++ + L+ C R+G + +AL
Sbjct: 153 INGNVPVLPCGLRDSRVAIADAMASLSLCFDEEIRRKMSDLLIEVYCTQFKRDGCY-LAL 211
Query: 195 EELGRLKDFGYKPSQTTYNALIQVFLRADKLDTAYLVKREMLSYAFVMDRYTLSCFAYSL 254
+ L + G PS+TT N L+ +RA++ +++ D Y + +
Sbjct: 212 DVFPVLANKGMFPSKTTCNILLTSLVRANEFQKC-CEAFDVVCKGVSPDVYLFTTAINAF 270
Query: 255 CKGGKCREAFDLIDEAED--FVPDTVFYNRMVSGLCEASLFEEAMDILHRMRSSSCIPNV 312
CKGGK EA L + E+ P+ V +N ++ GL ++EA +M P +
Sbjct: 271 CKGGKVEEAVKLFSKMEEAGVAPNVVTFNTVIDGLGMCGRYDEAFMFKEKMVERGMEPTL 330
Query: 313 VTYRILLSGCLRKGQLGRCKRILSMMITEGCYPNREIFNSLIHAYCKSRDYSYAYKLFKK 372
+TY IL+ G R ++G +L M +G PN ++N+LI ++ ++ + A ++
Sbjct: 331 ITYSILVKGLTRAKRIGDAYFVLKEMTKKGFPPNVIVYNNLIDSFIEAGSLNKAIEIKDL 390
Query: 373 MIKCGCQPGYLVYNIFIGSVCSNEEQPSSDILDLVEKAYSEMLDLGVVLNKVNVSNFARC 432
M+ G YN I C N + D E+ EML +G +N+ + ++
Sbjct: 391 MVSKGLSLTSSTYNTLIKGYCKNGQA------DNAERLLKEMLSIGFNVNQGSFTSVICL 444
Query: 433 LCGAGKFDQAFKIICEMMGKGFVPDDSTYSKVIGFLCDASKVEKAFSLFEEMKRNGIVPS 492
LC FD A + + EM+ + P + +I LC K KA L+ + G V
Sbjct: 445 LCSHLMFDSALRFVGEMLLRNMSPGGGLLTTLISGLCKHGKHSKALELWFQFLNKGFVVD 504
Query: 493 VYTYTILIDSFCKAGLIQQARKWFDEMLHKGCTPNVVTYTALIHAYLKAKQMPVADELFE 552
T L+ C+AG + +A + E+L +GC + V+Y LI K++ A +
Sbjct: 505 TRTSNALLHGLCEAGKLDEAFRIQKEILGRGCVMDRVSYNTLISGCCGKKKLDEAFMFLD 564
Query: 553 MMLLEGCKPNVVTYTALIDGHCKAGQIEKACQIYARMRGDIESSDMDKYFKLDHNNCEGP 612
M+ G KP+ TY+ LI G ++E+A Q + + N P
Sbjct: 565 EMVKRGLKPDNYTYSILICGLFNMNKVEEAIQFWDDCK----------------RNGMLP 608
Query: 613 NVITYGALVDGLCKANRVKEAHELLDTMLAHGCEPNQIVYDAVIDGFCKIGKLQDAQEVF 672
+V TY ++DG CKA R +E E D M++ +PN +VY+ +I +C+ G+L A E+
Sbjct: 609 DVYTYSVMIDGCCKAERTEEGQEFFDEMMSKNVQPNTVVYNHLIRAYCRSGRLSMALELR 668
Query: 673 TKMSERGYSPNLYTYSSFIDCLFKDNRLDLVLKVLSKMLENSCTPNVVIYTEMVDGLCKI 732
M +G SPN TY+S I + +R++ + +M PNV YT ++DG K+
Sbjct: 669 EDMKHKGISPNSATYTSLIKGMSIISRVEEAKLLFEEMRMEGLEPNVFHYTALIDGYGKL 728
Query: 733 GKTDEAYKLMLKMEEKGCNPNVVTYTAMIDGFGKSGKIEQCLELFRDMCSKGCAPNFITY 792
G+ + L+ +M K +PN +TYT MI G+ + G + + L +M KG P+ ITY
Sbjct: 729 GQMVKVECLLREMHSKNVHPNKITYTVMIGGYARDGNVTEASRLLNEMREKGIVPDSITY 788
Query: 793 RVLINHCCSNGLLDEAYKLLDE 814
+ I G + EA+K DE
Sbjct: 789 KEFIYGYLKQGGVLEAFKGSDE 810
Score = 202 bits (515), Expect = 6e-52
Identities = 176/686 (25%), Positives = 303/686 (43%), Gaps = 77/686 (11%)
Query: 264 FDLIDEAEDFVPDTVFYNRMVSGLCEASLFEEAMDILHRMRSSSCIPNVVTYRILLSGCL 323
F L ++ F Y ++ L +A+L A +L R+ + + +P +L L
Sbjct: 113 FRLASDSFSFSFSLRSYCLLIGLLLDANLLSAARVVLIRLINGN-VP-------VLPCGL 164
Query: 324 RKGQLGRCKRILSMMITEGCYPNREIFNSLIHAYCKS--RDYSY-AYKLFKKMIKCGCQP 380
R ++ + S+ + R++ + LI YC RD Y A +F + G P
Sbjct: 165 RDSRVAIADAMASLSLCFDEEIRRKMSDLLIEVYCTQFKRDGCYLALDVFPVLANKGMFP 224
Query: 381 GYLVYNIFIGS-VCSNEEQPSSDILDLVEKAYSEMLDLGVVLNKVNVSNFARCLCGAGKF 439
NI + S V +NE Q + D+V K S D+ + +N C GK
Sbjct: 225 SKTTCNILLTSLVRANEFQKCCEAFDVVCKGVSP--DVYLFTTAINA------FCKGGKV 276
Query: 440 DQAFKIICEMMGKGFVPDDSTYSKVIGFLCDASKVEKAFSLFEEMKRNGIVPSVYTYTIL 499
++A K+ +M G P+ T++ VI L + ++AF E+M G+ P++ TY+IL
Sbjct: 277 EEAVKLFSKMEEAGVAPNVVTFNTVIDGLGMCGRYDEAFMFKEKMVERGMEPTLITYSIL 336
Query: 500 IDSFCKAGLIQQARKWFDEMLHKGCTPNVVTYTALIHAYLKAKQMPVADELFEMMLLEGC 559
+ +A I A EM KG PNV+ Y LI ++++A + A E+ ++M+ +G
Sbjct: 337 VKGLTRAKRIGDAYFVLKEMTKKGFPPNVIVYNNLIDSFIEAGSLNKAIEIKDLMVSKGL 396
Query: 560 KPNVVTYTALIDGHCKAGQIEKACQIYARMRGDIESSDMDKYFKLDHNNCEGPNVITYGA 619
TY LI G+CK GQ + A ++ M + N +G ++ +
Sbjct: 397 SLTSSTYNTLIKGYCKNGQADNAERLLKEM------------LSIGFNVNQG----SFTS 440
Query: 620 LVDGLCKANRVKEAHELLDTMLAHGCEPNQIVYDAVIDGFCKIGKLQDAQEVFTKMSERG 679
++ LC A + ML P + +I G CK GK A E++ + +G
Sbjct: 441 VICLLCSHLMFDSALRFVGEMLLRNMSPGGGLLTTLISGLCKHGKHSKALELWFQFLNKG 500
Query: 680 YSPNLYTYSSFIDCLFKDNRLDLVLKVLSKMLENSCTPNVVIYTEMVDGLCKIGKTDEAY 739
+ + T ++ + L + +LD ++ ++L C + V Y ++ G C K DEA+
Sbjct: 501 FVVDTRTSNALLHGLCEAGKLDEAFRIQKEILGRGCVMDRVSYNTLISGCCGKKKLDEAF 560
Query: 740 KLMLKMEEKGCNPNVVTYTAMIDGFGKSGKIEQCLELFRDMCSKGCAPNFITYRVLINHC 799
+ +M ++G P+ TY+ +I G K+E+ ++ + D G P+ TY V+I+ C
Sbjct: 561 MFLDEMVKRGLKPDNYTYSILICGLFNMNKVEEAIQFWDDCKRNGMLPDVYTYSVMIDGC 620
Query: 800 CSNGLLDEAYKLLDEMKQTYWPKHILSHRKIIEGFSQEFITSIGLLDELSENESVPVDSL 859
C +E + DEM +S+N P +
Sbjct: 621 CKAERTEEGQEFFDEM--------------------------------MSKNVQ-PNTVV 647
Query: 860 YRILIDNYIKAGRLEVALDLLEEISS---SPSHAVSNKYLYASLIENLSHASKVDKALEL 916
Y LI Y ++GRL +AL+L E++ SP+ A Y SLI+ +S S+V++A L
Sbjct: 648 YNHLIRAYCRSGRLSMALELREDMKHKGISPNSAT-----YTSLIKGMSIISRVEEAKLL 702
Query: 917 YASMISKNVVPELSILVHLIKGLIKV 942
+ M + + P + LI G K+
Sbjct: 703 FEEMRMEGLEPNVFHYTALIDGYGKL 728
Score = 117 bits (293), Expect = 3e-26
Identities = 93/383 (24%), Positives = 161/383 (41%), Gaps = 39/383 (10%)
Query: 628 NRVKEAHELLDTMLAHGCEPNQIVYDAVIDGFCKIGKLQD---AQEVFTKMSERGYSPNL 684
+RV A + L E + + D +I+ +C K A +VF ++ +G P+
Sbjct: 167 SRVAIADAMASLSLCFDEEIRRKMSDLLIEVYCTQFKRDGCYLALDVFPVLANKGMFPSK 226
Query: 685 YTYSSFIDCLFKDNRLDLVLKVLSKMLENSCTPNVVIYTEMVDGLCKIGKTDEAYKLMLK 744
T + + L + N + + + +P+V ++T ++ CK GK +EA KL K
Sbjct: 227 TTCNILLTSLVRANEFQKCCEAFDVVCKG-VSPDVYLFTTAINAFCKGGKVEEAVKLFSK 285
Query: 745 MEEKGCNPNVVTYTAMIDGFGKSGKIEQCLELFRDMCSKGCAPNFITYRVLINHCCSNGL 804
MEE G PNVVT+ +IDG G G+ ++ M +G P ITY +L+
Sbjct: 286 MEEAGVAPNVVTFNTVIDGLGMCGRYDEAFMFKEKMVERGMEPTLITYSILVKGLTRAKR 345
Query: 805 LDEAYKLLDEMKQTYWPKHILSHRKIIEGFSQ--EFITSIGLLDELSENESVPVDSLYRI 862
+ +AY +L EM + +P +++ + +I+ F + +I + D + S Y
Sbjct: 346 IGDAYFVLKEMTKKGFPPNVIVYNNLIDSFIEAGSLNKAIEIKDLMVSKGLSLTSSTYNT 405
Query: 863 LIDNYIKAGRLEVALDLLEEISSSPSHAVSNKY--------------------------- 895
LI Y K G+ + A LL+E+ S + +
Sbjct: 406 LIKGYCKNGQADNAERLLKEMLSIGFNVNQGSFTSVICLLCSHLMFDSALRFVGEMLLRN 465
Query: 896 ------LYASLIENLSHASKVDKALELYASMISKNVVPELSILVHLIKGLIKVDKWQEAL 949
L +LI L K KALEL+ ++K V + L+ GL + K EA
Sbjct: 466 MSPGGGLLTTLISGLCKHGKHSKALELWFQFLNKGFVVDTRTSNALLHGLCEAGKLDEAF 525
Query: 950 QLSDSICQMVCLTLSQTFQPLVN 972
++ I C+ ++ L++
Sbjct: 526 RIQKEILGRGCVMDRVSYNTLIS 548
>At5g59900 putative protein
Length = 907
Score = 274 bits (700), Expect = 2e-73
Identities = 190/649 (29%), Positives = 297/649 (45%), Gaps = 42/649 (6%)
Query: 178 NFLVRKCCRNGWWNMALEELGRLKDFG---YKPSQTTYNALIQVFLRADKLDTAYLVKRE 234
N L+ C+ E +G KD KP TY L+ + + + + E
Sbjct: 266 NVLIDGLCKK---QKVWEAVGIKKDLAGKDLKPDVVTYCTLVYGLCKVQEFEIGLEMMDE 322
Query: 235 MLSYAFVMDRYTLSCFAYSLCKGGKCREAFDLIDEAEDF--VPDTVFYNRMVSGLCEASL 292
ML F +S L K GK EA +L+ DF P+ YN ++ LC+
Sbjct: 323 MLCLRFSPSEAAVSSLVEGLRKRGKIEEALNLVKRVVDFGVSPNLFVYNALIDSLCKGRK 382
Query: 293 FEEAMDILHRMRSSSCIPNVVTYRILLSGCLRKGQLGRCKRILSMMITEGCYPNREIFNS 352
F EA + RM PN VTY IL+ R+G+L L M+ G + +NS
Sbjct: 383 FHEAELLFDRMGKIGLRPNDVTYSILIDMFCRRGKLDTALSFLGEMVDTGLKLSVYPYNS 442
Query: 353 LIHAYCKSRDYSYAYKLFKKMIKCGCQPGYLVYNIFIGSVCSNEEQPSSDILDLVEKAYS 412
LI+ +CK D S A +MI +P + Y +G CS + ++ + Y
Sbjct: 443 LINGHCKFGDISAAEGFMAEMINKKLEPTVVTYTSLMGGYCSKGK------INKALRLYH 496
Query: 413 EMLDLGVVLNKVNVSNFARCLCGAGKFDQAFKIICEMMGKGFVPDDSTYSKVIGFLCDAS 472
EM G+ + + L AG A K+ EM P+ TY+ +I C+
Sbjct: 497 EMTGKGIAPSIYTFTTLLSGLFRAGLIRDAVKLFNEMAEWNVKPNRVTYNVMIEGYCEEG 556
Query: 473 KVEKAFSLFEEMKRNGIVPSVYTYTILIDSFCKAGLIQQARKWFDEMLHKG-CTPNVVTY 531
+ KAF +EM GIVP Y+Y LI C G +A K F + LHKG C N + Y
Sbjct: 557 DMSKAFEFLKEMTEKGIVPDTYSYRPLIHGLCLTGQASEA-KVFVDGLHKGNCELNEICY 615
Query: 532 TALIHAYLKAKQMPVADELFEMMLLEGCKPNVVTYTALIDGHCKAGQIEKACQIYARMRG 591
T L+H + + ++ A + + M+ G ++V Y LIDG K K +++ +
Sbjct: 616 TGLLHGFCREGKLEEALSVCQEMVQRGVDLDLVCYGVLIDGSLK----HKDRKLFFGL-- 669
Query: 592 DIESSDMDKYFKLDHNNCEGPNVITYGALVDGLCKANRVKEAHELLDTMLAHGCEPNQIV 651
K H+ P+ + Y +++D K KEA + D M+ GC PN++
Sbjct: 670 ----------LKEMHDRGLKPDDVIYTSMIDAKSKTGDFKEAFGIWDLMINEGCVPNEVT 719
Query: 652 YDAVIDGFCKIGKLQDAQEVFTKMSERGYSPNLYTYSSFIDCLFK-----DNRLDLVLKV 706
Y AVI+G CK G + +A+ + +KM PN TY F+D L K ++L +
Sbjct: 720 YTAVINGLCKAGFVNEAEVLCSKMQPVSSVPNQVTYGCFLDILTKGEVDMQKAVELHNAI 779
Query: 707 LSKMLENSCTPNVVIYTEMVDGLCKIGKTDEAYKLMLKMEEKGCNPNVVTYTAMIDGFGK 766
L +L N+ T Y ++ G C+ G+ +EA +L+ +M G +P+ +TYT MI+ +
Sbjct: 780 LKGLLANTAT-----YNMLIRGFCRQGRIEEASELITRMIGDGVSPDCITYTTMINELCR 834
Query: 767 SGKIEQCLELFRDMCSKGCAPNFITYRVLINHCCSNGLLDEAYKLLDEM 815
+++ +EL+ M KG P+ + Y LI+ CC G + +A +L +EM
Sbjct: 835 RNDVKKAIELWNSMTEKGIRPDRVAYNTLIHGCCVAGEMGKATELRNEM 883
Score = 274 bits (700), Expect = 2e-73
Identities = 226/881 (25%), Positives = 391/881 (43%), Gaps = 87/881 (9%)
Query: 110 VMNNVKNPELCVKFFLWAGRQIGYSHTPQVFDKLLDLLGCNVNADDRVPLKFLMEIKDDD 169
++ + +P+L ++FF + G G+ H+ F L+ L V A+ P L++
Sbjct: 77 LIGTIDDPKLGLRFFNFLGLHRGFDHSTASFCILIHAL---VKANLFWPASSLLQ----- 128
Query: 170 HELLRRLLNFLVRKCCRNGWWNMALEELGRLKDFGYKPSQTTYNALIQVFLRADKLDTAY 229
L+R + +N+ + K S ++++ LIQ ++R+ ++
Sbjct: 129 --------TLLLRALKPSDVFNVLFSCYEKCK----LSSSSSFDLLIQHYVRSRRVLDGV 176
Query: 230 LVKREMLSYAFVMDRY-TLSCFAYSLCKGGKCREAFDLIDE--AEDFVPDTVFYNRMVSG 286
LV + M++ ++ TLS + L K A +L ++ + PD Y ++
Sbjct: 177 LVFKMMITKVSLLPEVRTLSALLHGLVKFRHFGLAMELFNDMVSVGIRPDVYIYTGVIRS 236
Query: 287 LCEASLFEEAMDILHRMRSSSCIPNVVTYRILLSGCLRKGQLGRCKRILSMMITEGCYPN 346
LCE A +++ M ++ C N+V Y +L+ G +K ++ I + + P+
Sbjct: 237 LCELKDLSRAKEMIAHMEATGCDVNIVPYNVLIDGLCKKQKVWEAVGIKKDLAGKDLKPD 296
Query: 347 REIFNSLIHAYCKSRDYSYAYKLFKKMIKCGCQPGYLVYNIFIGSVCSNEEQPSSDILDL 406
+ +L++ CK +++ ++ +M+ P + + + + + L+L
Sbjct: 297 VVTYCTLVYGLCKVQEFEIGLEMMDEMLCLRFSPSEAAVSSLVEGL--RKRGKIEEALNL 354
Query: 407 VEKAYSEMLDLGVVLNKVNVSNFARCLCGAGKFDQAFKIICEMMGK-GFVPDDSTYSKVI 465
V++ ++D GV N + LC KF +A +++ + MGK G P+D TYS +I
Sbjct: 355 VKR----VVDFGVSPNLFVYNALIDSLCKGRKFHEA-ELLFDRMGKIGLRPNDVTYSILI 409
Query: 466 GFLCDASKVEKAFSLFEEMKRNGIVPSVYTYTILIDSFCKAGLIQQARKWFDEMLHKGCT 525
C K++ A S EM G+ SVY Y LI+ CK G I A + EM++K
Sbjct: 410 DMFCRRGKLDTALSFLGEMVDTGLKLSVYPYNSLINGHCKFGDISAAEGFMAEMINKKLE 469
Query: 526 PNVVTYTALIHAYLKAKQMPVADELFEMMLLEGCKPNVVTYTALIDGHCKAGQIEKACQI 585
P VVTYT+L+ Y ++ A L+ M +G P++ T+T L+ G +AG I A ++
Sbjct: 470 PTVVTYTSLMGGYCSKGKINKALRLYHEMTGKGIAPSIYTFTTLLSGLFRAGLIRDAVKL 529
Query: 586 YARM----------------RGDIESSDMDKYFKLDHNNCEG---PNVITYGALVDGLCK 626
+ M G E DM K F+ E P+ +Y L+ GLC
Sbjct: 530 FNEMAEWNVKPNRVTYNVMIEGYCEEGDMSKAFEFLKEMTEKGIVPDTYSYRPLIHGLCL 589
Query: 627 ANRVKEAHELLDTMLAHGCEPNQIVYDAVIDGFCKIGKLQDAQEVFTKMSERGYSPNLYT 686
+ EA +D + CE N+I Y ++ GFC+ GKL++A V +M +RG +L
Sbjct: 590 TGQASEAKVFVDGLHKGNCELNEICYTGLLHGFCREGKLEEALSVCQEMVQRGVDLDLVC 649
Query: 687 YSSFIDCLFKDNRLDLVLKVLSKMLENSCTPNVVIYTEMVDGLCKIGKTDEAYKLMLKME 746
Y ID K L +L +M + P+ VIYT M+D K G EA+ + M
Sbjct: 650 YGVLIDGSLKHKDRKLFFGLLKEMHDRGLKPDDVIYTSMIDAKSKTGDFKEAFGIWDLMI 709
Query: 747 EKGCNPNVVTYTAMIDGFGKSGKIEQCLELFRDMCSKGCAPNFITYRVLINHCCSNGLLD 806
+GC PN VTYTA+I+G K+G + + L M PN +TY ++ + G +D
Sbjct: 710 NEGCVPNEVTYTAVINGLCKAGFVNEAEVLCSKMQPVSSVPNQVTYGCFLD-ILTKGEVD 768
Query: 807 EAYKLLDEMKQTYWPKHILSHRKIIEGFSQEFITSIGLLDELSENESVPVDSLYRILIDN 866
K + H I++G T Y +LI
Sbjct: 769 -------------MQKAVELHNAILKGLLANTAT-------------------YNMLIRG 796
Query: 867 YIKAGRLEVALDLLEEISSSPSHAVSNKYL-YASLIENLSHASKVDKALELYASMISKNV 925
+ + GR+E A +L+ + VS + Y ++I L + V KA+EL+ SM K +
Sbjct: 797 FCRQGRIEEASELITRMIGD---GVSPDCITYTTMINELCRRNDVKKAIELWNSMTEKGI 853
Query: 926 VPELSILVHLIKGLIKVDKWQEALQLSDSICQMVCLTLSQT 966
P+ LI G + +A +L + + + + ++T
Sbjct: 854 RPDRVAYNTLIHGCCVAGEMGKATELRNEMLRQGLIPNNKT 894
Score = 258 bits (660), Expect = 9e-69
Identities = 184/719 (25%), Positives = 306/719 (41%), Gaps = 97/719 (13%)
Query: 175 RLLNFLVRKCCRNGWWNMALEELGRLKDFGYKPSQTTYNALIQVFLRADKLDTAYLVKRE 234
R L+ L+ + + +A+E + G +P Y +I+ L A +
Sbjct: 193 RTLSALLHGLVKFRHFGLAMELFNDMVSVGIRPDVYIYTGVIRSLCELKDLSRAKEMIAH 252
Query: 235 MLSYAFVMDRYTLSCFAYSLCKGGKCREAFDLIDE--AEDFVPDTVFYNRMVSGLCEASL 292
M + ++ + LCK K EA + + +D PD V Y +V GLC+
Sbjct: 253 MEATGCDVNIVPYNVLIDGLCKKQKVWEAVGIKKDLAGKDLKPDVVTYCTLVYGLCKVQE 312
Query: 293 FEEAMDILHRMRSSSCIPNVVTYRILLSGCLRKGQLGRCKRILSMMITEGCYPNREIFNS 352
FE ++++ M P+ L+ G ++G++ ++ ++ G PN ++N+
Sbjct: 313 FEIGLEMMDEMLCLRFSPSEAAVSSLVEGLRKRGKIEEALNLVKRVVDFGVSPNLFVYNA 372
Query: 353 LIHAYCKSRDYSYAYKLFKKMIKCGCQPGYLVYNIFIGSVCSNEEQPSSDILDLVEKAYS 412
LI + CK R + A LF +M K G +P + Y+I I C + LD
Sbjct: 373 LIDSLCKGRKFHEAELLFDRMGKIGLRPNDVTYSILIDMFCRRGK------LDTALSFLG 426
Query: 413 EMLDLGVVLNKVNVSNFARCLCGAGKFDQAFKIICEMMGKGFVPDDSTYSKVIGFLCDAS 472
EM+D G+ L+ ++ C G A + EM+ K P TY+ ++G C
Sbjct: 427 EMVDTGLKLSVYPYNSLINGHCKFGDISAAEGFMAEMINKKLEPTVVTYTSLMGGYCSKG 486
Query: 473 KVEKAFSLFEEMKRNGIVPSVYTYTILIDSFCKAGLIQQARKWFDEMLHKGCTPNVVTYT 532
K+ KA L+ EM GI PS+YT+T L+ +AGLI+ A K F+EM PN VTY
Sbjct: 487 KINKALRLYHEMTGKGIAPSIYTFTTLLSGLFRAGLIRDAVKLFNEMAEWNVKPNRVTYN 546
Query: 533 ALIHAYLKAKQMPVADELFEMMLLEG---------------------------------- 558
+I Y + M A E + M +G
Sbjct: 547 VMIEGYCEEGDMSKAFEFLKEMTEKGIVPDTYSYRPLIHGLCLTGQASEAKVFVDGLHKG 606
Query: 559 -CKPNVVTYTALIDGHCKAGQIEKACQIYARM----------------RGDIESSDMDKY 601
C+ N + YT L+ G C+ G++E+A + M G ++ D +
Sbjct: 607 NCELNEICYTGLLHGFCREGKLEEALSVCQEMVQRGVDLDLVCYGVLIDGSLKHKDRKLF 666
Query: 602 FKL---DHNNCEGPNVITYGALVDGLCKANRVKEAHELLDTMLAHGCEPNQIVYDAVIDG 658
F L H+ P+ + Y +++D K KEA + D M+ GC PN++ Y AVI+G
Sbjct: 667 FGLLKEMHDRGLKPDDVIYTSMIDAKSKTGDFKEAFGIWDLMINEGCVPNEVTYTAVING 726
Query: 659 FCKIGKLQDAQEVFTKMSERGYSPNLYTYSSFIDCLFK---------------------- 696
CK G + +A+ + +KM PN TY F+D L K
Sbjct: 727 LCKAGFVNEAEVLCSKMQPVSSVPNQVTYGCFLDILTKGEVDMQKAVELHNAILKGLLAN 786
Query: 697 -------------DNRLDLVLKVLSKMLENSCTPNVVIYTEMVDGLCKIGKTDEAYKLML 743
R++ +++++M+ + +P+ + YT M++ LC+ +A +L
Sbjct: 787 TATYNMLIRGFCRQGRIEEASELITRMIGDGVSPDCITYTTMINELCRRNDVKKAIELWN 846
Query: 744 KMEEKGCNPNVVTYTAMIDGFGKSGKIEQCLELFRDMCSKGCAPNFITYRVLINHCCSN 802
M EKG P+ V Y +I G +G++ + EL +M +G PN T R ++ S+
Sbjct: 847 SMTEKGIRPDRVAYNTLIHGCCVAGEMGKATELRNEMLRQGLIPNNKTSRTTTSNDTSS 905
Score = 159 bits (401), Expect = 1e-38
Identities = 124/510 (24%), Positives = 229/510 (44%), Gaps = 38/510 (7%)
Query: 488 GIVPSVYTYTILIDSFCKAGLIQQARKWFDEMLHKGCTPNVV-----------------T 530
G S ++ ILI + KA L A +L + P+ V +
Sbjct: 99 GFDHSTASFCILIHALVKANLFWPASSLLQTLLLRALKPSDVFNVLFSCYEKCKLSSSSS 158
Query: 531 YTALIHAYLKAKQMPVADELFEMMLLE-GCKPNVVTYTALIDGHCKAGQIEKACQIYARM 589
+ LI Y++++++ +F+MM+ + P V T +AL+ G K A +++
Sbjct: 159 FDLLIQHYVRSRRVLDGVLVFKMMITKVSLLPEVRTLSALLHGLVKFRHFGLAMELF--- 215
Query: 590 RGDIESSDMDKYFKLDHNNCEGPNVITYGALVDGLCKANRVKEAHELLDTMLAHGCEPNQ 649
D+ S + P+V Y ++ LC+ + A E++ M A GC+ N
Sbjct: 216 -NDMVSVGIR------------PDVYIYTGVIRSLCELKDLSRAKEMIAHMEATGCDVNI 262
Query: 650 IVYDAVIDGFCKIGKLQDAQEVFTKMSERGYSPNLYTYSSFIDCLFKDNRLDLVLKVLSK 709
+ Y+ +IDG CK K+ +A + ++ + P++ TY + + L K ++ L+++ +
Sbjct: 263 VPYNVLIDGLCKKQKVWEAVGIKKDLAGKDLKPDVVTYCTLVYGLCKVQEFEIGLEMMDE 322
Query: 710 MLENSCTPNVVIYTEMVDGLCKIGKTDEAYKLMLKMEEKGCNPNVVTYTAMIDGFGKSGK 769
ML +P+ + +V+GL K GK +EA L+ ++ + G +PN+ Y A+ID K K
Sbjct: 323 MLCLRFSPSEAAVSSLVEGLRKRGKIEEALNLVKRVVDFGVSPNLFVYNALIDSLCKGRK 382
Query: 770 IEQCLELFRDMCSKGCAPNFITYRVLINHCCSNGLLDEAYKLLDEMKQTYWPKHILSHRK 829
+ LF M G PN +TY +LI+ C G LD A L EM T + +
Sbjct: 383 FHEAELLFDRMGKIGLRPNDVTYSILIDMFCRRGKLDTALSFLGEMVDTGLKLSVYPYNS 442
Query: 830 IIEGFSQ--EFITSIGLLDELSENESVPVDSLYRILIDNYIKAGRLEVALDLLEEISSSP 887
+I G + + + G + E+ + P Y L+ Y G++ AL L E++
Sbjct: 443 LINGHCKFGDISAAEGFMAEMINKKLEPTVVTYTSLMGGYCSKGKINKALRLYHEMTGKG 502
Query: 888 SHAVSNKYLYASLIENLSHASKVDKALELYASMISKNVVPELSILVHLIKGLIKVDKWQE 947
+ Y + +L+ L A + A++L+ M NV P +I+G + +
Sbjct: 503 --IAPSIYTFTTLLSGLFRAGLIRDAVKLFNEMAEWNVKPNRVTYNVMIEGYCEEGDMSK 560
Query: 948 ALQLSDSICQMVCLTLSQTFQPLVNLNLLT 977
A + + + + + +++PL++ LT
Sbjct: 561 AFEFLKEMTEKGIVPDTYSYRPLIHGLCLT 590
>At5g61990 unknown protein
Length = 974
Score = 273 bits (699), Expect = 3e-73
Identities = 182/667 (27%), Positives = 309/667 (46%), Gaps = 47/667 (7%)
Query: 157 VPLKFLMEIKDDDHELLRRLLNFLVRKCCRNGWWNMALEELGRLKDFGYKPSQTTYNALI 216
VPLK+ ++ D ++RL + A L + G TY+ LI
Sbjct: 274 VPLKYTYDVLIDGLCKIKRLED--------------AKSLLVEMDSLGVSLDNHTYSLLI 319
Query: 217 QVFLRADKLDTAYLVKREMLSYAFVMDRYTLSCFAYSLCKGGKCREAFDLIDE--AEDFV 274
L+ D A + EM+S+ + Y C + K G +A L D A +
Sbjct: 320 DGLLKGRNADAAKGLVHEMVSHGINIKPYMYDCCICVMSKEGVMEKAKALFDGMIASGLI 379
Query: 275 PDTVFYNRMVSGLCEASLFEEAMDILHRMRSSSCIPNVVTYRILLSGCLRKGQLGRCKRI 334
P Y ++ G C + ++L M+ + + + TY ++ G G L I
Sbjct: 380 PQAQAYASLIEGYCREKNVRQGYELLVEMKKRNIVISPYTYGTVVKGMCSSGDLDGAYNI 439
Query: 335 LSMMITEGCYPNREIFNSLIHAYCKSRDYSYAYKLFKKMIKCGCQPGYLVYNIFIGSVCS 394
+ MI GC PN I+ +LI + ++ + A ++ K+M + G P YN I +
Sbjct: 440 VKEMIASGCRPNVVIYTTLIKTFLQNSRFGDAMRVLKEMKEQGIAPDIFCYNSLIIGLSK 499
Query: 395 NEEQPSSDILDLVEKAYSEMLDLGVVLNKVNVSNFARCLCGAGKFDQAFKIICEMMGKGF 454
+ +D EM++ G+ N F A +F A K + EM G
Sbjct: 500 AKR------MDEARSFLVEMVENGLKPNAFTYGAFISGYIEASEFASADKYVKEMRECGV 553
Query: 455 VPDDSTYSKVIGFLCDASKVEKAFSLFEEMKRNGIVPSVYTYTILIDSFCKAGLIQQARK 514
+P+ + +I C KV +A S + M GI+ TYT+L++ K + A +
Sbjct: 554 LPNKVLCTGLINEYCKKGKVIEACSAYRSMVDQGILGDAKTYTVLMNGLFKNDKVDDAEE 613
Query: 515 WFDEMLHKGCTPNVVTYTALIHAYLKAKQMPVADELFEMMLLEGCKPNVVTYTALIDGHC 574
F EM KG P+V +Y LI+ + K M A +F+ M+ EG PNV+ Y L+ G C
Sbjct: 614 IFREMRGKGIAPDVFSYGVLINGFSKLGNMQKASSIFDEMVEEGLTPNVIIYNMLLGGFC 673
Query: 575 KAGQIEKACQIYARMRGDIESSDMDKYFKLDHNNCEG--PNVITYGALVDGLCKANRVKE 632
++G+IEKA ++ LD + +G PN +TY ++DG CK+ + E
Sbjct: 674 RSGEIEKAKEL------------------LDEMSVKGLHPNAVTYCTIIDGYCKSGDLAE 715
Query: 633 AHELLDTMLAHGCEPNQIVYDAVIDGFCKIGKLQDAQEVFTKMSERGYSPNLYTYSSFID 692
A L D M G P+ VY ++DG C++ ++ A +F +++G + + +++ I+
Sbjct: 716 AFRLFDEMKLKGLVPDSFVYTTLVDGCCRLNDVERAITIFGT-NKKGCASSTAPFNALIN 774
Query: 693 CLFKDNRLDLVLKVLSKMLENSCT----PNVVIYTEMVDGLCKIGKTDEAYKLMLKMEEK 748
+FK + +L +VL+++++ S PN V Y M+D LCK G + A +L +M+
Sbjct: 775 WVFKFGKTELKTEVLNRLMDGSFDRFGKPNDVTYNIMIDYLCKEGNLEAAKELFHQMQNA 834
Query: 749 GCNPNVVTYTAMIDGFGKSGKIEQCLELFRDMCSKGCAPNFITYRVLINHCCSNGLLDEA 808
P V+TYT++++G+ K G+ + +F + + G P+ I Y V+IN G+ +A
Sbjct: 835 NLMPTVITYTSLLNGYDKMGRRAEMFPVFDEAIAAGIEPDHIMYSVIINAFLKEGMTTKA 894
Query: 809 YKLLDEM 815
L+D+M
Sbjct: 895 LVLVDQM 901
Score = 266 bits (680), Expect = 4e-71
Identities = 189/784 (24%), Positives = 343/784 (43%), Gaps = 64/784 (8%)
Query: 207 PSQTTYNALIQVFLRADKLDTAYLVKREMLSYAFVMDRYTLSCFAYSLCKGGKCREAFDL 266
P + L+ LR ++LD + V + M+ V D T + C+ G + D+
Sbjct: 184 PRLSRCKVLLDALLRWNRLDLFWDVYKGMVERNVVFDVKTYHMLIIAHCRAGNVQLGKDV 243
Query: 267 IDEAE-----------------------DFVPDTVFYNRMVSGLCEASLFEEAMDILHRM 303
+ + E VP Y+ ++ GLC+ E+A +L M
Sbjct: 244 LFKTEKEFRTATLNVDGALKLKESMICKGLVPLKYTYDVLIDGLCKIKRLEDAKSLLVEM 303
Query: 304 RSSSCIPNVVTYRILLSGCLRKGQLGRCKRILSMMITEGCYPNREIFNSLIHAYCKSRDY 363
S + TY +L+ G L+ K ++ M++ G +++ I K
Sbjct: 304 DSLGVSLDNHTYSLLIDGLLKGRNADAAKGLVHEMVSHGINIKPYMYDCCICVMSKEGVM 363
Query: 364 SYAYKLFKKMIKCGCQPGYLVYNIFIGSVCSNEEQPSSDILDLVEKAYS---EMLDLGVV 420
A LF MI G P Y I C + V + Y EM +V
Sbjct: 364 EKAKALFDGMIASGLIPQAQAYASLIEGYCREKN---------VRQGYELLVEMKKRNIV 414
Query: 421 LNKVNVSNFARCLCGAGKFDQAFKIICEMMGKGFVPDDSTYSKVIGFLCDASKVEKAFSL 480
++ + +C +G D A+ I+ EM+ G P+ Y+ +I S+ A +
Sbjct: 415 ISPYTYGTVVKGMCSSGDLDGAYNIVKEMIASGCRPNVVIYTTLIKTFLQNSRFGDAMRV 474
Query: 481 FEEMKRNGIVPSVYTYTILIDSFCKAGLIQQARKWFDEMLHKGCTPNVVTYTALIHAYLK 540
+EMK GI P ++ Y LI KA + +AR + EM+ G PN TY A I Y++
Sbjct: 475 LKEMKEQGIAPDIFCYNSLIIGLSKAKRMDEARSFLVEMVENGLKPNAFTYGAFISGYIE 534
Query: 541 AKQMPVADELFEMMLLEGCKPNVVTYTALIDGHCKAGQIEKACQIYARM-----RGDIES 595
A + AD+ + M G PN V T LI+ +CK G++ +AC Y M GD ++
Sbjct: 535 ASEFASADKYVKEMRECGVLPNKVLCTGLINEYCKKGKVIEACSAYRSMVDQGILGDAKT 594
Query: 596 --------------SDMDKYFKLDHNNCEGPNVITYGALVDGLCKANRVKEAHELLDTML 641
D ++ F+ P+V +YG L++G K +++A + D M+
Sbjct: 595 YTVLMNGLFKNDKVDDAEEIFREMRGKGIAPDVFSYGVLINGFSKLGNMQKASSIFDEMV 654
Query: 642 AHGCEPNQIVYDAVIDGFCKIGKLQDAQEVFTKMSERGYSPNLYTYSSFIDCLFKDNRLD 701
G PN I+Y+ ++ GFC+ G+++ A+E+ +MS +G PN TY + ID K L
Sbjct: 655 EEGLTPNVIIYNMLLGGFCRSGEIEKAKELLDEMSVKGLHPNAVTYCTIIDGYCKSGDLA 714
Query: 702 LVLKVLSKMLENSCTPNVVIYTEMVDGLCKIGKTDEAYKLMLKMEEKGCNPNVVTYTAMI 761
++ +M P+ +YT +VDG C++ + A + +KGC + + A+I
Sbjct: 715 EAFRLFDEMKLKGLVPDSFVYTTLVDGCCRLNDVERAITI-FGTNKKGCASSTAPFNALI 773
Query: 762 DGFGKSGKIEQCLELFRDMC----SKGCAPNFITYRVLINHCCSNGLLDEAYKLLDEMKQ 817
+ K GK E E+ + + PN +TY ++I++ C G L+ A +L +M+
Sbjct: 774 NWVFKFGKTELKTEVLNRLMDGSFDRFGKPNDVTYNIMIDYLCKEGNLEAAKELFHQMQN 833
Query: 818 TYWPKHILSHRKIIEGFSQ--EFITSIGLLDELSENESVPVDSLYRILIDNYIKAGRLEV 875
++++ ++ G+ + + DE P +Y ++I+ ++K G
Sbjct: 834 ANLMPTVITYTSLLNGYDKMGRRAEMFPVFDEAIAAGIEPDHIMYSVIINAFLKEGMTTK 893
Query: 876 ALDLLEEISSSPSHAVSNKYLYA---SLIENLSHASKVDKALELYASMISKNVVPELSIL 932
AL L++++ + + K + +L+ + +++ A ++ +M+ +P+ + +
Sbjct: 894 ALVLVDQMFAKNAVDDGCKLSISTCRALLSGFAKVGEMEVAEKVMENMVRLQYIPDSATV 953
Query: 933 VHLI 936
+ LI
Sbjct: 954 IELI 957
Score = 236 bits (601), Expect = 6e-62
Identities = 174/687 (25%), Positives = 303/687 (43%), Gaps = 60/687 (8%)
Query: 298 DILHRMRSSSCIPNVVTYRILLSGCLRKG--QLGR-------------------CKRILS 336
D+ M + + +V TY +L+ R G QLG+ ++
Sbjct: 207 DVYKGMVERNVVFDVKTYHMLIIAHCRAGNVQLGKDVLFKTEKEFRTATLNVDGALKLKE 266
Query: 337 MMITEGCYPNREIFNSLIHAYCKSRDYSYAYKLFKKMIKCGCQPGYLVYNIFIGSVCSNE 396
MI +G P + ++ LI CK + A L +M G Y++ I +
Sbjct: 267 SMICKGLVPLKYTYDVLIDGLCKIKRLEDAKSLLVEMDSLGVSLDNHTYSLLIDGLLKGR 326
Query: 397 EQPSSDILDLVEKAYSEMLDLGVVLNKVNVSNFARCLCGAGK---FDQAFKIICEMMGKG 453
D + EM+ G+ + + C+C K ++A + M+ G
Sbjct: 327 NA------DAAKGLVHEMVSHGI---NIKPYMYDCCICVMSKEGVMEKAKALFDGMIASG 377
Query: 454 FVPDDSTYSKVIGFLCDASKVEKAFSLFEEMKRNGIVPSVYTYTILIDSFCKAGLIQQAR 513
+P Y+ +I C V + + L EMK+ IV S YTY ++ C +G + A
Sbjct: 378 LIPQAQAYASLIEGYCREKNVRQGYELLVEMKKRNIVISPYTYGTVVKGMCSSGDLDGAY 437
Query: 514 KWFDEMLHKGCTPNVVTYTALIHAYLKAKQMPVADELFEMMLLEGCKPNVVTYTALIDGH 573
EM+ GC PNVV YT LI +L+ + A + + M +G P++ Y +LI G
Sbjct: 438 NIVKEMIASGCRPNVVIYTTLIKTFLQNSRFGDAMRVLKEMKEQGIAPDIFCYNSLIIGL 497
Query: 574 CKAGQIEKACQIYARMRGDIESSDMDKYFKLDHNNCEGPNVITYGALVDGLCKANRVKEA 633
KA ++++A M N PN TYGA + G +A+ A
Sbjct: 498 SKAKRMDEARSFLVEMV----------------ENGLKPNAFTYGAFISGYIEASEFASA 541
Query: 634 HELLDTMLAHGCEPNQIVYDAVIDGFCKIGKLQDAQEVFTKMSERGYSPNLYTYSSFIDC 693
+ + M G PN+++ +I+ +CK GK+ +A + M ++G + TY+ ++
Sbjct: 542 DKYVKEMRECGVLPNKVLCTGLINEYCKKGKVIEACSAYRSMVDQGILGDAKTYTVLMNG 601
Query: 694 LFKDNRLDLVLKVLSKMLENSCTPNVVIYTEMVDGLCKIGKTDEAYKLMLKMEEKGCNPN 753
LFK++++D ++ +M P+V Y +++G K+G +A + +M E+G PN
Sbjct: 602 LFKNDKVDDAEEIFREMRGKGIAPDVFSYGVLINGFSKLGNMQKASSIFDEMVEEGLTPN 661
Query: 754 VVTYTAMIDGFGKSGKIEQCLELFRDMCSKGCAPNFITYRVLINHCCSNGLLDEAYKLLD 813
V+ Y ++ GF +SG+IE+ EL +M KG PN +TY +I+ C +G L EA++L D
Sbjct: 662 VIIYNMLLGGFCRSGEIEKAKELLDEMSVKGLHPNAVTYCTIIDGYCKSGDLAEAFRLFD 721
Query: 814 EMKQTYWPKHILSHRKIIEGFS-----QEFITSIGLLDELSENESVPVDSLYRILIDNYI 868
EMK + +++G + IT G + + + P + LI+
Sbjct: 722 EMKLKGLVPDSFVYTTLVDGCCRLNDVERAITIFGTNKKGCASSTAP----FNALINWVF 777
Query: 869 KAGRLEVALDLLEEI--SSSPSHAVSNKYLYASLIENLSHASKVDKALELYASMISKNVV 926
K G+ E+ ++L + S N Y +I+ L ++ A EL+ M + N++
Sbjct: 778 KFGKTELKTEVLNRLMDGSFDRFGKPNDVTYNIMIDYLCKEGNLEAAKELFHQMQNANLM 837
Query: 927 PELSILVHLIKGLIKVDKWQEALQLSD 953
P + L+ G K+ + E + D
Sbjct: 838 PTVITYTSLLNGYDKMGRRAEMFPVFD 864
Score = 167 bits (422), Expect = 4e-41
Identities = 126/510 (24%), Positives = 224/510 (43%), Gaps = 28/510 (5%)
Query: 468 LCDASKVEKAFSLFEEM-KRNGIVPSVYT------------------YTILIDSFCKAGL 508
LC+ EKA S+ E M +RN V V++ + IL D + G
Sbjct: 107 LCNFGSFEKALSVVERMIERNWPVAEVWSSIVRCSQEFVGKSDDGVLFGILFDGYIAKGY 166
Query: 509 IQQARKWFDEMLHKGCTPNVVTYTALIHAYLKAKQMPVADELFEMMLLEGCKPNVVTYTA 568
I++A F + P + L+ A L+ ++ + ++++ M+ +V TY
Sbjct: 167 IEEAVFVFSSSMGLELVPRLSRCKVLLDALLRWNRLDLFWDVYKGMVERNVVFDVKTYHM 226
Query: 569 LIDGHCKAGQIEKACQIYARMRGDIESS--DMDKYFKLDHNN-CEG--PNVITYGALVDG 623
LI HC+AG ++ + + + ++ ++D KL + C+G P TY L+DG
Sbjct: 227 LIIAHCRAGNVQLGKDVLFKTEKEFRTATLNVDGALKLKESMICKGLVPLKYTYDVLIDG 286
Query: 624 LCKANRVKEAHELLDTMLAHGCEPNQIVYDAVIDGFCKIGKLQDAQEVFTKMSERGYSPN 683
LCK R+++A LL M + G + Y +IDG K A+ + +M G +
Sbjct: 287 LCKIKRLEDAKSLLVEMDSLGVSLDNHTYSLLIDGLLKGRNADAAKGLVHEMVSHGINIK 346
Query: 684 LYTYSSFIDCLFKDNRLDLVLKVLSKMLENSCTPNVVIYTEMVDGLCKIGKTDEAYKLML 743
Y Y I + K+ ++ + M+ + P Y +++G C+ + Y+L++
Sbjct: 347 PYMYDCCICVMSKEGVMEKAKALFDGMIASGLIPQAQAYASLIEGYCREKNVRQGYELLV 406
Query: 744 KMEEKGCNPNVVTYTAMIDGFGKSGKIEQCLELFRDMCSKGCAPNFITYRVLINHCCSNG 803
+M+++ + TY ++ G SG ++ + ++M + GC PN + Y LI N
Sbjct: 407 EMKKRNIVISPYTYGTVVKGMCSSGDLDGAYNIVKEMIASGCRPNVVIYTTLIKTFLQNS 466
Query: 804 LLDEAYKLLDEMKQTYWPKHILSHRKIIEGFS--QEFITSIGLLDELSENESVPVDSLYR 861
+A ++L EMK+ I + +I G S + + L E+ EN P Y
Sbjct: 467 RFGDAMRVLKEMKEQGIAPDIFCYNSLIIGLSKAKRMDEARSFLVEMVENGLKPNAFTYG 526
Query: 862 ILIDNYIKAGRLEVALDLLEEISSSPSHAVSNKYLYASLIENLSHASKVDKALELYASMI 921
I YI+A A ++E+ + NK L LI KV +A Y SM+
Sbjct: 527 AFISGYIEASEFASADKYVKEMRE--CGVLPNKVLCTGLINEYCKKGKVIEACSAYRSMV 584
Query: 922 SKNVVPELSILVHLIKGLIKVDKWQEALQL 951
+ ++ + L+ GL K DK +A ++
Sbjct: 585 DQGILGDAKTYTVLMNGLFKNDKVDDAEEI 614
Score = 159 bits (402), Expect = 7e-39
Identities = 105/402 (26%), Positives = 188/402 (46%), Gaps = 20/402 (4%)
Query: 200 LKDFGYKPSQTTYNALIQVFLRADKLDTAYLVKREMLSYAFVMDRYTLSCFAYSLCKGGK 259
+++ G P++ LI + + K+ A R M+ + D T + L K K
Sbjct: 548 MRECGVLPNKVLCTGLINEYCKKGKVIEACSAYRSMVDQGILGDAKTYTVLMNGLFKNDK 607
Query: 260 CREAFDLIDE--AEDFVPDTVFYNRMVSGLCEASLFEEAMDILHRMRSSSCIPNVVTYRI 317
+A ++ E + PD Y +++G + ++A I M PNV+ Y +
Sbjct: 608 VDDAEEIFREMRGKGIAPDVFSYGVLINGFSKLGNMQKASSIFDEMVEEGLTPNVIIYNM 667
Query: 318 LLSGCLRKGQLGRCKRILSMMITEGCYPNREIFNSLIHAYCKSRDYSYAYKLFKKMIKCG 377
LL G R G++ + K +L M +G +PN + ++I YCKS D + A++LF +M G
Sbjct: 668 LLGGFCRSGEIEKAKELLDEMSVKGLHPNAVTYCTIIDGYCKSGDLAEAFRLFDEMKLKG 727
Query: 378 CQPGYLVYNIFIGSVCS-NEEQPSSDILDLVEKAYSEMLDLGVVLNKVNVSNFARCLCGA 436
P VY + C N+ + + I +K G + + +
Sbjct: 728 LVPDSFVYTTLVDGCCRLNDVERAITIFGTNKK--------GCASSTAPFNALINWVFKF 779
Query: 437 GKFDQAFKIICEMMGKGF----VPDDSTYSKVIGFLCDASKVEKAFSLFEEMKRNGIVPS 492
GK + +++ +M F P+D TY+ +I +LC +E A LF +M+ ++P+
Sbjct: 780 GKTELKTEVLNRLMDGSFDRFGKPNDVTYNIMIDYLCKEGNLEAAKELFHQMQNANLMPT 839
Query: 493 VYTYTILIDSFCKAGLIQQARKWFDEMLHKGCTPNVVTYTALIHAYLK----AKQMPVAD 548
V TYT L++ + K G + FDE + G P+ + Y+ +I+A+LK K + + D
Sbjct: 840 VITYTSLLNGYDKMGRRAEMFPVFDEAIAAGIEPDHIMYSVIINAFLKEGMTTKALVLVD 899
Query: 549 ELFEMMLL-EGCKPNVVTYTALIDGHCKAGQIEKACQIYARM 589
++F + +GCK ++ T AL+ G K G++E A ++ M
Sbjct: 900 QMFAKNAVDDGCKLSISTCRALLSGFAKVGEMEVAEKVMENM 941
>At5g55840 putative protein
Length = 1274
Score = 273 bits (699), Expect = 3e-73
Identities = 193/760 (25%), Positives = 353/760 (46%), Gaps = 30/760 (3%)
Query: 207 PSQTTYNALIQVFLRADKLDTAYLVKREMLSYAFVMDRYTLSCFAYSLCKGGKCREAFDL 266
P T+N LI V + + + ++M + T + + CK G+ + A +L
Sbjct: 191 PDVATFNILINVLCAEGSFEKSSYLMQKMEKSGYAPTIVTYNTVLHWYCKKGRFKAAIEL 250
Query: 267 ID--EAEDFVPDTVFYNRMVSGLCEASLFEEAMDILHRMRSSSCIPNVVTYRILLSGCLR 324
+D +++ D YN ++ LC ++ + +L MR PN VTY L++G
Sbjct: 251 LDHMKSKGVDADVCTYNMLIHDLCRSNRIAKGYLLLRDMRKRMIHPNEVTYNTLINGFSN 310
Query: 325 KGQLGRCKRILSMMITEGCYPNREIFNSLIHAYCKSRDYSYAYKLFKKMIKCGCQPGYLV 384
+G++ ++L+ M++ G PN FN+LI + ++ A K+F M G P +
Sbjct: 311 EGKVLIASQLLNEMLSFGLSPNHVTFNALIDGHISEGNFKEALKMFYMMEAKGLTPSEVS 370
Query: 385 YNIFIGSVCSNEEQPSSDILDLVEKAYSEMLDLGVVLNKVNVSNFARCLCGAGKFDQAFK 444
Y + + +C N E DL Y M GV + ++ + LC G D+A
Sbjct: 371 YGVLLDGLCKNAE------FDLARGFYMRMKRNGVCVGRITYTGMIDGLCKNGFLDEAVV 424
Query: 445 IICEMMGKGFVPDDSTYSKVIGFLCDASKVEKAFSLFEEMKRNGIVPSVYTYTILIDSFC 504
++ EM G PD TYS +I C + + A + + R G+ P+ Y+ LI + C
Sbjct: 425 LLNEMSKDGIDPDIVTYSALINGFCKVGRFKTAKEIVCRIYRVGLSPNGIIYSTLIYNCC 484
Query: 505 KAGLIQQARKWFDEMLHKGCTPNVVTYTALIHAYLKAKQMPVADELFEMMLLEGCKPNVV 564
+ G +++A + ++ M+ +G T + T+ L+ + KA ++ A+E M +G PN V
Sbjct: 485 RMGCLKEAIRIYEAMILEGHTRDHFTFNVLVTSLCKAGKVAEAEEFMRCMTSDGILPNTV 544
Query: 565 TYTALIDGHCKAGQIEKACQIYARMRGDIESSDMDKYFKLDHNNCEGPNVITYGALVDGL 624
++ LI+G+ +G+ KA ++ D+ K+ H+ P TYG+L+ GL
Sbjct: 545 SFDCLINGYGNSGEGLKAFSVF------------DEMTKVGHH----PTFFTYGSLLKGL 588
Query: 625 CKANRVKEAHELLDTMLAHGCEPNQIVYDAVIDGFCKIGKLQDAQEVFTKMSERGYSPNL 684
CK ++EA + L ++ A + ++Y+ ++ CK G L A +F +M +R P+
Sbjct: 589 CKGGHLREAEKFLKSLHAVPAAVDTVMYNTLLTAMCKSGNLAKAVSLFGEMVQRSILPDS 648
Query: 685 YTYSSFIDCLFKDNRLDL-VLKVLSKMLENSCTPNVVIYTEMVDGLCKIGKTDEAYKLML 743
YTY+S I L + + + +L + PN V+YT VDG+ K G+
Sbjct: 649 YTYTSLISGLCRKGKTVIAILFAKEAEARGNVLPNKVMYTCFVDGMFKAGQWKAGIYFRE 708
Query: 744 KMEEKGCNPNVVTYTAMIDGFGKSGKIEQCLELFRDMCSKGCAPNFITYRVLINHCCSNG 803
+M+ G P++VT AMIDG+ + GKIE+ +L +M ++ PN TY +L++
Sbjct: 709 QMDNLGHTPDIVTTNAMIDGYSRMGKIEKTNDLLPEMGNQNGGPNLTTYNILLHGYSKRK 768
Query: 804 LLDEAYKLLDEMKQTYWPKHILSHRKIIEGFSQEFITSIGL-LDELSENESVPVDS-LYR 861
+ ++ L + L+ ++ G + + IGL + + V VD +
Sbjct: 769 DVSTSFLLYRSIILNGILPDKLTCHSLVLGICESNMLEIGLKILKAFICRGVEVDRYTFN 828
Query: 862 ILIDNYIKAGRLEVALDLLEEISSSPSHAVSNKYLYASLIENLSHASKVDKALELYASMI 921
+LI G + A DL++ ++S +K +++ L+ + ++ + M
Sbjct: 829 MLISKCCANGEINWAFDLVKVMTSLGISL--DKDTCDAMVSVLNRNHRFQESRMVLHEMS 886
Query: 922 SKNVVPELSILVHLIKGLIKVDKWQEALQLSDS-ICQMVC 960
+ + PE + LI GL +V + A + + I +C
Sbjct: 887 KQGISPESRKYIGLINGLCRVGDIKTAFVVKEEMIAHKIC 926
Score = 264 bits (675), Expect = 2e-70
Identities = 202/803 (25%), Positives = 353/803 (43%), Gaps = 64/803 (7%)
Query: 210 TTYNALIQVFLRADKLDTAYLVKREMLSYAFVMDRYTLSCFAYSLCKGGKCREAFDLIDE 269
+ Y+ LI+V+LR + + + R M Y F YT + S+ K G+ + + E
Sbjct: 124 SVYDILIRVYLREGMIQDSLEIFRLMGLYGFNPSVYTCNAILGSVVKSGEDVSVWSFLKE 183
Query: 270 --AEDFVPDTVFYNRMVSGLCEASLFEEAMDILHRMRSSSCIPNVVTYRILLSGCLRKGQ 327
PD +N +++ LC FE++ ++ +M S P +VTY +L +KG+
Sbjct: 184 MLKRKICPDVATFNILINVLCAEGSFEKSSYLMQKMEKSGYAPTIVTYNTVLHWYCKKGR 243
Query: 328 LGRCKRILSMMITEGCYPNREIFNSLIHAYCKSRDYSYAYKLFKKMIKCGCQPGYLVYNI 387
+L M ++G + +N LIH C+S + Y L + M K P + YN
Sbjct: 244 FKAAIELLDHMKSKGVDADVCTYNMLIHDLCRSNRIAKGYLLLRDMRKRMIHPNEVTYNT 303
Query: 388 FIGSVCSNEEQPSSDILDLVEKAYSEMLDLGVVLNKVNVSNFARCLCGAGKFDQAFKIIC 447
I SNE + + + + +EML G+ N V + G F +A K+
Sbjct: 304 LINGF-SNEGK-----VLIASQLLNEMLSFGLSPNHVTFNALIDGHISEGNFKEALKMFY 357
Query: 448 EMMGKGFVPDDSTYSKVIGFLCDASKVEKAFSLFEEMKRNGIVPSVYTYTILIDSFCKAG 507
M KG P + +Y ++ LC ++ + A + MKRNG+ TYT +ID CK G
Sbjct: 358 MMEAKGLTPSEVSYGVLLDGLCKNAEFDLARGFYMRMKRNGVCVGRITYTGMIDGLCKNG 417
Query: 508 LIQQARKWFDEMLHKGCTPNVVTYTALIHAYLKAKQMPVADELFEMMLLEGCKPNVVTYT 567
+ +A +EM G P++VTY+ALI+ + K + A E+ + G PN + Y+
Sbjct: 418 FLDEAVVLLNEMSKDGIDPDIVTYSALINGFCKVGRFKTAKEIVCRIYRVGLSPNGIIYS 477
Query: 568 ALIDGHCKAGQIEKACQIYARMRGDIESSDMDKYFKLDHNNCEGPNVITYGALVDGLCKA 627
LI C+ G +++A +IY M +E D + T+ LV LCKA
Sbjct: 478 TLIYNCCRMGCLKEAIRIYEAM--ILEGHTRDHF--------------TFNVLVTSLCKA 521
Query: 628 NRVKEAHELLDTMLAHGCEPNQIVYDAVIDGFCKIGKLQDAQEVFTKMSERGYSPNLYTY 687
+V EA E + M + G PN + +D +I+G+ G+ A VF +M++ G+ P +TY
Sbjct: 522 GKVAEAEEFMRCMTSDGILPNTVSFDCLINGYGNSGEGLKAFSVFDEMTKVGHHPTFFTY 581
Query: 688 SSFIDCLFKDNRL-----------------DLVL------------------KVLSKMLE 712
S + L K L D V+ + +M++
Sbjct: 582 GSLLKGLCKGGHLREAEKFLKSLHAVPAAVDTVMYNTLLTAMCKSGNLAKAVSLFGEMVQ 641
Query: 713 NSCTPNVVIYTEMVDGLCKIGKTDEAYKLMLKMEEKG-CNPNVVTYTAMIDGFGKSGKIE 771
S P+ YT ++ GLC+ GKT A + E +G PN V YT +DG K+G+ +
Sbjct: 642 RSILPDSYTYTSLISGLCRKGKTVIAILFAKEAEARGNVLPNKVMYTCFVDGMFKAGQWK 701
Query: 772 QCLELFRDMCSKGCAPNFITYRVLINHCCSNGLLDEAYKLLDEMKQTYWPKHILSHRKII 831
+ M + G P+ +T +I+ G +++ LL EM ++ ++ ++
Sbjct: 702 AGIYFREQMDNLGHTPDIVTTNAMIDGYSRMGKIEKTNDLLPEMGNQNGGPNLTTYNILL 761
Query: 832 EGFS--QEFITSIGLLDELSENESVPVDSLYRILIDNYIKAGRLEVALDLLEEISSSPSH 889
G+S ++ TS L + N +P L+ ++ LE+ L +L+
Sbjct: 762 HGYSKRKDVSTSFLLYRSIILNGILPDKLTCHSLVLGICESNMLEIGLKILKAFICRGVE 821
Query: 890 AVSNKYLYASLIENLSHASKVDKALELYASMISKNVVPELSILVHLIKGLIKVDKWQEAL 949
++Y + LI +++ A +L M S + + ++ L + ++QE+
Sbjct: 822 V--DRYTFNMLISKCCANGEINWAFDLVKVMTSLGISLDKDTCDAMVSVLNRNHRFQESR 879
Query: 950 QLSDSICQMVCLTLSQTFQPLVN 972
+ + + S+ + L+N
Sbjct: 880 MVLHEMSKQGISPESRKYIGLIN 902
Score = 233 bits (593), Expect = 5e-61
Identities = 165/660 (25%), Positives = 307/660 (46%), Gaps = 38/660 (5%)
Query: 308 CIPNVVTYRILLSGCLRKGQLGRCKRILSMMITEGCYPNREIFNSLIHAYCKSRDYSYAY 367
C N Y IL+ LR+G + I +M G P+ N+++ + KS + +
Sbjct: 119 CNSNPSVYDILIRVYLREGMIQDSLEIFRLMGLYGFNPSVYTCNAILGSVVKSGEDVSVW 178
Query: 368 KLFKKMIKCGCQPGYLVYNIFIGSVCSNEE-QPSSDILDLVEKAYSEMLDLGVVLNKVNV 426
K+M+K P +NI I +C+ + SS ++ +EK+ G V
Sbjct: 179 SFLKEMLKRKICPDVATFNILINVLCAEGSFEKSSYLMQKMEKS-------GYAPTIVTY 231
Query: 427 SNFARCLCGAGKFDQAFKIICEMMGKGFVPDDSTYSKVIGFLCDASKVEKAFSLFEEMKR 486
+ C G+F A +++ M KG D TY+ +I LC ++++ K + L +M++
Sbjct: 232 NTVLHWYCKKGRFKAAIELLDHMKSKGVDADVCTYNMLIHDLCRSNRIAKGYLLLRDMRK 291
Query: 487 NGIVPSVYTYTILIDSFCKAGLIQQARKWFDEMLHKGCTPNVVTYTALIHAYLKAKQMPV 546
I P+ TY LI+ F G + A + +EML G +PN VT+ ALI ++
Sbjct: 292 RMIHPNEVTYNTLINGFSNEGKVLIASQLLNEMLSFGLSPNHVTFNALIDGHISEGNFKE 351
Query: 547 ADELFEMMLLEGCKPNVVTYTALIDGHCKAGQIEKACQIYARMR------GDIESSDM-- 598
A ++F MM +G P+ V+Y L+DG CK + + A Y RM+ G I + M
Sbjct: 352 ALKMFYMMEAKGLTPSEVSYGVLLDGLCKNAEFDLARGFYMRMKRNGVCVGRITYTGMID 411
Query: 599 ---------DKYFKLDHNNCEG--PNVITYGALVDGLCKANRVKEAHELLDTMLAHGCEP 647
+ L+ + +G P+++TY AL++G CK R K A E++ + G P
Sbjct: 412 GLCKNGFLDEAVVLLNEMSKDGIDPDIVTYSALINGFCKVGRFKTAKEIVCRIYRVGLSP 471
Query: 648 NQIVYDAVIDGFCKIGKLQDAQEVFTKMSERGYSPNLYTYSSFIDCLFKDNRLDLVLKVL 707
N I+Y +I C++G L++A ++ M G++ + +T++ + L K ++ + +
Sbjct: 472 NGIIYSTLIYNCCRMGCLKEAIRIYEAMILEGHTRDHFTFNVLVTSLCKAGKVAEAEEFM 531
Query: 708 SKMLENSCTPNVVIYTEMVDGLCKIGKTDEAYKLMLKMEEKGCNPNVVTYTAMIDGFGKS 767
M + PN V + +++G G+ +A+ + +M + G +P TY +++ G K
Sbjct: 532 RCMTSDGILPNTVSFDCLINGYGNSGEGLKAFSVFDEMTKVGHHPTFFTYGSLLKGLCKG 591
Query: 768 GKIEQCLELFRDMCSKGCAPNFITYRVLINHCCSNGLLDEAYKLLDEMKQTYWPKHILSH 827
G + + + + + + A + + Y L+ C +G L +A L EM Q ++
Sbjct: 592 GHLREAEKFLKSLHAVPAAVDTVMYNTLLTAMCKSGNLAKAVSLFGEMVQRSILPDSYTY 651
Query: 828 RKIIEGFSQEFITSIGLL---DELSENESVPVDSLYRILIDNYIKAGRLEVAL---DLLE 881
+I G ++ T I +L + + +P +Y +D KAG+ + + + ++
Sbjct: 652 TSLISGLCRKGKTVIAILFAKEAEARGNVLPNKVMYTCFVDGMFKAGQWKAGIYFREQMD 711
Query: 882 EISSSPSHAVSNKYLYASLIENLSHASKVDKALELYASMISKNVVPELSILVHLIKGLIK 941
+ +P +N ++I+ S K++K +L M ++N P L+ L+ G K
Sbjct: 712 NLGHTPDIVTTN-----AMIDGYSRMGKIEKTNDLLPEMGNQNGGPNLTTYNILLHGYSK 766
Score = 233 bits (593), Expect = 5e-61
Identities = 189/813 (23%), Positives = 330/813 (40%), Gaps = 97/813 (11%)
Query: 178 NFLVRKCCRNGWWNMALEELGRLKDFGYKPSQTTYNALIQVFLRADKLDTAYLVKREMLS 237
N ++ C+ G + A+E L +K G TYN LI R++++ YL+ R+M
Sbjct: 232 NTVLHWYCKKGRFKAAIELLDHMKSKGVDADVCTYNMLIHDLCRSNRIAKGYLLLRDM-- 289
Query: 238 YAFVMDRYTLSCFAYSLCKGGKCREAFDLIDEAEDFVPDTVFYNRMVSGLCEASLFEEAM 297
P+ V YN +++G A
Sbjct: 290 -------------------------------RKRMIHPNEVTYNTLINGFSNEGKVLIAS 318
Query: 298 DILHRMRSSSCIPNVVTYRILLSGCLRKGQLGRCKRILSMMITEGCYPNREIFNSLIHAY 357
+L+ M S PN VT+ L+ G + +G ++ MM +G P+ + L+
Sbjct: 319 QLLNEMLSFGLSPNHVTFNALIDGHISEGNFKEALKMFYMMEAKGLTPSEVSYGVLLDGL 378
Query: 358 CKSRDYSYAYKLFKKMIKCGCQPGYLVYNIFIGSVCSNEEQPSSDILDLVEKAYSEMLDL 417
CK+ ++ A + +M + G G + Y I +C N LD +EM
Sbjct: 379 CKNAEFDLARGFYMRMKRNGVCVGRITYTGMIDGLCKN------GFLDEAVVLLNEMSKD 432
Query: 418 GVVLNKVNVSNFARCLCGAGKFDQAFKIICEMMGKGFVPDDSTYSKVIGFLCDASKVEKA 477
G+ + V S C G+F A +I+C + G P+ YS +I C +++A
Sbjct: 433 GIDPDIVTYSALINGFCKVGRFKTAKEIVCRIYRVGLSPNGIIYSTLIYNCCRMGCLKEA 492
Query: 478 FSLFEEMKRNGIVPSVYTYTILIDSFCKAGLIQQARKWFDEMLHKGCTPNVVTYTALIHA 537
++E M G +T+ +L+ S CKAG + +A ++ M G PN V++ LI+
Sbjct: 493 IRIYEAMILEGHTRDHFTFNVLVTSLCKAGKVAEAEEFMRCMTSDGILPNTVSFDCLING 552
Query: 538 YLKAKQMPVADELFEMMLLEGCKPNVVTYTALIDGHCKAGQIEKACQIYARMRGDIESSD 597
Y + + A +F+ M G P TY +L+ G CK G + +A
Sbjct: 553 YGNSGEGLKAFSVFDEMTKVGHHPTFFTYGSLLKGLCKGGHLREA--------------- 597
Query: 598 MDKYFKLDHNNCEGPNVITYGALVDGLCKANRVKEAHELLDTMLAHGCEPNQIVYDAVID 657
+K+ K H + + Y L+ +CK+ + +A L M+ P+ Y ++I
Sbjct: 598 -EKFLKSLHAVPAAVDTVMYNTLLTAMCKSGNLAKAVSLFGEMVQRSILPDSYTYTSLIS 656
Query: 658 GFCKIGKLQDAQEVFTKMSERG-YSPNLYTYSSFIDCLFKDNRLDLVLKVLSKMLENSCT 716
G C+ GK A + RG PN Y+ F+D +FK + + +M T
Sbjct: 657 GLCRKGKTVIAILFAKEAEARGNVLPNKVMYTCFVDGMFKAGQWKAGIYFREQMDNLGHT 716
Query: 717 PNVVIYTEMVDGLCKIGKTDEAYKLMLKMEEKGCNPNVVTYTAMIDGFGKSGKIEQCLEL 776
P++V M+DG ++GK ++ L+ +M + PN+ TY ++ G+ K + L
Sbjct: 717 PDIVTTNAMIDGYSRMGKIEKTNDLLPEMGNQNGGPNLTTYNILLHGYSKRKDVSTSFLL 776
Query: 777 FRDMCSKGCAPNFI-----------------------------------TYRVLINHCCS 801
+R + G P+ + T+ +LI+ CC+
Sbjct: 777 YRSIILNGILPDKLTCHSLVLGICESNMLEIGLKILKAFICRGVEVDRYTFNMLISKCCA 836
Query: 802 NGLLDEAYKLLDEMKQ--TYWPKHILSHRKIIEGFSQEFITSIGLLDELSENESVPVDSL 859
NG ++ A+ L+ M K + + F S +L E+S+ P
Sbjct: 837 NGEINWAFDLVKVMTSLGISLDKDTCDAMVSVLNRNHRFQESRMVLHEMSKQGISPESRK 896
Query: 860 YRILIDNYIKAGRLEVALDLLEEISSSPSHAVSNKYLYAS-LIENLSHASKVDKALELYA 918
Y LI+ + G ++ A + EE+ +H + + S ++ L+ K D+A L
Sbjct: 897 YIGLINGLCRVGDIKTAFVVKEEMI---AHKICPPNVAESAMVRALAKCGKADEATLLLR 953
Query: 919 SMISKNVVPELSILVHLIKGLIKVDKWQEALQL 951
M+ +VP ++ L+ K EAL+L
Sbjct: 954 FMLKMKLVPTIASFTTLMHLCCKNGNVIEALEL 986
Score = 219 bits (558), Expect = 6e-57
Identities = 177/712 (24%), Positives = 305/712 (41%), Gaps = 32/712 (4%)
Query: 178 NFLVRKCCRNGWWNMALEELGRLKDFGYKPSQTTYNALIQVFLRADKLDTAYLVKREMLS 237
N L+ G + AL+ ++ G PS+ +Y L+ + + D A M
Sbjct: 337 NALIDGHISEGNFKEALKMFYMMEAKGLTPSEVSYGVLLDGLCKNAEFDLARGFYMRMKR 396
Query: 238 YAFVMDRYTLSCFAYSLCKGGKCREAFDLIDEA--EDFVPDTVFYNRMVSGLCEASLFEE 295
+ R T + LCK G EA L++E + PD V Y+ +++G C+ F+
Sbjct: 397 NGVCVGRITYTGMIDGLCKNGFLDEAVVLLNEMSKDGIDPDIVTYSALINGFCKVGRFKT 456
Query: 296 AMDILHRMRSSSCIPNVVTYRILLSGCLRKGQLGRCKRILSMMITEGCYPNREIFNSLIH 355
A +I+ R+ PN + Y L+ C R G L RI MI EG + FN L+
Sbjct: 457 AKEIVCRIYRVGLSPNGIIYSTLIYNCCRMGCLKEAIRIYEAMILEGHTRDHFTFNVLVT 516
Query: 356 AYCKSRDYSYAYKLFKKMIKCGCQPGYLVYNIFIGSVCSNEEQPSSDILDLVEKAYSEML 415
+ CK+ + A + + M G P + ++ I ++ E + + EM
Sbjct: 517 SLCKAGKVAEAEEFMRCMTSDGILPNTVSFDCLINGYGNSGEGLKAF------SVFDEMT 570
Query: 416 DLGVVLNKVNVSNFARCLCGAGKFDQAFKIICEMMGKGFVPDDSTYSKVIGFLCDASKVE 475
+G + + LC G +A K + + D Y+ ++ +C + +
Sbjct: 571 KVGHHPTFFTYGSLLKGLCKGGHLREAEKFLKSLHAVPAAVDTVMYNTLLTAMCKSGNLA 630
Query: 476 KAFSLFEEMKRNGIVPSVYTYTILIDSFCKAGLIQQARKWFDEMLHKG-CTPNVVTYTAL 534
KA SLF EM + I+P YTYT LI C+ G A + E +G PN V YT
Sbjct: 631 KAVSLFGEMVQRSILPDSYTYTSLISGLCRKGKTVIAILFAKEAEARGNVLPNKVMYTCF 690
Query: 535 IHAYLKAKQMPVADELFEMMLLEGCKPNVVTYTALIDGHCKAGQIEKACQIYARMRGDIE 594
+ KA Q E M G P++VT A+IDG+ + G+IEK + M
Sbjct: 691 VDGMFKAGQWKAGIYFREQMDNLGHTPDIVTTNAMIDGYSRMGKIEKTNDLLPEMGNQNG 750
Query: 595 SSDMDKYFKLDHNNCEG-------------------PNVITYGALVDGLCKANRVKEAHE 635
++ Y L H + P+ +T +LV G+C++N ++ +
Sbjct: 751 GPNLTTYNILLHGYSKRKDVSTSFLLYRSIILNGILPDKLTCHSLVLGICESNMLEIGLK 810
Query: 636 LLDTMLAHGCEPNQIVYDAVIDGFCKIGKLQDAQEVFTKMSERGYSPNLYTYSSFIDCLF 695
+L + G E ++ ++ +I C G++ A ++ M+ G S + T + + L
Sbjct: 811 ILKAFICRGVEVDRYTFNMLISKCCANGEINWAFDLVKVMTSLGISLDKDTCDAMVSVLN 870
Query: 696 KDNRLDLVLKVLSKMLENSCTPNVVIYTEMVDGLCKIGKTDEAYKLMLKM-EEKGCNPNV 754
+++R VL +M + +P Y +++GLC++G A+ + +M K C PNV
Sbjct: 871 RNHRFQESRMVLHEMSKQGISPESRKYIGLINGLCRVGDIKTAFVVKEEMIAHKICPPNV 930
Query: 755 VTYTAMIDGFGKSGKIEQCLELFRDMCSKGCAPNFITYRVLINHCCSNGLLDEAYKLLDE 814
+AM+ K GK ++ L R M P ++ L++ CC NG + EA +L
Sbjct: 931 AE-SAMVRALAKCGKADEATLLLRFMLKMKLVPTIASFTTLMHLCCKNGNVIEALELRVV 989
Query: 815 MKQTYWPKHILSHRKIIEGF--SQEFITSIGLLDELSENESVPVDSLYRILI 864
M ++S+ +I G + + L +E+ + + + Y+ LI
Sbjct: 990 MSNCGLKLDLVSYNVLITGLCAKGDMALAFELYEEMKGDGFLANATTYKALI 1041
Score = 208 bits (529), Expect = 1e-53
Identities = 179/792 (22%), Positives = 336/792 (41%), Gaps = 42/792 (5%)
Query: 178 NFLVRKCCRNGWWNMALEELGRLKDFGYKPSQTTYNALIQVFLRADKLDTAYLVKREMLS 237
N L+ CR+ L ++ P++ TYN LI F K+ A + EMLS
Sbjct: 267 NMLIHDLCRSNRIAKGYLLLRDMRKRMIHPNEVTYNTLINGFSNEGKVLIASQLLNEMLS 326
Query: 238 YAFVMDRYTLSCFAYSLCKGGKCREAFDL--IDEAEDFVPDTVFYNRMVSGLCEASLFEE 295
+ + T + G +EA + + EA+ P V Y ++ GLC+ + F+
Sbjct: 327 FGLSPNHVTFNALIDGHISEGNFKEALKMFYMMEAKGLTPSEVSYGVLLDGLCKNAEFDL 386
Query: 296 AMDILHRMRSSSCIPNVVTYRILLSGCLRKGQLGRCKRILSMMITEGCYPNREIFNSLIH 355
A RM+ + +TY ++ G + G L +L+ M +G P+ +++LI+
Sbjct: 387 ARGFYMRMKRNGVCVGRITYTGMIDGLCKNGFLDEAVVLLNEMSKDGIDPDIVTYSALIN 446
Query: 356 AYCKSRDYSYAYKLFKKMIKCGCQPGYLVYNIFIGSVCSNEEQPSSDILDLVEKAYSEML 415
+CK + A ++ ++ + G P ++Y+ I + C L + Y M+
Sbjct: 447 GFCKVGRFKTAKEIVCRIYRVGLSPNGIIYSTLIYNCCR------MGCLKEAIRIYEAMI 500
Query: 416 DLGVVLNKVNVSNFARCLCGAGKFDQAFKIICEMMGKGFVPDDSTYSKVIGFLCDASKVE 475
G + + LC AGK +A + + M G +P+ ++ +I ++ +
Sbjct: 501 LEGHTRDHFTFNVLVTSLCKAGKVAEAEEFMRCMTSDGILPNTVSFDCLINGYGNSGEGL 560
Query: 476 KAFSLFEEMKRNGIVPSVYTYTILIDSFCKAGLIQQARKWFDEMLHKGCTPNVVTYTALI 535
KAFS+F+EM + G P+ +TY L+ CK G +++A K+ + + V Y L+
Sbjct: 561 KAFSVFDEMTKVGHHPTFFTYGSLLKGLCKGGHLREAEKFLKSLHAVPAAVDTVMYNTLL 620
Query: 536 HAYLKAKQMPVADELFEMMLLEGCKPNVVTYTALIDGHCKAGQIEKACQIY--ARMRGDI 593
A K+ + A LF M+ P+ TYT+LI G C+ G+ A A RG++
Sbjct: 621 TAMCKSGNLAKAVSLFGEMVQRSILPDSYTYTSLISGLCRKGKTVIAILFAKEAEARGNV 680
Query: 594 ESSDM-----------------DKYFKLDHNNC-EGPNVITYGALVDGLCKANRVKEAHE 635
+ + YF+ +N P+++T A++DG + ++++ ++
Sbjct: 681 LPNKVMYTCFVDGMFKAGQWKAGIYFREQMDNLGHTPDIVTTNAMIDGYSRMGKIEKTND 740
Query: 636 LLDTMLAHGCEPNQIVYDAVIDGFCKIGKLQDAQEVFTKMSERGYSPNLYTYSSFIDCLF 695
LL M PN Y+ ++ G+ K + + ++ + G P+ T S + +
Sbjct: 741 LLPEMGNQNGGPNLTTYNILLHGYSKRKDVSTSFLLYRSIILNGILPDKLTCHSLVLGIC 800
Query: 696 KDNRLDLVLKVLSKMLENSCTPNVVIYTEMVDGLCKIGKTDEAYKLMLKMEEKGCNPNVV 755
+ N L++ LK+L + + + ++ C G+ + A+ L+ M G + +
Sbjct: 801 ESNMLEIGLKILKAFICRGVEVDRYTFNMLISKCCANGEINWAFDLVKVMTSLGISLDKD 860
Query: 756 TYTAMIDGFGKSGKIEQCLELFRDMCSKGCAPNFITYRVLINHCCSNGLLDEAYKLLDEM 815
T AM+ ++ + ++ + +M +G +P Y LIN C G + A+ + +EM
Sbjct: 861 TCDAMVSVLNRNHRFQESRMVLHEMSKQGISPESRKYIGLINGLCRVGDIKTAFVVKEEM 920
Query: 816 KQTYWPKHILSHRKIIEGFSQEFITSIGLLDELS-------ENESVPVDSLYRILIDNYI 868
H + + E + G DE + + + VP + + L+
Sbjct: 921 IA-----HKICPPNVAESAMVRALAKCGKADEATLLLRFMLKMKLVPTIASFTTLMHLCC 975
Query: 869 KAGRLEVALDLLEEISSSPSHAVSNKYLYASLIENLSHASKVDKALELYASMISKNVVPE 928
K G + AL+L + S + Y LI L + A ELY M +
Sbjct: 976 KNGNVIEALEL--RVVMSNCGLKLDLVSYNVLITGLCAKGDMALAFELYEEMKGDGFLAN 1033
Query: 929 LSILVHLIKGLI 940
+ LI+GL+
Sbjct: 1034 ATTYKALIRGLL 1045
Score = 127 bits (319), Expect = 3e-29
Identities = 87/312 (27%), Positives = 150/312 (47%), Gaps = 10/312 (3%)
Query: 645 CEPNQIVYDAVIDGFCKIGKLQDAQEVFTKMSERGYSPNLYTYSSFIDCLFKDNRLDLVL 704
C N VYD +I + + G +QD+ E+F M G++P++YT ++ + + K V
Sbjct: 119 CNSNPSVYDILIRVYLREGMIQDSLEIFRLMGLYGFNPSVYTCNAILGSVVKSGEDVSVW 178
Query: 705 KVLSKMLENSCTPNVVIYTEMVDGLCKIGKTDEAYKLMLKMEEKGCNPNVVTYTAMIDGF 764
L +ML+ P+V + +++ LC G +++ LM KME+ G P +VTY ++ +
Sbjct: 179 SFLKEMLKRKICPDVATFNILINVLCAEGSFEKSSYLMQKMEKSGYAPTIVTYNTVLHWY 238
Query: 765 GKSGKIEQCLELFRDMCSKGCAPNFITYRVLINHCCSNGLLDEAYKLLDEMKQTYWPKHI 824
K G+ + +EL M SKG + TY +LI+ C + + + Y LL +M++ +
Sbjct: 239 CKKGRFKAAIELLDHMKSKGVDADVCTYNMLIHDLCRSNRIAKGYLLLRDMRKRMIHPNE 298
Query: 825 LSHRKIIEGFSQE--FITSIGLLDELSENESVPVDSLYRILIDNYIKAGRLEVALD---L 879
+++ +I GFS E + + LL+E+ P + LID +I G + AL +
Sbjct: 299 VTYNTLINGFSNEGKVLIASQLLNEMLSFGLSPNHVTFNALIDGHISEGNFKEALKMFYM 358
Query: 880 LEEISSSPSHAVSNKYLYASLIENLSHASKVDKALELYASMISKNVVPELSILVHLIKGL 939
+E +PS + Y L++ L ++ D A Y M V +I GL
Sbjct: 359 MEAKGLTPS-----EVSYGVLLDGLCKNAEFDLARGFYMRMKRNGVCVGRITYTGMIDGL 413
Query: 940 IKVDKWQEALQL 951
K EA+ L
Sbjct: 414 CKNGFLDEAVVL 425
>At1g12300 hypothetical protein
Length = 637
Score = 273 bits (699), Expect = 3e-73
Identities = 162/561 (28%), Positives = 279/561 (48%), Gaps = 27/561 (4%)
Query: 262 EAFDLIDEAEDFV-----PDTVFYNRMVSGLCEASLFEEAMDILHRMRSSSCIPNVVTYR 316
+A D ID D + P + ++R+ S + + ++ + + +M N+ T
Sbjct: 68 KADDAIDLFRDMIHSRPLPTVIDFSRLFSAIAKTKQYDLVLALCKQMELKGIAHNLYTLS 127
Query: 317 ILLSGCLRKGQLGRCKRILSMMITEGCYPNREIFNSLIHAYCKSRDYSYAYKLFKKMIKC 376
I+++ R +L + +I G PN F++LI+ C S A +L +M++
Sbjct: 128 IMINCFCRCRKLCLAFSAMGKIIKLGYEPNTITFSTLINGLCLEGRVSEALELVDRMVEM 187
Query: 377 GCQPGYLVYNIFIGSVCSNEEQPSSDILDLVEKAYSEMLDLGVVLNKVNVSNFARCLCGA 436
G +P + N + +C + ++ + +L +M++ G N V +C +
Sbjct: 188 GHKPDLITINTLVNGLCLSGKEAEAMLL------IDKMVEYGCQPNAVTYGPVLNVMCKS 241
Query: 437 GKFDQAFKIICEMMGKGFVPDDSTYSKVIGFLCDASKVEKAFSLFEEMKRNGIVPSVYTY 496
G+ A +++ +M + D YS +I LC ++ AF+LF EM+ GI ++ TY
Sbjct: 242 GQTALAMELLRKMEERNIKLDAVKYSIIIDGLCKHGSLDNAFNLFNEMEMKGITTNIITY 301
Query: 497 TILIDSFCKAGLIQQARKWFDEMLHKGCTPNVVTYTALIHAYLKAKQMPVADELFEMMLL 556
ILI FC AG K +M+ + PNVVT++ LI +++K ++ A+EL + M+
Sbjct: 302 NILIGGFCNAGRWDDGAKLLRDMIKRKINPNVVTFSVLIDSFVKEGKLREAEELHKEMIH 361
Query: 557 EGCKPNVVTYTALIDGHCKAGQIEKACQIYARMRGDIESSDMDKYFKLDHNNCEGPNVIT 616
G P+ +TYT+LIDG CK ++KA Q+ M C+ PN+ T
Sbjct: 362 RGIAPDTITYTSLIDGFCKENHLDKANQMVDLMVS---------------KGCD-PNIRT 405
Query: 617 YGALVDGLCKANRVKEAHELLDTMLAHGCEPNQIVYDAVIDGFCKIGKLQDAQEVFTKMS 676
+ L++G CKANR+ + EL M G + + Y+ +I GFC++GKL A+E+F +M
Sbjct: 406 FNILINGYCKANRIDDGLELFRKMSLRGVVADTVTYNTLIQGFCELGKLNVAKELFQEMV 465
Query: 677 ERGYSPNLYTYSSFIDCLFKDNRLDLVLKVLSKMLENSCTPNVVIYTEMVDGLCKIGKTD 736
R PN+ TY +D L + + L++ K+ ++ ++ IY ++ G+C K D
Sbjct: 466 SRKVPPNIVTYKILLDGLCDNGESEKALEIFEKIEKSKMELDIGIYNIIIHGMCNASKVD 525
Query: 737 EAYKLMLKMEEKGCNPNVVTYTAMIDGFGKSGKIEQCLELFRDMCSKGCAPNFITYRVLI 796
+A+ L + KG P V TY MI G K G + + LFR M G AP+ TY +LI
Sbjct: 526 DAWDLFCSLPLKGVKPGVKTYNIMIGGLCKKGPLSEAELLFRKMEEDGHAPDGWTYNILI 585
Query: 797 NHCCSNGLLDEAYKLLDEMKQ 817
+G ++ KL++E+K+
Sbjct: 586 RAHLGDGDATKSVKLIEELKR 606
Score = 242 bits (617), Expect = 9e-64
Identities = 166/605 (27%), Positives = 280/605 (45%), Gaps = 31/605 (5%)
Query: 304 RSSSCIPNVVTYRILLSGCLRKGQLGRCKRILSMMITEG--CYPNREI-FNSLIHAYCKS 360
R SS + V R+L +G LR + C LS G + +R + + + +
Sbjct: 8 RLSSQVSKFVQPRLLETGTLRIALIN-CPNELSFCCERGFSAFSDRNLSYRERLRSGLVD 66
Query: 361 RDYSYAYKLFKKMIKCGCQPGYLVYNIFIGSVCSNEEQPSSDILDLVEKAYSEMLDLGVV 420
A LF+ MI P + ++ ++ ++ DLV +M G+
Sbjct: 67 IKADDAIDLFRDMIHSRPLPTVIDFSRLFSAIAKTKQY------DLVLALCKQMELKGIA 120
Query: 421 LNKVNVSNFARCLCGAGKFDQAFKIICEMMGKGFVPDDSTYSKVIGFLCDASKVEKAFSL 480
N +S C C K AF + +++ G+ P+ T+S +I LC +V +A L
Sbjct: 121 HNLYTLSIMINCFCRCRKLCLAFSAMGKIIKLGYEPNTITFSTLINGLCLEGRVSEALEL 180
Query: 481 FEEMKRNGIVPSVYTYTILIDSFCKAGLIQQARKWFDEMLHKGCTPNVVTYTALIHAYLK 540
+ M G P + T L++ C +G +A D+M+ GC PN VTY +++ K
Sbjct: 181 VDRMVEMGHKPDLITINTLVNGLCLSGKEAEAMLLIDKMVEYGCQPNAVTYGPVLNVMCK 240
Query: 541 AKQMPVADELFEMMLLEGCKPNVVTYTALIDGHCKAGQIEKACQIYARMRGDIESSDMDK 600
+ Q +A EL M K + V Y+ +IDG CK G ++ A ++ M ++
Sbjct: 241 SGQTALAMELLRKMEERNIKLDAVKYSIIIDGLCKHGSLDNAFNLFNEMEMKGITT---- 296
Query: 601 YFKLDHNNCEGPNVITYGALVDGLCKANRVKEAHELLDTMLAHGCEPNQIVYDAVIDGFC 660
N+ITY L+ G C A R + +LL M+ PN + + +ID F
Sbjct: 297 ------------NIITYNILIGGFCNAGRWDDGAKLLRDMIKRKINPNVVTFSVLIDSFV 344
Query: 661 KIGKLQDAQEVFTKMSERGYSPNLYTYSSFIDCLFKDNRLDLVLKVLSKMLENSCTPNVV 720
K GKL++A+E+ +M RG +P+ TY+S ID K+N LD +++ M+ C PN+
Sbjct: 345 KEGKLREAEELHKEMIHRGIAPDTITYTSLIDGFCKENHLDKANQMVDLMVSKGCDPNIR 404
Query: 721 IYTEMVDGLCKIGKTDEAYKLMLKMEEKGCNPNVVTYTAMIDGFGKSGKIEQCLELFRDM 780
+ +++G CK + D+ +L KM +G + VTY +I GF + GK+ ELF++M
Sbjct: 405 TFNILINGYCKANRIDDGLELFRKMSLRGVVADTVTYNTLIQGFCELGKLNVAKELFQEM 464
Query: 781 CSKGCAPNFITYRVLINHCCSNGLLDEAYKLLDEMKQTYWPKHILSHRKIIEGF--SQEF 838
S+ PN +TY++L++ C NG ++A ++ ++++++ I + II G + +
Sbjct: 465 VSRKVPPNIVTYKILLDGLCDNGESEKALEIFEKIEKSKMELDIGIYNIIIHGMCNASKV 524
Query: 839 ITSIGLLDELSENESVPVDSLYRILIDNYIKAGRL---EVALDLLEEISSSPSHAVSNKY 895
+ L L P Y I+I K G L E+ +EE +P N
Sbjct: 525 DDAWDLFCSLPLKGVKPGVKTYNIMIGGLCKKGPLSEAELLFRKMEEDGHAPDGWTYNIL 584
Query: 896 LYASL 900
+ A L
Sbjct: 585 IRAHL 589
Score = 220 bits (560), Expect = 4e-57
Identities = 154/573 (26%), Positives = 259/573 (44%), Gaps = 61/573 (10%)
Query: 140 FDKLLDLLGCNVNADDRVPLKFLMEIKDDDHELLRRLLNFLVRKCCRNGWWNMALEELGR 199
F +L + D + L ME+K H L L+ ++ CR +A +G+
Sbjct: 91 FSRLFSAIAKTKQYDLVLALCKQMELKGIAHNLYT--LSIMINCFCRCRKLCLAFSAMGK 148
Query: 200 LKDFGYKPSQTTYNALIQVFLRADKLDTAYLVKREMLSYAFVMDRYTLSCFAYSLCKGGK 259
+ GY+P+ T++ LI LC G+
Sbjct: 149 IIKLGYEPNTITFSTLIN-----------------------------------GLCLEGR 173
Query: 260 CREAFDLIDEAEDF--VPDTVFYNRMVSGLCEASLFEEAMDILHRMRSSSCIPNVVTYRI 317
EA +L+D + PD + N +V+GLC + EAM ++ +M C PN VTY
Sbjct: 174 VSEALELVDRMVEMGHKPDLITINTLVNGLCLSGKEAEAMLLIDKMVEYGCQPNAVTYGP 233
Query: 318 LLSGCLRKGQLGRCKRILSMMITEGCYPNREIFNSLIHAYCKSRDYSYAYKLFKKMIKCG 377
+L+ + GQ +L M + ++ +I CK A+ LF +M G
Sbjct: 234 VLNVMCKSGQTALAMELLRKMEERNIKLDAVKYSIIIDGLCKHGSLDNAFNLFNEMEMKG 293
Query: 378 CQPGYLVYNIFIGSVCSNEEQPSSDILDLVEKAYSEMLDLGVVLNKVNVSNFARCLCGAG 437
+ YNI IG C+ D L+ ++ VV V + +F + G
Sbjct: 294 ITTNIITYNILIGGFCNAGRW--DDGAKLLRDMIKRKINPNVVTFSVLIDSFVK----EG 347
Query: 438 KFDQAFKIICEMMGKGFVPDDSTYSKVIGFLCDASKVEKAFSLFEEMKRNGIVPSVYTYT 497
K +A ++ EM+ +G PD TY+ +I C + ++KA + + M G P++ T+
Sbjct: 348 KLREAEELHKEMIHRGIAPDTITYTSLIDGFCKENHLDKANQMVDLMVSKGCDPNIRTFN 407
Query: 498 ILIDSFCKAGLIQQARKWFDEMLHKGCTPNVVTYTALIHAYLKAKQMPVADELFEMMLLE 557
ILI+ +CKA I + F +M +G + VTY LI + + ++ VA ELF+ M+
Sbjct: 408 ILINGYCKANRIDDGLELFRKMSLRGVVADTVTYNTLIQGFCELGKLNVAKELFQEMVSR 467
Query: 558 GCKPNVVTYTALIDGHCKAGQIEKACQIYARMRGDIESSDMDKYFKLDHNNCEGPNVITY 617
PN+VTY L+DG C G+ EKA +I+ + IE S M+ ++ Y
Sbjct: 468 KVPPNIVTYKILLDGLCDNGESEKALEIFEK----IEKSKMEL------------DIGIY 511
Query: 618 GALVDGLCKANRVKEAHELLDTMLAHGCEPNQIVYDAVIDGFCKIGKLQDAQEVFTKMSE 677
++ G+C A++V +A +L ++ G +P Y+ +I G CK G L +A+ +F KM E
Sbjct: 512 NIIIHGMCNASKVDDAWDLFCSLPLKGVKPGVKTYNIMIGGLCKKGPLSEAELLFRKMEE 571
Query: 678 RGYSPNLYTYSSFIDCLFKDNRLDLVLKVLSKM 710
G++P+ +TY+ I D +K++ ++
Sbjct: 572 DGHAPDGWTYNILIRAHLGDGDATKSVKLIEEL 604
Score = 218 bits (554), Expect = 2e-56
Identities = 137/487 (28%), Positives = 242/487 (49%), Gaps = 24/487 (4%)
Query: 473 KVEKAFSLFEEMKRNGIVPSVYTYTILIDSFCKAGLIQQARKWFDEMLHKGCTPNVVTYT 532
K + A LF +M + +P+V ++ L + K +M KG N+ T +
Sbjct: 68 KADDAIDLFRDMIHSRPLPTVIDFSRLFSAIAKTKQYDLVLALCKQMELKGIAHNLYTLS 127
Query: 533 ALIHAYLKAKQMPVADELFEMMLLEGCKPNVVTYTALIDGHCKAGQIEKACQIYARMRGD 592
+I+ + + +++ +A ++ G +PN +T++ LI+G C G++ +A ++
Sbjct: 128 IMINCFCRCRKLCLAFSAMGKIIKLGYEPNTITFSTLINGLCLEGRVSEALEL------- 180
Query: 593 IESSDMDKYFKLDHNNCEGPNVITYGALVDGLCKANRVKEAHELLDTMLAHGCEPNQIVY 652
+D+ ++ H P++IT LV+GLC + + EA L+D M+ +GC+PN + Y
Sbjct: 181 -----VDRMVEMGHK----PDLITINTLVNGLCLSGKEAEAMLLIDKMVEYGCQPNAVTY 231
Query: 653 DAVIDGFCKIGKLQDAQEVFTKMSERGYSPNLYTYSSFIDCLFKDNRLDLVLKVLSKMLE 712
V++ CK G+ A E+ KM ER + YS ID L K LD + ++M
Sbjct: 232 GPVLNVMCKSGQTALAMELLRKMEERNIKLDAVKYSIIIDGLCKHGSLDNAFNLFNEMEM 291
Query: 713 NSCTPNVVIYTEMVDGLCKIGKTDEAYKLMLKMEEKGCNPNVVTYTAMIDGFGKSGKIEQ 772
T N++ Y ++ G C G+ D+ KL+ M ++ NPNVVT++ +ID F K GK+ +
Sbjct: 292 KGITTNIITYNILIGGFCNAGRWDDGAKLLRDMIKRKINPNVVTFSVLIDSFVKEGKLRE 351
Query: 773 CLELFRDMCSKGCAPNFITYRVLINHCCSNGLLDEAYKLLDEMKQTYWPKHILSHRKIIE 832
EL ++M +G AP+ ITY LI+ C LD+A +++D M +I + +I
Sbjct: 352 AEELHKEMIHRGIAPDTITYTSLIDGFCKENHLDKANQMVDLMVSKGCDPNIRTFNILIN 411
Query: 833 GF--SQEFITSIGLLDELSENESVPVDSLYRILIDNYIKAGRLEVALDLLEEISSS--PS 888
G+ + + L ++S V Y LI + + G+L VA +L +E+ S P
Sbjct: 412 GYCKANRIDDGLELFRKMSLRGVVADTVTYNTLIQGFCELGKLNVAKELFQEMVSRKVPP 471
Query: 889 HAVSNKYLYASLIENLSHASKVDKALELYASMISKNVVPELSILVHLIKGLIKVDKWQEA 948
+ V+ K L L +N + +KALE++ + + ++ I +I G+ K +A
Sbjct: 472 NIVTYKILLDGLCDN----GESEKALEIFEKIEKSKMELDIGIYNIIIHGMCNASKVDDA 527
Query: 949 LQLSDSI 955
L S+
Sbjct: 528 WDLFCSL 534
Score = 174 bits (440), Expect = 3e-43
Identities = 120/463 (25%), Positives = 207/463 (43%), Gaps = 10/463 (2%)
Query: 130 QIGYSHTPQVFDKLLDLLGCNVNADDRVPLKFLMEIKDDDHELLRRLLNFLVRKCCRNGW 189
++GY F L++ L + + L M +L+ +N LV C +G
Sbjct: 151 KLGYEPNTITFSTLINGLCLEGRVSEALELVDRMVEMGHKPDLIT--INTLVNGLCLSGK 208
Query: 190 WNMALEELGRLKDFGYKPSQTTYNALIQVFLRADKLDTAYLVKREMLSYAFVMDRYTLSC 249
A+ + ++ ++G +P+ TY ++ V ++ + A + R+M +D S
Sbjct: 209 EAEAMLLIDKMVEYGCQPNAVTYGPVLNVMCKSGQTALAMELLRKMEERNIKLDAVKYSI 268
Query: 250 FAYSLCKGGKCREAFDLIDEAE--DFVPDTVFYNRMVSGLCEASLFEEAMDILHRMRSSS 307
LCK G AF+L +E E + + YN ++ G C A +++ +L M
Sbjct: 269 IIDGLCKHGSLDNAFNLFNEMEMKGITTNIITYNILIGGFCNAGRWDDGAKLLRDMIKRK 328
Query: 308 CIPNVVTYRILLSGCLRKGQLGRCKRILSMMITEGCYPNREIFNSLIHAYCKSRDYSYAY 367
PNVVT+ +L+ +++G+L + + MI G P+ + SLI +CK A
Sbjct: 329 INPNVVTFSVLIDSFVKEGKLREAEELHKEMIHRGIAPDTITYTSLIDGFCKENHLDKAN 388
Query: 368 KLFKKMIKCGCQPGYLVYNIFIGSVCSNEEQPSSDILDLVEKAYSEMLDLGVVLNKVNVS 427
++ M+ GC P +NI I C + D L+L K M GVV + V +
Sbjct: 389 QMVDLMVSKGCDPNIRTFNILINGYC--KANRIDDGLELFRK----MSLRGVVADTVTYN 442
Query: 428 NFARCLCGAGKFDQAFKIICEMMGKGFVPDDSTYSKVIGFLCDASKVEKAFSLFEEMKRN 487
+ C GK + A ++ EM+ + P+ TY ++ LCD + EKA +FE+++++
Sbjct: 443 TLIQGFCELGKLNVAKELFQEMVSRKVPPNIVTYKILLDGLCDNGESEKALEIFEKIEKS 502
Query: 488 GIVPSVYTYTILIDSFCKAGLIQQARKWFDEMLHKGCTPNVVTYTALIHAYLKAKQMPVA 547
+ + Y I+I C A + A F + KG P V TY +I K + A
Sbjct: 503 KMELDIGIYNIIIHGMCNASKVDDAWDLFCSLPLKGVKPGVKTYNIMIGGLCKKGPLSEA 562
Query: 548 DELFEMMLLEGCKPNVVTYTALIDGHCKAGQIEKACQIYARMR 590
+ LF M +G P+ TY LI H G K+ ++ ++
Sbjct: 563 ELLFRKMEEDGHAPDGWTYNILIRAHLGDGDATKSVKLIEELK 605
Score = 141 bits (355), Expect = 2e-33
Identities = 103/381 (27%), Positives = 171/381 (44%), Gaps = 35/381 (9%)
Query: 592 DIESSDMDKYFKLDHNNCEGPNVITYGALVDGLCKANRVKEAHELLDTMLAHGCEPNQIV 651
DI++ D F+ ++ P VI + L + K + L M G N
Sbjct: 66 DIKADDAIDLFRDMIHSRPLPTVIDFSRLFSAIAKTKQYDLVLALCKQMELKGIAHNLYT 125
Query: 652 YDAVIDGFCKIGKLQDAQEVFTKMSERGYSPNLYTYSSFIDCLFKDNRLDLVLKVLSKML 711
+I+ FC+ KL A K+ + GY PN T+S+ I+ L + R+ L+++ +M+
Sbjct: 126 LSIMINCFCRCRKLCLAFSAMGKIIKLGYEPNTITFSTLINGLCLEGRVSEALELVDRMV 185
Query: 712 ENSCTPNVVIYTEMVDGLCKIGKTDEAYKLMLKMEEKGCNPNVVTYTAMIDGFGKSGKIE 771
E P+++ +V+GLC GK EA L+ KM E GC PN VTY +++ KSG+
Sbjct: 186 EMGHKPDLITINTLVNGLCLSGKEAEAMLLIDKMVEYGCQPNAVTYGPVLNVMCKSGQTA 245
Query: 772 QCLELFRDMCSKGCAPNFITYRVLINHCCSNGLLDEAYKLLDEMKQTYWPKHILSHRKII 831
+EL R M + + + Y ++I+ C +G LD A+ L +EM+ +
Sbjct: 246 LAMELLRKMEERNIKLDAVKYSIIIDGLCKHGSLDNAFNLFNEME--------------M 291
Query: 832 EGFSQEFITSIGLLDELSENESVPVDSLYRILIDNYIKAGRLEVALDLLEEISSSPSHAV 891
+G + IT Y ILI + AGR + LL ++ +
Sbjct: 292 KGITTNIIT-------------------YNILIGGFCNAGRWDDGAKLLRDMIKRKINP- 331
Query: 892 SNKYLYASLIENLSHASKVDKALELYASMISKNVVPELSILVHLIKGLIKVDKWQEALQL 951
N ++ LI++ K+ +A EL+ MI + + P+ LI G K + +A Q+
Sbjct: 332 -NVVTFSVLIDSFVKEGKLREAEELHKEMIHRGIAPDTITYTSLIDGFCKENHLDKANQM 390
Query: 952 SDSICQMVCLTLSQTFQPLVN 972
D + C +TF L+N
Sbjct: 391 VDLMVSKGCDPNIRTFNILIN 411
Score = 87.4 bits (215), Expect = 4e-17
Identities = 56/205 (27%), Positives = 94/205 (45%), Gaps = 2/205 (0%)
Query: 175 RLLNFLVRKCCRNGWWNMALEELGRLKDFGYKPSQTTYNALIQVFLRADKLDTAYLVKRE 234
R N L+ C+ + LE ++ G TYN LIQ F KL+ A + +E
Sbjct: 404 RTFNILINGYCKANRIDDGLELFRKMSLRGVVADTVTYNTLIQGFCELGKLNVAKELFQE 463
Query: 235 MLSYAFVMDRYTLSCFAYSLCKGGKCREAFDLIDEAED--FVPDTVFYNRMVSGLCEASL 292
M+S + T LC G+ +A ++ ++ E D YN ++ G+C AS
Sbjct: 464 MVSRKVPPNIVTYKILLDGLCDNGESEKALEIFEKIEKSKMELDIGIYNIIIHGMCNASK 523
Query: 293 FEEAMDILHRMRSSSCIPNVVTYRILLSGCLRKGQLGRCKRILSMMITEGCYPNREIFNS 352
++A D+ + P V TY I++ G +KG L + + M +G P+ +N
Sbjct: 524 VDDAWDLFCSLPLKGVKPGVKTYNIMIGGLCKKGPLSEAELLFRKMEEDGHAPDGWTYNI 583
Query: 353 LIHAYCKSRDYSYAYKLFKKMIKCG 377
LI A+ D + + KL +++ +CG
Sbjct: 584 LIRAHLGDGDATKSVKLIEELKRCG 608
Score = 41.2 bits (95), Expect = 0.003
Identities = 24/81 (29%), Positives = 41/81 (49%)
Query: 893 NKYLYASLIENLSHASKVDKALELYASMISKNVVPELSILVHLIKGLIKVDKWQEALQLS 952
N +++LI L +V +ALEL M+ P+L + L+ GL K EA+ L
Sbjct: 157 NTITFSTLINGLCLEGRVSEALELVDRMVEMGHKPDLITINTLVNGLCLSGKEAEAMLLI 216
Query: 953 DSICQMVCLTLSQTFQPLVNL 973
D + + C + T+ P++N+
Sbjct: 217 DKMVEYGCQPNAVTYGPVLNV 237
>At3g53700 putative protein
Length = 754
Score = 273 bits (698), Expect = 3e-73
Identities = 175/635 (27%), Positives = 303/635 (47%), Gaps = 28/635 (4%)
Query: 205 YKPSQTTYNALIQVFLRADKLDTAYLVKREMLSYAFVMDRYTLSCFAYSLCKGGKCREAF 264
+ P Y ++ R+ D + +M S M T S + E
Sbjct: 79 FSPEPALYEEILLRLGRSGSFDDMKKILEDMKSSRCEMGTSTFLILIESYAQFELQDEIL 138
Query: 265 DLIDEAED---FVPDTVFYNRMVSGLCEASLFEEAMDILHRMRSSSCI-PNVVTYRILLS 320
++D D PDT FYNRM++ L + + + ++I H S I P+V T+ +L+
Sbjct: 139 SVVDWMIDEFGLKPDTHFYNRMLNLLVDGNSLK-LVEISHAKMSVWGIKPDVSTFNVLIK 197
Query: 321 GCLRKGQLGRCKRILSMMITEGCYPNREIFNSLIHAYCKSRDYSYAYKLFKKMIKCGCQP 380
R QL +L M + G P+ + F +++ Y + D A ++ ++M++ GC
Sbjct: 198 ALCRAHQLRPAILMLEDMPSYGLVPDEKTFTTVMQGYIEEGDLDGALRIREQMVEFGCSW 257
Query: 381 GYLVYNIFIGSVCSNEEQPSSDILDLVEKAYSEMLDLGVVLNKVNVSNFARCLCGAGKFD 440
+ N+ + C +E D L+ +++ ++ G ++ + LC AG
Sbjct: 258 SNVSVNVIVHGFC--KEGRVEDALNFIQEMSNQD---GFFPDQYTFNTLVNGLCKAGHVK 312
Query: 441 QAFKIICEMMGKGFVPDDSTYSKVIGFLCDASKVEKAFSLFEEMKRNGIVPSVYTYTILI 500
A +I+ M+ +G+ PD TY+ VI LC +V++A + ++M P+ TY LI
Sbjct: 313 HAIEIMDVMLQEGYDPDVYTYNSVISGLCKLGEVKEAVEVLDQMITRDCSPNTVTYNTLI 372
Query: 501 DSFCKAGLIQQARKWFDEMLHKGCTPNVVTYTALIHAYLKAKQMPVADELFEMMLLEGCK 560
+ CK +++A + + KG P+V T+ +LI + VA ELFE M +GC+
Sbjct: 373 STLCKENQVEEATELARVLTSKGILPDVCTFNSLIQGLCLTRNHRVAMELFEEMRSKGCE 432
Query: 561 PNVVTYTALIDGHCKAGQIEKACQIYARMRGDIESSDMDKYFKLDHNNCEGPNVITYGAL 620
P+ TY LID C G++++A + +M + + C +VITY L
Sbjct: 433 PDEFTYNMLIDSLCSKGKLDEALNMLKQM---------------ELSGC-ARSVITYNTL 476
Query: 621 VDGLCKANRVKEAHELLDTMLAHGCEPNQIVYDAVIDGFCKIGKLQDAQEVFTKMSERGY 680
+DG CKAN+ +EA E+ D M HG N + Y+ +IDG CK +++DA ++ +M G
Sbjct: 477 IDGFCKANKTREAEEIFDEMEVHGVSRNSVTYNTLIDGLCKSRRVEDAAQLMDQMIMEGQ 536
Query: 681 SPNLYTYSSFIDCLFKDNRLDLVLKVLSKMLENSCTPNVVIYTEMVDGLCKIGKTDEAYK 740
P+ YTY+S + + + ++ M N C P++V Y ++ GLCK G+ + A K
Sbjct: 537 KPDKYTYNSLLTHFCRGGDIKKAADIVQAMTSNGCEPDIVTYGTLISGLCKAGRVEVASK 596
Query: 741 LMLKMEEKGCNPNVVTYTAMIDGFGKSGKIEQCLELFRDMCSKGCA-PNFITYRVLINHC 799
L+ ++ KG N Y +I G + K + + LFR+M + A P+ ++YR++
Sbjct: 597 LLRSIQMKGINLTPHAYNPVIQGLFRKRKTTEAINLFREMLEQNEAPPDAVSYRIVFRGL 656
Query: 800 CS-NGLLDEAYKLLDEMKQTYWPKHILSHRKIIEG 833
C+ G + EA L E+ + + S + EG
Sbjct: 657 CNGGGPIREAVDFLVELLEKGFVPEFSSLYMLAEG 691
Score = 250 bits (639), Expect = 2e-66
Identities = 167/667 (25%), Positives = 314/667 (47%), Gaps = 32/667 (4%)
Query: 104 DSLVVEVMNNVKNPELCVKFFLWAGRQIGYSHTPQVFDKLLDLLGCNVNADDRVPLKFLM 163
D +++ + + + ++ F A ++ +S P +++++L LG + + DD K L
Sbjct: 50 DVKLLDSLRSQPDDSAALRLFNLASKKPNFSPEPALYEEILLRLGRSGSFDDMK--KILE 107
Query: 164 EIKDDDHELLRRLLNFLVRKCCRNGWWNMALEELGRLKD-FGYKPSQTTYNALIQVFLRA 222
++K E+ L+ + + L + + D FG KP YN ++ + +
Sbjct: 108 DMKSSRCEMGTSTFLILIESYAQFELQDEILSVVDWMIDEFGLKPDTHFYNRMLNLLVDG 167
Query: 223 DKLDTAYLVKREMLSYAFVMDRYTLSCFAYSLCKGGKCREAFDLIDEAEDF--VPDTVFY 280
+ L + +M + D T + +LC+ + R A ++++ + VPD +
Sbjct: 168 NSLKLVEISHAKMSVWGIKPDVSTFNVLIKALCRAHQLRPAILMLEDMPSYGLVPDEKTF 227
Query: 281 NRMVSGLCEASLFEEAMDILHRMRSSSCIPNVVTYRILLSGCLRKGQLGRCKRILSMMIT 340
++ G E + A+ I +M C + V+ +++ G ++G++ + M
Sbjct: 228 TTVMQGYIEEGDLDGALRIREQMVEFGCSWSNVSVNVIVHGFCKEGRVEDALNFIQEMSN 287
Query: 341 -EGCYPNREIFNSLIHAYCKSRDYSYAYKLFKKMIKCGCQPGYLVYNIFIGSVCS-NEEQ 398
+G +P++ FN+L++ CK+ +A ++ M++ G P YN I +C E +
Sbjct: 288 QDGFFPDQYTFNTLVNGLCKAGHVKHAIEIMDVMLQEGYDPDVYTYNSVISGLCKLGEVK 347
Query: 399 PSSDILDLVEKAYSEMLDLGVVLNKVNVSNFARCLCGAGKFDQAFKIICEMMGKGFVPDD 458
+ ++LD +M+ N V + LC + ++A ++ + KG +PD
Sbjct: 348 EAVEVLD-------QMITRDCSPNTVTYNTLISTLCKENQVEEATELARVLTSKGILPDV 400
Query: 459 STYSKVIGFLCDASKVEKAFSLFEEMKRNGIVPSVYTYTILIDSFCKAGLIQQARKWFDE 518
T++ +I LC A LFEEM+ G P +TY +LIDS C G + +A +
Sbjct: 401 CTFNSLIQGLCLTRNHRVAMELFEEMRSKGCEPDEFTYNMLIDSLCSKGKLDEALNMLKQ 460
Query: 519 MLHKGCTPNVVTYTALIHAYLKAKQMPVADELFEMMLLEGCKPNVVTYTALIDGHCKAGQ 578
M GC +V+TY LI + KA + A+E+F+ M + G N VTY LIDG CK+ +
Sbjct: 461 MELSGCARSVITYNTLIDGFCKANKTREAEEIFDEMEVHGVSRNSVTYNTLIDGLCKSRR 520
Query: 579 IEKACQIYARMRGDIESSDMDKYFKLDHNNCEGPNVITYGALVDGLCKANRVKEAHELLD 638
+E A Q+ +M +E DKY TY +L+ C+ +K+A +++
Sbjct: 521 VEDAAQLMDQM--IMEGQKPDKY--------------TYNSLLTHFCRGGDIKKAADIVQ 564
Query: 639 TMLAHGCEPNQIVYDAVIDGFCKIGKLQDAQEVFTKMSERGYSPNLYTYSSFIDCLFKDN 698
M ++GCEP+ + Y +I G CK G+++ A ++ + +G + + Y+ I LF+
Sbjct: 565 AMTSNGCEPDIVTYGTLISGLCKAGRVEVASKLLRSIQMKGINLTPHAYNPVIQGLFRKR 624
Query: 699 RLDLVLKVLSKMLE-NSCTPNVVIYTEMVDGLCK-IGKTDEAYKLMLKMEEKGCNPNVVT 756
+ + + +MLE N P+ V Y + GLC G EA ++++ EKG P +
Sbjct: 625 KTTEAINLFREMLEQNEAPPDAVSYRIVFRGLCNGGGPIREAVDFLVELLEKGFVPEFSS 684
Query: 757 YTAMIDG 763
+ +G
Sbjct: 685 LYMLAEG 691
Score = 250 bits (639), Expect = 2e-66
Identities = 178/704 (25%), Positives = 322/704 (45%), Gaps = 73/704 (10%)
Query: 264 FDLIDEAEDFVPDTVFYNRMVSGLCEASLFEEAMDILHRMRSSSCIPNVVTYRILLSGCL 323
F+L + +F P+ Y ++ L + F++ IL M+SS C T+ IL+
Sbjct: 70 FNLASKKPNFSPEPALYEEILLRLGRSGSFDDMKKILEDMKSSRCEMGTSTFLILIESYA 129
Query: 324 RKGQLGRCKRILSMMITE-GCYPNREIFNSLIHAYCKSRDYSYAYKLFKKMIKCGCQPGY 382
+ ++ MI E G P+ +N ++
Sbjct: 130 QFELQDEILSVVDWMIDEFGLKPDTHFYNRML---------------------------- 161
Query: 383 LVYNIFIGSVCSNEEQPSSDILDLVEKAYSEMLDLGVVLNKVNVSNF---ARCLCGAGKF 439
N+ + + L LVE ++++M G+ K +VS F + LC A +
Sbjct: 162 ---NLLV----------DGNSLKLVEISHAKMSVWGI---KPDVSTFNVLIKALCRAHQL 205
Query: 440 DQAFKIICEMMGKGFVPDDSTYSKVIGFLCDASKVEKAFSLFEEMKRNGIVPSVYTYTIL 499
A ++ +M G VPD+ T++ V+ + ++ A + E+M G S + ++
Sbjct: 206 RPAILMLEDMPSYGLVPDEKTFTTVMQGYIEEGDLDGALRIREQMVEFGCSWSNVSVNVI 265
Query: 500 IDSFCKAGLIQQARKWFDEMLHK-GCTPNVVTYTALIHAYLKAKQMPVADELFEMMLLEG 558
+ FCK G ++ A + EM ++ G P+ T+ L++ KA + A E+ ++ML EG
Sbjct: 266 VHGFCKEGRVEDALNFIQEMSNQDGFFPDQYTFNTLVNGLCKAGHVKHAIEIMDVMLQEG 325
Query: 559 CKPNVVTYTALIDGHCKAGQIEKACQIYARMRGDIESSDMDKYFKLDHNNCEGPNVITYG 618
P+V TY ++I G CK G++++A ++ +D+ D + PN +TY
Sbjct: 326 YDPDVYTYNSVISGLCKLGEVKEAVEV------------LDQMITRDCS----PNTVTYN 369
Query: 619 ALVDGLCKANRVKEAHELLDTMLAHGCEPNQIVYDAVIDGFCKIGKLQDAQEVFTKMSER 678
L+ LCK N+V+EA EL + + G P+ ++++I G C + A E+F +M +
Sbjct: 370 TLISTLCKENQVEEATELARVLTSKGILPDVCTFNSLIQGLCLTRNHRVAMELFEEMRSK 429
Query: 679 GYSPNLYTYSSFIDCLFKDNRLDLVLKVLSKMLENSCTPNVVIYTEMVDGLCKIGKTDEA 738
G P+ +TY+ ID L +LD L +L +M + C +V+ Y ++DG CK KT EA
Sbjct: 430 GCEPDEFTYNMLIDSLCSKGKLDEALNMLKQMELSGCARSVITYNTLIDGFCKANKTREA 489
Query: 739 YKLMLKMEEKGCNPNVVTYTAMIDGFGKSGKIEQCLELFRDMCSKGCAPNFITYRVLINH 798
++ +ME G + N VTY +IDG KS ++E +L M +G P+ TY L+ H
Sbjct: 490 EEIFDEMEVHGVSRNSVTYNTLIDGLCKSRRVEDAAQLMDQMIMEGQKPDKYTYNSLLTH 549
Query: 799 CCSNGLLDEAYKLLDEMKQTYWPKHILSHRKIIEGFSQEFITSIG--LLDELSENESVPV 856
C G + +A ++ M I+++ +I G + + LL +
Sbjct: 550 FCRGGDIKKAADIVQAMTSNGCEPDIVTYGTLISGLCKAGRVEVASKLLRSIQMKGINLT 609
Query: 857 DSLYRILIDNYIKAGRLEVALDLLEEI---SSSPSHAVSNKYLYASLIENLSHASKVDKA 913
Y +I + + A++L E+ + +P AVS + ++ L + +A
Sbjct: 610 PHAYNPVIQGLFRKRKTTEAINLFREMLEQNEAPPDAVSYRIVFRGLCNG---GGPIREA 666
Query: 914 LELYASMISKNVVPELSILVHLIKGLIKVDKWQEALQLSDSICQ 957
++ ++ K VPE S L L +GL+ + + ++L + + Q
Sbjct: 667 VDFLVELLEKGFVPEFSSLYMLAEGLLTLSMEETLVKLVNMVMQ 710
Score = 218 bits (555), Expect = 1e-56
Identities = 148/543 (27%), Positives = 261/543 (47%), Gaps = 29/543 (5%)
Query: 454 FVPDDSTYSKVIGFLCDASKVEKAFSLFEEMKRNGIVPSVYTYTILIDSFCKAGLIQQAR 513
F P+ + Y +++ L + + + E+MK + T+ ILI+S+ + L +
Sbjct: 79 FSPEPALYEEILLRLGRSGSFDDMKKILEDMKSSRCEMGTSTFLILIESYAQFELQDEIL 138
Query: 514 KWFDEMLHK-GCTPNVVTYTALIHAYLKAKQMPVADELFEMMLLEGCKPNVVTYTALIDG 572
D M+ + G P+ Y +++ + + + + M + G KP+V T+ LI
Sbjct: 139 SVVDWMIDEFGLKPDTHFYNRMLNLLVDGNSLKLVEISHAKMSVWGIKPDVSTFNVLIKA 198
Query: 573 HCKAGQIEKACQIYARM----------------RGDIESSDMDKYFKLDHN----NCEGP 612
C+A Q+ A + M +G IE D+D ++ C
Sbjct: 199 LCRAHQLRPAILMLEDMPSYGLVPDEKTFTTVMQGYIEEGDLDGALRIREQMVEFGCSWS 258
Query: 613 NVITYGALVDGLCKANRVKEAHELLDTMLAH-GCEPNQIVYDAVIDGFCKIGKLQDAQEV 671
NV + +V G CK RV++A + M G P+Q ++ +++G CK G ++ A E+
Sbjct: 259 NV-SVNVIVHGFCKEGRVEDALNFIQEMSNQDGFFPDQYTFNTLVNGLCKAGHVKHAIEI 317
Query: 672 FTKMSERGYSPNLYTYSSFIDCLFKDNRLDLVLKVLSKMLENSCTPNVVIYTEMVDGLCK 731
M + GY P++YTY+S I L K + ++VL +M+ C+PN V Y ++ LCK
Sbjct: 318 MDVMLQEGYDPDVYTYNSVISGLCKLGEVKEAVEVLDQMITRDCSPNTVTYNTLISTLCK 377
Query: 732 IGKTDEAYKLMLKMEEKGCNPNVVTYTAMIDGFGKSGKIEQCLELFRDMCSKGCAPNFIT 791
+ +EA +L + KG P+V T+ ++I G + +ELF +M SKGC P+ T
Sbjct: 378 ENQVEEATELARVLTSKGILPDVCTFNSLIQGLCLTRNHRVAMELFEEMRSKGCEPDEFT 437
Query: 792 YRVLINHCCSNGLLDEAYKLLDEMKQTYWPKHILSHRKIIEGFSQEFITSIG--LLDELS 849
Y +LI+ CS G LDEA +L +M+ + + ++++ +I+GF + T + DE+
Sbjct: 438 YNMLIDSLCSKGKLDEALNMLKQMELSGCARSVITYNTLIDGFCKANKTREAEEIFDEME 497
Query: 850 ENESVPVDSLYRILIDNYIKAGRLEVALDLLEEISSSPSHAVSNKYLYASLIENLSHASK 909
+ Y LID K+ R+E A L++++ +KY Y SL+ +
Sbjct: 498 VHGVSRNSVTYNTLIDGLCKSRRVEDAAQLMDQMIMEGQKP--DKYTYNSLLTHFCRGGD 555
Query: 910 VDKALELYASMISKNVVPELSILVHLIKGLIKVDKWQEALQLSDSICQMVCLTLS-QTFQ 968
+ KA ++ +M S P++ LI GL K + + A +L SI QM + L+ +
Sbjct: 556 IKKAADIVQAMTSNGCEPDIVTYGTLISGLCKAGRVEVASKLLRSI-QMKGINLTPHAYN 614
Query: 969 PLV 971
P++
Sbjct: 615 PVI 617
Score = 171 bits (433), Expect = 2e-42
Identities = 128/498 (25%), Positives = 220/498 (43%), Gaps = 31/498 (6%)
Query: 177 LNFLVRKCCRNGWWNMALEELGRLKDF-GYKPSQTTYNALIQVFLRADKLDTAYLVKREM 235
+N +V C+ G AL + + + G+ P Q T+N L+ +A + A + M
Sbjct: 262 VNVIVHGFCKEGRVEDALNFIQEMSNQDGFFPDQYTFNTLVNGLCKAGHVKHAIEIMDVM 321
Query: 236 LSYAFVMDRYTLSCFAYSLCKGGKCREAFDLIDE--AEDFVPDTVFYNRMVSGLCEASLF 293
L + D YT + LCK G+ +EA +++D+ D P+TV YN ++S LC+ +
Sbjct: 322 LQEGYDPDVYTYNSVISGLCKLGEVKEAVEVLDQMITRDCSPNTVTYNTLISTLCKENQV 381
Query: 294 EEAMDILHRMRSSSCIPNVVTYRILLSGCLRKGQLGRCKRILSMMITEGCYPNREIFNSL 353
EEA ++ + S +P+V T+ L+ G + M ++GC P+ +N L
Sbjct: 382 EEATELARVLTSKGILPDVCTFNSLIQGLCLTRNHRVAMELFEEMRSKGCEPDEFTYNML 441
Query: 354 IHAYCKSRDYSYAYKLFKKMIKCGCQPGYLVYNIFIGSVC-SNEEQPSSDILDLVEKAYS 412
I + C A + K+M GC + YN I C +N+ + + +I D
Sbjct: 442 IDSLCSKGKLDEALNMLKQMELSGCARSVITYNTLIDGFCKANKTREAEEIFD------- 494
Query: 413 EMLDLGVVLNKVNVSNFARCLCGAGKFDQAFKIICEMMGKGFVPDDSTYSKVIGFLCDAS 472
EM GV N V + LC + + + A +++ +M+ +G PD TY+ ++ C
Sbjct: 495 EMEVHGVSRNSVTYNTLIDGLCKSRRVEDAAQLMDQMIMEGQKPDKYTYNSLLTHFCRGG 554
Query: 473 KVEKAFSLFEEMKRNGIVPSVYTYTILIDSFCKAGLIQQARKWFDEMLHKGCTPNVVTYT 532
++KA + + M NG P + TY LI CKAG ++ A K + KG Y
Sbjct: 555 DIKKAADIVQAMTSNGCEPDIVTYGTLISGLCKAGRVEVASKLLRSIQMKGINLTPHAYN 614
Query: 533 ALIHAYLKAKQMPVADELFEMMLLEG-CKPNVVTYTALIDGHCK-AGQIEKACQIYARMR 590
+I + ++ A LF ML + P+ V+Y + G C G I +A +
Sbjct: 615 PVIQGLFRKRKTTEAINLFREMLEQNEAPPDAVSYRIVFRGLCNGGGPIREAVDFLVEL- 673
Query: 591 GDIESSDMDKYFKLDHNNCEGPNVITYGALVDGLCKANRVKEAHELLDTMLAHGCEPNQI 650
++K F P + L +GL + + +L++ ++ +
Sbjct: 674 -------LEKGFV--------PEFSSLYMLAEGLLTLSMEETLVKLVNMVMQKARFSEEE 718
Query: 651 VYDAVIDGFCKIGKLQDA 668
V +++ G KI K QDA
Sbjct: 719 V--SMVKGLLKIRKFQDA 734
Score = 108 bits (269), Expect = 2e-23
Identities = 91/394 (23%), Positives = 175/394 (44%), Gaps = 8/394 (2%)
Query: 584 QIYARMRGDIESSDMDKYFK-LDHNNCEGPNVITYGALVDGLCKANRVKEAHELLDTMLA 642
+I R+ DM K + + + CE T+ L++ + E ++D M+
Sbjct: 88 EILLRLGRSGSFDDMKKILEDMKSSRCE-MGTSTFLILIESYAQFELQDEILSVVDWMID 146
Query: 643 H-GCEPNQIVYDAVIDGFCKIGKLQDAQEVFTKMSERGYSPNLYTYSSFIDCLFKDNRLD 701
G +P+ Y+ +++ L+ + KMS G P++ T++ I L + ++L
Sbjct: 147 EFGLKPDTHFYNRMLNLLVDGNSLKLVEISHAKMSVWGIKPDVSTFNVLIKALCRAHQLR 206
Query: 702 LVLKVLSKMLENSCTPNVVIYTEMVDGLCKIGKTDEAYKLMLKMEEKGCNPNVVTYTAMI 761
+ +L M P+ +T ++ G + G D A ++ +M E GC+ + V+ ++
Sbjct: 207 PAILMLEDMPSYGLVPDEKTFTTVMQGYIEEGDLDGALRIREQMVEFGCSWSNVSVNVIV 266
Query: 762 DGFGKSGKIEQCLELFRDMCSK-GCAPNFITYRVLINHCCSNGLLDEAYKLLDEMKQTYW 820
GF K G++E L ++M ++ G P+ T+ L+N C G + A +++D M Q +
Sbjct: 267 HGFCKEGRVEDALNFIQEMSNQDGFFPDQYTFNTLVNGLCKAGHVKHAIEIMDVMLQEGY 326
Query: 821 PKHILSHRKIIEGFSQ--EFITSIGLLDELSENESVPVDSLYRILIDNYIKAGRLEVALD 878
+ ++ +I G + E ++ +LD++ + P Y LI K ++E A +
Sbjct: 327 DPDVYTYNSVISGLCKLGEVKEAVEVLDQMITRDCSPNTVTYNTLISTLCKENQVEEATE 386
Query: 879 LLEEISSSPSHAVSNKYLYASLIENLSHASKVDKALELYASMISKNVVPELSILVHLIKG 938
L ++S + + + SLI+ L A+EL+ M SK P+ LI
Sbjct: 387 LARVLTSKG--ILPDVCTFNSLIQGLCLTRNHRVAMELFEEMRSKGCEPDEFTYNMLIDS 444
Query: 939 LIKVDKWQEALQLSDSICQMVCLTLSQTFQPLVN 972
L K EAL + + C T+ L++
Sbjct: 445 LCSKGKLDEALNMLKQMELSGCARSVITYNTLID 478
Score = 95.1 bits (235), Expect = 2e-19
Identities = 81/324 (25%), Positives = 142/324 (43%), Gaps = 9/324 (2%)
Query: 148 GCNVNADDRVPLKFLMEIKDDDHELLRRLLNFLVRKCCRNGWWNMALEELGRLKDFGYKP 207
G + + RV ++ E++ E N L+ C G + AL L +++ G
Sbjct: 409 GLCLTRNHRVAMELFEEMRSKGCEPDEFTYNMLIDSLCSKGKLDEALNMLKQMELSGCAR 468
Query: 208 SQTTYNALIQVFLRADKLDTAYLVKREMLSYAFVMDRYTLSCFAYSLCKGGKCREAFDLI 267
S TYN LI F +A+K A + EM + + T + LCK + +A L+
Sbjct: 469 SVITYNTLIDGFCKANKTREAEEIFDEMEVHGVSRNSVTYNTLIDGLCKSRRVEDAAQLM 528
Query: 268 DE--AEDFVPDTVFYNRMVSGLCEASLFEEAMDILHRMRSSSCIPNVVTYRILLSGCLRK 325
D+ E PD YN +++ C ++A DI+ M S+ C P++VTY L+SG +
Sbjct: 529 DQMIMEGQKPDKYTYNSLLTHFCRGGDIKKAADIVQAMTSNGCEPDIVTYGTLISGLCKA 588
Query: 326 GQLGRCKRILSMMITEGCYPNREIFNSLIHAYCKSRDYSYAYKLFKKMIKCG-CQPGYLV 384
G++ ++L + +G +N +I + R + A LF++M++ P +
Sbjct: 589 GRVEVASKLLRSIQMKGINLTPHAYNPVIQGLFRKRKTTEAINLFREMLEQNEAPPDAVS 648
Query: 385 YNIFIGSVCSNEEQPSSDILDLVEKAYSEMLDLGVVLNKVNVSNFARCLCGAGKFDQAFK 444
Y I +C N P + +D + E+L+ G V ++ A L + K
Sbjct: 649 YRIVFRGLC-NGGGPIREAVDFL----VELLEKGFVPEFSSLYMLAEGLLTLSMEETLVK 703
Query: 445 IICEMMGKGFVPDDSTYSKVIGFL 468
++ +M K ++ S V G L
Sbjct: 704 LVNMVMQKARFSEEEV-SMVKGLL 726
Score = 85.5 bits (210), Expect = 1e-16
Identities = 77/368 (20%), Positives = 156/368 (41%), Gaps = 37/368 (10%)
Query: 612 PNVITYGALVDGLCKANRVKEAHELLDTMLAHGCEPNQIVYDAVIDGFCKIGKLQDA--Q 669
P Y ++ L ++ + ++L+ M + CE + +I+ + + +LQD
Sbjct: 81 PEPALYEEILLRLGRSGSFDDMKKILEDMKSSRCEMGTSTFLILIESYAQF-ELQDEILS 139
Query: 670 EVFTKMSERGYSPNLYTYSSFIDCLFKDNRLDLVLKVLSKMLENSCTPNVVIYTEMVDGL 729
V + E G P+ + Y+ ++ L N L LV +KM P+V + ++ L
Sbjct: 140 VVDWMIDEFGLKPDTHFYNRMLNLLVDGNSLKLVEISHAKMSVWGIKPDVSTFNVLIKAL 199
Query: 730 CKIGKTDEAYKLMLKMEEKGCNPNVVTYTAMIDGFGKSGKIEQCLELFRDMCSKGCAPNF 789
C+ + A ++ M G P+ T+T ++ G+ + G ++ L + M GC+ +
Sbjct: 200 CRAHQLRPAILMLEDMPSYGLVPDEKTFTTVMQGYIEEGDLDGALRIREQMVEFGCSWSN 259
Query: 790 ITYRVLINHCCSNGLLDEAYKLLDEMKQTYWPKHILSHRKIIEGFSQEFITSIGLLDELS 849
++ V+++ C G +++A + EM +GF + T
Sbjct: 260 VSVNVIVHGFCKEGRVEDALNFIQEMSNQ-------------DGFFPDQYT--------- 297
Query: 850 ENESVPVDSLYRILIDNYIKAGRLEVALDLLEEISSSPSHAVSNKYLYASLIENLSHASK 909
+ L++ KAG ++ A+++++ + + Y Y S+I L +
Sbjct: 298 ----------FNTLVNGLCKAGHVKHAIEIMDVMLQEGYDP--DVYTYNSVISGLCKLGE 345
Query: 910 VDKALELYASMISKNVVPELSILVHLIKGLIKVDKWQEALQLSDSICQMVCLTLSQTFQP 969
V +A+E+ MI+++ P LI L K ++ +EA +L+ + L TF
Sbjct: 346 VKEAVEVLDQMITRDCSPNTVTYNTLISTLCKENQVEEATELARVLTSKGILPDVCTFNS 405
Query: 970 LVNLNLLT 977
L+ LT
Sbjct: 406 LIQGLCLT 413
>At3g54980 unknown protein (At3g54980)
Length = 851
Score = 271 bits (692), Expect = 2e-72
Identities = 186/632 (29%), Positives = 310/632 (48%), Gaps = 30/632 (4%)
Query: 346 NREIFNSLIHAYCKSRDYSYAYKLFKKMIKCGCQPGYLVYNIFIGSVCSNEEQPSSDILD 405
N FN L++AY K R +A + +M++ P + N + ++ L
Sbjct: 162 NSRAFNYLLNAYSKDRQTDHAVDIVNQMLELDVIPFFPYVNRTLSALVQRNS------LT 215
Query: 406 LVEKAYSEMLDLGVVLNKVNVSNFARCLCGAGKFDQAFKIICEMMGKGFVPDDSTYSKVI 465
++ YS M+ +GV + V R K +A +++ + +G PD YS +
Sbjct: 216 EAKELYSRMVAIGVDGDNVTTQLLMRASLREEKPAEALEVLSRAIERGAEPDSLLYSLAV 275
Query: 466 GFLCDASKVEKAFSLFEEMKRNGI-VPSVYTYTILIDSFCKAGLIQQARKWFDEMLHKGC 524
C + A SL EMK + VPS TYT +I + K G + A + DEML G
Sbjct: 276 QACCKTLDLAMANSLLREMKEKKLCVPSQETYTSVILASVKQGNMDDAIRLKDEMLSDGI 335
Query: 525 TPNVVTYTALIHAYLKAKQMPVADELFEMMLLEGCKPNVVTYTALIDGHCKAGQIEKACQ 584
+ NVV T+LI + K + A LF+ M EG PN VT++ LI+ K G++EKA +
Sbjct: 336 SMNVVAATSLITGHCKNNDLVSALVLFDKMEKEGPSPNSVTFSVLIEWFRKNGEMEKALE 395
Query: 585 IYARM----------------RGDIESSDMDKYFKLDHNNCEG--PNVITYGALVDGLCK 626
Y +M +G ++ ++ KL + E NV ++ LCK
Sbjct: 396 FYKKMEVLGLTPSVFHVHTIIQGWLKGQKHEEALKLFDESFETGLANVFVCNTILSWLCK 455
Query: 627 ANRVKEAHELLDTMLAHGCEPNQIVYDAVIDGFCKIGKLQDAQEVFTKMSERGYSPNLYT 686
+ EA ELL M + G PN + Y+ V+ G C+ + A+ VF+ + E+G PN YT
Sbjct: 456 QGKTDEATELLSKMESRGIGPNVVSYNNVMLGHCRQKNMDLARIVFSNILEKGLKPNNYT 515
Query: 687 YSSFIDCLFKDNRLDLVLKVLSKMLENSCTPNVVIYTEMVDGLCKIGKTDEAYKLMLKM- 745
YS ID F+++ L+V++ M ++ N V+Y +++GLCK+G+T +A +L+ M
Sbjct: 516 YSILIDGCFRNHDEQNALEVVNHMTSSNIEVNGVVYQTIINGLCKVGQTSKARELLANMI 575
Query: 746 EEKGCNPNVVTYTAMIDGFGKSGKIEQCLELFRDMCSKGCAPNFITYRVLINHCCSNGLL 805
EEK + ++Y ++IDGF K G+++ + + +MC G +PN ITY L+N C N +
Sbjct: 576 EEKRLCVSCMSYNSIIDGFFKEGEMDSAVAAYEEMCGNGISPNVITYTSLMNGLCKNNRM 635
Query: 806 DEAYKLLDEMKQTYWPKHILSHRKIIEGFSQE--FITSIGLLDELSENESVPVDSLYRIL 863
D+A ++ DEMK I ++ +I+GF + ++ L EL E P +Y L
Sbjct: 636 DQALEMRDEMKNKGVKLDIPAYGALIDGFCKRSNMESASALFSELLEEGLNPSQPIYNSL 695
Query: 864 IDNYIKAGRLEVALDLLEEISSSPSHAVSNKYLYASLIENLSHASKVDKALELYASMISK 923
I + G + ALDL +++ + Y +LI+ L + A ELY M +
Sbjct: 696 ISGFRNLGNMVAALDLYKKMLKDGLRC--DLGTYTTLIDGLLKDGNLILASELYTEMQAV 753
Query: 924 NVVPELSILVHLIKGLIKVDKWQEALQLSDSI 955
+VP+ I ++ GL K ++ + +++ + +
Sbjct: 754 GLVPDEIIYTVIVNGLSKKGQFVKVVKMFEEM 785
Score = 257 bits (657), Expect = 2e-68
Identities = 197/762 (25%), Positives = 341/762 (43%), Gaps = 57/762 (7%)
Query: 101 QLNDSLVVEVM-NNVKNPELCVKFFLWAGRQIGYSH--------------TPQVFDKLLD 145
Q +D+ V++V+ N NPE ++F+ WA G +P+ + + D
Sbjct: 69 QKDDASVIDVLLNRRNNPEAALRFYNWARPWRGSFEDGDVFWVLIHILVSSPETYGRASD 128
Query: 146 LLGCNVNADDRVPL------KFLMEIKDDDHELLRRLLNFLVRKCCRNGWWNMALEELGR 199
LL V+ + P+ K + K E+ R N+L+ ++ + A++ + +
Sbjct: 129 LLIRYVSTSNPTPMASVLVSKLVDSAKSFGFEVNSRAFNYLLNAYSKDRQTDHAVDIVNQ 188
Query: 200 LKDFGYKPSQTTYNALIQVFLRADKLDTAYLVKREMLSYAFVMDRYTLSCFAYSLCKGGK 259
+ + P N + ++ + L A + M++ D T + + K
Sbjct: 189 MLELDVIPFFPYVNRTLSALVQRNSLTEAKELYSRMVAIGVDGDNVTTQLLMRASLREEK 248
Query: 260 CREAFDLIDEAED--FVPDTVFYNRMVSGLCEASLFEEAMDILHRMRSSS-CIPNVVTYR 316
EA +++ A + PD++ Y+ V C+ A +L M+ C+P+ TY
Sbjct: 249 PAEALEVLSRAIERGAEPDSLLYSLAVQACCKTLDLAMANSLLREMKEKKLCVPSQETYT 308
Query: 317 ILLSGCLRKGQLGRCKRILSMMITEGCYPNREIFNSLIHAYCKSRDYSYAYKLFKKMIKC 376
++ +++G + R+ M+++G N SLI +CK+ D A LF KM K
Sbjct: 309 SVILASVKQGNMDDAIRLKDEMLSDGISMNVVAATSLITGHCKNNDLVSALVLFDKMEKE 368
Query: 377 GCQPGYLVYNIFIGSVCSNEEQPSSDILDLVEKA---YSEMLDLGVVLNKVNVSNFARCL 433
G P + +++ I N E +EKA Y +M LG+ + +V +
Sbjct: 369 GPSPNSVTFSVLIEWFRKNGE---------MEKALEFYKKMEVLGLTPSVFHVHTIIQGW 419
Query: 434 CGAGKFDQAFKIICEMMGKGFVPDDSTYSKVIGFLCDASKVEKAFSLFEEMKRNGIVPSV 493
K ++A K+ E G + + ++ +LC K ++A L +M+ GI P+V
Sbjct: 420 LKGQKHEEALKLFDESFETGLA-NVFVCNTILSWLCKQGKTDEATELLSKMESRGIGPNV 478
Query: 494 YTYTILIDSFCKAGLIQQARKWFDEMLHKGCTPNVVTYTALIHAYLKAKQMPVADELFEM 553
+Y ++ C+ + AR F +L KG PN TY+ LI + A E+
Sbjct: 479 VSYNNVMLGHCRQKNMDLARIVFSNILEKGLKPNNYTYSILIDGCFRNHDEQNALEVVNH 538
Query: 554 MLLEGCKPNVVTYTALIDGHCKAGQIEKACQIYARM-----------------RGDIESS 596
M + N V Y +I+G CK GQ KA ++ A M G +
Sbjct: 539 MTSSNIEVNGVVYQTIINGLCKVGQTSKARELLANMIEEKRLCVSCMSYNSIIDGFFKEG 598
Query: 597 DMDKYFKLDHNNCEG---PNVITYGALVDGLCKANRVKEAHELLDTMLAHGCEPNQIVYD 653
+MD C PNVITY +L++GLCK NR+ +A E+ D M G + + Y
Sbjct: 599 EMDSAVAAYEEMCGNGISPNVITYTSLMNGLCKNNRMDQALEMRDEMKNKGVKLDIPAYG 658
Query: 654 AVIDGFCKIGKLQDAQEVFTKMSERGYSPNLYTYSSFIDCLFKDNRLDLVLKVLSKMLEN 713
A+IDGFCK ++ A +F+++ E G +P+ Y+S I + L + KML++
Sbjct: 659 ALIDGFCKRSNMESASALFSELLEEGLNPSQPIYNSLISGFRNLGNMVAALDLYKKMLKD 718
Query: 714 SCTPNVVIYTEMVDGLCKIGKTDEAYKLMLKMEEKGCNPNVVTYTAMIDGFGKSGKIEQC 773
++ YT ++DGL K G A +L +M+ G P+ + YT +++G K G+ +
Sbjct: 719 GLRCDLGTYTTLIDGLLKDGNLILASELYTEMQAVGLVPDEIIYTVIVNGLSKKGQFVKV 778
Query: 774 LELFRDMCSKGCAPNFITYRVLINHCCSNGLLDEAYKLLDEM 815
+++F +M PN + Y +I G LDEA++L DEM
Sbjct: 779 VKMFEEMKKNNVTPNVLIYNAVIAGHYREGNLDEAFRLHDEM 820
Score = 174 bits (441), Expect = 2e-43
Identities = 141/576 (24%), Positives = 256/576 (43%), Gaps = 25/576 (4%)
Query: 386 NIFIGSVCSNEEQPSSDILDLVEKAYSEMLDLGVVLNKVNVSNFARCLCGAGKFDQAFKI 445
++ I V ++ P + +L V K G +N + + D A I
Sbjct: 128 DLLIRYVSTSNPTPMASVL--VSKLVDSAKSFGFEVNSRAFNYLLNAYSKDRQTDHAVDI 185
Query: 446 ICEMMGKGFVPDDSTYSKVIGFLCDASKVEKAFSLFEEMKRNGIVPSVYTYTILIDSFCK 505
+ +M+ +P ++ + L + + +A L+ M G+ T +L+ + +
Sbjct: 186 VNQMLELDVIPFFPYVNRTLSALVQRNSLTEAKELYSRMVAIGVDGDNVTTQLLMRASLR 245
Query: 506 AGLIQQARKWFDEMLHKGCTPNVVTYTALIHAYLKAKQMPVADELF-EMMLLEGCKPNVV 564
+A + + +G P+ + Y+ + A K + +A+ L EM + C P+
Sbjct: 246 EEKPAEALEVLSRAIERGAEPDSLLYSLAVQACCKTLDLAMANSLLREMKEKKLCVPSQE 305
Query: 565 TYTALIDGHCKAGQIEKACQIYARMRGDIESSDMDKYFKLDHNNCEGPNVITYGALVDGL 624
TYT++I K G ++ A ++ M D S NV+ +L+ G
Sbjct: 306 TYTSVILASVKQGNMDDAIRLKDEMLSDGISM----------------NVVAATSLITGH 349
Query: 625 CKANRVKEAHELLDTMLAHGCEPNQIVYDAVIDGFCKIGKLQDAQEVFTKMSERGYSPNL 684
CK N + A L D M G PN + + +I+ F K G+++ A E + KM G +P++
Sbjct: 350 CKNNDLVSALVLFDKMEKEGPSPNSVTFSVLIEWFRKNGEMEKALEFYKKMEVLGLTPSV 409
Query: 685 YTYSSFIDCLFKDNRLDLVLKVLSKMLENSCTPNVVIYTEMVDGLCKIGKTDEAYKLMLK 744
+ + I K + + LK+ + E NV + ++ LCK GKTDEA +L+ K
Sbjct: 410 FHVHTIIQGWLKGQKHEEALKLFDESFETGLA-NVFVCNTILSWLCKQGKTDEATELLSK 468
Query: 745 MEEKGCNPNVVTYTAMIDGFGKSGKIEQCLELFRDMCSKGCAPNFITYRVLINHCCSNGL 804
ME +G PNVV+Y ++ G + ++ +F ++ KG PN TY +LI+ C N
Sbjct: 469 MESRGIGPNVVSYNNVMLGHCRQKNMDLARIVFSNILEKGLKPNNYTYSILIDGCFRNHD 528
Query: 805 LDEAYKLLDEMKQTYWPKHILSHRKIIEGFSQEFITSIG--LLDELSENESVPVDSL-YR 861
A ++++ M + + + ++ II G + TS LL + E + + V + Y
Sbjct: 529 EQNALEVVNHMTSSNIEVNGVVYQTIINGLCKVGQTSKARELLANMIEEKRLCVSCMSYN 588
Query: 862 ILIDNYIKAGRLEVALDLLEEISSSPSHAVSNKYLYASLIENLSHASKVDKALELYASMI 921
+ID + K G ++ A+ EE+ + N Y SL+ L +++D+ALE+ M
Sbjct: 589 SIIDGFFKEGEMDSAVAAYEEMCGNGISP--NVITYTSLMNGLCKNNRMDQALEMRDEMK 646
Query: 922 SKNVVPELSILVHLIKGLIKVDKWQEALQLSDSICQ 957
+K V ++ LI G K + A L + +
Sbjct: 647 NKGVKLDIPAYGALIDGFCKRSNMESASALFSELLE 682
Score = 73.9 bits (180), Expect = 4e-13
Identities = 85/373 (22%), Positives = 155/373 (40%), Gaps = 48/373 (12%)
Query: 590 RGDIESSDMDKYFKLDHNNCEGPNVITYGALVDGLCK----ANRVKEAH----ELLDTML 641
RG E D+ ++ L H P TYG D L + +N A +L+D+
Sbjct: 100 RGSFEDGDV--FWVLIHILVSSPE--TYGRASDLLIRYVSTSNPTPMASVLVSKLVDSAK 155
Query: 642 AHGCEPNQIVYDAVIDGFCKIGKLQDAQEVFTKMSERGYSPNLYTYSSFIDCLFKDNRLD 701
+ G E N ++ +++ + K + A ++ +M E P + + L + N L
Sbjct: 156 SFGFEVNSRAFNYLLNAYSKDRQTDHAVDIVNQMLELDVIPFFPYVNRTLSALVQRNSLT 215
Query: 702 LVLKVLSKMLENSCTPNVVIYTEMVDGLCKIGKTDEAYKLMLKMEEKGCNPNVVTYTAMI 761
++ S+M+ + V ++ + K EA +++ + E+G P+ + Y+ +
Sbjct: 216 EAKELYSRMVAIGVDGDNVTTQLLMRASLREEKPAEALEVLSRAIERGAEPDSLLYSLAV 275
Query: 762 DGFGKSGKIEQCLELFRDMCSKG-CAPNFITYRVLINHCCSNGLLDEAYKLLDEMKQTYW 820
K+ + L R+M K C P+ TY +I G +D+A +L DEM
Sbjct: 276 QACCKTLDLAMANSLLREMKEKKLCVPSQETYTSVILASVKQGNMDDAIRLKDEM----- 330
Query: 821 PKHILSHRKIIEGFSQEFITSIGLLDELSENESVPVDSLYRILIDNYIKAGRLEVALDLL 880
+ +G S + + L+ +N + V +L V D +
Sbjct: 331 ---------LSDGISMNVVAATSLITGHCKNNDL-VSAL---------------VLFDKM 365
Query: 881 EEISSSPSHAVSNKYLYASLIENLSHASKVDKALELYASMISKNVVPELSILVHLIKGLI 940
E+ SP N ++ LIE +++KALE Y M + P + + +I+G +
Sbjct: 366 EKEGPSP-----NSVTFSVLIEWFRKNGEMEKALEFYKKMEVLGLTPSVFHVHTIIQGWL 420
Query: 941 KVDKWQEALQLSD 953
K K +EAL+L D
Sbjct: 421 KGQKHEEALKLFD 433
Score = 64.7 bits (156), Expect = 2e-10
Identities = 46/218 (21%), Positives = 92/218 (42%), Gaps = 2/218 (0%)
Query: 180 LVRKCCRNGWWNMALEELGRLKDFGYKPSQTTYNALIQVFLRADKLDTAYLVKREMLSYA 239
L+ C+N + ALE +K+ G K Y ALI F + +++A + E+L
Sbjct: 625 LMNGLCKNNRMDQALEMRDEMKNKGVKLDIPAYGALIDGFCKRSNMESASALFSELLEEG 684
Query: 240 FVMDRYTLSCFAYSLCKGGKCREAFDLIDEA--EDFVPDTVFYNRMVSGLCEASLFEEAM 297
+ + G A DL + + D Y ++ GL + A
Sbjct: 685 LNPSQPIYNSLISGFRNLGNMVAALDLYKKMLKDGLRCDLGTYTTLIDGLLKDGNLILAS 744
Query: 298 DILHRMRSSSCIPNVVTYRILLSGCLRKGQLGRCKRILSMMITEGCYPNREIFNSLIHAY 357
++ M++ +P+ + Y ++++G +KGQ + ++ M PN I+N++I +
Sbjct: 745 ELYTEMQAVGLVPDEIIYTVIVNGLSKKGQFVKVVKMFEEMKKNNVTPNVLIYNAVIAGH 804
Query: 358 CKSRDYSYAYKLFKKMIKCGCQPGYLVYNIFIGSVCSN 395
+ + A++L +M+ G P ++I + N
Sbjct: 805 YREGNLDEAFRLHDEMLDKGILPDGATFDILVSGQVGN 842
>At1g12620 putative protein
Length = 621
Score = 268 bits (684), Expect = 1e-71
Identities = 158/543 (29%), Positives = 274/543 (50%), Gaps = 22/543 (4%)
Query: 275 PDTVFYNRMVSGLCEASLFEEAMDILHRMRSSSCIPNVVTYRILLSGCLRKGQLGRCKRI 334
P + ++R+ S + ++ +D+ +M N+ T I+++ C R +L
Sbjct: 70 PRLIDFSRLFSVVARTKQYDLVLDLCKQMELKGIAHNLYTLSIMINCCCRCRKLSLAFSA 129
Query: 335 LSMMITEGCYPNREIFNSLIHAYCKSRDYSYAYKLFKKMIKCGCQPGYLVYNIFIGSVCS 394
+ +I G P+ F++LI+ C S A +L +M++ G +P + N + +C
Sbjct: 130 MGKIIKLGYEPDTVTFSTLINGLCLEGRVSEALELVDRMVEMGHKPTLITLNALVNGLCL 189
Query: 395 NEEQPSSDILDLVEKAYSEMLDLGVVLNKVNVSNFARCLCGAGKFDQAFKIICEMMGKGF 454
N + SD + L+++ M++ G N+V + +C +G+ A +++ +M +
Sbjct: 190 NGKV--SDAVLLIDR----MVETGFQPNEVTYGPVLKVMCKSGQTALAMELLRKMEERKI 243
Query: 455 VPDDSTYSKVIGFLCDASKVEKAFSLFEEMKRNGIVPSVYTYTILIDSFCKAGLIQQARK 514
D YS +I LC ++ AF+LF EM+ G + YT LI FC AG K
Sbjct: 244 KLDAVKYSIIIDGLCKDGSLDNAFNLFNEMEIKGFKADIIIYTTLIRGFCYAGRWDDGAK 303
Query: 515 WFDEMLHKGCTPNVVTYTALIHAYLKAKQMPVADELFEMMLLEGCKPNVVTYTALIDGHC 574
+M+ + TP+VV ++ALI ++K ++ A+EL + M+ G P+ VTYT+LIDG C
Sbjct: 304 LLRDMIKRKITPDVVAFSALIDCFVKEGKLREAEELHKEMIQRGISPDTVTYTSLIDGFC 363
Query: 575 KAGQIEKACQIYARMRGDIESSDMDKYFKLDHNNCEGPNVITYGALVDGLCKANRVKEAH 634
K Q++KA + M C GPN+ T+ L++G CKAN + +
Sbjct: 364 KENQLDKANHMLDLMVS---------------KGC-GPNIRTFNILINGYCKANLIDDGL 407
Query: 635 ELLDTMLAHGCEPNQIVYDAVIDGFCKIGKLQDAQEVFTKMSERGYSPNLYTYSSFIDCL 694
EL M G + + Y+ +I GFC++GKL+ A+E+F +M R P++ +Y +D L
Sbjct: 408 ELFRKMSLRGVVADTVTYNTLIQGFCELGKLEVAKELFQEMVSRRVRPDIVSYKILLDGL 467
Query: 695 FKDNRLDLVLKVLSKMLENSCTPNVVIYTEMVDGLCKIGKTDEAYKLMLKMEEKGCNPNV 754
+ + L++ K+ ++ ++ IY ++ G+C K D+A+ L + KG P+V
Sbjct: 468 CDNGEPEKALEIFEKIEKSKMELDIGIYNIIIHGMCNASKVDDAWDLFCSLPLKGVKPDV 527
Query: 755 VTYTAMIDGFGKSGKIEQCLELFRDMCSKGCAPNFITYRVLINHCCSNGLLDEAYKLLDE 814
TY MI G K G + + LFR M G +PN TY +LI G ++ KL++E
Sbjct: 528 KTYNIMIGGLCKKGSLSEADLLFRKMEEDGHSPNGCTYNILIRAHLGEGDATKSAKLIEE 587
Query: 815 MKQ 817
+K+
Sbjct: 588 IKR 590
Score = 236 bits (601), Expect = 6e-62
Identities = 164/556 (29%), Positives = 265/556 (47%), Gaps = 31/556 (5%)
Query: 366 AYKLFKKMIKCGCQPGYLVYNIFIGSVCSNEEQPSSDILDLVEKAYSEMLDLGVVLNKVN 425
A LF++M + +P + ++ V ++ DLV +M G+ N
Sbjct: 56 AVDLFQEMTRSRPRPRLIDFSRLFSVVARTKQY------DLVLDLCKQMELKGIAHNLYT 109
Query: 426 VSNFARCLCGAGKFDQAFKIICEMMGKGFVPDDSTYSKVIGFLCDASKVEKAFSLFEEMK 485
+S C C K AF + +++ G+ PD T+S +I LC +V +A L + M
Sbjct: 110 LSIMINCCCRCRKLSLAFSAMGKIIKLGYEPDTVTFSTLINGLCLEGRVSEALELVDRMV 169
Query: 486 RNGIVPSVYTYTILIDSFCKAGLIQQARKWFDEMLHKGCTPNVVTYTALIHAYLKAKQMP 545
G P++ T L++ C G + A D M+ G PN VTY ++ K+ Q
Sbjct: 170 EMGHKPTLITLNALVNGLCLNGKVSDAVLLIDRMVETGFQPNEVTYGPVLKVMCKSGQTA 229
Query: 546 VADELFEMMLLEGCKPNVVTYTALIDGHCKAGQIEKACQIYARMRGDIESSDMDKYFKLD 605
+A EL M K + V Y+ +IDG CK G ++ A ++ M K FK D
Sbjct: 230 LAMELLRKMEERKIKLDAVKYSIIIDGLCKDGSLDNAFNLFNEME--------IKGFKAD 281
Query: 606 HNNCEGPNVITYGALVDGLCKANRVKEAHELLDTMLAHGCEPNQIVYDAVIDGFCKIGKL 665
+I Y L+ G C A R + +LL M+ P+ + + A+ID F K GKL
Sbjct: 282 --------IIIYTTLIRGFCYAGRWDDGAKLLRDMIKRKITPDVVAFSALIDCFVKEGKL 333
Query: 666 QDAQEVFTKMSERGYSPNLYTYSSFIDCLFKDNRLDLVLKVLSKMLENSCTPNVVIYTEM 725
++A+E+ +M +RG SP+ TY+S ID K+N+LD +L M+ C PN+ + +
Sbjct: 334 REAEELHKEMIQRGISPDTVTYTSLIDGFCKENQLDKANHMLDLMVSKGCGPNIRTFNIL 393
Query: 726 VDGLCKIGKTDEAYKLMLKMEEKGCNPNVVTYTAMIDGFGKSGKIEQCLELFRDMCSKGC 785
++G CK D+ +L KM +G + VTY +I GF + GK+E ELF++M S+
Sbjct: 394 INGYCKANLIDDGLELFRKMSLRGVVADTVTYNTLIQGFCELGKLEVAKELFQEMVSRRV 453
Query: 786 APNFITYRVLINHCCSNGLLDEAYKLLDEMKQTYWPKHILSHRKIIEGF--SQEFITSIG 843
P+ ++Y++L++ C NG ++A ++ ++++++ I + II G + + +
Sbjct: 454 RPDIVSYKILLDGLCDNGEPEKALEIFEKIEKSKMELDIGIYNIIIHGMCNASKVDDAWD 513
Query: 844 LLDELSENESVPVDSLYRILIDNYIKAGRLEVALDLL----EEISSSPSHAVSNKYLYAS 899
L L P Y I+I K G L A DLL EE SP+ N + A
Sbjct: 514 LFCSLPLKGVKPDVKTYNIMIGGLCKKGSLSEA-DLLFRKMEEDGHSPNGCTYNILIRAH 572
Query: 900 LIENLSHASKVDKALE 915
L E A+K K +E
Sbjct: 573 LGE--GDATKSAKLIE 586
Score = 222 bits (566), Expect = 7e-58
Identities = 146/513 (28%), Positives = 254/513 (49%), Gaps = 20/513 (3%)
Query: 438 KFDQAFKIICEMMGKGFVPDDSTYSKVIGFLCDASKVEKAFSLFEEMKRNGIVPSVYTYT 497
K D A + EM P +S++ + + + L ++M+ GI ++YT +
Sbjct: 52 KEDDAVDLFQEMTRSRPRPRLIDFSRLFSVVARTKQYDLVLDLCKQMELKGIAHNLYTLS 111
Query: 498 ILIDSFCKAGLIQQARKWFDEMLHKGCTPNVVTYTALIHAYLKAKQMPVADELFEMMLLE 557
I+I+ C+ + A +++ G P+ VT++ LI+ ++ A EL + M+
Sbjct: 112 IMINCCCRCRKLSLAFSAMGKIIKLGYEPDTVTFSTLINGLCLEGRVSEALELVDRMVEM 171
Query: 558 GCKPNVVTYTALIDGHCKAGQIEKACQIYARMRGDIESSDMDKYFKLDHNNCEGPNVITY 617
G KP ++T AL++G C G++ A + RM +E+ PN +TY
Sbjct: 172 GHKPTLITLNALVNGLCLNGKVSDAVLLIDRM---VETGFQ-------------PNEVTY 215
Query: 618 GALVDGLCKANRVKEAHELLDTMLAHGCEPNQIVYDAVIDGFCKIGKLQDAQEVFTKMSE 677
G ++ +CK+ + A ELL M + + + Y +IDG CK G L +A +F +M
Sbjct: 216 GPVLKVMCKSGQTALAMELLRKMEERKIKLDAVKYSIIIDGLCKDGSLDNAFNLFNEMEI 275
Query: 678 RGYSPNLYTYSSFIDCLFKDNRLDLVLKVLSKMLENSCTPNVVIYTEMVDGLCKIGKTDE 737
+G+ ++ Y++ I R D K+L M++ TP+VV ++ ++D K GK E
Sbjct: 276 KGFKADIIIYTTLIRGFCYAGRWDDGAKLLRDMIKRKITPDVVAFSALIDCFVKEGKLRE 335
Query: 738 AYKLMLKMEEKGCNPNVVTYTAMIDGFGKSGKIEQCLELFRDMCSKGCAPNFITYRVLIN 797
A +L +M ++G +P+ VTYT++IDGF K ++++ + M SKGC PN T+ +LIN
Sbjct: 336 AEELHKEMIQRGISPDTVTYTSLIDGFCKENQLDKANHMLDLMVSKGCGPNIRTFNILIN 395
Query: 798 HCCSNGLLDEAYKLLDEMKQTYWPKHILSHRKIIEGFSQ--EFITSIGLLDELSENESVP 855
C L+D+ +L +M +++ +I+GF + + + L E+ P
Sbjct: 396 GYCKANLIDDGLELFRKMSLRGVVADTVTYNTLIQGFCELGKLEVAKELFQEMVSRRVRP 455
Query: 856 VDSLYRILIDNYIKAGRLEVALDLLEEISSSPSHAVSNKYLYASLIENLSHASKVDKALE 915
Y+IL+D G E AL++ E+I S + +Y +I + +ASKVD A +
Sbjct: 456 DIVSYKILLDGLCDNGEPEKALEIFEKIEKSKMEL--DIGIYNIIIHGMCNASKVDDAWD 513
Query: 916 LYASMISKNVVPELSILVHLIKGLIKVDKWQEA 948
L+ S+ K V P++ +I GL K EA
Sbjct: 514 LFCSLPLKGVKPDVKTYNIMIGGLCKKGSLSEA 546
Score = 216 bits (551), Expect = 4e-56
Identities = 150/554 (27%), Positives = 257/554 (46%), Gaps = 61/554 (11%)
Query: 140 FDKLLDLLGCNVNADDRVPLKFLMEIKDDDHELLRRLLNFLVRKCCRNGWWNMALEELGR 199
F +L ++ D + L ME+K H L L+ ++ CCR ++A +G+
Sbjct: 75 FSRLFSVVARTKQYDLVLDLCKQMELKGIAHNLYT--LSIMINCCCRCRKLSLAFSAMGK 132
Query: 200 LKDFGYKPSQTTYNALIQVFLRADKLDTAYLVKREMLSYAFVMDRYTLSCFAYSLCKGGK 259
+ GY+P T++ LI ++ A + M+ TL+ LC GK
Sbjct: 133 IIKLGYEPDTVTFSTLINGLCLEGRVSEALELVDRMVEMGHKPTLITLNALVNGLCLNGK 192
Query: 260 CREAFDLIDEAED--FVPDTVFYNRMVSGLCEASLFEEAMDILHRMRSSSCIPNVVTYRI 317
+A LID + F P+ V Y ++ +C++ AM++L +M + V Y I
Sbjct: 193 VSDAVLLIDRMVETGFQPNEVTYGPVLKVMCKSGQTALAMELLRKMEERKIKLDAVKYSI 252
Query: 318 LLSGCLRKGQLGRCKRILSMMITEGCYPNREIFNSLIHAYCKSRDYSYAYKLFKKMIKCG 377
++ G + G L + + M +G + I+ +LI +C + + KL + MIK
Sbjct: 253 IIDGLCKDGSLDNAFNLFNEMEIKGFKADIIIYTTLIRGFCYAGRWDDGAKLLRDMIKRK 312
Query: 378 CQPGYLVYNIFIGSVCSNEEQPSSDILDLVEKAYSEMLDLGVVLNKVNVSNFARCLCGAG 437
P D+V A+S ++D C G
Sbjct: 313 ITP------------------------DVV--AFSALID---------------CFVKEG 331
Query: 438 KFDQAFKIICEMMGKGFVPDDSTYSKVIGFLCDASKVEKAFSLFEEMKRNGIVPSVYTYT 497
K +A ++ EM+ +G PD TY+ +I C ++++KA + + M G P++ T+
Sbjct: 332 KLREAEELHKEMIQRGISPDTVTYTSLIDGFCKENQLDKANHMLDLMVSKGCGPNIRTFN 391
Query: 498 ILIDSFCKAGLIQQARKWFDEMLHKGCTPNVVTYTALIHAYLKAKQMPVADELFEMMLLE 557
ILI+ +CKA LI + F +M +G + VTY LI + + ++ VA ELF+ M+
Sbjct: 392 ILINGYCKANLIDDGLELFRKMSLRGVVADTVTYNTLIQGFCELGKLEVAKELFQEMVSR 451
Query: 558 GCKPNVVTYTALIDGHCKAGQIEKACQIYARMRGDIESSDMDKYFKLDHNNCEGPNVITY 617
+P++V+Y L+DG C G+ EKA +I+ + IE S M+ ++ Y
Sbjct: 452 RVRPDIVSYKILLDGLCDNGEPEKALEIFEK----IEKSKMEL------------DIGIY 495
Query: 618 GALVDGLCKANRVKEAHELLDTMLAHGCEPNQIVYDAVIDGFCKIGKLQDAQEVFTKMSE 677
++ G+C A++V +A +L ++ G +P+ Y+ +I G CK G L +A +F KM E
Sbjct: 496 NIIIHGMCNASKVDDAWDLFCSLPLKGVKPDVKTYNIMIGGLCKKGSLSEADLLFRKMEE 555
Query: 678 RGYSPNLYTYSSFI 691
G+SPN TY+ I
Sbjct: 556 DGHSPNGCTYNILI 569
Score = 202 bits (514), Expect = 8e-52
Identities = 134/485 (27%), Positives = 237/485 (48%), Gaps = 20/485 (4%)
Query: 473 KVEKAFSLFEEMKRNGIVPSVYTYTILIDSFCKAGLIQQARKWFDEMLHKGCTPNVVTYT 532
K + A LF+EM R+ P + ++ L + +M KG N+ T +
Sbjct: 52 KEDDAVDLFQEMTRSRPRPRLIDFSRLFSVVARTKQYDLVLDLCKQMELKGIAHNLYTLS 111
Query: 533 ALIHAYLKAKQMPVADELFEMMLLEGCKPNVVTYTALIDGHCKAGQIEKACQIYARMRGD 592
+I+ + +++ +A ++ G +P+ VT++ LI+G C G++ +A ++
Sbjct: 112 IMINCCCRCRKLSLAFSAMGKIIKLGYEPDTVTFSTLINGLCLEGRVSEALEL------- 164
Query: 593 IESSDMDKYFKLDHNNCEGPNVITYGALVDGLCKANRVKEAHELLDTMLAHGCEPNQIVY 652
+D+ ++ H P +IT ALV+GLC +V +A L+D M+ G +PN++ Y
Sbjct: 165 -----VDRMVEMGHK----PTLITLNALVNGLCLNGKVSDAVLLIDRMVETGFQPNEVTY 215
Query: 653 DAVIDGFCKIGKLQDAQEVFTKMSERGYSPNLYTYSSFIDCLFKDNRLDLVLKVLSKMLE 712
V+ CK G+ A E+ KM ER + YS ID L KD LD + ++M
Sbjct: 216 GPVLKVMCKSGQTALAMELLRKMEERKIKLDAVKYSIIIDGLCKDGSLDNAFNLFNEMEI 275
Query: 713 NSCTPNVVIYTEMVDGLCKIGKTDEAYKLMLKMEEKGCNPNVVTYTAMIDGFGKSGKIEQ 772
+++IYT ++ G C G+ D+ KL+ M ++ P+VV ++A+ID F K GK+ +
Sbjct: 276 KGFKADIIIYTTLIRGFCYAGRWDDGAKLLRDMIKRKITPDVVAFSALIDCFVKEGKLRE 335
Query: 773 CLELFRDMCSKGCAPNFITYRVLINHCCSNGLLDEAYKLLDEMKQTYWPKHILSHRKIIE 832
EL ++M +G +P+ +TY LI+ C LD+A +LD M +I + +I
Sbjct: 336 AEELHKEMIQRGISPDTVTYTSLIDGFCKENQLDKANHMLDLMVSKGCGPNIRTFNILIN 395
Query: 833 GFSQEFITSIGL--LDELSENESVPVDSLYRILIDNYIKAGRLEVALDLLEEISSSPSHA 890
G+ + + GL ++S V Y LI + + G+LEVA +L +E+ S
Sbjct: 396 GYCKANLIDDGLELFRKMSLRGVVADTVTYNTLIQGFCELGKLEVAKELFQEMVSR--RV 453
Query: 891 VSNKYLYASLIENLSHASKVDKALELYASMISKNVVPELSILVHLIKGLIKVDKWQEALQ 950
+ Y L++ L + +KALE++ + + ++ I +I G+ K +A
Sbjct: 454 RPDIVSYKILLDGLCDNGEPEKALEIFEKIEKSKMELDIGIYNIIIHGMCNASKVDDAWD 513
Query: 951 LSDSI 955
L S+
Sbjct: 514 LFCSL 518
Score = 184 bits (467), Expect = 2e-46
Identities = 120/463 (25%), Positives = 211/463 (44%), Gaps = 10/463 (2%)
Query: 130 QIGYSHTPQVFDKLLDLLGCNVNADDRVPLKFLMEIKDDDHELLRRLLNFLVRKCCRNGW 189
++GY F L++ L + L+ + + + H+ LN LV C NG
Sbjct: 135 KLGYEPDTVTFSTLINGLCLEGRVSEA--LELVDRMVEMGHKPTLITLNALVNGLCLNGK 192
Query: 190 WNMALEELGRLKDFGYKPSQTTYNALIQVFLRADKLDTAYLVKREMLSYAFVMDRYTLSC 249
+ A+ + R+ + G++P++ TY +++V ++ + A + R+M +D S
Sbjct: 193 VSDAVLLIDRMVETGFQPNEVTYGPVLKVMCKSGQTALAMELLRKMEERKIKLDAVKYSI 252
Query: 250 FAYSLCKGGKCREAFDLIDEAE--DFVPDTVFYNRMVSGLCEASLFEEAMDILHRMRSSS 307
LCK G AF+L +E E F D + Y ++ G C A +++ +L M
Sbjct: 253 IIDGLCKDGSLDNAFNLFNEMEIKGFKADIIIYTTLIRGFCYAGRWDDGAKLLRDMIKRK 312
Query: 308 CIPNVVTYRILLSGCLRKGQLGRCKRILSMMITEGCYPNREIFNSLIHAYCKSRDYSYAY 367
P+VV + L+ +++G+L + + MI G P+ + SLI +CK A
Sbjct: 313 ITPDVVAFSALIDCFVKEGKLREAEELHKEMIQRGISPDTVTYTSLIDGFCKENQLDKAN 372
Query: 368 KLFKKMIKCGCQPGYLVYNIFIGSVCSNEEQPSSDILDLVEKAYSEMLDLGVVLNKVNVS 427
+ M+ GC P +NI I C ++++D + + +M GVV + V +
Sbjct: 373 HMLDLMVSKGCGPNIRTFNILINGYC------KANLIDDGLELFRKMSLRGVVADTVTYN 426
Query: 428 NFARCLCGAGKFDQAFKIICEMMGKGFVPDDSTYSKVIGFLCDASKVEKAFSLFEEMKRN 487
+ C GK + A ++ EM+ + PD +Y ++ LCD + EKA +FE+++++
Sbjct: 427 TLIQGFCELGKLEVAKELFQEMVSRRVRPDIVSYKILLDGLCDNGEPEKALEIFEKIEKS 486
Query: 488 GIVPSVYTYTILIDSFCKAGLIQQARKWFDEMLHKGCTPNVVTYTALIHAYLKAKQMPVA 547
+ + Y I+I C A + A F + KG P+V TY +I K + A
Sbjct: 487 KMELDIGIYNIIIHGMCNASKVDDAWDLFCSLPLKGVKPDVKTYNIMIGGLCKKGSLSEA 546
Query: 548 DELFEMMLLEGCKPNVVTYTALIDGHCKAGQIEKACQIYARMR 590
D LF M +G PN TY LI H G K+ ++ ++
Sbjct: 547 DLLFRKMEEDGHSPNGCTYNILIRAHLGEGDATKSAKLIEEIK 589
Score = 134 bits (338), Expect = 2e-31
Identities = 97/365 (26%), Positives = 171/365 (46%), Gaps = 10/365 (2%)
Query: 592 DIESSDMDKYFKLDHNNCEGPNVITYGALVDGLCKANRVKEAHELLDTMLAHGCEPNQIV 651
DI+ D F+ + P +I + L + + + +L M G N
Sbjct: 50 DIKEDDAVDLFQEMTRSRPRPRLIDFSRLFSVVARTKQYDLVLDLCKQMELKGIAHNLYT 109
Query: 652 YDAVIDGFCKIGKLQDAQEVFTKMSERGYSPNLYTYSSFIDCLFKDNRLDLVLKVLSKML 711
+I+ C+ KL A K+ + GY P+ T+S+ I+ L + R+ L+++ +M+
Sbjct: 110 LSIMINCCCRCRKLSLAFSAMGKIIKLGYEPDTVTFSTLINGLCLEGRVSEALELVDRMV 169
Query: 712 ENSCTPNVVIYTEMVDGLCKIGKTDEAYKLMLKMEEKGCNPNVVTYTAMIDGFGKSGKIE 771
E P ++ +V+GLC GK +A L+ +M E G PN VTY ++ KSG+
Sbjct: 170 EMGHKPTLITLNALVNGLCLNGKVSDAVLLIDRMVETGFQPNEVTYGPVLKVMCKSGQTA 229
Query: 772 QCLELFRDMCSKGCAPNFITYRVLINHCCSNGLLDEAYKLLDEMKQTYWPKHILSHRKII 831
+EL R M + + + Y ++I+ C +G LD A+ L +EM+ + I+ + +I
Sbjct: 230 LAMELLRKMEERKIKLDAVKYSIIIDGLCKDGSLDNAFNLFNEMEIKGFKADIIIYTTLI 289
Query: 832 EGF--SQEFITSIGLLDELSENESVPVDSLYRILIDNYIKAGRLEVALDLLEEI---SSS 886
GF + + LL ++ + + P + LID ++K G+L A +L +E+ S
Sbjct: 290 RGFCYAGRWDDGAKLLRDMIKRKITPDVVAFSALIDCFVKEGKLREAEELHKEMIQRGIS 349
Query: 887 PSHAVSNKYLYASLIENLSHASKVDKALELYASMISKNVVPELSILVHLIKGLIKVDKWQ 946
P + Y SLI+ +++DKA + M+SK P + LI G K +
Sbjct: 350 P-----DTVTYTSLIDGFCKENQLDKANHMLDLMVSKGCGPNIRTFNILINGYCKANLID 404
Query: 947 EALQL 951
+ L+L
Sbjct: 405 DGLEL 409
Score = 88.2 bits (217), Expect = 2e-17
Identities = 57/205 (27%), Positives = 94/205 (45%), Gaps = 2/205 (0%)
Query: 175 RLLNFLVRKCCRNGWWNMALEELGRLKDFGYKPSQTTYNALIQVFLRADKLDTAYLVKRE 234
R N L+ C+ + LE ++ G TYN LIQ F KL+ A + +E
Sbjct: 388 RTFNILINGYCKANLIDDGLELFRKMSLRGVVADTVTYNTLIQGFCELGKLEVAKELFQE 447
Query: 235 MLSYAFVMDRYTLSCFAYSLCKGGKCREAFDLIDEAED--FVPDTVFYNRMVSGLCEASL 292
M+S D + LC G+ +A ++ ++ E D YN ++ G+C AS
Sbjct: 448 MVSRRVRPDIVSYKILLDGLCDNGEPEKALEIFEKIEKSKMELDIGIYNIIIHGMCNASK 507
Query: 293 FEEAMDILHRMRSSSCIPNVVTYRILLSGCLRKGQLGRCKRILSMMITEGCYPNREIFNS 352
++A D+ + P+V TY I++ G +KG L + M +G PN +N
Sbjct: 508 VDDAWDLFCSLPLKGVKPDVKTYNIMIGGLCKKGSLSEADLLFRKMEEDGHSPNGCTYNI 567
Query: 353 LIHAYCKSRDYSYAYKLFKKMIKCG 377
LI A+ D + + KL +++ +CG
Sbjct: 568 LIRAHLGEGDATKSAKLIEEIKRCG 592
Score = 29.6 bits (65), Expect = 8.8
Identities = 22/91 (24%), Positives = 42/91 (45%), Gaps = 2/91 (2%)
Query: 178 NFLVRKCCRNGWWNMALEELGRLKDFGYKPSQTTYNALIQVFL-RADKLDTAYLVKREML 236
N ++ C+ G + A ++++ G+ P+ TYN LI+ L D +A L++ E+
Sbjct: 531 NIMIGGLCKKGSLSEADLLFRKMEEDGHSPNGCTYNILIRAHLGEGDATKSAKLIE-EIK 589
Query: 237 SYAFVMDRYTLSCFAYSLCKGGKCREAFDLI 267
F +D T+ L G + D++
Sbjct: 590 RCGFSVDASTVKMVVDMLSDGRLKKSFLDML 620
>At1g62930 unknown protein
Length = 659
Score = 267 bits (683), Expect = 2e-71
Identities = 166/561 (29%), Positives = 276/561 (48%), Gaps = 59/561 (10%)
Query: 259 KCREAFDLIDEAEDF--VPDTVFYNRMVSGLCEASLFEEAMDILHRMRSSSCIPNVVTYR 316
K +A DL E +P V +N+++S + + + F+ + + RM++ ++ +Y
Sbjct: 60 KLDDAVDLFGEMVQSRPLPSIVEFNKLLSAIAKMNKFDLVISLGERMQNLRISYDLYSYN 119
Query: 317 ILLSGCLRKGQLGRCKRILSMMITEGCYPNREIFNSLIHAYCKSRDYSYAYKLFKKMIKC 376
IL++ R+ QL +L M+ G P+ +SL++ YC + S A L +M
Sbjct: 120 ILINCFCRRSQLPLALAVLGKMMKLGYEPDIVTLSSLLNGYCHGKRISEAVALVDQMFVM 179
Query: 377 GCQPGYLVYNIFIGSVCSNEEQPSSDILDLVEKAYSEMLDLGVVLNKVNVSNFARCLCGA 436
QP + +N I G+ L+
Sbjct: 180 EYQPNTVTFNTLIH---------------------------GLFLHN------------- 199
Query: 437 GKFDQAFKIICEMMGKGFVPDDSTYSKVIGFLCDASKVEKAFSLFEEMKRNGIVPSVYTY 496
K +A +I M+ +G PD TY V+ LC ++ A SL ++M++ I V Y
Sbjct: 200 -KASEAVALIDRMVARGCQPDLFTYGTVVNGLCKRGDIDLALSLLKKMEKGKIEADVVIY 258
Query: 497 TILIDSFCKAGLIQQARKWFDEMLHKGCTPNVVTYTALIHAYLKAKQMPVADELFEMMLL 556
T +ID+ C + A F EM +KG PNVVTY +LI + A L M+
Sbjct: 259 TTIIDALCNYKNVNDALNLFTEMDNKGIRPNVVTYNSLIRCLCNYGRWSDASRLLSDMIE 318
Query: 557 EGCKPNVVTYTALIDGHCKAGQIEKACQIYARMRGDIESSDMDKYFKLDHNNCEGPNVIT 616
PNVVT++ALID K G++ +A ++Y M I+ S +D P++ T
Sbjct: 319 RKINPNVVTFSALIDAFVKEGKLVEAEKLYDEM---IKRS-------ID------PDIFT 362
Query: 617 YGALVDGLCKANRVKEAHELLDTMLAHGCEPNQIVYDAVIDGFCKIGKLQDAQEVFTKMS 676
Y +L++G C +R+ EA + + M++ C PN + Y+ +I GFCK ++++ E+F +MS
Sbjct: 363 YSSLINGFCMHDRLDEAKHMFELMISKDCFPNVVTYNTLIKGFCKAKRVEEGMELFREMS 422
Query: 677 ERGYSPNLYTYSSFIDCLFKDNRLDLVLKVLSKMLENSCTPNVVIYTEMVDGLCKIGKTD 736
+RG N TY++ I LF+ D+ K+ KM+ + P+++ Y+ ++DGLCK GK +
Sbjct: 423 QRGLVGNTVTYNTLIQGLFQAGDCDMAQKIFKKMVSDGVPPDIITYSILLDGLCKYGKLE 482
Query: 737 EAYKLMLKMEEKGCNPNVVTYTAMIDGFGKSGKIEQCLELFRDMCSKGCAPNFITYRVLI 796
+A + +++ P++ TY MI+G K+GK+E +LF + KG PN I Y +I
Sbjct: 483 KALVVFEYLQKSKMEPDIYTYNIMIEGMCKAGKVEDGWDLFCSLSLKGVKPNVIIYTTMI 542
Query: 797 NHCCSNGLLDEAYKLLDEMKQ 817
+ C GL +EA L EMK+
Sbjct: 543 SGFCRKGLKEEADALFREMKE 563
Score = 246 bits (628), Expect = 5e-65
Identities = 159/605 (26%), Positives = 277/605 (45%), Gaps = 59/605 (9%)
Query: 193 ALEELGRLKDFGYKPSQTTYNALIQVFLRADKLDTAYLVKREMLSYAFVMDRYTLSCFAY 252
A++ G + PS +N L+ + +K D + M + D Y+ +
Sbjct: 64 AVDLFGEMVQSRPLPSIVEFNKLLSAIAKMNKFDLVISLGERMQNLRISYDLYSYNILIN 123
Query: 253 SLCKGGKCREAFDLIDEAED--FVPDTVFYNRMVSGLCEASLFEEAMDILHRMRSSSCIP 310
C+ + A ++ + + PD V + +++G C EA+ ++ +M P
Sbjct: 124 CFCRRSQLPLALAVLGKMMKLGYEPDIVTLSSLLNGYCHGKRISEAVALVDQMFVMEYQP 183
Query: 311 NVVTYRILLSGCLRKGQLGRCKRILSMMITEGCYPNREIFNSLIHAYCKSRDYSYAYKLF 370
N VT+ L+ G + ++ M+ GC P+ + ++++ CK D A L
Sbjct: 184 NTVTFNTLIHGLFLHNKASEAVALIDRMVARGCQPDLFTYGTVVNGLCKRGDIDLALSLL 243
Query: 371 KKMIKCGCQPGYLVYNIFIGSVCSNEEQPSSDILDLVEKAYSEMLDLGVVLNKVNVSNFA 430
KKM K + ++Y I ++C+ + +D L+L ++EM + G+ N V ++
Sbjct: 244 KKMEKGKIEADVVIYTTIIDALCNYKN--VNDALNL----FTEMDNKGIRPNVVTYNSLI 297
Query: 431 RCLCGAGKFDQAFKIICEMMGKGFVPDDSTYSKVIGFLCDASKVEKAFSLFEEMKRNGIV 490
RCLC G++ A +++ +M+ + P+ T+S +I K+ +A L++EM + I
Sbjct: 298 RCLCNYGRWSDASRLLSDMIERKINPNVVTFSALIDAFVKEGKLVEAEKLYDEMIKRSID 357
Query: 491 PSVYTYTILIDSFCKAGLIQQARKWFDEMLHKGCTPNVVTYTALIHAYLKAKQMPVADEL 550
P ++TY+ LI+ FC + +A+ F+ M+ K C PNVVTY LI + KAK++ EL
Sbjct: 358 PDIFTYSSLINGFCMHDRLDEAKHMFELMISKDCFPNVVTYNTLIKGFCKAKRVEEGMEL 417
Query: 551 FEMMLLEGCKPNVVTYTALIDGHCKAGQIEKACQIYARMRGDIESSDMDKYFKLDHNNCE 610
F M G N VTY LI G +AG + A +I+ +M D
Sbjct: 418 FREMSQRGLVGNTVTYNTLIQGLFQAGDCDMAQKIFKKMVSD----------------GV 461
Query: 611 GPNVITYGALVDGLCKANRVKEAHELLDTMLAHGCEPNQIVYDAVIDGFCKIGKLQDAQE 670
P++ITY L+DGLCK ++++A + + + EP+ Y+ +I+G CK GK++D +
Sbjct: 462 PPDIITYSILLDGLCKYGKLEKALVVFEYLQKSKMEPDIYTYNIMIEGMCKAGKVEDGWD 521
Query: 671 VFTKMSERGYSPNLYTYSSFIDCLFKDNRLDLVLKVLSKMLENSCTPNVVIYTEMVDGLC 730
+F +S +G PN+ Y++ I G C
Sbjct: 522 LFCSLSLKGVKPNVIIYTTMI-----------------------------------SGFC 546
Query: 731 KIGKTDEAYKLMLKMEEKGCNPNVVTYTAMIDGFGKSGKIEQCLELFRDMCSKGCAPNFI 790
+ G +EA L +M+E G PN TY +I + G EL ++M S G +
Sbjct: 547 RKGLKEEADALFREMKEDGTLPNSGTYNTLIRARLRDGDKAASAELIKEMRSCGFVGDAS 606
Query: 791 TYRVL 795
T +L
Sbjct: 607 TISML 611
Score = 240 bits (612), Expect = 3e-63
Identities = 157/540 (29%), Positives = 263/540 (48%), Gaps = 20/540 (3%)
Query: 438 KFDQAFKIICEMMGKGFVPDDSTYSKVIGFLCDASKVEKAFSLFEEMKRNGIVPSVYTYT 497
K D A + EM+ +P ++K++ + +K + SL E M+ I +Y+Y
Sbjct: 60 KLDDAVDLFGEMVQSRPLPSIVEFNKLLSAIAKMNKFDLVISLGERMQNLRISYDLYSYN 119
Query: 498 ILIDSFCKAGLIQQARKWFDEMLHKGCTPNVVTYTALIHAYLKAKQMPVADELFEMMLLE 557
ILI+ FC+ + A +M+ G P++VT ++L++ Y K++ A L + M +
Sbjct: 120 ILINCFCRRSQLPLALAVLGKMMKLGYEPDIVTLSSLLNGYCHGKRISEAVALVDQMFVM 179
Query: 558 GCKPNVVTYTALIDGHCKAGQIEKACQIYARMRGDIESSDMDKYFKLDHNNCEGPNVITY 617
+PN VT+ LI G + +A + RM C+ P++ TY
Sbjct: 180 EYQPNTVTFNTLIHGLFLHNKASEAVALIDRMVA---------------RGCQ-PDLFTY 223
Query: 618 GALVDGLCKANRVKEAHELLDTMLAHGCEPNQIVYDAVIDGFCKIGKLQDAQEVFTKMSE 677
G +V+GLCK + A LL M E + ++Y +ID C + DA +FT+M
Sbjct: 224 GTVVNGLCKRGDIDLALSLLKKMEKGKIEADVVIYTTIIDALCNYKNVNDALNLFTEMDN 283
Query: 678 RGYSPNLYTYSSFIDCLFKDNRLDLVLKVLSKMLENSCTPNVVIYTEMVDGLCKIGKTDE 737
+G PN+ TY+S I CL R ++LS M+E PNVV ++ ++D K GK E
Sbjct: 284 KGIRPNVVTYNSLIRCLCNYGRWSDASRLLSDMIERKINPNVVTFSALIDAFVKEGKLVE 343
Query: 738 AYKLMLKMEEKGCNPNVVTYTAMIDGFGKSGKIEQCLELFRDMCSKGCAPNFITYRVLIN 797
A KL +M ++ +P++ TY+++I+GF ++++ +F M SK C PN +TY LI
Sbjct: 344 AEKLYDEMIKRSIDPDIFTYSSLINGFCMHDRLDEAKHMFELMISKDCFPNVVTYNTLIK 403
Query: 798 HCCSNGLLDEAYKLLDEMKQTYWPKHILSHRKIIEGFSQEFITSIG-LLDELSENESVPV 856
C ++E +L EM Q + +++ +I+G Q + + + ++ VP
Sbjct: 404 GFCKAKRVEEGMELFREMSQRGLVGNTVTYNTLIQGLFQAGDCDMAQKIFKKMVSDGVPP 463
Query: 857 DSL-YRILIDNYIKAGRLEVALDLLEEISSSPSHAVSNKYLYASLIENLSHASKVDKALE 915
D + Y IL+D K G+LE AL + E + S + Y Y +IE + A KV+ +
Sbjct: 464 DIITYSILLDGLCKYGKLEKALVVFEYLQKSKME--PDIYTYNIMIEGMCKAGKVEDGWD 521
Query: 916 LYASMISKNVVPELSILVHLIKGLIKVDKWQEALQLSDSICQMVCLTLSQTFQPLVNLNL 975
L+ S+ K V P + I +I G + +EA L + + L S T+ L+ L
Sbjct: 522 LFCSLSLKGVKPNVIIYTTMISGFCRKGLKEEADALFREMKEDGTLPNSGTYNTLIRARL 581
Score = 232 bits (591), Expect = 9e-61
Identities = 144/522 (27%), Positives = 248/522 (46%), Gaps = 59/522 (11%)
Query: 178 NFLVRKCCRNGWWNMALEELGRLKDFGYKPSQTTYNALIQVFLRADKLDTAYLVKREMLS 237
N L+ CR +AL LG++ GY+P
Sbjct: 119 NILINCFCRRSQLPLALAVLGKMMKLGYEP------------------------------ 148
Query: 238 YAFVMDRYTLSCFAYSLCKGGKCREAFDLIDE--AEDFVPDTVFYNRMVSGLCEASLFEE 295
D TLS C G + EA L+D+ ++ P+TV +N ++ GL + E
Sbjct: 149 -----DIVTLSSLLNGYCHGKRISEAVALVDQMFVMEYQPNTVTFNTLIHGLFLHNKASE 203
Query: 296 AMDILHRMRSSSCIPNVVTYRILLSGCLRKGQLGRCKRILSMMITEGCYPNREIFNSLIH 355
A+ ++ RM + C P++ TY +++G ++G + +L M + I+ ++I
Sbjct: 204 AVALIDRMVARGCQPDLFTYGTVVNGLCKRGDIDLALSLLKKMEKGKIEADVVIYTTIID 263
Query: 356 AYCKSRDYSYAYKLFKKMIKCGCQPGYLVYNIFIGSVCSNEEQPSSDILDLVEKAYSEML 415
A C ++ + A LF +M G +P + YN I +C+ + + S+M+
Sbjct: 264 ALCNYKNVNDALNLFTEMDNKGIRPNVVTYNSLIRCLCNYGRWSDA------SRLLSDMI 317
Query: 416 DLGVVLNKVNVSNFARCLCGAGKFDQAFKIICEMMGKGFVPDDSTYSKVIGFLCDASKVE 475
+ + N V S GK +A K+ EM+ + PD TYS +I C +++
Sbjct: 318 ERKINPNVVTFSALIDAFVKEGKLVEAEKLYDEMIKRSIDPDIFTYSSLINGFCMHDRLD 377
Query: 476 KAFSLFEEMKRNGIVPSVYTYTILIDSFCKAGLIQQARKWFDEMLHKGCTPNVVTYTALI 535
+A +FE M P+V TY LI FCKA +++ + F EM +G N VTY LI
Sbjct: 378 EAKHMFELMISKDCFPNVVTYNTLIKGFCKAKRVEEGMELFREMSQRGLVGNTVTYNTLI 437
Query: 536 HAYLKAKQMPVADELFEMMLLEGCKPNVVTYTALIDGHCKAGQIEKACQIYARMRGDIES 595
+A +A ++F+ M+ +G P+++TY+ L+DG CK G++EKA ++ ++
Sbjct: 438 QGLFQAGDCDMAQKIFKKMVSDGVPPDIITYSILLDGLCKYGKLEKALVVFEY----LQK 493
Query: 596 SDMDKYFKLDHNNCEGPNVITYGALVDGLCKANRVKEAHELLDTMLAHGCEPNQIVYDAV 655
S M+ P++ TY +++G+CKA +V++ +L ++ G +PN I+Y +
Sbjct: 494 SKME------------PDIYTYNIMIEGMCKAGKVEDGWDLFCSLSLKGVKPNVIIYTTM 541
Query: 656 IDGFCKIGKLQDAQEVFTKMSERGYSPNLYTYSSFIDCLFKD 697
I GFC+ G ++A +F +M E G PN TY++ I +D
Sbjct: 542 ISGFCRKGLKEEADALFREMKEDGTLPNSGTYNTLIRARLRD 583
Score = 223 bits (568), Expect = 4e-58
Identities = 154/564 (27%), Positives = 257/564 (45%), Gaps = 37/564 (6%)
Query: 404 LDLVEKAYSEMLDLGVVLNKVNVSNFARCLCGAGKFDQAFKIICEMMGKGFVPDDSTYSK 463
LD + EM+ + + V + + KFD + M D +Y+
Sbjct: 61 LDDAVDLFGEMVQSRPLPSIVEFNKLLSAIAKMNKFDLVISLGERMQNLRISYDLYSYNI 120
Query: 464 VIGFLCDASKVEKAFSLFEEMKRNGIVPSVYTYTILIDSFCKAGLIQQARKWFDEMLHKG 523
+I C S++ A ++ +M + G P + T + L++ +C I +A D+M
Sbjct: 121 LINCFCRRSQLPLALAVLGKMMKLGYEPDIVTLSSLLNGYCHGKRISEAVALVDQMFVME 180
Query: 524 CTPNVVTYTALIHAYLKAKQMPVADELFEMMLLEGCKPNVVTYTALIDGHCKAGQIEKAC 583
PN VT+ LIH + A L + M+ GC+P++ TY +++G CK G I+ A
Sbjct: 181 YQPNTVTFNTLIHGLFLHNKASEAVALIDRMVARGCQPDLFTYGTVVNGLCKRGDIDLAL 240
Query: 584 QIYARM-RGDIES------------------SDMDKYFKLDHNNCEGPNVITYGALVDGL 624
+ +M +G IE+ +D F N PNV+TY +L+ L
Sbjct: 241 SLLKKMEKGKIEADVVIYTTIIDALCNYKNVNDALNLFTEMDNKGIRPNVVTYNSLIRCL 300
Query: 625 CKANRVKEAHELLDTMLAHGCEPNQIVYDAVIDGFCKIGKLQDAQEVFTKMSERGYSPNL 684
C R +A LL M+ PN + + A+ID F K GKL +A++++ +M +R P++
Sbjct: 301 CNYGRWSDASRLLSDMIERKINPNVVTFSALIDAFVKEGKLVEAEKLYDEMIKRSIDPDI 360
Query: 685 YTYSSFIDCLFKDNRLDLVLKVLSKMLENSCTPNVVIYTEMVDGLCKIGKTDEAYKLMLK 744
+TYSS I+ +RLD + M+ C PNVV Y ++ G CK + +E +L +
Sbjct: 361 FTYSSLINGFCMHDRLDEAKHMFELMISKDCFPNVVTYNTLIKGFCKAKRVEEGMELFRE 420
Query: 745 MEEKGCNPNVVTYTAMIDGFGKSGKIEQCLELFRDMCSKGCAPNFITYRVLINHCCSNGL 804
M ++G N VTY +I G ++G + ++F+ M S G P+ ITY +L++ C G
Sbjct: 421 MSQRGLVGNTVTYNTLIQGLFQAGDCDMAQKIFKKMVSDGVPPDIITYSILLDGLCKYGK 480
Query: 805 LDEAYKLLDEMKQTYWPKHILSHRKIIEGFSQEFITSIG--LLDELSENESVPVDSLYRI 862
L++A + + ++++ I ++ +IEG + G L LS P +Y
Sbjct: 481 LEKALVVFEYLQKSKMEPDIYTYNIMIEGMCKAGKVEDGWDLFCSLSLKGVKPNVIIYTT 540
Query: 863 LIDNYIKAGRLEVALDLLEEISSS---PSHAVSN-----------KYLYASLIENLSHAS 908
+I + + G E A L E+ P+ N K A LI+ +
Sbjct: 541 MISGFCRKGLKEEADALFREMKEDGTLPNSGTYNTLIRARLRDGDKAASAELIKEMRSCG 600
Query: 909 KVDKALELYASMISKNVVPELSIL 932
V A + SM+S N V + SIL
Sbjct: 601 FVGDASTI--SMLSGNAVLKQSIL 622
Score = 96.3 bits (238), Expect = 8e-20
Identities = 66/247 (26%), Positives = 108/247 (43%), Gaps = 10/247 (4%)
Query: 178 NFLVRKCCRNGWWNMALEELGRLKDFGYKPSQTTYNALIQVFLRADKLDTAYLVKREMLS 237
N L++ C+ +E + G + TYN LIQ +A D A + ++M+S
Sbjct: 399 NTLIKGFCKAKRVEEGMELFREMSQRGLVGNTVTYNTLIQGLFQAGDCDMAQKIFKKMVS 458
Query: 238 YAFVMDRYTLSCFAYSLCKGGKCREAFDLID--EAEDFVPDTVFYNRMVSGLCEASLFEE 295
D T S LCK GK +A + + + PD YN M+ G+C+A E+
Sbjct: 459 DGVPPDIITYSILLDGLCKYGKLEKALVVFEYLQKSKMEPDIYTYNIMIEGMCKAGKVED 518
Query: 296 AMDILHRMRSSSCIPNVVTYRILLSGCLRKGQLGRCKRILSMMITEGCYPNREIFNSLIH 355
D+ + PNV+ Y ++SG RKG + M +G PN +N+LI
Sbjct: 519 GWDLFCSLSLKGVKPNVIIYTTMISGFCRKGLKEEADALFREMKEDGTLPNSGTYNTLIR 578
Query: 356 AYCKSRDYSYAYKLFKKMIKCGCQPGYLVYNIFIGSVCSNE--------EQPSSDILDLV 407
A + D + + +L K+M CG ++ G+ + EQPSS ++ V
Sbjct: 579 ARLRDGDKAASAELIKEMRSCGFVGDASTISMLSGNAVLKQSILEKRIGEQPSSLVIAYV 638
Query: 408 EKAYSEM 414
++ +M
Sbjct: 639 SVSFVDM 645
Score = 89.7 bits (221), Expect = 7e-18
Identities = 62/285 (21%), Positives = 133/285 (45%), Gaps = 15/285 (5%)
Query: 697 DNRLDLVLKVLSKMLENSCTPNVVIYTEMVDGLCKIGKTDEAYKLMLKMEEKGCNPNVVT 756
D +LD + + +M+++ P++V + +++ + K+ K D L +M+ + ++ +
Sbjct: 58 DLKLDDAVDLFGEMVQSRPLPSIVEFNKLLSAIAKMNKFDLVISLGERMQNLRISYDLYS 117
Query: 757 YTAMIDGFGKSGKIEQCLELFRDMCSKGCAPNFITYRVLINHCCSNGLLDEAYKLLDEMK 816
Y +I+ F + ++ L + M G P+ +T L+N C + EA L+D+M
Sbjct: 118 YNILINCFCRRSQLPLALAVLGKMMKLGYEPDIVTLSSLLNGYCHGKRISEAVALVDQMF 177
Query: 817 QTYWPKHILSHRKIIEG--FSQEFITSIGLLDELSENESVPVDSLYRILIDNYIKAGRLE 874
+ + ++ +I G + ++ L+D + P Y +++ K G ++
Sbjct: 178 VMEYQPNTVTFNTLIHGLFLHNKASEAVALIDRMVARGCQPDLFTYGTVVNGLCKRGDID 237
Query: 875 VALDLLEEISSSPSHAVSNKYLYASLIENLSHASKVDKALELYASMISKNVVPELSILVH 934
+AL LL+++ A + +Y ++I+ L + V+ AL L+ M +K + P +
Sbjct: 238 LALSLLKKMEKGKIEA--DVVIYTTIIDALCNYKNVNDALNLFTEMDNKGIRPNVVTYNS 295
Query: 935 LIKGLIKVDKWQEALQLSDSICQMVCLTLSQTFQPLVNLNLLTLS 979
LI+ L +W +A +L LS + +N N++T S
Sbjct: 296 LIRCLCNYGRWSDASRL-----------LSDMIERKINPNVVTFS 329
>At1g63130 unknown protein
Length = 630
Score = 266 bits (681), Expect = 3e-71
Identities = 162/545 (29%), Positives = 273/545 (49%), Gaps = 28/545 (5%)
Query: 275 PDTVFYNRMVSGLCEASLFEEAMDILHRMRSSSCIPNVVTYRILLSGCLRKGQLGRCKRI 334
P V +++++S + + + F+ + + +M++ N+ TY IL++ R+ QL +
Sbjct: 79 PSIVEFSKLLSAIAKMNKFDLVISLGEQMQNLGISHNLYTYSILINCFCRRSQLSLALAV 138
Query: 335 LSMMITEGCYPNREIFNSLIHAYCKSRDYSYAYKLFKKMIKCGCQPGYLVYNIFI-GSVC 393
L+ M+ G P+ NSL++ +C S A L +M++ G QP +N I G
Sbjct: 139 LAKMMKLGYEPDIVTLNSLLNGFCHGNRISDAVSLVGQMVEMGYQPDSFTFNTLIHGLFR 198
Query: 394 SNEEQPSSDILD--LVEKAYSEMLDLGVVLNKVNVSNFARCLCGAGKFDQAFKIICEMMG 451
N + ++D +V+ +++ G+V+N LC G D A ++ +M
Sbjct: 199 HNRASEAVALVDRMVVKGCQPDLVTYGIVVNG---------LCKRGDIDLALSLLKKMEQ 249
Query: 452 KGFVPDDSTYSKVIGFLCDASKVEKAFSLFEEMKRNGIVPSVYTYTILIDSFCKAGLIQQ 511
P Y+ +I LC+ V A +LF EM GI P+V TY LI C G
Sbjct: 250 GKIEPGVVIYNTIIDALCNYKNVNDALNLFTEMDNKGIRPNVVTYNSLIRCLCNYGRWSD 309
Query: 512 ARKWFDEMLHKGCTPNVVTYTALIHAYLKAKQMPVADELFEMMLLEGCKPNVVTYTALID 571
A + +M+ + PNVVT++ALI A++K ++ A++L++ M+ P++ TY++LI+
Sbjct: 310 ASRLLSDMIERKINPNVVTFSALIDAFVKEGKLVEAEKLYDEMIKRSIDPDIFTYSSLIN 369
Query: 572 GHCKAGQIEKACQIYARMRGDIESSDMDKYFKLDHNNCEGPNVITYGALVDGLCKANRVK 631
G C ++++A ++ M +C PNV+TY L+ G CKA RV
Sbjct: 370 GFCMHDRLDEAKHMFELMIS---------------KDCF-PNVVTYNTLIKGFCKAKRVD 413
Query: 632 EAHELLDTMLAHGCEPNQIVYDAVIDGFCKIGKLQDAQEVFTKMSERGYSPNLYTYSSFI 691
E EL M G N + Y +I GF + + +AQ VF +M G P++ TYS +
Sbjct: 414 EGMELFREMSQRGLVGNTVTYTTLIHGFFQARECDNAQIVFKQMVSDGVLPDIMTYSILL 473
Query: 692 DCLFKDNRLDLVLKVLSKMLENSCTPNVVIYTEMVDGLCKIGKTDEAYKLMLKMEEKGCN 751
D L + +++ L V + + P++ Y M++G+CK GK ++ + L + KG
Sbjct: 474 DGLCNNGKVETALVVFEYLQRSKMEPDIYTYNIMIEGMCKAGKVEDGWDLFCSLSLKGVK 533
Query: 752 PNVVTYTAMIDGFGKSGKIEQCLELFRDMCSKGCAPNFITYRVLINHCCSNGLLDEAYKL 811
PNVVTYT M+ GF + G E+ LFR+M +G P+ TY LI +G + +L
Sbjct: 534 PNVVTYTTMMSGFCRKGLKEEADALFREMKEEGPLPDSGTYNTLIRAHLRDGDKAASAEL 593
Query: 812 LDEMK 816
+ EM+
Sbjct: 594 IREMR 598
Score = 259 bits (663), Expect = 4e-69
Identities = 155/524 (29%), Positives = 260/524 (49%), Gaps = 22/524 (4%)
Query: 294 EEAMDILHRMRSSSCIPNVVTYRILLSGCLRKGQLGRCKRILSMMITEGCYPNREIFNSL 353
++A+++ M S P++V + LLS + + + M G N ++ L
Sbjct: 63 DDAVNLFGDMVKSRPFPSIVEFSKLLSAIAKMNKFDLVISLGEQMQNLGISHNLYTYSIL 122
Query: 354 IHAYCKSRDYSYAYKLFKKMIKCGCQPGYLVYNIFIGSVCSNEEQPSSDILDLVEKAYSE 413
I+ +C+ S A + KM+K G +P + N + C SD + LV +
Sbjct: 123 INCFCRRSQLSLALAVLAKMMKLGYEPDIVTLNSLLNGFCHGNR--ISDAVSLV----GQ 176
Query: 414 MLDLGVVLNKVNVSNFARCLCGAGKFDQAFKIICEMMGKGFVPDDSTYSKVIGFLCDASK 473
M+++G + + L + +A ++ M+ KG PD TY V+ LC
Sbjct: 177 MVEMGYQPDSFTFNTLIHGLFRHNRASEAVALVDRMVVKGCQPDLVTYGIVVNGLCKRGD 236
Query: 474 VEKAFSLFEEMKRNGIVPSVYTYTILIDSFCKAGLIQQARKWFDEMLHKGCTPNVVTYTA 533
++ A SL ++M++ I P V Y +ID+ C + A F EM +KG PNVVTY +
Sbjct: 237 IDLALSLLKKMEQGKIEPGVVIYNTIIDALCNYKNVNDALNLFTEMDNKGIRPNVVTYNS 296
Query: 534 LIHAYLKAKQMPVADELFEMMLLEGCKPNVVTYTALIDGHCKAGQIEKACQIYARMRGDI 593
LI + A L M+ PNVVT++ALID K G++ +A ++Y M I
Sbjct: 297 LIRCLCNYGRWSDASRLLSDMIERKINPNVVTFSALIDAFVKEGKLVEAEKLYDEM---I 353
Query: 594 ESSDMDKYFKLDHNNCEGPNVITYGALVDGLCKANRVKEAHELLDTMLAHGCEPNQIVYD 653
+ S +D P++ TY +L++G C +R+ EA + + M++ C PN + Y+
Sbjct: 354 KRS-------ID------PDIFTYSSLINGFCMHDRLDEAKHMFELMISKDCFPNVVTYN 400
Query: 654 AVIDGFCKIGKLQDAQEVFTKMSERGYSPNLYTYSSFIDCLFKDNRLDLVLKVLSKMLEN 713
+I GFCK ++ + E+F +MS+RG N TY++ I F+ D V +M+ +
Sbjct: 401 TLIKGFCKAKRVDEGMELFREMSQRGLVGNTVTYTTLIHGFFQARECDNAQIVFKQMVSD 460
Query: 714 SCTPNVVIYTEMVDGLCKIGKTDEAYKLMLKMEEKGCNPNVVTYTAMIDGFGKSGKIEQC 773
P+++ Y+ ++DGLC GK + A + ++ P++ TY MI+G K+GK+E
Sbjct: 461 GVLPDIMTYSILLDGLCNNGKVETALVVFEYLQRSKMEPDIYTYNIMIEGMCKAGKVEDG 520
Query: 774 LELFRDMCSKGCAPNFITYRVLINHCCSNGLLDEAYKLLDEMKQ 817
+LF + KG PN +TY +++ C GL +EA L EMK+
Sbjct: 521 WDLFCSLSLKGVKPNVVTYTTMMSGFCRKGLKEEADALFREMKE 564
Score = 250 bits (639), Expect = 2e-66
Identities = 160/605 (26%), Positives = 284/605 (46%), Gaps = 60/605 (9%)
Query: 207 PSQTTYNALIQVFLRADKLDTAYLVKREMLSYAFVMDRYTLSCFAYSLCKGGKCREAFDL 266
PS ++ L+ + +K D + +M + + YT S C+ + A +
Sbjct: 79 PSIVEFSKLLSAIAKMNKFDLVISLGEQMQNLGISHNLYTYSILINCFCRRSQLSLALAV 138
Query: 267 IDEAED--FVPDTVFYNRMVSGLCEASLFEEAMDILHRMRSSSCIPNVVTYRILLSGCLR 324
+ + + PD V N +++G C + +A+ ++ +M P+ T+ L+ G R
Sbjct: 139 LAKMMKLGYEPDIVTLNSLLNGFCHGNRISDAVSLVGQMVEMGYQPDSFTFNTLIHGLFR 198
Query: 325 KGQLGRCKRILSMMITEGCYPNREIFNSLIHAYCKSRDYSYAYKLFKKMIKCGCQPGYLV 384
+ ++ M+ +GC P+ + +++ CK D A L KKM + +PG ++
Sbjct: 199 HNRASEAVALVDRMVVKGCQPDLVTYGIVVNGLCKRGDIDLALSLLKKMEQGKIEPGVVI 258
Query: 385 YNIFIGSVCSNEEQPSSDILDLVEKAYSEMLDLGVVLNKVNVSNFARCLCGAGKFDQAFK 444
YN I ++C+ + +D L+L ++EM + G+ N V ++ RCLC G++ A +
Sbjct: 259 YNTIIDALCNYKN--VNDALNL----FTEMDNKGIRPNVVTYNSLIRCLCNYGRWSDASR 312
Query: 445 IICEMMGKGFVPDDSTYSKVIGFLCDASKVEKAFSLFEEMKRNGIVPSVYTYTILIDSFC 504
++ +M+ + P+ T+S +I K+ +A L++EM + I P ++TY+ LI+ FC
Sbjct: 313 LLSDMIERKINPNVVTFSALIDAFVKEGKLVEAEKLYDEMIKRSIDPDIFTYSSLINGFC 372
Query: 505 KAGLIQQARKWFDEMLHKGCTPNVVTYTALIHAYLKAKQMPVADELFEMMLLEGCKPNVV 564
+ +A+ F+ M+ K C PNVVTY LI + KAK++ ELF M G N V
Sbjct: 373 MHDRLDEAKHMFELMISKDCFPNVVTYNTLIKGFCKAKRVDEGMELFREMSQRGLVGNTV 432
Query: 565 TYTALIDGHCKAGQIEKACQIYARMRGDIESSDMDKYFKLDHNNCEGPNVITYGALVDGL 624
TYT LI G +A + + A ++ +M D P+++TY L+DGL
Sbjct: 433 TYTTLIHGFFQARECDNAQIVFKQMVSDGVL----------------PDIMTYSILLDGL 476
Query: 625 CKANRVKEAHELLDTMLAHGCEPNQIVYDAVIDGFCKIGKLQDAQEVFTKMSERGYSPNL 684
C +V+ A + + + EP+ Y+ +I+G CK GK++D ++F +S +G PN+
Sbjct: 477 CNNGKVETALVVFEYLQRSKMEPDIYTYNIMIEGMCKAGKVEDGWDLFCSLSLKGVKPNV 536
Query: 685 YTYSSFIDCLFKDNRLDLVLKVLSKMLENSCTPNVVIYTEMVDGLCKIGKTDEAYKLMLK 744
TY T M+ G C+ G +EA L +
Sbjct: 537 VTY-----------------------------------TTMMSGFCRKGLKEEADALFRE 561
Query: 745 MEEKGCNPNVVTYTAMIDGFGKSGKIEQCLELFRDMCSKGCAPNFITYRVLINHCCSNGL 804
M+E+G P+ TY +I + G EL R+M S + T L+ + +G
Sbjct: 562 MKEEGPLPDSGTYNTLIRAHLRDGDKAASAELIREMRSCRFVGDASTIG-LVTNMLHDGR 620
Query: 805 LDEAY 809
LD+++
Sbjct: 621 LDKSF 625
Score = 239 bits (610), Expect = 6e-63
Identities = 154/540 (28%), Positives = 264/540 (48%), Gaps = 20/540 (3%)
Query: 438 KFDQAFKIICEMMGKGFVPDDSTYSKVIGFLCDASKVEKAFSLFEEMKRNGIVPSVYTYT 497
K D A + +M+ P +SK++ + +K + SL E+M+ GI ++YTY+
Sbjct: 61 KLDDAVNLFGDMVKSRPFPSIVEFSKLLSAIAKMNKFDLVISLGEQMQNLGISHNLYTYS 120
Query: 498 ILIDSFCKAGLIQQARKWFDEMLHKGCTPNVVTYTALIHAYLKAKQMPVADELFEMMLLE 557
ILI+ FC+ + A +M+ G P++VT +L++ + ++ A L M+
Sbjct: 121 ILINCFCRRSQLSLALAVLAKMMKLGYEPDIVTLNSLLNGFCHGNRISDAVSLVGQMVEM 180
Query: 558 GCKPNVVTYTALIDGHCKAGQIEKACQIYARMRGDIESSDMDKYFKLDHNNCEGPNVITY 617
G +P+ T+ LI G + + +A + RM C+ P+++TY
Sbjct: 181 GYQPDSFTFNTLIHGLFRHNRASEAVALVDRMVV---------------KGCQ-PDLVTY 224
Query: 618 GALVDGLCKANRVKEAHELLDTMLAHGCEPNQIVYDAVIDGFCKIGKLQDAQEVFTKMSE 677
G +V+GLCK + A LL M EP ++Y+ +ID C + DA +FT+M
Sbjct: 225 GIVVNGLCKRGDIDLALSLLKKMEQGKIEPGVVIYNTIIDALCNYKNVNDALNLFTEMDN 284
Query: 678 RGYSPNLYTYSSFIDCLFKDNRLDLVLKVLSKMLENSCTPNVVIYTEMVDGLCKIGKTDE 737
+G PN+ TY+S I CL R ++LS M+E PNVV ++ ++D K GK E
Sbjct: 285 KGIRPNVVTYNSLIRCLCNYGRWSDASRLLSDMIERKINPNVVTFSALIDAFVKEGKLVE 344
Query: 738 AYKLMLKMEEKGCNPNVVTYTAMIDGFGKSGKIEQCLELFRDMCSKGCAPNFITYRVLIN 797
A KL +M ++ +P++ TY+++I+GF ++++ +F M SK C PN +TY LI
Sbjct: 345 AEKLYDEMIKRSIDPDIFTYSSLINGFCMHDRLDEAKHMFELMISKDCFPNVVTYNTLIK 404
Query: 798 HCCSNGLLDEAYKLLDEMKQTYWPKHILSHRKIIEGFSQ--EFITSIGLLDELSENESVP 855
C +DE +L EM Q + +++ +I GF Q E + + ++ + +P
Sbjct: 405 GFCKAKRVDEGMELFREMSQRGLVGNTVTYTTLIHGFFQARECDNAQIVFKQMVSDGVLP 464
Query: 856 VDSLYRILIDNYIKAGRLEVALDLLEEISSSPSHAVSNKYLYASLIENLSHASKVDKALE 915
Y IL+D G++E AL + E + S + Y Y +IE + A KV+ +
Sbjct: 465 DIMTYSILLDGLCNNGKVETALVVFEYLQRSKME--PDIYTYNIMIEGMCKAGKVEDGWD 522
Query: 916 LYASMISKNVVPELSILVHLIKGLIKVDKWQEALQLSDSICQMVCLTLSQTFQPLVNLNL 975
L+ S+ K V P + ++ G + +EA L + + L S T+ L+ +L
Sbjct: 523 LFCSLSLKGVKPNVVTYTTMMSGFCRKGLKEEADALFREMKEEGPLPDSGTYNTLIRAHL 582
Score = 228 bits (581), Expect = 1e-59
Identities = 148/560 (26%), Positives = 257/560 (45%), Gaps = 61/560 (10%)
Query: 140 FDKLLDLLGCNVNADDRVPLKFLMEIKDDDHELLRRLLNFLVRKCCRNGWWNMALEELGR 199
F KLL + D + L M+ H L + L+ CR ++AL L +
Sbjct: 84 FSKLLSAIAKMNKFDLVISLGEQMQNLGISHNLYT--YSILINCFCRRSQLSLALAVLAK 141
Query: 200 LKDFGYKPSQTTYNALIQVFLRADKLDTAYLVKREMLSYAFVMDRYTLSCFAYSLCKGGK 259
+ GY+P T N+L+ F +++ A + +M+ + D +T + + L + +
Sbjct: 142 MMKLGYEPDIVTLNSLLNGFCHGNRISDAVSLVGQMVEMGYQPDSFTFNTLIHGLFRHNR 201
Query: 260 CREAFDLIDE--AEDFVPDTVFYNRMVSGLCEASLFEEAMDILHRMRSSSCIPNVVTYRI 317
EA L+D + PD V Y +V+GLC+ + A+ +L +M P VV Y
Sbjct: 202 ASEAVALVDRMVVKGCQPDLVTYGIVVNGLCKRGDIDLALSLLKKMEQGKIEPGVVIY-- 259
Query: 318 LLSGCLRKGQLGRCKRILSMMITEGCYPNREIFNSLIHAYCKSRDYSYAYKLFKKMIKCG 377
N++I A C ++ + A LF +M G
Sbjct: 260 ---------------------------------NTIIDALCNYKNVNDALNLFTEMDNKG 286
Query: 378 CQPGYLVYNIFIGSVCSNEEQPSSDILDLVEKAYSEMLDLGVVLNKVNVSNFARCLCGAG 437
+P + YN I +C+ + + S+M++ + N V S G
Sbjct: 287 IRPNVVTYNSLIRCLCNYGRWSDA------SRLLSDMIERKINPNVVTFSALIDAFVKEG 340
Query: 438 KFDQAFKIICEMMGKGFVPDDSTYSKVIGFLCDASKVEKAFSLFEEMKRNGIVPSVYTYT 497
K +A K+ EM+ + PD TYS +I C ++++A +FE M P+V TY
Sbjct: 341 KLVEAEKLYDEMIKRSIDPDIFTYSSLINGFCMHDRLDEAKHMFELMISKDCFPNVVTYN 400
Query: 498 ILIDSFCKAGLIQQARKWFDEMLHKGCTPNVVTYTALIHAYLKAKQMPVADELFEMMLLE 557
LI FCKA + + + F EM +G N VTYT LIH + +A++ A +F+ M+ +
Sbjct: 401 TLIKGFCKAKRVDEGMELFREMSQRGLVGNTVTYTTLIHGFFQARECDNAQIVFKQMVSD 460
Query: 558 GCKPNVVTYTALIDGHCKAGQIEKACQIYARMRGDIESSDMDKYFKLDHNNCEGPNVITY 617
G P+++TY+ L+DG C G++E A ++ ++ S M+ P++ TY
Sbjct: 461 GVLPDIMTYSILLDGLCNNGKVETALVVFEY----LQRSKME------------PDIYTY 504
Query: 618 GALVDGLCKANRVKEAHELLDTMLAHGCEPNQIVYDAVIDGFCKIGKLQDAQEVFTKMSE 677
+++G+CKA +V++ +L ++ G +PN + Y ++ GFC+ G ++A +F +M E
Sbjct: 505 NIMIEGMCKAGKVEDGWDLFCSLSLKGVKPNVVTYTTMMSGFCRKGLKEEADALFREMKE 564
Query: 678 RGYSPNLYTYSSFIDCLFKD 697
G P+ TY++ I +D
Sbjct: 565 EGPLPDSGTYNTLIRAHLRD 584
Score = 227 bits (579), Expect = 2e-59
Identities = 151/553 (27%), Positives = 261/553 (46%), Gaps = 29/553 (5%)
Query: 404 LDLVEKAYSEMLDLGVVLNKVNVSNFARCLCGAGKFDQAFKIICEMMGKGFVPDDSTYSK 463
LD + +M+ + V S + KFD + +M G + TYS
Sbjct: 62 LDDAVNLFGDMVKSRPFPSIVEFSKLLSAIAKMNKFDLVISLGEQMQNLGISHNLYTYSI 121
Query: 464 VIGFLCDASKVEKAFSLFEEMKRNGIVPSVYTYTILIDSFCKAGLIQQARKWFDEMLHKG 523
+I C S++ A ++ +M + G P + T L++ FC I A +M+ G
Sbjct: 122 LINCFCRRSQLSLALAVLAKMMKLGYEPDIVTLNSLLNGFCHGNRISDAVSLVGQMVEMG 181
Query: 524 CTPNVVTYTALIHAYLKAKQMPVADELFEMMLLEGCKPNVVTYTALIDGHCKAGQIEKAC 583
P+ T+ LIH + + A L + M+++GC+P++VTY +++G CK G I+ A
Sbjct: 182 YQPDSFTFNTLIHGLFRHNRASEAVALVDRMVVKGCQPDLVTYGIVVNGLCKRGDIDLAL 241
Query: 584 QIYARM-RGDIE-------------------SSDMDKYFKLDHNNCEGPNVITYGALVDG 623
+ +M +G IE + ++ + ++D+ PNV+TY +L+
Sbjct: 242 SLLKKMEQGKIEPGVVIYNTIIDALCNYKNVNDALNLFTEMDNKGIR-PNVVTYNSLIRC 300
Query: 624 LCKANRVKEAHELLDTMLAHGCEPNQIVYDAVIDGFCKIGKLQDAQEVFTKMSERGYSPN 683
LC R +A LL M+ PN + + A+ID F K GKL +A++++ +M +R P+
Sbjct: 301 LCNYGRWSDASRLLSDMIERKINPNVVTFSALIDAFVKEGKLVEAEKLYDEMIKRSIDPD 360
Query: 684 LYTYSSFIDCLFKDNRLDLVLKVLSKMLENSCTPNVVIYTEMVDGLCKIGKTDEAYKLML 743
++TYSS I+ +RLD + M+ C PNVV Y ++ G CK + DE +L
Sbjct: 361 IFTYSSLINGFCMHDRLDEAKHMFELMISKDCFPNVVTYNTLIKGFCKAKRVDEGMELFR 420
Query: 744 KMEEKGCNPNVVTYTAMIDGFGKSGKIEQCLELFRDMCSKGCAPNFITYRVLINHCCSNG 803
+M ++G N VTYT +I GF ++ + + +F+ M S G P+ +TY +L++ C+NG
Sbjct: 421 EMSQRGLVGNTVTYTTLIHGFFQARECDNAQIVFKQMVSDGVLPDIMTYSILLDGLCNNG 480
Query: 804 LLDEAYKLLDEMKQTYWPKHILSHRKIIEGFSQEFITSIG--LLDELSENESVPVDSLYR 861
++ A + + ++++ I ++ +IEG + G L LS P Y
Sbjct: 481 KVETALVVFEYLQRSKMEPDIYTYNIMIEGMCKAGKVEDGWDLFCSLSLKGVKPNVVTYT 540
Query: 862 ILIDNYIKAGRLEVALDLLEEISSSPSHAVSNKYLYASLIENLSHASKVDKA--LELYAS 919
++ + + G E A L E+ + + Y +LI +H DKA EL
Sbjct: 541 TMMSGFCRKGLKEEADALFREMKE--EGPLPDSGTYNTLIR--AHLRDGDKAASAELIRE 596
Query: 920 MISKNVVPELSIL 932
M S V + S +
Sbjct: 597 MRSCRFVGDASTI 609
Score = 95.5 bits (236), Expect = 1e-19
Identities = 73/321 (22%), Positives = 143/321 (43%), Gaps = 17/321 (5%)
Query: 663 GKLQDAQEVFTKMS--ERGYSPNLYTYSSFIDCLFKDNRLDLVLKVLSKMLENSCTPNVV 720
GK A F+ S R +S Y Y D +LD + + M+++ P++V
Sbjct: 23 GKCGTAPPSFSHCSFWVRDFSGVRYDYRKISINRLNDLKLDDAVNLFGDMVKSRPFPSIV 82
Query: 721 IYTEMVDGLCKIGKTDEAYKLMLKMEEKGCNPNVVTYTAMIDGFGKSGKIEQCLELFRDM 780
+++++ + K+ K D L +M+ G + N+ TY+ +I+ F + ++ L + M
Sbjct: 83 EFSKLLSAIAKMNKFDLVISLGEQMQNLGISHNLYTYSILINCFCRRSQLSLALAVLAKM 142
Query: 781 CSKGCAPNFITYRVLINHCCSNGLLDEAYKLLDEMKQTYWPKHILSHRKIIEGFSQEFIT 840
G P+ +T L+N C + +A L+ +M + + + +I G +
Sbjct: 143 MKLGYEPDIVTLNSLLNGFCHGNRISDAVSLVGQMVEMGYQPDSFTFNTLIHGLFRHNRA 202
Query: 841 S--IGLLDELSENESVPVDSLYRILIDNYIKAGRLEVALDLLEEISSSPSHAVSNKYLYA 898
S + L+D + P Y I+++ K G +++AL LL+++ +Y
Sbjct: 203 SEAVALVDRMVVKGCQPDLVTYGIVVNGLCKRGDIDLALSLLKKMEQGKIE--PGVVIYN 260
Query: 899 SLIENLSHASKVDKALELYASMISKNVVPELSILVHLIKGLIKVDKWQEALQLSDSICQM 958
++I+ L + V+ AL L+ M +K + P + LI+ L +W +A +L
Sbjct: 261 TIIDALCNYKNVNDALNLFTEMDNKGIRPNVVTYNSLIRCLCNYGRWSDASRL------- 313
Query: 959 VCLTLSQTFQPLVNLNLLTLS 979
LS + +N N++T S
Sbjct: 314 ----LSDMIERKINPNVVTFS 330
>At1g62670 PPR-repeat protein
Length = 630
Score = 266 bits (679), Expect = 6e-71
Identities = 158/545 (28%), Positives = 275/545 (49%), Gaps = 28/545 (5%)
Query: 275 PDTVFYNRMVSGLCEASLFEEAMDILHRMRSSSCIPNVVTYRILLSGCLRKGQLGRCKRI 334
P + +++++S + + + F+ + + +M++ N TY IL++ R+ QL +
Sbjct: 79 PSIIEFSKLLSAIAKMNKFDVVISLGEQMQNLGIPHNHYTYSILINCFCRRSQLPLALAV 138
Query: 335 LSMMITEGCYPNREIFNSLIHAYCKSRDYSYAYKLFKKMIKCGCQPGYLVYNIFI-GSVC 393
L M+ G PN +SL++ YC S+ S A L +M G QP + +N I G
Sbjct: 139 LGKMMKLGYEPNIVTLSSLLNGYCHSKRISEAVALVDQMFVTGYQPNTVTFNTLIHGLFL 198
Query: 394 SNEEQPSSDILD--LVEKAYSEMLDLGVVLNKVNVSNFARCLCGAGKFDQAFKIICEMMG 451
N+ + ++D + + +++ GVV+N LC G D AF ++ +M
Sbjct: 199 HNKASEAMALIDRMVAKGCQPDLVTYGVVVNG---------LCKRGDTDLAFNLLNKMEQ 249
Query: 452 KGFVPDDSTYSKVIGFLCDASKVEKAFSLFEEMKRNGIVPSVYTYTILIDSFCKAGLIQQ 511
P Y+ +I LC ++ A +LF+EM+ GI P+V TY+ LI C G
Sbjct: 250 GKLEPGVLIYNTIIDGLCKYKHMDDALNLFKEMETKGIRPNVVTYSSLISCLCNYGRWSD 309
Query: 512 ARKWFDEMLHKGCTPNVVTYTALIHAYLKAKQMPVADELFEMMLLEGCKPNVVTYTALID 571
A + +M+ + P+V T++ALI A++K ++ A++L++ M+ P++VTY++LI+
Sbjct: 310 ASRLLSDMIERKINPDVFTFSALIDAFVKEGKLVEAEKLYDEMVKRSIDPSIVTYSSLIN 369
Query: 572 GHCKAGQIEKACQIYARMRGDIESSDMDKYFKLDHNNCEGPNVITYGALVDGLCKANRVK 631
G C ++++A Q++ M +C P+V+TY L+ G CK RV+
Sbjct: 370 GFCMHDRLDEAKQMFEFMVS---------------KHCF-PDVVTYNTLIKGFCKYKRVE 413
Query: 632 EAHELLDTMLAHGCEPNQIVYDAVIDGFCKIGKLQDAQEVFTKMSERGYSPNLYTYSSFI 691
E E+ M G N + Y+ +I G + G AQE+F +M G PN+ TY++ +
Sbjct: 414 EGMEVFREMSQRGLVGNTVTYNILIQGLFQAGDCDMAQEIFKEMVSDGVPPNIMTYNTLL 473
Query: 692 DCLFKDNRLDLVLKVLSKMLENSCTPNVVIYTEMVDGLCKIGKTDEAYKLMLKMEEKGCN 751
D L K+ +L+ + V + + P + Y M++G+CK GK ++ + L + KG
Sbjct: 474 DGLCKNGKLEKAMVVFEYLQRSKMEPTIYTYNIMIEGMCKAGKVEDGWDLFCNLSLKGVK 533
Query: 752 PNVVTYTAMIDGFGKSGKIEQCLELFRDMCSKGCAPNFITYRVLINHCCSNGLLDEAYKL 811
P+VV Y MI GF + G E+ LF++M G PN Y LI +G + + +L
Sbjct: 534 PDVVAYNTMISGFCRKGSKEEADALFKEMKEDGTLPNSGCYNTLIRARLRDGDREASAEL 593
Query: 812 LDEMK 816
+ EM+
Sbjct: 594 IKEMR 598
Score = 256 bits (655), Expect = 3e-68
Identities = 167/635 (26%), Positives = 287/635 (44%), Gaps = 68/635 (10%)
Query: 183 KCCRNGWWNMALEE----LGRLKDFGYKPSQTTYNALIQVFLRADKLDTAYLVKREMLSY 238
K RNG + L++ G + PS ++ L+ + +K D + +M +
Sbjct: 51 KLSRNGLSELKLDDAVALFGEMVKSRPFPSIIEFSKLLSAIAKMNKFDVVISLGEQMQNL 110
Query: 239 AFVMDRYTLSCFAYSLCKGGKCREAFDLIDEAED--FVPDTVFYNRMVSGLCEASLFEEA 296
+ YT S C+ + A ++ + + P+ V + +++G C + EA
Sbjct: 111 GIPHNHYTYSILINCFCRRSQLPLALAVLGKMMKLGYEPNIVTLSSLLNGYCHSKRISEA 170
Query: 297 MDILHRMRSSSCIPNVVTYRILLSGCLRKGQLGRCKRILSMMITEGCYPNREIFNSLIHA 356
+ ++ +M + PN VT+ L+ G + ++ M+ +GC P+ + +++
Sbjct: 171 VALVDQMFVTGYQPNTVTFNTLIHGLFLHNKASEAMALIDRMVAKGCQPDLVTYGVVVNG 230
Query: 357 YCKSRDYSYAYKLFKKMIKCGCQPGYLVYNIFIGSVCSNEEQPSSDILDLVEKAYSEMLD 416
CK D A+ L KM + +PG L+YN I +C Y M
Sbjct: 231 LCKRGDTDLAFNLLNKMEQGKLEPGVLIYNTIIDGLCK----------------YKHM-- 272
Query: 417 LGVVLNKVNVSNFARCLCGAGKFDQAFKIICEMMGKGFVPDDSTYSKVIGFLCDASKVEK 476
D A + EM KG P+ TYS +I LC+ +
Sbjct: 273 -----------------------DDALNLFKEMETKGIRPNVVTYSSLISCLCNYGRWSD 309
Query: 477 AFSLFEEMKRNGIVPSVYTYTILIDSFCKAGLIQQARKWFDEMLHKGCTPNVVTYTALIH 536
A L +M I P V+T++ LID+F K G + +A K +DEM+ + P++VTY++LI+
Sbjct: 310 ASRLLSDMIERKINPDVFTFSALIDAFVKEGKLVEAEKLYDEMVKRSIDPSIVTYSSLIN 369
Query: 537 AYLKAKQMPVADELFEMMLLEGCKPNVVTYTALIDGHCKAGQIEKACQIYARM--RGDIE 594
+ ++ A ++FE M+ + C P+VVTY LI G CK ++E+ +++ M RG +
Sbjct: 370 GFCMHDRLDEAKQMFEFMVSKHCFPDVVTYNTLIKGFCKYKRVEEGMEVFREMSQRGLVG 429
Query: 595 SSDMDKYFKLDHNNCEGPNVITYGALVDGLCKANRVKEAHELLDTMLAHGCEPNQIVYDA 654
N +TY L+ GL +A A E+ M++ G PN + Y+
Sbjct: 430 ------------------NTVTYNILIQGLFQAGDCDMAQEIFKEMVSDGVPPNIMTYNT 471
Query: 655 VIDGFCKIGKLQDAQEVFTKMSERGYSPNLYTYSSFIDCLFKDNRLDLVLKVLSKMLENS 714
++DG CK GKL+ A VF + P +YTY+ I+ + K +++ + +
Sbjct: 472 LLDGLCKNGKLEKAMVVFEYLQRSKMEPTIYTYNIMIEGMCKAGKVEDGWDLFCNLSLKG 531
Query: 715 CTPNVVIYTEMVDGLCKIGKTDEAYKLMLKMEEKGCNPNVVTYTAMIDGFGKSGKIEQCL 774
P+VV Y M+ G C+ G +EA L +M+E G PN Y +I + G E
Sbjct: 532 VKPDVVAYNTMISGFCRKGSKEEADALFKEMKEDGTLPNSGCYNTLIRARLRDGDREASA 591
Query: 775 ELFRDMCSKGCAPNFITYRVLINHCCSNGLLDEAY 809
EL ++M S G A + T L+ + +G LD+++
Sbjct: 592 ELIKEMRSCGFAGDASTIG-LVTNMLHDGRLDKSF 625
Score = 247 bits (631), Expect = 2e-65
Identities = 156/540 (28%), Positives = 265/540 (48%), Gaps = 20/540 (3%)
Query: 438 KFDQAFKIICEMMGKGFVPDDSTYSKVIGFLCDASKVEKAFSLFEEMKRNGIVPSVYTYT 497
K D A + EM+ P +SK++ + +K + SL E+M+ GI + YTY+
Sbjct: 61 KLDDAVALFGEMVKSRPFPSIIEFSKLLSAIAKMNKFDVVISLGEQMQNLGIPHNHYTYS 120
Query: 498 ILIDSFCKAGLIQQARKWFDEMLHKGCTPNVVTYTALIHAYLKAKQMPVADELFEMMLLE 557
ILI+ FC+ + A +M+ G PN+VT ++L++ Y +K++ A L + M +
Sbjct: 121 ILINCFCRRSQLPLALAVLGKMMKLGYEPNIVTLSSLLNGYCHSKRISEAVALVDQMFVT 180
Query: 558 GCKPNVVTYTALIDGHCKAGQIEKACQIYARMRGDIESSDMDKYFKLDHNNCEGPNVITY 617
G +PN VT+ LI G + +A + RM C+ P+++TY
Sbjct: 181 GYQPNTVTFNTLIHGLFLHNKASEAMALIDRMVA---------------KGCQ-PDLVTY 224
Query: 618 GALVDGLCKANRVKEAHELLDTMLAHGCEPNQIVYDAVIDGFCKIGKLQDAQEVFTKMSE 677
G +V+GLCK A LL+ M EP ++Y+ +IDG CK + DA +F +M
Sbjct: 225 GVVVNGLCKRGDTDLAFNLLNKMEQGKLEPGVLIYNTIIDGLCKYKHMDDALNLFKEMET 284
Query: 678 RGYSPNLYTYSSFIDCLFKDNRLDLVLKVLSKMLENSCTPNVVIYTEMVDGLCKIGKTDE 737
+G PN+ TYSS I CL R ++LS M+E P+V ++ ++D K GK E
Sbjct: 285 KGIRPNVVTYSSLISCLCNYGRWSDASRLLSDMIERKINPDVFTFSALIDAFVKEGKLVE 344
Query: 738 AYKLMLKMEEKGCNPNVVTYTAMIDGFGKSGKIEQCLELFRDMCSKGCAPNFITYRVLIN 797
A KL +M ++ +P++VTY+++I+GF ++++ ++F M SK C P+ +TY LI
Sbjct: 345 AEKLYDEMVKRSIDPSIVTYSSLINGFCMHDRLDEAKQMFEFMVSKHCFPDVVTYNTLIK 404
Query: 798 HCCSNGLLDEAYKLLDEMKQTYWPKHILSHRKIIEGFSQEFITSIG--LLDELSENESVP 855
C ++E ++ EM Q + +++ +I+G Q + + E+ + P
Sbjct: 405 GFCKYKRVEEGMEVFREMSQRGLVGNTVTYNILIQGLFQAGDCDMAQEIFKEMVSDGVPP 464
Query: 856 VDSLYRILIDNYIKAGRLEVALDLLEEISSSPSHAVSNKYLYASLIENLSHASKVDKALE 915
Y L+D K G+LE A+ + E + S Y Y +IE + A KV+ +
Sbjct: 465 NIMTYNTLLDGLCKNGKLEKAMVVFEYLQRSKMEPTI--YTYNIMIEGMCKAGKVEDGWD 522
Query: 916 LYASMISKNVVPELSILVHLIKGLIKVDKWQEALQLSDSICQMVCLTLSQTFQPLVNLNL 975
L+ ++ K V P++ +I G + +EA L + + L S + L+ L
Sbjct: 523 LFCNLSLKGVKPDVVAYNTMISGFCRKGSKEEADALFKEMKEDGTLPNSGCYNTLIRARL 582
Score = 201 bits (511), Expect = 2e-51
Identities = 135/528 (25%), Positives = 237/528 (44%), Gaps = 29/528 (5%)
Query: 178 NFLVRKCCRNGWWNMALEELGRLKDFGYKPSQTTYNALIQVFLRADKLDTAYLVKREMLS 237
+ L+ CR +AL LG++ GY+P+ T ++L+ + + ++ A + +M
Sbjct: 120 SILINCFCRRSQLPLALAVLGKMMKLGYEPNIVTLSSLLNGYCHSKRISEAVALVDQMFV 179
Query: 238 YAFVMDRYTLSCFAYSLCKGGKCREAFDLIDE--AEDFVPDTVFYNRMVSGLCEASLFEE 295
+ + T + + L K EA LID A+ PD V Y +V+GLC+ +
Sbjct: 180 TGYQPNTVTFNTLIHGLFLHNKASEAMALIDRMVAKGCQPDLVTYGVVVNGLCKRGDTDL 239
Query: 296 AMDILHRMRSSSCIPNVVTYRILLSGCLRKGQLGRCKRILSMMITEGCYPNREIFNSLIH 355
A ++L++M P V+ Y ++ G + + + M T+G PN ++SLI
Sbjct: 240 AFNLLNKMEQGKLEPGVLIYNTIIDGLCKYKHMDDALNLFKEMETKGIRPNVVTYSSLIS 299
Query: 356 AYCKSRDYSYAYKLFKKMIKCGCQPGYLVYNIFIGSVCSNEEQPSSDILDLVEKAYSEML 415
C +S A +L MI+ P ++ I + + L EK Y EM+
Sbjct: 300 CLCNYGRWSDASRLLSDMIERKINPDVFTFSALIDAFVKEGK------LVEAEKLYDEMV 353
Query: 416 DLGVVLNKVNVSNFARCLCGAGKFDQAFKIICEMMGKGFVPDDSTYSKVIGFLCDASKVE 475
+ + V S+ C + D+A ++ M+ K PD TY+ +I C +VE
Sbjct: 354 KRSIDPSIVTYSSLINGFCMHDRLDEAKQMFEFMVSKHCFPDVVTYNTLIKGFCKYKRVE 413
Query: 476 KAFSLFEEMKRNGIVPSVYTYTILIDSFCKAGLIQQARKWFDEMLHKGCTPNVVTYTALI 535
+ +F EM + G+V + TY ILI +AG A++ F EM+ G PN++TY L+
Sbjct: 414 EGMEVFREMSQRGLVGNTVTYNILIQGLFQAGDCDMAQEIFKEMVSDGVPPNIMTYNTLL 473
Query: 536 HAYLKAKQMPVADELFEMMLLEGCKPNVVTYTALIDGHCKAGQIEKACQIYARMRGDIES 595
K ++ A +FE + +P + TY +I+G CKAG++E ++ +
Sbjct: 474 DGLCKNGKLEKAMVVFEYLQRSKMEPTIYTYNIMIEGMCKAGKVEDGWDLFCNL------ 527
Query: 596 SDMDKYFKLDHNNCEG--PNVITYGALVDGLCKANRVKEAHELLDTMLAHGCEPNQIVYD 653
+ +G P+V+ Y ++ G C+ +EA L M G PN Y+
Sbjct: 528 ------------SLKGVKPDVVAYNTMISGFCRKGSKEEADALFKEMKEDGTLPNSGCYN 575
Query: 654 AVIDGFCKIGKLQDAQEVFTKMSERGYSPNLYTYSSFIDCLFKDNRLD 701
+I + G + + E+ +M G++ + T + + D RLD
Sbjct: 576 TLIRARLRDGDREASAELIKEMRSCGFAGDASTI-GLVTNMLHDGRLD 622
Score = 181 bits (458), Expect = 2e-45
Identities = 114/418 (27%), Positives = 199/418 (47%), Gaps = 8/418 (1%)
Query: 177 LNFLVRKCCRNGWWNMALEELGRLKDFGYKPSQTTYNALIQVFLRADKLDTAYLVKREML 236
L+ L+ C + + A+ + ++ GY+P+ T+N LI +K A + M+
Sbjct: 154 LSSLLNGYCHSKRISEAVALVDQMFVTGYQPNTVTFNTLIHGLFLHNKASEAMALIDRMV 213
Query: 237 SYAFVMDRYTLSCFAYSLCKGGKCREAFDLIDEAED--FVPDTVFYNRMVSGLCEASLFE 294
+ D T LCK G AF+L+++ E P + YN ++ GLC+ +
Sbjct: 214 AKGCQPDLVTYGVVVNGLCKRGDTDLAFNLLNKMEQGKLEPGVLIYNTIIDGLCKYKHMD 273
Query: 295 EAMDILHRMRSSSCIPNVVTYRILLSGCLRKGQLGRCKRILSMMITEGCYPNREIFNSLI 354
+A+++ M + PNVVTY L+S G+ R+LS MI P+ F++LI
Sbjct: 274 DALNLFKEMETKGIRPNVVTYSSLISCLCNYGRWSDASRLLSDMIERKINPDVFTFSALI 333
Query: 355 HAYCKSRDYSYAYKLFKKMIKCGCQPGYLVYNIFIGSVCSNEEQPSSDILDLVEKAYSEM 414
A+ K A KL+ +M+K P + Y+ I C + D LD ++ + M
Sbjct: 334 DAFVKEGKLVEAEKLYDEMVKRSIDPSIVTYSSLINGFCMH------DRLDEAKQMFEFM 387
Query: 415 LDLGVVLNKVNVSNFARCLCGAGKFDQAFKIICEMMGKGFVPDDSTYSKVIGFLCDASKV 474
+ + V + + C + ++ ++ EM +G V + TY+ +I L A
Sbjct: 388 VSKHCFPDVVTYNTLIKGFCKYKRVEEGMEVFREMSQRGLVGNTVTYNILIQGLFQAGDC 447
Query: 475 EKAFSLFEEMKRNGIVPSVYTYTILIDSFCKAGLIQQARKWFDEMLHKGCTPNVVTYTAL 534
+ A +F+EM +G+ P++ TY L+D CK G +++A F+ + P + TY +
Sbjct: 448 DMAQEIFKEMVSDGVPPNIMTYNTLLDGLCKNGKLEKAMVVFEYLQRSKMEPTIYTYNIM 507
Query: 535 IHAYLKAKQMPVADELFEMMLLEGCKPNVVTYTALIDGHCKAGQIEKACQIYARMRGD 592
I KA ++ +LF + L+G KP+VV Y +I G C+ G E+A ++ M+ D
Sbjct: 508 IEGMCKAGKVEDGWDLFCNLSLKGVKPDVVAYNTMISGFCRKGSKEEADALFKEMKED 565
Score = 32.3 bits (72), Expect = 1.4
Identities = 27/91 (29%), Positives = 40/91 (43%), Gaps = 2/91 (2%)
Query: 178 NFLVRKCCRNGWWNMALEELGRLKDFGYKPSQTTYNALIQVFLR-ADKLDTAYLVKREML 236
N ++ CR G A +K+ G P+ YN LI+ LR D+ +A L+K EM
Sbjct: 540 NTMISGFCRKGSKEEADALFKEMKEDGTLPNSGCYNTLIRARLRDGDREASAELIK-EMR 598
Query: 237 SYAFVMDRYTLSCFAYSLCKGGKCREAFDLI 267
S F D T+ L G + D++
Sbjct: 599 SCGFAGDASTIGLVTNMLHDGRLDKSFLDML 629
>At3g22470 hypothetical protein
Length = 619
Score = 264 bits (675), Expect = 2e-70
Identities = 178/612 (29%), Positives = 288/612 (46%), Gaps = 82/612 (13%)
Query: 279 FYNRMVSGLCEASLFEEAMDILHRMRSSSCIPNVVTYRILLSGCLRKGQ----LGRCKRI 334
+ R+ +G+ + + +A+D+ M S +P + + L S R Q LG CK
Sbjct: 38 YKERLRNGIVDIKV-NDAIDLFESMIQSRPLPTPIDFNRLCSAVARTKQYDLVLGFCKG- 95
Query: 335 LSMMITEGCYPNREIFNSLIHAYCKSRDYSYAYKLFKKMIKCGCQPGYLVYNIFIGSVCS 394
M G + +I+ YC+ + +A+ + + K G +P + ++ + C
Sbjct: 96 ---MELNGIEHDMYTMTIMINCYCRKKKLLFAFSVLGRAWKLGYEPDTITFSTLVNGFCL 152
Query: 395 NEEQPSSDILDLVEKAYSEMLDLGVVLNKVNVSNFARCLCGAGKFDQAFKIICEMMGKGF 454
E S+ + LV++ M+++ + V VS LC G+ +A +I M+ GF
Sbjct: 153 --EGRVSEAVALVDR----MVEMKQRPDLVTVSTLINGLCLKGRVSEALVLIDRMVEYGF 206
Query: 455 VPDDSTYSKVIGFLCDASKVEKAFSLFEEMKRNGIVPSVYTYTILIDSFCKAGLIQQARK 514
PD+ TY V+ LC + A LF +M+ I SV Y+I+IDS CK G A
Sbjct: 207 QPDEVTYGPVLNRLCKSGNSALALDLFRKMEERNIKASVVQYSIVIDSLCKDGSFDDALS 266
Query: 515 WFDEMLHKGCT-----------------------------------PNVVTYTALIHAYL 539
F+EM KG P+VVT++ALI ++
Sbjct: 267 LFNEMEMKGIKADVVTYSSLIGGLCNDGKWDDGAKMLREMIGRNIIPDVVTFSALIDVFV 326
Query: 540 KAKQMPVADELFEMMLLEGCKPNVVTYTALIDGHCKAGQIEKACQIYARMRGDIESSDMD 599
K ++ A EL+ M+ G P+ +TY +LIDG CK + +A Q++ M
Sbjct: 327 KEGKLLEAKELYNEMITRGIAPDTITYNSLIDGFCKENCLHEANQMFDLMVS-------- 378
Query: 600 KYFKLDHNNCEGPNVITYGALVDGLCKANRVKEAHELLDTMLAHGCEPNQIVYDAVIDGF 659
CE P+++TY L++ CKA RV + L + + G PN I Y+ ++ GF
Sbjct: 379 -------KGCE-PDIVTYSILINSYCKAKRVDDGMRLFREISSKGLIPNTITYNTLVLGF 430
Query: 660 CKIGKLQDAQEVFTKMSERGYSPNLYTYSSFIDCLFKDNRLDLVLKVLSKMLENSCTPNV 719
C+ GKL A+E+F +M RG P++ TY +D L + L+ L++ KM ++ T +
Sbjct: 431 CQSGKLNAAKELFQEMVSRGVPPSVVTYGILLDGLCDNGELNKALEIFEKMQKSRMTLGI 490
Query: 720 VIYTEMVDGLCKIGKTDEAYKLMLKMEEKGCNPNVVTYTAMIDGFGKSGKIEQCLELFRD 779
IY ++ G+C K D+A+ L + +KG P+VVTY MI G K G + + LFR
Sbjct: 491 GIYNIIIHGMCNASKVDDAWSLFCSLSDKGVKPDVVTYNVMIGGLCKKGSLSEADMLFRK 550
Query: 780 MCSKGCAPNFITYRVLIN-HCCSNGLLDEAYKLLDEMKQTYWPKHILSHRKIIEGFSQEF 838
M GC P+ TY +LI H +GL+ + +L++EMK + GFS +
Sbjct: 551 MKEDGCTPDDFTYNILIRAHLGGSGLI-SSVELIEEMK--------------VCGFSADS 595
Query: 839 ITSIGLLDELSE 850
T ++D LS+
Sbjct: 596 STIKMVIDMLSD 607
Score = 224 bits (571), Expect = 2e-58
Identities = 151/557 (27%), Positives = 262/557 (46%), Gaps = 26/557 (4%)
Query: 366 AYKLFKKMIKCGCQPGYLVYNIFIGSVCSNEEQPSSDILDLVEKAYSEMLDLGVVLNKVN 425
A LF+ MI+ P + +N +V ++ DLV M G+ +
Sbjct: 54 AIDLFESMIQSRPLPTPIDFNRLCSAVARTKQY------DLVLGFCKGMELNGIEHDMYT 107
Query: 426 VSNFARCLCGAGKFDQAFKIICEMMGKGFVPDDSTYSKVIGFLCDASKVEKAFSLFEEMK 485
++ C C K AF ++ G+ PD T+S ++ C +V +A +L + M
Sbjct: 108 MTIMINCYCRKKKLLFAFSVLGRAWKLGYEPDTITFSTLVNGFCLEGRVSEAVALVDRMV 167
Query: 486 RNGIVPSVYTYTILIDSFCKAGLIQQARKWFDEMLHKGCTPNVVTYTALIHAYLKAKQMP 545
P + T + LI+ C G + +A D M+ G P+ VTY +++ K+
Sbjct: 168 EMKQRPDLVTVSTLINGLCLKGRVSEALVLIDRMVEYGFQPDEVTYGPVLNRLCKSGNSA 227
Query: 546 VADELFEMMLLEGCKPNVVTYTALIDGHCKAGQIEKACQIYARMRGDIESSDMDKYFKLD 605
+A +LF M K +VV Y+ +ID CK G + A ++ M K K D
Sbjct: 228 LALDLFRKMEERNIKASVVQYSIVIDSLCKDGSFDDALSLFNEMEM--------KGIKAD 279
Query: 606 HNNCEGPNVITYGALVDGLCKANRVKEAHELLDTMLAHGCEPNQIVYDAVIDGFCKIGKL 665
V+TY +L+ GLC + + ++L M+ P+ + + A+ID F K GKL
Sbjct: 280 --------VVTYSSLIGGLCNDGKWDDGAKMLREMIGRNIIPDVVTFSALIDVFVKEGKL 331
Query: 666 QDAQEVFTKMSERGYSPNLYTYSSFIDCLFKDNRLDLVLKVLSKMLENSCTPNVVIYTEM 725
+A+E++ +M RG +P+ TY+S ID K+N L ++ M+ C P++V Y+ +
Sbjct: 332 LEAKELYNEMITRGIAPDTITYNSLIDGFCKENCLHEANQMFDLMVSKGCEPDIVTYSIL 391
Query: 726 VDGLCKIGKTDEAYKLMLKMEEKGCNPNVVTYTAMIDGFGKSGKIEQCLELFRDMCSKGC 785
++ CK + D+ +L ++ KG PN +TY ++ GF +SGK+ ELF++M S+G
Sbjct: 392 INSYCKAKRVDDGMRLFREISSKGLIPNTITYNTLVLGFCQSGKLNAAKELFQEMVSRGV 451
Query: 786 APNFITYRVLINHCCSNGLLDEAYKLLDEMKQTYWPKHILSHRKIIEGF--SQEFITSIG 843
P+ +TY +L++ C NG L++A ++ ++M+++ I + II G + + +
Sbjct: 452 PPSVVTYGILLDGLCDNGELNKALEIFEKMQKSRMTLGIGIYNIIIHGMCNASKVDDAWS 511
Query: 844 LLDELSENESVPVDSLYRILIDNYIKAGRLEVALDLLEEISSSPSHAVSNKYLYASLIEN 903
L LS+ P Y ++I K G L A L ++ + + Y LI
Sbjct: 512 LFCSLSDKGVKPDVVTYNVMIGGLCKKGSLSEADMLFRKMKE--DGCTPDDFTYNILIRA 569
Query: 904 LSHASKVDKALELYASM 920
S + ++EL M
Sbjct: 570 HLGGSGLISSVELIEEM 586
Score = 214 bits (545), Expect = 2e-55
Identities = 143/531 (26%), Positives = 242/531 (44%), Gaps = 61/531 (11%)
Query: 163 MEIKDDDHELLRRLLNFLVRKCCRNGWWNMALEELGRLKDFGYKPSQTTYNALIQVFLRA 222
ME+ +H++ + ++ CR A LGR GY+P T++ L+ F
Sbjct: 96 MELNGIEHDMYT--MTIMINCYCRKKKLLFAFSVLGRAWKLGYEPDTITFSTLVNGFCLE 153
Query: 223 DKLDTAYLVKREMLSYAFVMDRYTLSCFAYSLCKGGKCREAFDLIDEAED--FVPDTVFY 280
++ A + M+ D T+S LC G+ EA LID + F PD V Y
Sbjct: 154 GRVSEAVALVDRMVEMKQRPDLVTVSTLINGLCLKGRVSEALVLIDRMVEYGFQPDEVTY 213
Query: 281 NRMVSGLCEASLFEEAMDILHRMRSSSCIPNVVTYRILLSGCLRKGQLGRCKRILSMMIT 340
+++ LC++ A+D+ +M + +VV Y I
Sbjct: 214 GPVLNRLCKSGNSALALDLFRKMEERNIKASVVQYSI----------------------- 250
Query: 341 EGCYPNREIFNSLIHAYCKSRDYSYAYKLFKKMIKCGCQPGYLVYNIFIGSVCSNEEQPS 400
+I + CK + A LF +M G + + Y+ IG +C++ +
Sbjct: 251 ------------VIDSLCKDGSFDDALSLFNEMEMKGIKADVVTYSSLIGGLCNDGK--- 295
Query: 401 SDILDLVEKAYSEMLDLGVVLNKVNVSNFARCLCGAGKFDQAFKIICEMMGKGFVPDDST 460
D K EM+ ++ + V S GK +A ++ EM+ +G PD T
Sbjct: 296 ---WDDGAKMLREMIGRNIIPDVVTFSALIDVFVKEGKLLEAKELYNEMITRGIAPDTIT 352
Query: 461 YSKVIGFLCDASKVEKAFSLFEEMKRNGIVPSVYTYTILIDSFCKAGLIQQARKWFDEML 520
Y+ +I C + + +A +F+ M G P + TY+ILI+S+CKA + + F E+
Sbjct: 353 YNSLIDGFCKENCLHEANQMFDLMVSKGCEPDIVTYSILINSYCKAKRVDDGMRLFREIS 412
Query: 521 HKGCTPNVVTYTALIHAYLKAKQMPVADELFEMMLLEGCKPNVVTYTALIDGHCKAGQIE 580
KG PN +TY L+ + ++ ++ A ELF+ M+ G P+VVTY L+DG C G++
Sbjct: 413 SKGLIPNTITYNTLVLGFCQSGKLNAAKELFQEMVSRGVPPSVVTYGILLDGLCDNGELN 472
Query: 581 KACQIYARMRGDIESSDMDKYFKLDHNNCEGPNVITYGALVDGLCKANRVKEAHELLDTM 640
KA +I+ +M + S M + Y ++ G+C A++V +A L ++
Sbjct: 473 KALEIFEKM----QKSRMTL------------GIGIYNIIIHGMCNASKVDDAWSLFCSL 516
Query: 641 LAHGCEPNQIVYDAVIDGFCKIGKLQDAQEVFTKMSERGYSPNLYTYSSFI 691
G +P+ + Y+ +I G CK G L +A +F KM E G +P+ +TY+ I
Sbjct: 517 SDKGVKPDVVTYNVMIGGLCKKGSLSEADMLFRKMKEDGCTPDDFTYNILI 567
Score = 204 bits (519), Expect = 2e-52
Identities = 134/483 (27%), Positives = 233/483 (47%), Gaps = 51/483 (10%)
Query: 473 KVEKAFSLFEEMKRNGIVPSVYTYTILIDSFCKAGLIQQARKWFDEMLHKGCTPNVVTYT 532
KV A LFE M ++ +P+ + L + + + M G ++ T T
Sbjct: 50 KVNDAIDLFESMIQSRPLPTPIDFNRLCSAVARTKQYDLVLGFCKGMELNGIEHDMYTMT 109
Query: 533 ALIHAYLKAKQMPVADELFEMMLLEGCKPNVVTYTALIDGHCKAGQIEKACQIYARMRGD 592
+I+ Y + K++ A + G +P+ +T++ L++G C G++ +A + RM
Sbjct: 110 IMINCYCRKKKLLFAFSVLGRAWKLGYEPDTITFSTLVNGFCLEGRVSEAVALVDRM--- 166
Query: 593 IESSDMDKYFKLDHNNCEGPNVITYGALVDGLCKANRVKEAHELLDTMLAHGCEPNQIVY 652
+E + P+++T L++GLC RV EA L+D M+ +G +P+++ Y
Sbjct: 167 VEMK-------------QRPDLVTVSTLINGLCLKGRVSEALVLIDRMVEYGFQPDEVTY 213
Query: 653 DAVIDGFCKIGKLQDAQEVFTKMSERGYSPNLYTYSSFIDCLFKDNRLDLVLKVLSKMLE 712
V++ CK G A ++F KM ER ++ YS ID L KD D L + ++M
Sbjct: 214 GPVLNRLCKSGNSALALDLFRKMEERNIKASVVQYSIVIDSLCKDGSFDDALSLFNEMEM 273
Query: 713 NSCTPNVVIYTEMVDGLCKIGKTDEAYKLMLKMEEKGCNPNVVTYTAMIDGFGKSGKIEQ 772
+VV Y+ ++ GLC GK D+ K++ +M + P+VVT++A+ID F K GK+ +
Sbjct: 274 KGIKADVVTYSSLIGGLCNDGKWDDGAKMLREMIGRNIIPDVVTFSALIDVFVKEGKLLE 333
Query: 773 CLELFRDMCSKGCAPNFITYRVLINHCCSNGLLDEAYKLLDEMKQTYWPKHILSHRKIIE 832
EL+ +M ++G AP+ ITY LI+ C L EA ++ D M + +
Sbjct: 334 AKELYNEMITRGIAPDTITYNSLIDGFCKENCLHEANQMFDLM--------------VSK 379
Query: 833 GFSQEFITSIGLLDELSENESVPVDSLYRILIDNYIKAGRLEVALDLLEEISSSPSHAVS 892
G + +T Y ILI++Y KA R++ + L EISS +
Sbjct: 380 GCEPDIVT-------------------YSILINSYCKAKRVDDGMRLFREISSKG--LIP 418
Query: 893 NKYLYASLIENLSHASKVDKALELYASMISKNVVPELSILVHLIKGLIKVDKWQEALQLS 952
N Y +L+ + K++ A EL+ M+S+ V P + L+ GL + +AL++
Sbjct: 419 NTITYNTLVLGFCQSGKLNAAKELFQEMVSRGVPPSVVTYGILLDGLCDNGELNKALEIF 478
Query: 953 DSI 955
+ +
Sbjct: 479 EKM 481
Score = 122 bits (307), Expect = 8e-28
Identities = 89/382 (23%), Positives = 175/382 (45%), Gaps = 4/382 (1%)
Query: 592 DIESSDMDKYFKLDHNNCEGPNVITYGALVDGLCKANRVKEAHELLDTMLAHGCEPNQIV 651
DI+ +D F+ + P I + L + + + M +G E +
Sbjct: 48 DIKVNDAIDLFESMIQSRPLPTPIDFNRLCSAVARTKQYDLVLGFCKGMELNGIEHDMYT 107
Query: 652 YDAVIDGFCKIGKLQDAQEVFTKMSERGYSPNLYTYSSFIDCLFKDNRLDLVLKVLSKML 711
+I+ +C+ KL A V + + GY P+ T+S+ ++ + R+ + ++ +M+
Sbjct: 108 MTIMINCYCRKKKLLFAFSVLGRAWKLGYEPDTITFSTLVNGFCLEGRVSEAVALVDRMV 167
Query: 712 ENSCTPNVVIYTEMVDGLCKIGKTDEAYKLMLKMEEKGCNPNVVTYTAMIDGFGKSGKIE 771
E P++V + +++GLC G+ EA L+ +M E G P+ VTY +++ KSG
Sbjct: 168 EMKQRPDLVTVSTLINGLCLKGRVSEALVLIDRMVEYGFQPDEVTYGPVLNRLCKSGNSA 227
Query: 772 QCLELFRDMCSKGCAPNFITYRVLINHCCSNGLLDEAYKLLDEMKQTYWPKHILSHRKII 831
L+LFR M + + + Y ++I+ C +G D+A L +EM+ ++++ +I
Sbjct: 228 LALDLFRKMEERNIKASVVQYSIVIDSLCKDGSFDDALSLFNEMEMKGIKADVVTYSSLI 287
Query: 832 EGFSQEFITSIG--LLDELSENESVPVDSLYRILIDNYIKAGRLEVALDLLEEISSSPSH 889
G + G +L E+ +P + LID ++K G+L A +L E+ +
Sbjct: 288 GGLCNDGKWDDGAKMLREMIGRNIIPDVVTFSALIDVFVKEGKLLEAKELYNEMIT--RG 345
Query: 890 AVSNKYLYASLIENLSHASKVDKALELYASMISKNVVPELSILVHLIKGLIKVDKWQEAL 949
+ Y SLI+ + + +A +++ M+SK P++ LI K + + +
Sbjct: 346 IAPDTITYNSLIDGFCKENCLHEANQMFDLMVSKGCEPDIVTYSILINSYCKAKRVDDGM 405
Query: 950 QLSDSICQMVCLTLSQTFQPLV 971
+L I + + T+ LV
Sbjct: 406 RLFREISSKGLIPNTITYNTLV 427
>At1g74580 hypothetical protein
Length = 763
Score = 263 bits (673), Expect = 3e-70
Identities = 174/642 (27%), Positives = 312/642 (48%), Gaps = 33/642 (5%)
Query: 201 KDFGYKPSQTTYNALIQ---VFLRADKLDTAYLVKREMLSYAFVMDRYTLSCFAYSLCKG 257
K+ G+K + +TY ++I+ + + + ++ + RE + + Y + Y +
Sbjct: 32 KEVGFKHTLSTYRSVIEKLGYYGKFEAMEEVLVDMRENVGNHMLEGVYVGAMKNYG--RK 89
Query: 258 GKCREAFDLIDEAE--DFVPDTVFYNRMVSGLCEASLFEEAMDILHRMRSSSCIPNVVTY 315
GK +EA ++ + + D P YN ++S L ++ F++A + RMR P+V ++
Sbjct: 90 GKVQEAVNVFERMDFYDCEPTVFSYNAIMSVLVDSGYFDQAHKVYMRMRDRGITPDVYSF 149
Query: 316 RILLSGCLRKGQLGRCKRILSMMITEGCYPNREIFNSLIHAYCKSRDYSYAYKLFKKMIK 375
I + + + R+L+ M ++GC N + +++ + + + Y+LF KM+
Sbjct: 150 TIRMKSFCKTSRPHAALRLLNNMSSQGCEMNVVAYCTVVGGFYEENFKAEGYELFGKMLA 209
Query: 376 CGCQPGYLVYNIFIGSVCSNEEQPSSDILDLVEKAYSEMLDLGVVLNKVNVSNFARCLCG 435
G +N + +C D+ + EK +++ GV+ N + F + LC
Sbjct: 210 SGVSLCLSTFNKLLRVLCK-----KGDVKE-CEKLLDKVIKRGVLPNLFTYNLFIQGLCQ 263
Query: 436 AGKFDQAFKIICEMMGKGFVPDDSTYSKVIGFLCDASKVEKAFSLFEEMKRNGIVPSVYT 495
G+ D A +++ ++ +G PD TY+ +I LC SK ++A +M G+ P YT
Sbjct: 264 RGELDGAVRMVGCLIEQGPKPDVITYNNLIYGLCKNSKFQEAEVYLGKMVNEGLEPDSYT 323
Query: 496 YTILIDSFCKAGLIQQARKWFDEMLHKGCTPNVVTYTALIHAYLKAKQMPVADELFEMML 555
Y LI +CK G++Q A + + + G P+ TY +LI + A LF L
Sbjct: 324 YNTLIAGYCKGGMVQLAERIVGDAVFNGFVPDQFTYRSLIDGLCHEGETNRALALFNEAL 383
Query: 556 LEGCKPNVVTYTALIDGHCKAGQIEKACQIYARM--RGDIES-----------------S 596
+G KPNV+ Y LI G G I +A Q+ M +G I S
Sbjct: 384 GKGIKPNVILYNTLIKGLSNQGMILEAAQLANEMSEKGLIPEVQTFNILVNGLCKMGCVS 443
Query: 597 DMDKYFKLDHNNCEGPNVITYGALVDGLCKANRVKEAHELLDTMLAHGCEPNQIVYDAVI 656
D D K+ + P++ T+ L+ G +++ A E+LD ML +G +P+ Y++++
Sbjct: 444 DADGLVKVMISKGYFPDIFTFNILIHGYSTQLKMENALEILDVMLDNGVDPDVYTYNSLL 503
Query: 657 DGFCKIGKLQDAQEVFTKMSERGYSPNLYTYSSFIDCLFKDNRLDLVLKVLSKMLENSCT 716
+G CK K +D E + M E+G +PNL+T++ ++ L + +LD L +L +M S
Sbjct: 504 NGLCKTSKFEDVMETYKTMVEKGCAPNLFTFNILLESLCRYRKLDEALGLLEEMKNKSVN 563
Query: 717 PNVVIYTEMVDGLCKIGKTDEAYKLMLKMEEK-GCNPNVVTYTAMIDGFGKSGKIEQCLE 775
P+ V + ++DG CK G D AY L KMEE + + TY +I F + + +
Sbjct: 564 PDAVTFGTLIDGFCKNGDLDGAYTLFRKMEEAYKVSSSTPTYNIIIHAFTEKLNVTMAEK 623
Query: 776 LFRDMCSKGCAPNFITYRVLINHCCSNGLLDEAYKLLDEMKQ 817
LF++M + P+ TYR++++ C G ++ YK L EM +
Sbjct: 624 LFQEMVDRCLGPDGYTYRLMVDGFCKTGNVNLGYKFLLEMME 665
Score = 258 bits (659), Expect = 1e-68
Identities = 168/641 (26%), Positives = 302/641 (46%), Gaps = 33/641 (5%)
Query: 294 EEAMDILHRMRSSSCIPNVVTYRILLSGCLRKGQLGRCKRILSMMITEGCYPNREIFNSL 353
+EA+++ RM C P V +Y ++S + G + ++ M G P+ F
Sbjct: 93 QEAVNVFERMDFYDCEPTVFSYNAIMSVLVDSGYFDQAHKVYMRMRDRGITPDVYSFTIR 152
Query: 354 IHAYCKSRDYSYAYKLFKKMIKCGCQPGYLVYNIFIGSVCSNEEQPSSDILDLVEKAYSE 413
+ ++CK+ A +L M GC+ + Y +G EE ++ +L + +
Sbjct: 153 MKSFCKTSRPHAALRLLNNMSSQGCEMNVVAYCTVVGGFY--EENFKAEGYEL----FGK 206
Query: 414 MLDLGVVLNKVNVSNFARCLCGAGKFDQAFKIICEMMGKGFVPDDSTYSKVIGFLCDASK 473
ML GV L + R LC G + K++ +++ +G +P+ TY+ I LC +
Sbjct: 207 MLASGVSLCLSTFNKLLRVLCKKGDVKECEKLLDKVIKRGVLPNLFTYNLFIQGLCQRGE 266
Query: 474 VEKAFSLFEEMKRNGIVPSVYTYTILIDSFCKAGLIQQARKWFDEMLHKGCTPNVVTYTA 533
++ A + + G P V TY LI CK Q+A + +M+++G P+ TY
Sbjct: 267 LDGAVRMVGCLIEQGPKPDVITYNNLIYGLCKNSKFQEAEVYLGKMVNEGLEPDSYTYNT 326
Query: 534 LIHAYLKAKQMPVADELFEMMLLEGCKPNVVTYTALIDGHCKAGQIEKACQIYARMRGDI 593
LI Y K + +A+ + + G P+ TY +LIDG C G+ +A ++ G
Sbjct: 327 LIAGYCKGGMVQLAERIVGDAVFNGFVPDQFTYRSLIDGLCHEGETNRALALFNEALG-- 384
Query: 594 ESSDMDKYFKLDHNNCEGPNVITYGALVDGLCKANRVKEAHELLDTMLAHGCEPNQIVYD 653
K K PNVI Y L+ GL + EA +L + M G P ++
Sbjct: 385 ------KGIK--------PNVILYNTLIKGLSNQGMILEAAQLANEMSEKGLIPEVQTFN 430
Query: 654 AVIDGFCKIGKLQDAQEVFTKMSERGYSPNLYTYSSFIDCLFKDNRLDLVLKVLSKMLEN 713
+++G CK+G + DA + M +GY P+++T++ I +++ L++L ML+N
Sbjct: 431 ILVNGLCKMGCVSDADGLVKVMISKGYFPDIFTFNILIHGYSTQLKMENALEILDVMLDN 490
Query: 714 SCTPNVVIYTEMVDGLCKIGKTDEAYKLMLKMEEKGCNPNVVTYTAMIDGFGKSGKIEQC 773
P+V Y +++GLCK K ++ + M EKGC PN+ T+ +++ + K+++
Sbjct: 491 GVDPDVYTYNSLLNGLCKTSKFEDVMETYKTMVEKGCAPNLFTFNILLESLCRYRKLDEA 550
Query: 774 LELFRDMCSKGCAPNFITYRVLINHCCSNGLLDEAYKLLDEMKQTY-WPKHILSHRKIIE 832
L L +M +K P+ +T+ LI+ C NG LD AY L +M++ Y ++ II
Sbjct: 551 LGLLEEMKNKSVNPDAVTFGTLIDGFCKNGDLDGAYTLFRKMEEAYKVSSSTPTYNIIIH 610
Query: 833 GFSQEFITSIG--LLDELSENESVPVDSLYRILIDNYIKAGRLEVALDLLEEISSS---P 887
F+++ ++ L E+ + P YR+++D + K G + + L E+ + P
Sbjct: 611 AFTEKLNVTMAEKLFQEMVDRCLGPDGYTYRLMVDGFCKTGNVNLGYKFLLEMMENGFIP 670
Query: 888 SHAVSNKYLYASLIENLSHASKVDKALELYASMISKNVVPE 928
S + + +E+ +V +A + M+ K +VPE
Sbjct: 671 SLTTLGRVINCLCVED-----RVYEAAGIIHRMVQKGLVPE 706
Score = 253 bits (646), Expect = 4e-67
Identities = 176/725 (24%), Positives = 321/725 (44%), Gaps = 67/725 (9%)
Query: 107 VVEVMNNVKNPELCVKFFLWAGRQIGYSHTPQVFDKLLDLLGC--NVNADDRVPLKFLME 164
V V+ K+P ++ F +++G+ HT + +++ LG A + V L++
Sbjct: 10 VTAVIKCQKDPMKALEMFNSMRKEVGFKHTLSTYRSVIEKLGYYGKFEAMEEV----LVD 65
Query: 165 IKDD-DHELLRRLLNFLVRKCCRNGWWNMALEELGRLKDFGYKPSQTTYNALIQVFLRAD 223
++++ + +L + ++ R G A+ R+ + +P+ +YNA++ V + +
Sbjct: 66 MRENVGNHMLEGVYVGAMKNYGRKGKVQEAVNVFERMDFYDCEPTVFSYNAIMSVLVDSG 125
Query: 224 KLDTAYLVKREMLSYAFVMDRYTLSCFAYSLCKGGKCREAFDLIDE-------------- 269
D A+ V M D Y+ + S CK + A L++
Sbjct: 126 YFDQAHKVYMRMRDRGITPDVYSFTIRMKSFCKTSRPHAALRLLNNMSSQGCEMNVVAYC 185
Query: 270 -------AEDFVPD----------------TVFYNRMVSGLCEASLFEEAMDILHRMRSS 306
E+F + +N+++ LC+ +E +L ++
Sbjct: 186 TVVGGFYEENFKAEGYELFGKMLASGVSLCLSTFNKLLRVLCKKGDVKECEKLLDKVIKR 245
Query: 307 SCIPNVVTYRILLSGCLRKGQLGRCKRILSMMITEGCYPNREIFNSLIHAYCKSRDYSYA 366
+PN+ TY + + G ++G+L R++ +I +G P+ +N+LI+ CK+ + A
Sbjct: 246 GVLPNLFTYNLFIQGLCQRGELDGAVRMVGCLIEQGPKPDVITYNNLIYGLCKNSKFQEA 305
Query: 367 YKLFKKMIKCGCQPGYLVYNIFIGSVCSNEEQPSSDILDLVEKAYSEMLDLGVVLNKVNV 426
KM+ G +P YN I C ++ L E+ + + G V ++
Sbjct: 306 EVYLGKMVNEGLEPDSYTYNTLIAGYC------KGGMVQLAERIVGDAVFNGFVPDQFTY 359
Query: 427 SNFARCLCGAGKFDQAFKIICEMMGKGFVPDDSTYSKVIGFLCDASKVEKAFSLFEEMKR 486
+ LC G+ ++A + E +GKG P+ Y+ +I L + + +A L EM
Sbjct: 360 RSLIDGLCHEGETNRALALFNEALGKGIKPNVILYNTLIKGLSNQGMILEAAQLANEMSE 419
Query: 487 NGIVPSVYTYTILIDSFCKAGLIQQARKWFDEMLHKGCTPNVVTYTALIHAYLKAKQMPV 546
G++P V T+ IL++ CK G + A M+ KG P++ T+ LIH Y +M
Sbjct: 420 KGLIPEVQTFNILVNGLCKMGCVSDADGLVKVMISKGYFPDIFTFNILIHGYSTQLKMEN 479
Query: 547 ADELFEMMLLEGCKPNVVTYTALIDGHCKAGQIEKACQIYARMRGDIESSDMDKYFKLDH 606
A E+ ++ML G P+V TY +L++G CK + E M+ Y +
Sbjct: 480 ALEILDVMLDNGVDPDVYTYNSLLNGLCKTSKFEDV---------------METYKTMVE 524
Query: 607 NNCEGPNVITYGALVDGLCKANRVKEAHELLDTMLAHGCEPNQIVYDAVIDGFCKIGKLQ 666
C PN+ T+ L++ LC+ ++ EA LL+ M P+ + + +IDGFCK G L
Sbjct: 525 KGC-APNLFTFNILLESLCRYRKLDEALGLLEEMKNKSVNPDAVTFGTLIDGFCKNGDLD 583
Query: 667 DAQEVFTKMSER-GYSPNLYTYSSFIDCLFKDNRLDLVLKVLSKMLENSCTPNVVIYTEM 725
A +F KM E S + TY+ I + + + K+ +M++ P+ Y M
Sbjct: 584 GAYTLFRKMEEAYKVSSSTPTYNIIIHAFTEKLNVTMAEKLFQEMVDRCLGPDGYTYRLM 643
Query: 726 VDGLCKIGKTDEAYKLMLKMEEKGCNPNVVTYTAMIDGFGKSGKIEQCLELFRDMCSKGC 785
VDG CK G + YK +L+M E G P++ T +I+ ++ + + M KG
Sbjct: 644 VDGFCKTGNVNLGYKFLLEMMENGFIPSLTTLGRVINCLCVEDRVYEAAGIIHRMVQKGL 703
Query: 786 APNFI 790
P +
Sbjct: 704 VPEAV 708
Score = 211 bits (537), Expect = 2e-54
Identities = 145/569 (25%), Positives = 269/569 (46%), Gaps = 35/569 (6%)
Query: 195 EELGRLKDFGYKPSQTTYNALIQVFLRADKLDTAYLVKREMLSYAFVMDRYTLSCFAYSL 254
E G++ G +T+N L++V + + + +++ + + +T + F L
Sbjct: 202 ELFGKMLASGVSLCLSTFNKLLRVLCKKGDVKECEKLLDKVIKRGVLPNLFTYNLFIQGL 261
Query: 255 CKGGKCREAFDLID--EAEDFVPDTVFYNRMVSGLCEASLFEEAMDILHRMRSSSCIPNV 312
C+ G+ A ++ + PD + YN ++ GLC+ S F+EA L +M + P+
Sbjct: 262 CQRGELDGAVRMVGCLIEQGPKPDVITYNNLIYGLCKNSKFQEAEVYLGKMVNEGLEPDS 321
Query: 313 VTYRILLSGCLRKGQLGRCKRILSMMITEGCYPNREIFNSLIHAYCKSRDYSYAYKLFKK 372
TY L++G + G + +RI+ + G P++ + SLI C + + A LF +
Sbjct: 322 YTYNTLIAGYCKGGMVQLAERIVGDAVFNGFVPDQFTYRSLIDGLCHEGETNRALALFNE 381
Query: 373 MIKCGCQPGYLVYNIFIGSVCSNEEQPSSDILDLVEKAYSEMLDLGVVLNKVNVSNFARC 432
+ G +P ++YN I + SN+ IL+ + A +EM + G++ +
Sbjct: 382 ALGKGIKPNVILYNTLIKGL-SNQGM----ILEAAQLA-NEMSEKGLIPEVQTFNILVNG 435
Query: 433 LCGAGKFDQAFKIICEMMGKGFVPDDSTYSKVIGFLCDASKVEKAFSLFEEMKRNGIVPS 492
LC G A ++ M+ KG+ PD T++ +I K+E A + + M NG+ P
Sbjct: 436 LCKMGCVSDADGLVKVMISKGYFPDIFTFNILIHGYSTQLKMENALEILDVMLDNGVDPD 495
Query: 493 VYTYTILIDSFCKAGLIQQARKWFDEMLHKGCTPNVVTYTALIHAYLKAKQMPVADELFE 552
VYTY L++ CK + + + M+ KGC PN+ T+ L+ + + +++ A L E
Sbjct: 496 VYTYNSLLNGLCKTSKFEDVMETYKTMVEKGCAPNLFTFNILLESLCRYRKLDEALGLLE 555
Query: 553 MMLLEGCKPNVVTYTALIDGHCKAGQIEKACQIYARMRGDIESSDMDKYFKLDHNNCEGP 612
M + P+ VT+ LIDG CK G ++ A ++ + M++ +K+
Sbjct: 556 EMKNKSVNPDAVTFGTLIDGFCKNGDLDGAYTLFRK---------MEEAYKV------SS 600
Query: 613 NVITYGALVDGLCKANRVKEAHELLDTMLAHGCEPNQIVYDAVIDGFCKIGKLQDAQEVF 672
+ TY ++ + V A +L M+ P+ Y ++DGFCK G + +
Sbjct: 601 STPTYNIIIHAFTEKLNVTMAEKLFQEMVDRCLGPDGYTYRLMVDGFCKTGNVNLGYKFL 660
Query: 673 TKMSERGYSPNLYTYSSFIDCLFKDNRLDLVLKVLSKMLENSCTPNVVIYTEMVDGLCKI 732
+M E G+ P+L T I+CL ++R+ ++ +M++ P E V+ +C +
Sbjct: 661 LEMMENGFIPSLTTLGRVINCLCVEDRVYEAAGIIHRMVQKGLVP------EAVNTICDV 714
Query: 733 GKTD-EAYKLMLK-MEEKGCNPNVVTYTA 759
K + A KL+L+ + +K C +TY A
Sbjct: 715 DKKEVAAPKLVLEDLLKKSC----ITYYA 739
Score = 211 bits (536), Expect = 2e-54
Identities = 145/542 (26%), Positives = 249/542 (45%), Gaps = 29/542 (5%)
Query: 403 ILDLVEKAYSEMLDLGVVLNKVNVSNFARCLCGAGKFDQAFKIICEMMGKGFVPDDSTYS 462
++D+ E + ML+ GV + + N+ R GK +A + M P +Y+
Sbjct: 63 LVDMRENVGNHMLE-GVYVGAMK--NYGR----KGKVQEAVNVFERMDFYDCEPTVFSYN 115
Query: 463 KVIGFLCDASKVEKAFSLFEEMKRNGIVPSVYTYTILIDSFCKAGLIQQARKWFDEMLHK 522
++ L D+ ++A ++ M+ GI P VY++TI + SFCK A + + M +
Sbjct: 116 AIMSVLVDSGYFDQAHKVYMRMRDRGITPDVYSFTIRMKSFCKTSRPHAALRLLNNMSSQ 175
Query: 523 GCTPNVVTYTALIHAYLKAKQMPVADELFEMMLLEGCKPNVVTYTALIDGHCKAGQIEKA 582
GC NVV Y ++ + + ELF ML G + T+ L+ CK G +++
Sbjct: 176 GCEMNVVAYCTVVGGFYEENFKAEGYELFGKMLASGVSLCLSTFNKLLRVLCKKGDVKEC 235
Query: 583 CQIYARMRGDIESSDMDKYFKLDHNNCEGPNVITYGALVDGLCKANRVKEAHELLDTMLA 642
++ +DK K PN+ TY + GLC+ + A ++ ++
Sbjct: 236 EKL------------LDKVIK----RGVLPNLFTYNLFIQGLCQRGELDGAVRMVGCLIE 279
Query: 643 HGCEPNQIVYDAVIDGFCKIGKLQDAQEVFTKMSERGYSPNLYTYSSFIDCLFKDNRLDL 702
G +P+ I Y+ +I G CK K Q+A+ KM G P+ YTY++ I K + L
Sbjct: 280 QGPKPDVITYNNLIYGLCKNSKFQEAEVYLGKMVNEGLEPDSYTYNTLIAGYCKGGMVQL 339
Query: 703 VLKVLSKMLENSCTPNVVIYTEMVDGLCKIGKTDEAYKLMLKMEEKGCNPNVVTYTAMID 762
+++ + N P+ Y ++DGLC G+T+ A L + KG PNV+ Y +I
Sbjct: 340 AERIVGDAVFNGFVPDQFTYRSLIDGLCHEGETNRALALFNEALGKGIKPNVILYNTLIK 399
Query: 763 GFGKSGKIEQCLELFRDMCSKGCAPNFITYRVLINHCCSNGLLDEAYKLLDEMKQTYWPK 822
G G I + +L +M KG P T+ +L+N C G + +A L+ M +
Sbjct: 400 GLSNQGMILEAAQLANEMSEKGLIPEVQTFNILVNGLCKMGCVSDADGLVKVMISKGYFP 459
Query: 823 HILSHRKIIEGFSQE--FITSIGLLDELSENESVPVDSLYRILIDNYIKAGRLEVALDLL 880
I + +I G+S + ++ +LD + +N P Y L++ K + E D++
Sbjct: 460 DIFTFNILIHGYSTQLKMENALEILDVMLDNGVDPDVYTYNSLLNGLCKTSKFE---DVM 516
Query: 881 EEISSSPSHAVS-NKYLYASLIENLSHASKVDKALELYASMISKNVVPELSILVHLIKGL 939
E + + N + + L+E+L K+D+AL L M +K+V P+ LI G
Sbjct: 517 ETYKTMVEKGCAPNLFTFNILLESLCRYRKLDEALGLLEEMKNKSVNPDAVTFGTLIDGF 576
Query: 940 IK 941
K
Sbjct: 577 CK 578
Score = 146 bits (368), Expect = 6e-35
Identities = 110/442 (24%), Positives = 208/442 (46%), Gaps = 21/442 (4%)
Query: 535 IHAYLKAKQMPV-ADELFEMMLLE-GCKPNVVTYTALIDGHCKAGQIEKACQIYARMRGD 592
+ A +K ++ P+ A E+F M E G K + TY ++I+ G+ E ++ MR +
Sbjct: 10 VTAVIKCQKDPMKALEMFNSMRKEVGFKHTLSTYRSVIEKLGYYGKFEAMEEVLVDMREN 69
Query: 593 IESSDMDKYFKLDHNNCEGPNVITYGALVDGLCKANRVKEAHELLDTMLAHGCEPNQIVY 652
+ + ++ + N YG + +V+EA + + M + CEP Y
Sbjct: 70 VGNHMLEGVYVGAMKN--------YG-------RKGKVQEAVNVFERMDFYDCEPTVFSY 114
Query: 653 DAVIDGFCKIGKLQDAQEVFTKMSERGYSPNLYTYSSFIDCLFKDNRLDLVLKVLSKMLE 712
+A++ G A +V+ +M +RG +P++Y+++ + K +R L++L+ M
Sbjct: 115 NAIMSVLVDSGYFDQAHKVYMRMRDRGITPDVYSFTIRMKSFCKTSRPHAALRLLNNMSS 174
Query: 713 NSCTPNVVIYTEMVDGLCKIGKTDEAYKLMLKMEEKGCNPNVVTYTAMIDGFGKSGKIEQ 772
C NVV Y +V G + E Y+L KM G + + T+ ++ K G +++
Sbjct: 175 QGCEMNVVAYCTVVGGFYEENFKAEGYELFGKMLASGVSLCLSTFNKLLRVLCKKGDVKE 234
Query: 773 CLELFRDMCSKGCAPNFITYRVLINHCCSNGLLDEAYKLLDEMKQTYWPKHILSHRKIIE 832
C +L + +G PN TY + I C G LD A +++ + + ++++ +I
Sbjct: 235 CEKLLDKVIKRGVLPNLFTYNLFIQGLCQRGELDGAVRMVGCLIEQGPKPDVITYNNLIY 294
Query: 833 GF--SQEFITSIGLLDELSENESVPVDSLYRILIDNYIKAGRLEVALDLLEEISSSPSHA 890
G + +F + L ++ P Y LI Y K G +++A ++ + + +
Sbjct: 295 GLCKNSKFQEAEVYLGKMVNEGLEPDSYTYNTLIAGYCKGGMVQLAERIVGD--AVFNGF 352
Query: 891 VSNKYLYASLIENLSHASKVDKALELYASMISKNVVPELSILVHLIKGLIKVDKWQEALQ 950
V +++ Y SLI+ L H + ++AL L+ + K + P + + LIKGL EA Q
Sbjct: 353 VPDQFTYRSLIDGLCHEGETNRALALFNEALGKGIKPNVILYNTLIKGLSNQGMILEAAQ 412
Query: 951 LSDSICQMVCLTLSQTFQPLVN 972
L++ + + + QTF LVN
Sbjct: 413 LANEMSEKGLIPEVQTFNILVN 434
Score = 124 bits (311), Expect = 3e-28
Identities = 92/363 (25%), Positives = 155/363 (42%), Gaps = 21/363 (5%)
Query: 176 LLNFLVRKCCRNGWWNMALEELGRLKDFGYKPSQTTYNALIQVFLRADKLDTAYLVKREM 235
L N L++ G A + + + G P T+N L+ + + A + + M
Sbjct: 393 LYNTLIKGLSNQGMILEAAQLANEMSEKGLIPEVQTFNILVNGLCKMGCVSDADGLVKVM 452
Query: 236 LSYAFVMDRYTLSCFAYSLCKGGKCREAFDLIDEAED--FVPDTVFYNRMVSGLCEASLF 293
+S + D +T + + K A +++D D PD YN +++GLC+ S F
Sbjct: 453 ISKGYFPDIFTFNILIHGYSTQLKMENALEILDVMLDNGVDPDVYTYNSLLNGLCKTSKF 512
Query: 294 EEAMDILHRMRSSSCIPNVVTYRILLSGCLRKGQLGRCKRILSMMITEGCYPNREIFNSL 353
E+ M+ M C PN+ T+ ILL R +L +L M + P+ F +L
Sbjct: 513 EDVMETYKTMVEKGCAPNLFTFNILLESLCRYRKLDEALGLLEEMKNKSVNPDAVTFGTL 572
Query: 354 IHAYCKSRDYSYAYKLFKKMIKC-GCQPGYLVYNIFIGSVCSNEEQPSSDILDLVEKAYS 412
I +CK+ D AY LF+KM + YNI I + + + EK +
Sbjct: 573 IDGFCKNGDLDGAYTLFRKMEEAYKVSSSTPTYNIIIHAFTEKLN------VTMAEKLFQ 626
Query: 413 EMLDLGVVLNKVNVSNFARCLCGAGKFDQAFKIICEMMGKGFVPDDSTYSKVIGFLCDAS 472
EM+D + + C G + +K + EMM GF+P +T +VI LC
Sbjct: 627 EMVDRCLGPDGYTYRLMVDGFCKTGNVNLGYKFLLEMMENGFIPSLTTLGRVINCLCVED 686
Query: 473 KVEKAFSLFEEMKRNGIVPSVYTYTILIDSFCKAGLIQQA--RKWFDEMLHKGCTPNVVT 530
+V +A + M + G+VP +++ C + A + +++L K C +T
Sbjct: 687 RVYEAAGIIHRMVQKGLVPEA------VNTICDVDKKEVAAPKLVLEDLLKKSC----IT 736
Query: 531 YTA 533
Y A
Sbjct: 737 YYA 739
>At1g63080 unknown protein
Length = 614
Score = 263 bits (672), Expect = 4e-70
Identities = 168/560 (30%), Positives = 271/560 (48%), Gaps = 24/560 (4%)
Query: 259 KCREAFDLIDEAEDF--VPDTVFYNRMVSGLCEASLFEEAMDILHRMRSSSCIPNVVTYR 316
K EA DL E P V +++++S + + F+ + +M N+ TY
Sbjct: 45 KLDEAVDLFGEMVKSRPFPSIVEFSKLLSAIAKMKKFDLVISFGEKMEILGVSHNLYTYN 104
Query: 317 ILLSGCLRKGQLGRCKRILSMMITEGCYPNREIFNSLIHAYCKSRDYSYAYKLFKKMIKC 376
I+++ R+ QL IL M+ G P+ NSL++ +C S A L +M++
Sbjct: 105 IMINCLCRRSQLSFALAILGKMMKLGYGPSIVTLNSLLNGFCHGNRISEAVALVDQMVEM 164
Query: 377 GCQPGYLVYNIFIGSVCSNEEQPSSDILDLVEKAYSEMLDLGVVLNKVNVSNFARCLCGA 436
G QP + + + + + + +S+ + LVE+ M+ G + V LC
Sbjct: 165 GYQPDTVTFTTLVHGLFQHNK--ASEAVALVER----MVVKGCQPDLVTYGAVINGLCKR 218
Query: 437 GKFDQAFKIICEMMGKGFVPDDSTYSKVIGFLCDASKVEKAFSLFEEMKRNGIVPSVYTY 496
G+ D A ++ +M D YS VI LC V+ A +LF EM GI P V+TY
Sbjct: 219 GEPDLALNLLNKMEKGKIEADVVIYSTVIDSLCKYRHVDDALNLFTEMDNKGIRPDVFTY 278
Query: 497 TILIDSFCKAGLIQQARKWFDEMLHKGCTPNVVTYTALIHAYLKAKQMPVADELFEMMLL 556
+ LI C G A + +ML + PNVVT+ +LI A+ K ++ A++LF+ M+
Sbjct: 279 SSLISCLCNYGRWSDASRLLSDMLERKINPNVVTFNSLIDAFAKEGKLIEAEKLFDEMIQ 338
Query: 557 EGCKPNVVTYTALIDGHCKAGQIEKACQIYARMRGDIESSDMDKYFKLDHNNCEGPNVIT 616
PN+VTY +LI+G C ++++A QI+ M +C P+V+T
Sbjct: 339 RSIDPNIVTYNSLINGFCMHDRLDEAQQIFTLMVS---------------KDCL-PDVVT 382
Query: 617 YGALVDGLCKANRVKEAHELLDTMLAHGCEPNQIVYDAVIDGFCKIGKLQDAQEVFTKMS 676
Y L++G CKA +V + EL M G N + Y +I GF + +AQ VF +M
Sbjct: 383 YNTLINGFCKAKKVVDGMELFRDMSRRGLVGNTVTYTTLIHGFFQASDCDNAQMVFKQMV 442
Query: 677 ERGYSPNLYTYSSFIDCLFKDNRLDLVLKVLSKMLENSCTPNVVIYTEMVDGLCKIGKTD 736
G PN+ TY++ +D L K+ +L+ + V + ++ P++ Y M +G+CK GK +
Sbjct: 443 SDGVHPNIMTYNTLLDGLCKNGKLEKAMVVFEYLQKSKMEPDIYTYNIMSEGMCKAGKVE 502
Query: 737 EAYKLMLKMEEKGCNPNVVTYTAMIDGFGKSGKIEQCLELFRDMCSKGCAPNFITYRVLI 796
+ + L + KG P+V+ Y MI GF K G E+ LF M G P+ TY LI
Sbjct: 503 DGWDLFCSLSLKGVKPDVIAYNTMISGFCKKGLKEEAYTLFIKMKEDGPLPDSGTYNTLI 562
Query: 797 NHCCSNGLLDEAYKLLDEMK 816
+G + +L+ EM+
Sbjct: 563 RAHLRDGDKAASAELIKEMR 582
Score = 258 bits (658), Expect = 2e-68
Identities = 158/532 (29%), Positives = 258/532 (47%), Gaps = 27/532 (5%)
Query: 280 YNRMVSGLCEASLFEEAMDILHRMRSSSCIPNVVTYRILLSGCLRKGQLGRCKRILSMMI 339
YN M++ LC S A+ IL +M P++VT LL+G ++ ++ M+
Sbjct: 103 YNIMINCLCRRSQLSFALAILGKMMKLGYGPSIVTLNSLLNGFCHGNRISEAVALVDQMV 162
Query: 340 TEGCYPNREIFNSLIHAYCKSRDYSYAYKLFKKMIKCGCQPGYLVYNIFIGSVCSNEEQP 399
G P+ F +L+H + S A L ++M+ GCQP + Y I +C E
Sbjct: 163 EMGYQPDTVTFTTLVHGLFQHNKASEAVALVERMVVKGCQPDLVTYGAVINGLCKRGEPD 222
Query: 400 SSDILDLVEKAYSEMLDLGVVLNKVNVSNFARCLCGAGKFDQAFKIICEMMGKGFVPDDS 459
+ L+L+ K ++ VV+ + + LC D A + EM KG PD
Sbjct: 223 LA--LNLLNKMEKGKIEADVVIYSTVIDS----LCKYRHVDDALNLFTEMDNKGIRPDVF 276
Query: 460 TYSKVIGFLCDASKVEKAFSLFEEMKRNGIVPSVYTYTILIDSFCKAGLIQQARKWFDEM 519
TYS +I LC+ + A L +M I P+V T+ LID+F K G + +A K FDEM
Sbjct: 277 TYSSLISCLCNYGRWSDASRLLSDMLERKINPNVVTFNSLIDAFAKEGKLIEAEKLFDEM 336
Query: 520 LHKGCTPNVVTYTALIHAYLKAKQMPVADELFEMMLLEGCKPNVVTYTALIDGHCKAGQI 579
+ + PN+VTY +LI+ + ++ A ++F +M+ + C P+VVTY LI+G CKA ++
Sbjct: 337 IQRSIDPNIVTYNSLINGFCMHDRLDEAQQIFTLMVSKDCLPDVVTYNTLINGFCKAKKV 396
Query: 580 EKACQIYARM--RGDIESSDMDKYFKLDHNNCEGPNVITYGALVDGLCKANRVKEAHELL 637
+++ M RG + N +TY L+ G +A+ A +
Sbjct: 397 VDGMELFRDMSRRGLVG------------------NTVTYTTLIHGFFQASDCDNAQMVF 438
Query: 638 DTMLAHGCEPNQIVYDAVIDGFCKIGKLQDAQEVFTKMSERGYSPNLYTYSSFIDCLFKD 697
M++ G PN + Y+ ++DG CK GKL+ A VF + + P++YTY+ + + K
Sbjct: 439 KQMVSDGVHPNIMTYNTLLDGLCKNGKLEKAMVVFEYLQKSKMEPDIYTYNIMSEGMCKA 498
Query: 698 NRLDLVLKVLSKMLENSCTPNVVIYTEMVDGLCKIGKTDEAYKLMLKMEEKGCNPNVVTY 757
+++ + + P+V+ Y M+ G CK G +EAY L +KM+E G P+ TY
Sbjct: 499 GKVEDGWDLFCSLSLKGVKPDVIAYNTMISGFCKKGLKEEAYTLFIKMKEDGPLPDSGTY 558
Query: 758 TAMIDGFGKSGKIEQCLELFRDMCSKGCAPNFITYRVLINHCCSNGLLDEAY 809
+I + G EL ++M S A + TY L+ +G LD+ +
Sbjct: 559 NTLIRAHLRDGDKAASAELIKEMRSCRFAGDASTYG-LVTDMLHDGRLDKGF 609
Score = 238 bits (606), Expect = 2e-62
Identities = 152/540 (28%), Positives = 265/540 (48%), Gaps = 20/540 (3%)
Query: 438 KFDQAFKIICEMMGKGFVPDDSTYSKVIGFLCDASKVEKAFSLFEEMKRNGIVPSVYTYT 497
K D+A + EM+ P +SK++ + K + S E+M+ G+ ++YTY
Sbjct: 45 KLDEAVDLFGEMVKSRPFPSIVEFSKLLSAIAKMKKFDLVISFGEKMEILGVSHNLYTYN 104
Query: 498 ILIDSFCKAGLIQQARKWFDEMLHKGCTPNVVTYTALIHAYLKAKQMPVADELFEMMLLE 557
I+I+ C+ + A +M+ G P++VT +L++ + ++ A L + M+
Sbjct: 105 IMINCLCRRSQLSFALAILGKMMKLGYGPSIVTLNSLLNGFCHGNRISEAVALVDQMVEM 164
Query: 558 GCKPNVVTYTALIDGHCKAGQIEKACQIYARMRGDIESSDMDKYFKLDHNNCEGPNVITY 617
G +P+ VT+T L+ G + + +A + RM C+ P+++TY
Sbjct: 165 GYQPDTVTFTTLVHGLFQHNKASEAVALVERMVV---------------KGCQ-PDLVTY 208
Query: 618 GALVDGLCKANRVKEAHELLDTMLAHGCEPNQIVYDAVIDGFCKIGKLQDAQEVFTKMSE 677
GA+++GLCK A LL+ M E + ++Y VID CK + DA +FT+M
Sbjct: 209 GAVINGLCKRGEPDLALNLLNKMEKGKIEADVVIYSTVIDSLCKYRHVDDALNLFTEMDN 268
Query: 678 RGYSPNLYTYSSFIDCLFKDNRLDLVLKVLSKMLENSCTPNVVIYTEMVDGLCKIGKTDE 737
+G P+++TYSS I CL R ++LS MLE PNVV + ++D K GK E
Sbjct: 269 KGIRPDVFTYSSLISCLCNYGRWSDASRLLSDMLERKINPNVVTFNSLIDAFAKEGKLIE 328
Query: 738 AYKLMLKMEEKGCNPNVVTYTAMIDGFGKSGKIEQCLELFRDMCSKGCAPNFITYRVLIN 797
A KL +M ++ +PN+VTY ++I+GF ++++ ++F M SK C P+ +TY LIN
Sbjct: 329 AEKLFDEMIQRSIDPNIVTYNSLINGFCMHDRLDEAQQIFTLMVSKDCLPDVVTYNTLIN 388
Query: 798 HCCSNGLLDEAYKLLDEMKQTYWPKHILSHRKIIEGFSQ--EFITSIGLLDELSENESVP 855
C + + +L +M + + +++ +I GF Q + + + ++ + P
Sbjct: 389 GFCKAKKVVDGMELFRDMSRRGLVGNTVTYTTLIHGFFQASDCDNAQMVFKQMVSDGVHP 448
Query: 856 VDSLYRILIDNYIKAGRLEVALDLLEEISSSPSHAVSNKYLYASLIENLSHASKVDKALE 915
Y L+D K G+LE A+ + E + S + Y Y + E + A KV+ +
Sbjct: 449 NIMTYNTLLDGLCKNGKLEKAMVVFEYLQKSKME--PDIYTYNIMSEGMCKAGKVEDGWD 506
Query: 916 LYASMISKNVVPELSILVHLIKGLIKVDKWQEALQLSDSICQMVCLTLSQTFQPLVNLNL 975
L+ S+ K V P++ +I G K +EA L + + L S T+ L+ +L
Sbjct: 507 LFCSLSLKGVKPDVIAYNTMISGFCKKGLKEEAYTLFIKMKEDGPLPDSGTYNTLIRAHL 566
Score = 206 bits (525), Expect = 4e-53
Identities = 149/574 (25%), Positives = 254/574 (43%), Gaps = 32/574 (5%)
Query: 140 FDKLLDLLGCNVNADDRVPLKFLMEIKDDDHELLRRLLNFLVRKCCRNGWWNMALEELGR 199
F KLL + D + MEI H L N ++ CR + AL LG+
Sbjct: 68 FSKLLSAIAKMKKFDLVISFGEKMEILGVSHNLYT--YNIMINCLCRRSQLSFALAILGK 125
Query: 200 LKDFGYKPSQTTYNALIQVFLRADKLDTAYLVKREMLSYAFVMDRYTLSCFAYSLCKGGK 259
+ GY PS T N+L+ F +++ A + +M+ + D T + + L + K
Sbjct: 126 MMKLGYGPSIVTLNSLLNGFCHGNRISEAVALVDQMVEMGYQPDTVTFTTLVHGLFQHNK 185
Query: 260 CREAFDLIDE--AEDFVPDTVFYNRMVSGLCEASLFEEAMDILHRMRSSSCIPNVVTYRI 317
EA L++ + PD V Y +++GLC+ + A+++L++M +VV Y
Sbjct: 186 ASEAVALVERMVVKGCQPDLVTYGAVINGLCKRGEPDLALNLLNKMEKGKIEADVVIYST 245
Query: 318 LLSGCLRKGQLGRCKRILSMMITEGCYPNREIFNSLIHAYCKSRDYSYAYKLFKKMIKCG 377
++ + + + + M +G P+ ++SLI C +S A +L M++
Sbjct: 246 VIDSLCKYRHVDDALNLFTEMDNKGIRPDVFTYSSLISCLCNYGRWSDASRLLSDMLERK 305
Query: 378 CQPGYLVYNIFIGSVCSNEEQPSSDILDLVEKAYSEMLDLGVVLNKVNVSNFARCLCGAG 437
P + +N I + + L EK + EM+ + N V ++ C
Sbjct: 306 INPNVVTFNSLIDAFAKEGK------LIEAEKLFDEMIQRSIDPNIVTYNSLINGFCMHD 359
Query: 438 KFDQAFKIICEMMGKGFVPDDSTYSKVIGFLCDASKVEKAFSLFEEMKRNGIVPSVYTYT 497
+ D+A +I M+ K +PD TY+ +I C A KV LF +M R G+V + TYT
Sbjct: 360 RLDEAQQIFTLMVSKDCLPDVVTYNTLINGFCKAKKVVDGMELFRDMSRRGLVGNTVTYT 419
Query: 498 ILIDSFCKAGLIQQARKWFDEMLHKGCTPNVVTYTALIHAYLKAKQMPVADELFEMMLLE 557
LI F +A A+ F +M+ G PN++TY L+ K ++ A +FE +
Sbjct: 420 TLIHGFFQASDCDNAQMVFKQMVSDGVHPNIMTYNTLLDGLCKNGKLEKAMVVFEYLQKS 479
Query: 558 GCKPNVVTYTALIDGHCKAGQIEKACQIYARMRGDIESSDMDKYFKLDHNNCEG--PNVI 615
+P++ TY + +G CKAG++E ++ + + +G P+VI
Sbjct: 480 KMEPDIYTYNIMSEGMCKAGKVEDGWDLFCSL------------------SLKGVKPDVI 521
Query: 616 TYGALVDGLCKANRVKEAHELLDTMLAHGCEPNQIVYDAVIDGFCKIGKLQDAQEVFTKM 675
Y ++ G CK +EA+ L M G P+ Y+ +I + G + E+ +M
Sbjct: 522 AYNTMISGFCKKGLKEEAYTLFIKMKEDGPLPDSGTYNTLIRAHLRDGDKAASAELIKEM 581
Query: 676 SERGYSPNLYTYSSFIDCLFKDNRLDL-VLKVLS 708
++ + TY D L D RLD L+VLS
Sbjct: 582 RSCRFAGDASTYGLVTDML-HDGRLDKGFLEVLS 614
>At1g09900 hypothetical protein
Length = 598
Score = 259 bits (663), Expect = 4e-69
Identities = 152/536 (28%), Positives = 259/536 (47%), Gaps = 54/536 (10%)
Query: 421 LNKVNVSNFARCLCGAGKFDQAFKIICEMMGKGFVPDDSTYSKVIGFLCDASKVEKAFSL 480
L V +N R + G+ ++ FK + M+ G VPD + +I C K KA +
Sbjct: 100 LEDVESNNHLRQMVRTGELEEGFKFLENMVYHGNVPDIIPCTTLIRGFCRLGKTRKAAKI 159
Query: 481 FEEMKRNGIVPSVYTYTILIDSFCKAGLIQQARKWFDEMLHKGCTPNVVTYTALIHAYLK 540
E ++ +G VP V TY ++I +CKAG I A D M +P+VVTY ++ +
Sbjct: 160 LEILEGSGAVPDVITYNVMISGYCKAGEINNALSVLDRM---SVSPDVVTYNTILRSLCD 216
Query: 541 AKQMPVADELFEMMLLEGCKPNVVTYTALIDGHCKAGQIEKACQIYARMRGDIESSDMDK 600
+ ++ A E+ + ML C P+V+TYT LI+ C+ + A ++ MR
Sbjct: 217 SGKLKQAMEVLDRMLQRDCYPDVITYTILIEATCRDSGVGHAMKLLDEMRD--------- 267
Query: 601 YFKLDHNNCEGPNVITYGALVDGLCKANRVKEAHELLDTMLAHGCEPNQIVYDAVIDGFC 660
C P+V+TY LV+G+CK R+ EA + L+ M + GC+PN I ++ ++ C
Sbjct: 268 ------RGCT-PDVVTYNVLVNGICKEGRLDEAIKFLNDMPSSGCQPNVITHNIILRSMC 320
Query: 661 KIGKLQDAQEVFTKMSERGYSPNLYTYSSFIDCLFKDNRLDLVLKVLSKMLENSCTPNVV 720
G+ DA+++ M +G+SP++ T++ I+ L + L + +L KM ++ C PN +
Sbjct: 321 STGRWMDAEKLLADMLRKGFSPSVVTFNILINFLCRKGLLGRAIDILEKMPQHGCQPNSL 380
Query: 721 IYTEMVDGLCKIGKTDEAYKLMLKMEEKGCNPNVVTYTAMIDGFGKSGKIEQCLELFRDM 780
Y ++ G CK K D A + + +M +GC P++VTY M+ K GK+E +E+ +
Sbjct: 381 SYNPLLHGFCKEKKMDRAIEYLERMVSRGCYPDIVTYNTMLTALCKDGKVEDAVEILNQL 440
Query: 781 CSKGCAPNFITYRVLINHCCSNGLLDEAYKLLDEMKQTYWPKHILSHRKIIEGFSQEFIT 840
SKGC+P ITY +I+ G +A KLLDEM+ +++ ++ G S+E
Sbjct: 441 SSKGCSPVLITYNTVIDGLAKAGKTGKAIKLLDEMRAKDLKPDTITYSSLVGGLSRE--- 497
Query: 841 SIGLLDELSENESVPVDSLYRILIDNYIKAGRLEVALDLLEEISSSPSHAVSNKYLYASL 900
G+++ A+ E N + S+
Sbjct: 498 ------------------------------GKVDEAIKFFHEFERMGIR--PNAVTFNSI 525
Query: 901 IENLSHASKVDKALELYASMISKNVVPELSILVHLIKGLIKVDKWQEALQLSDSIC 956
+ L + + D+A++ MI++ P + LI+GL +EAL+L + +C
Sbjct: 526 MLGLCKSRQTDRAIDFLVFMINRGCKPNETSYTILIEGLAYEGMAKEALELLNELC 581
Score = 242 bits (618), Expect = 7e-64
Identities = 140/498 (28%), Positives = 240/498 (48%), Gaps = 27/498 (5%)
Query: 254 LCKGGKCREAFDLIDEA--EDFVPDTVFYNRMVSGLCEASLFEEAMDILHRMRSSSCIPN 311
+ + G+ E F ++ VPD + ++ G C +A IL + S +P+
Sbjct: 112 MVRTGELEEGFKFLENMVYHGNVPDIIPCTTLIRGFCRLGKTRKAAKILEILEGSGAVPD 171
Query: 312 VVTYRILLSGCLRKGQLGRCKRILSMMITEGCYPNREIFNSLIHAYCKSRDYSYAYKLFK 371
V+TY +++SG + G++ +L M P+ +N+++ + C S A ++
Sbjct: 172 VITYNVMISGYCKAGEINNALSVLDRMSVS---PDVVTYNTILRSLCDSGKLKQAMEVLD 228
Query: 372 KMIKCGCQPGYLVYNIFIGSVCSNEEQPSSDILDLVEKAYSEMLDLGVVLNKVNVSNFAR 431
+M++ C P + Y I I + C + + K EM D G + V +
Sbjct: 229 RMLQRDCYPDVITYTILIEATCRDSG------VGHAMKLLDEMRDRGCTPDVVTYNVLVN 282
Query: 432 CLCGAGKFDQAFKIICEMMGKGFVPDDSTYSKVIGFLCDASKVEKAFSLFEEMKRNGIVP 491
+C G+ D+A K + +M G P+ T++ ++ +C + A L +M R G P
Sbjct: 283 GICKEGRLDEAIKFLNDMPSSGCQPNVITHNIILRSMCSTGRWMDAEKLLADMLRKGFSP 342
Query: 492 SVYTYTILIDSFCKAGLIQQARKWFDEMLHKGCTPNVVTYTALIHAYLKAKQMPVADELF 551
SV T+ ILI+ C+ GL+ +A ++M GC PN ++Y L+H + K K+M A E
Sbjct: 343 SVVTFNILINFLCRKGLLGRAIDILEKMPQHGCQPNSLSYNPLLHGFCKEKKMDRAIEYL 402
Query: 552 EMMLLEGCKPNVVTYTALIDGHCKAGQIEKACQIYARMRGDIESSDMDKYFKLDHNNCEG 611
E M+ GC P++VTY ++ CK G++E A +I +L C
Sbjct: 403 ERMVSRGCYPDIVTYNTMLTALCKDGKVEDAVEILN---------------QLSSKGC-S 446
Query: 612 PNVITYGALVDGLCKANRVKEAHELLDTMLAHGCEPNQIVYDAVIDGFCKIGKLQDAQEV 671
P +ITY ++DGL KA + +A +LLD M A +P+ I Y +++ G + GK+ +A +
Sbjct: 447 PVLITYNTVIDGLAKAGKTGKAIKLLDEMRAKDLKPDTITYSSLVGGLSREGKVDEAIKF 506
Query: 672 FTKMSERGYSPNLYTYSSFIDCLFKDNRLDLVLKVLSKMLENSCTPNVVIYTEMVDGLCK 731
F + G PN T++S + L K + D + L M+ C PN YT +++GL
Sbjct: 507 FHEFERMGIRPNAVTFNSIMLGLCKSRQTDRAIDFLVFMINRGCKPNETSYTILIEGLAY 566
Query: 732 IGKTDEAYKLMLKMEEKG 749
G EA +L+ ++ KG
Sbjct: 567 EGMAKEALELLNELCNKG 584
Score = 209 bits (532), Expect = 6e-54
Identities = 124/425 (29%), Positives = 214/425 (50%), Gaps = 23/425 (5%)
Query: 549 ELFEMMLLEGCKPNVVTYTALIDGHCKAGQIEKACQIYARMRGDIESSDMDKYFKLDHNN 608
+ E M+ G P+++ T LI G C+ G+ KA +I + G +
Sbjct: 123 KFLENMVYHGNVPDIIPCTTLIRGFCRLGKTRKAAKILEILEG----------------S 166
Query: 609 CEGPNVITYGALVDGLCKANRVKEAHELLDTMLAHGCEPNQIVYDAVIDGFCKIGKLQDA 668
P+VITY ++ G CKA + A +LD M P+ + Y+ ++ C GKL+ A
Sbjct: 167 GAVPDVITYNVMISGYCKAGEINNALSVLDRM---SVSPDVVTYNTILRSLCDSGKLKQA 223
Query: 669 QEVFTKMSERGYSPNLYTYSSFIDCLFKDNRLDLVLKVLSKMLENSCTPNVVIYTEMVDG 728
EV +M +R P++ TY+ I+ +D+ + +K+L +M + CTP+VV Y +V+G
Sbjct: 224 MEVLDRMLQRDCYPDVITYTILIEATCRDSGVGHAMKLLDEMRDRGCTPDVVTYNVLVNG 283
Query: 729 LCKIGKTDEAYKLMLKMEEKGCNPNVVTYTAMIDGFGKSGKIEQCLELFRDMCSKGCAPN 788
+CK G+ DEA K + M GC PNV+T+ ++ +G+ +L DM KG +P+
Sbjct: 284 ICKEGRLDEAIKFLNDMPSSGCQPNVITHNIILRSMCSTGRWMDAEKLLADMLRKGFSPS 343
Query: 789 FITYRVLINHCCSNGLLDEAYKLLDEMKQTYWPKHILSHRKIIEGFSQE--FITSIGLLD 846
+T+ +LIN C GLL A +L++M Q + LS+ ++ GF +E +I L+
Sbjct: 344 VVTFNILINFLCRKGLLGRAIDILEKMPQHGCQPNSLSYNPLLHGFCKEKKMDRAIEYLE 403
Query: 847 ELSENESVPVDSLYRILIDNYIKAGRLEVALDLLEEISSSPSHAVSNKYLYASLIENLSH 906
+ P Y ++ K G++E A+++L ++SS V Y ++I+ L+
Sbjct: 404 RMVSRGCYPDIVTYNTMLTALCKDGKVEDAVEILNQLSSKGCSPVL--ITYNTVIDGLAK 461
Query: 907 ASKVDKALELYASMISKNVVPELSILVHLIKGLIKVDKWQEALQLSDSICQMVCLTLSQT 966
A K KA++L M +K++ P+ L+ GL + K EA++ +M + T
Sbjct: 462 AGKTGKAIKLLDEMRAKDLKPDTITYSSLVGGLSREGKVDEAIKFFHEFERMGIRPNAVT 521
Query: 967 FQPLV 971
F ++
Sbjct: 522 FNSIM 526
Score = 203 bits (516), Expect = 4e-52
Identities = 134/467 (28%), Positives = 221/467 (46%), Gaps = 27/467 (5%)
Query: 180 LVRKCCRNGWWNMALEELGRLKDFGYKPSQTTYNALIQVFLRADKLDTAYLVKREMLSYA 239
L+R CR G A + L L+ G P TYN +I + +A +++ A V M +
Sbjct: 143 LIRGFCRLGKTRKAAKILEILEGSGAVPDVITYNVMISGYCKAGEINNALSVLDRM---S 199
Query: 240 FVMDRYTLSCFAYSLCKGGKCREAFDLIDE--AEDFVPDTVFYNRMVSGLCEASLFEEAM 297
D T + SLC GK ++A +++D D PD + Y ++ C S AM
Sbjct: 200 VSPDVVTYNTILRSLCDSGKLKQAMEVLDRMLQRDCYPDVITYTILIEATCRDSGVGHAM 259
Query: 298 DILHRMRSSSCIPNVVTYRILLSGCLRKGQLGRCKRILSMMITEGCYPNREIFNSLIHAY 357
+L MR C P+VVTY +L++G ++G+L + L+ M + GC PN N ++ +
Sbjct: 260 KLLDEMRDRGCTPDVVTYNVLVNGICKEGRLDEAIKFLNDMPSSGCQPNVITHNIILRSM 319
Query: 358 CKSRDYSYAYKLFKKMIKCGCQPGYLVYNIFIGSVCSNEEQPSSDILDLVEKAYSEMLDL 417
C + + A KL M++ G P + +NI I +C + +D++EK M
Sbjct: 320 CSTGRWMDAEKLLADMLRKGFSPSVVTFNILINFLC--RKGLLGRAIDILEK----MPQH 373
Query: 418 GVVLNKVNVSNFARCLCGAGKFDQAFKIICEMMGKGFVPDDSTYSKVIGFLCDASKVEKA 477
G N ++ + C K D+A + + M+ +G PD TY+ ++ LC KVE A
Sbjct: 374 GCQPNSLSYNPLLHGFCKEKKMDRAIEYLERMVSRGCYPDIVTYNTMLTALCKDGKVEDA 433
Query: 478 FSLFEEMKRNGIVPSVYTYTILIDSFCKAGLIQQARKWFDEMLHKGCTPNVVTYTALIHA 537
+ ++ G P + TY +ID KAG +A K DEM K P+ +TY++L+
Sbjct: 434 VEILNQLSSKGCSPVLITYNTVIDGLAKAGKTGKAIKLLDEMRAKDLKPDTITYSSLVGG 493
Query: 538 YLKAKQMPVADELFEMMLLEGCKPNVVTYTALIDGHCKAGQIEKACQIYARMRGDIESSD 597
+ ++ A + F G +PN VT+ +++ G CK+ Q ++A
Sbjct: 494 LSREGKVDEAIKFFHEFERMGIRPNAVTFNSIMLGLCKSRQTDRA--------------- 538
Query: 598 MDKYFKLDHNNCEGPNVITYGALVDGLCKANRVKEAHELLDTMLAHG 644
+D + + C+ PN +Y L++GL KEA ELL+ + G
Sbjct: 539 IDFLVFMINRGCK-PNETSYTILIEGLAYEGMAKEALELLNELCNKG 584
Score = 180 bits (456), Expect = 4e-45
Identities = 102/388 (26%), Positives = 197/388 (50%), Gaps = 43/388 (11%)
Query: 589 MRGDIESSDMDKYFKLDHN---NCEGPNVITYGALVDGLCKANRVKEAHELLDTMLAHGC 645
+R + + ++++ FK N + P++I L+ G C+ + ++A ++L+ + G
Sbjct: 109 LRQMVRTGELEEGFKFLENMVYHGNVPDIIPCTTLIRGFCRLGKTRKAAKILEILEGSGA 168
Query: 646 EPNQIVYDAVIDGFCKIGKLQDAQEVFTKMSERGYSPNLYTYSSFIDCLFKDNRLDLVLK 705
P+ I Y+ +I G+CK G++ +A V +MS SP++ TY++ + L +L ++
Sbjct: 169 VPDVITYNVMISGYCKAGEINNALSVLDRMS---VSPDVVTYNTILRSLCDSGKLKQAME 225
Query: 706 VLSKMLENSCTPNVVIYTEMVDGLCKIGKTDEAYKLMLKMEEKGCNPNVVTYTAMIDGFG 765
VL +ML+ C P+V+ YT +++ C+ A KL+ +M ++GC P+VVTY +++G
Sbjct: 226 VLDRMLQRDCYPDVITYTILIEATCRDSGVGHAMKLLDEMRDRGCTPDVVTYNVLVNGIC 285
Query: 766 KSGKIEQCLELFRDMCSKGCAPNFITYRVLINHCCSNGLLDEAYKLLDEMKQTYWPKHIL 825
K G++++ ++ DM S GC PN IT+ +++ CS G +A KLL +M +
Sbjct: 286 KEGRLDEAIKFLNDMPSSGCQPNVITHNIILRSMCSTGRWMDAEKLLADMLR-------- 337
Query: 826 SHRKIIEGFSQEFITSIGLLDELSENESVPVDSLYRILIDNYIKAGRLEVALDLLEEISS 885
+GFS +T + ILI+ + G L A+D+LE++
Sbjct: 338 ------KGFSPSVVT-------------------FNILINFLCRKGLLGRAIDILEKM-- 370
Query: 886 SPSHAVS-NKYLYASLIENLSHASKVDKALELYASMISKNVVPELSILVHLIKGLIKVDK 944
P H N Y L+ K+D+A+E M+S+ P++ ++ L K K
Sbjct: 371 -PQHGCQPNSLSYNPLLHGFCKEKKMDRAIEYLERMVSRGCYPDIVTYNTMLTALCKDGK 429
Query: 945 WQEALQLSDSICQMVCLTLSQTFQPLVN 972
++A+++ + + C + T+ +++
Sbjct: 430 VEDAVEILNQLSSKGCSPVLITYNTVID 457
Score = 177 bits (449), Expect = 3e-44
Identities = 111/410 (27%), Positives = 181/410 (44%), Gaps = 43/410 (10%)
Query: 178 NFLVRKCCRNGWWNMALEELGRLKDFGYKPSQTTYNALIQVFLRADKLDTAYLVKREMLS 237
N ++R C +G A+E L R+ P TY LI+ R + A + EM
Sbjct: 208 NTILRSLCDSGKLKQAMEVLDRMLQRDCYPDVITYTILIEATCRDSGVGHAMKLLDEMRD 267
Query: 238 YAFVMDRYTLSCFAYSLCKGGKCREAFDLIDE--AEDFVPDTVFYNRMVSGLCEASLFEE 295
D T + +CK G+ EA +++ + P+ + +N ++ +C + +
Sbjct: 268 RGCTPDVVTYNVLVNGICKEGRLDEAIKFLNDMPSSGCQPNVITHNIILRSMCSTGRWMD 327
Query: 296 AMDILHRMRSSSCIPNVVTYRILLSGCLRKGQLGRCKRILSMMITEGCYPNREIFNSLIH 355
A +L M P+VVT+ IL++ RKG LGR IL M GC PN +N L+H
Sbjct: 328 AEKLLADMLRKGFSPSVVTFNILINFLCRKGLLGRAIDILEKMPQHGCQPNSLSYNPLLH 387
Query: 356 AYCKSRDYSYAYKLFKKMIKCGCQPGYLVYNIFIGSVCSNEEQPSSDILDLVEKAYSEML 415
+CK + A + ++M+ GC P + YN + ++C +
Sbjct: 388 GFCKEKKMDRAIEYLERMVSRGCYPDIVTYNTMLTALCKD-------------------- 427
Query: 416 DLGVVLNKVNVSNFARCLCGAGKFDQAFKIICEMMGKGFVPDDSTYSKVIGFLCDASKVE 475
GK + A +I+ ++ KG P TY+ VI L A K
Sbjct: 428 ---------------------GKVEDAVEILNQLSSKGCSPVLITYNTVIDGLAKAGKTG 466
Query: 476 KAFSLFEEMKRNGIVPSVYTYTILIDSFCKAGLIQQARKWFDEMLHKGCTPNVVTYTALI 535
KA L +EM+ + P TY+ L+ + G + +A K+F E G PN VT+ +++
Sbjct: 467 KAIKLLDEMRAKDLKPDTITYSSLVGGLSREGKVDEAIKFFHEFERMGIRPNAVTFNSIM 526
Query: 536 HAYLKAKQMPVADELFEMMLLEGCKPNVVTYTALIDGHCKAGQIEKACQI 585
K++Q A + M+ GCKPN +YT LI+G G ++A ++
Sbjct: 527 LGLCKSRQTDRAIDFLVFMINRGCKPNETSYTILIEGLAYEGMAKEALEL 576
Score = 156 bits (395), Expect = 5e-38
Identities = 110/413 (26%), Positives = 176/413 (41%), Gaps = 53/413 (12%)
Query: 155 DRVPLKFLMEIKDDDHELLRR-------LLNFLVRKCCRNGWWNMALEELGRLKDFGYKP 207
D LK ME+ D +L+R L+ CR+ A++ L ++D G P
Sbjct: 216 DSGKLKQAMEVLD---RMLQRDCYPDVITYTILIEATCRDSGVGHAMKLLDEMRDRGCTP 272
Query: 208 SQTTYNALIQVFLRADKLDTAYLVKREMLSYAFVMDRYTLSCFAYSLCKGGKCREAFDLI 267
TYN L+ + +LD A +M S + T + S+C G+ +A L+
Sbjct: 273 DVVTYNVLVNGICKEGRLDEAIKFLNDMPSSGCQPNVITHNIILRSMCSTGRWMDAEKLL 332
Query: 268 DE--AEDFVPDTVFYNRMVSGLCEASLFEEAMDILHRMRSSSCIPNVVTYRILLSGCLRK 325
+ + F P V +N +++ LC L A+DIL +M C PN ++Y LL G ++
Sbjct: 333 ADMLRKGFSPSVVTFNILINFLCRKGLLGRAIDILEKMPQHGCQPNSLSYNPLLHGFCKE 392
Query: 326 GQLGRCKRILSMMITEGCYPNREIFNSLIHAYCKSRDYSYAYKLFKKMIKCGCQPGYLVY 385
++ R L M++ GCYP+ +N+++ A CK A ++ ++ GC P + Y
Sbjct: 393 KKMDRAIEYLERMVSRGCYPDIVTYNTMLTALCKDGKVEDAVEILNQLSSKGCSPVLITY 452
Query: 386 NIFIGSVCSNEEQPSSDILDLVEKAYSEMLDLGVVLNKVNVSNFARCLCGAGKFDQAFKI 445
N I L AGK +A K+
Sbjct: 453 NTVIDG-----------------------------------------LAKAGKTGKAIKL 471
Query: 446 ICEMMGKGFVPDDSTYSKVIGFLCDASKVEKAFSLFEEMKRNGIVPSVYTYTILIDSFCK 505
+ EM K PD TYS ++G L KV++A F E +R GI P+ T+ ++ CK
Sbjct: 472 LDEMRAKDLKPDTITYSSLVGGLSREGKVDEAIKFFHEFERMGIRPNAVTFNSIMLGLCK 531
Query: 506 AGLIQQARKWFDEMLHKGCTPNVVTYTALIHAYLKAKQMPVADELFEMMLLEG 558
+ +A + M+++GC PN +YT LI A EL + +G
Sbjct: 532 SRQTDRAIDFLVFMINRGCKPNETSYTILIEGLAYEGMAKEALELLNELCNKG 584
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.322 0.138 0.412
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 22,410,113
Number of Sequences: 26719
Number of extensions: 1010142
Number of successful extensions: 22282
Number of sequences better than 10.0: 479
Number of HSP's better than 10.0 without gapping: 450
Number of HSP's successfully gapped in prelim test: 30
Number of HSP's that attempted gapping in prelim test: 2500
Number of HSP's gapped (non-prelim): 3890
length of query: 987
length of database: 11,318,596
effective HSP length: 109
effective length of query: 878
effective length of database: 8,406,225
effective search space: 7380665550
effective search space used: 7380665550
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.9 bits)
S2: 65 (29.6 bits)
Medicago: description of AC126792.3


