

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC126787.10 - phase: 0
(306 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
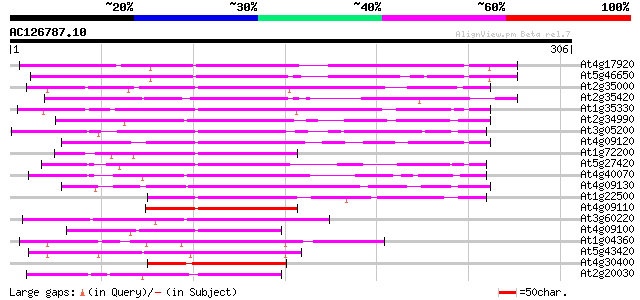
Score E
Sequences producing significant alignments: (bits) Value
At4g17920 putative protein 207 6e-54
At5g46650 putative protein 163 9e-41
At2g35000 putative RING zinc finger protein 140 7e-34
At2g35420 unknown protein 126 1e-29
At1g35330 hypothetical protein 126 1e-29
At2g34990 putative RING zinc finger protein 116 2e-26
At3g05200 RING-H2 zinc finger protein ATL6 (ATL6) 115 3e-26
At4g09120 putative protein 114 7e-26
At1g72200 RING-H2 zinc finger protein ATL3, putative 114 9e-26
At5g27420 RING-H2 zinc finger protein-like 112 3e-25
At4g40070 unknown protein 111 6e-25
At4g09130 putative protein 105 3e-23
At1g22500 Similar to zinc finger protein 104 7e-23
At4g09110 putative protein 92 5e-19
At3g60220 RING-H2 zinc finger protein ATL4 92 5e-19
At4g09100 putative protein 90 2e-18
At1g04360 unknown protein (At1g04360) 87 9e-18
At5g43420 unknown protein 86 3e-17
At4g30400 unknown protein 86 3e-17
At2g20030 putative RING zinc finger protein 85 4e-17
>At4g17920 putative protein
Length = 289
Score = 207 bits (527), Expect = 6e-54
Identities = 121/280 (43%), Positives = 149/280 (53%), Gaps = 28/280 (10%)
Query: 6 PKYDTPPAPTYKTPPALVAFTLTVLILCFVAFSVVYVCKYCFAGFFHTWALQRTTSGSLV 65
P PP Y TPP V T+ +L+ F+ F +Y CK W L G V
Sbjct: 8 PLAPQPPQQHYVTPPLTVILTVILLVFFFIGFFTLYFCKCFLDTMVQAWRLHH--GGDTV 65
Query: 66 RLSPDRSPSR-----GLDNTLLEKFPTFLYSSVKDLRKEKSYSLECAICLLEFDDDSMLR 120
+P + P GL+ ++ FPTF YSSVKDLR+EK Y LECAICLLEFD D +LR
Sbjct: 66 SDNPLQQPEAPPVNPGLELRIINSFPTFPYSSVKDLREEK-YGLECAICLLEFDGDHVLR 124
Query: 121 LLTICCHVFHQECIDLWLESHKTCPVCRTDLDSPPNQMSKHGEGNHNNNNNNLNVQEGMT 180
LLT C HVFHQECIDLW ESH+TCPVCR DLD PP N + +
Sbjct: 125 LLTTCYHVFHQECIDLWFESHRTCPVCRRDLDPPPPPE---------------NTKPTVD 169
Query: 181 SLPCDDIHIDVRGEESDNVGEITRAQVHEGDQHDHHVGMSMQQQEDHRFSRSHSTGHSIV 240
+ D I EE D+ + T Q+ + +Q +FSRSHSTGHSIV
Sbjct: 170 EMIIDVIQETSDDEEDDHHRQQTTTQIDTWPSSGQTSSIKKEQNLPEKFSRSHSTGHSIV 229
Query: 241 TIRGEEKDHEKYTLRLPEHV---IRGGHNYTRSCTNYNEM 277
+ EE+D KYTLRLPEHV + GH+ T SC + E+
Sbjct: 230 RNKPEEED--KYTLRLPEHVKIKVTRGHSQTESCVTFAEL 267
>At5g46650 putative protein
Length = 289
Score = 163 bits (413), Expect = 9e-41
Identities = 100/277 (36%), Positives = 152/277 (54%), Gaps = 36/277 (12%)
Query: 12 PAPTYKTPPALVAFTLTVLILCFVAFSVVYVCKYCFAGFFHTWALQRTTSGSLVRLSPDR 71
P Y PP ++ T+ +L++ F+ F +Y CK + W ++ +
Sbjct: 16 PPQHYSKPPLVIILTVILLVVFFIGFFAIYFCKCFYHTLTEAWNHHYHNGLPENQIQAQQ 75
Query: 72 SPSR-----GLDNTLLEKFPTFLYSSVKDLRKEKSYSLECAICLLEFDDDS-MLRLLTIC 125
P + GL+ +++ +P F +SSVKDLR++K Y LECAICLLEF+++ +LRLLT C
Sbjct: 76 EPVQPPVNPGLEPHIIQSYPLFPFSSVKDLREDK-YGLECAICLLEFEEEHILLRLLTTC 134
Query: 126 CHVFHQECIDLWLESHKTCPVCRTDLDSPPNQMSKHGEGNHNNNNNNLNVQEGMTSLPCD 185
HVFHQECID WLES+KTCPVCR +LD PN N++E + + +
Sbjct: 135 YHVFHQECIDQWLESNKTCPVCRRNLD--PNAPE--------------NIKELIIEVIQE 178
Query: 186 DIHIDVRGEESDNVGEITRAQVHEGDQHDHHVGMSMQQQEDHRFSRSHSTGHSIVTIRGE 245
+ H + E++ E+ ++ G+ ++ D +FSRS +TGHSIV + E
Sbjct: 179 NAHENRDQEQTSTSNEVMLSRQSSGNNE-----RKIETLPD-KFSRSKTTGHSIVRNKPE 232
Query: 246 EKDHEKYTLRLPEHV-----IRGGHNYTRSCTNYNEM 277
E+D +YTLRLP+HV R +N T SC ++ E+
Sbjct: 233 EED--RYTLRLPDHVKIKVTRRHNNNQTESCISFGEL 267
>At2g35000 putative RING zinc finger protein
Length = 378
Score = 140 bits (354), Expect = 7e-34
Identities = 89/263 (33%), Positives = 136/263 (50%), Gaps = 28/263 (10%)
Query: 10 TPPAPTYKTP--PALVAFTLTVLILCFVAFSVVYVCKYCFAGFFHTWALQRTTSGS---L 64
TPP T P P +V T+ L++ F+ F ++ C+ A F + + T + + +
Sbjct: 38 TPPGTTKTKPNDPVVVVITVLFLVIFFMVFGSIF-CRRSNAQFSRSSIFRSTDADAESQV 96
Query: 65 VRLSPDRSPSRGLDNTLLEKFPTFLYSSVKDLRKEKSYSLECAICLLEFDDDSMLRLLTI 124
VR+ R +RGLD +E FPTFLYS VK +R K +ECA+CL EF+DD LRL+
Sbjct: 97 VRIR--RLTARGLDAEAIETFPTFLYSEVKAVRIGKG-GVECAVCLCEFEDDETLRLMPP 153
Query: 125 CCHVFHQECIDLWLESHKTCPVCRTDL-----DSPPNQMSKHGEGNHNNNNNNLNVQEGM 179
CCHVFH +C+D+WL H TCP+CR DL + + + +++ + + GM
Sbjct: 154 CCHVFHADCVDVWLSEHSTCPLCRADLVLNQQGDDDDSTESYSGTDPGTISSSTDPERGM 213
Query: 180 TSLPCDDIHIDVRGEESDNVGEITRAQVHEGDQHDHHVGMSMQQQEDHRFSRSHSTGHSI 239
D +D + N+ +++ G+S Q F RSHSTGHS+
Sbjct: 214 VLESSDAHLLDAVTWSNSNITPRSKS-----------TGLSSWQITGILFPRSHSTGHSL 262
Query: 240 VTIRGEEKDHEKYTLRLPEHVIR 262
+ G + +++TLRLP+ V R
Sbjct: 263 IQPAG---NLDRFTLRLPDDVRR 282
>At2g35420 unknown protein
Length = 234
Score = 126 bits (317), Expect = 1e-29
Identities = 85/261 (32%), Positives = 120/261 (45%), Gaps = 53/261 (20%)
Query: 20 PALVAFTLTVLILCFVAFSVVYVCKYCFAGFFHTWALQRTTSGSLVRLSPDRSPSRGLDN 79
P V T +L + F F +++ ++ F TW LQRT G L+ ++ + GLD
Sbjct: 2 PVTVVLTGVLLFVIFAGFFSLFLWQFLLNRLFTTWNLQRTPYGDLIHVATPPE-NTGLDP 60
Query: 80 TLLEKFPTFLYSSVKDLRKEKSYSLECAICLLEFDDDSMLRLLTICCHVFHQECIDLWLE 139
++ FP F YSS +K++ ECAICL EF D+ +RL+T+C H FH CIDLW E
Sbjct: 61 FIIRSFPVFHYSSAT----KKNHGTECAICLSEFSDEDTVRLITVCRHPFHSNCIDLWFE 116
Query: 140 SHKTCPVCRTDLDSPPNQMSKHGEGNHNNNNNNLNVQEGMTSLPCDDIHIDVRGEESDNV 199
HKTCPVCR +LD P + G G R E N
Sbjct: 117 LHKTCPVCRCELD--PGMI---GSG---------------------------RLESFHNT 144
Query: 200 GEITRAQVHEGDQHDHHVGMSMQ--QQEDHRFSRSHSTGHSIV-TIRGEEKDHEKYTLRL 256
IT ++ +++ G S + + RFSRSHSTGH +V T K +
Sbjct: 145 VTITIQDINHDEENPPTAGSSKRLIEASAWRFSRSHSTGHFMVKTTDANVKSKRR----- 199
Query: 257 PEHVIRGGHNYTRSCTNYNEM 277
H T SC +++E+
Sbjct: 200 --------HYQTGSCVSFDEL 212
>At1g35330 hypothetical protein
Length = 327
Score = 126 bits (317), Expect = 1e-29
Identities = 92/265 (34%), Positives = 127/265 (47%), Gaps = 22/265 (8%)
Query: 5 EPKYDTPPAPTYK---TPPALVAFTLTVLILCFVAFSVVYVCKYCFAGFFHTWALQRTTS 61
+P+ PP + P ++A + L+ +A V Y KY T + T
Sbjct: 29 QPRTTAPPYIAQRPNQVPAVIIAMLMFTLLFSMLACCVCY--KYTNTSPHGTSS--DTEE 84
Query: 62 GSLVRLSPDRSPSRGLDNTLLEKFPTFLYSSVKDLRKEKSYSLECAICLLEFDDDSMLRL 121
G ++ R SRGL ++ FP+FLYS VK L+ K +ECAICL EF+D+ LRL
Sbjct: 85 GGHGEVAFTRRTSRGLGKDVINSFPSFLYSQVKGLKIGKG-GVECAICLNEFEDEETLRL 143
Query: 122 LTICCHVFHQECIDLWLESHKTCPVCRTDLDSPP----NQMSKHGEGNHNNNNNNLNVQE 177
+ C H FH CID+WL S TCPVCR L P N + + N + + NV
Sbjct: 144 MPPCSHAFHASCIDVWLSSRSTCPVCRASLPPKPGSDQNSLYPFIRPHDNQDMDLENVTA 203
Query: 178 GMTSLPCDDIHIDVRGEESDNVGEITRAQVHEGDQHDHHVGMSMQQQEDHRFSRSHSTGH 237
+ L D+ + R S+N G T G+S + + F RSHSTGH
Sbjct: 204 RRSVLESPDVRLLDRLSWSNNTGANT-------PPRSRSTGLSNWRITELLFPRSHSTGH 256
Query: 238 SIVTIRGEEKDHEKYTLRLPEHVIR 262
S+V R E D ++TL+LPE V R
Sbjct: 257 SLVP-RVENLD--RFTLQLPEEVRR 278
>At2g34990 putative RING zinc finger protein
Length = 302
Score = 116 bits (290), Expect = 2e-26
Identities = 80/241 (33%), Positives = 113/241 (46%), Gaps = 33/241 (13%)
Query: 26 TLTVLILCFVAFSVVYVCKYCFAGFFHTWALQRTTS---GSLVRLSPDRSPSRGLDNTLL 82
TL V+ + A +V + CF + Q + + S V + RGLD ++
Sbjct: 14 TLLVITIILFAIFIVGLASVCFRWTSRQFYSQESINPFTDSDVESRTSITAVRGLDEAII 73
Query: 83 EKFPTFLYSSVKDLRKEKSYSLECAICLLEFDDDSMLRLLTICCHVFHQECIDLWLESHK 142
FPTFLYS VK+ R+ +ECA+C+ EF+D LRL+ CCHVFH +C+ +WL H
Sbjct: 74 NSFPTFLYSEVKE-RRIGIGGVECAVCICEFEDHETLRLMPECCHVFHADCVSVWLSDHS 132
Query: 143 TCPVCRTDLDSPPNQMSKHGEGNHNNNNNNLNVQEGMTSLPCDDIHIDVRGEESDNVGEI 202
TCP+CR DL P GE ++ N P D+ D V
Sbjct: 133 TCPLCRVDLCLQP------GERSYLN--------------PEPDLVESTNSHLFDGVTWT 172
Query: 203 TRAQVHEGDQHDHHVGMSMQQQEDHRFSRSHSTGHSIVTIRGEEKDH-EKYTLRLPEHVI 261
R + +S + SRSHSTGHS+V + D+ +++TLRLPE V
Sbjct: 173 NRNR----PSRSWSTRLSQCRVSQILISRSHSTGHSVV----QPLDNLDRFTLRLPEEVR 224
Query: 262 R 262
R
Sbjct: 225 R 225
>At3g05200 RING-H2 zinc finger protein ATL6 (ATL6)
Length = 398
Score = 115 bits (288), Expect = 3e-26
Identities = 92/264 (34%), Positives = 128/264 (47%), Gaps = 34/264 (12%)
Query: 2 SSSEPKYDTPPAPTYKTPPAL-VAFTLTVLILCFVAFSVVYVCKYCF----AGFFHTWAL 56
S S+P P + PA+ V + + L F+ F +Y ++C AG
Sbjct: 29 SQSQPGPTNQPYNYGRLSPAMAVIVVILIAALFFMGFFSIYF-RHCSGVPDAGV------ 81
Query: 57 QRTTSGSLVRLSPDRSPSRGLDNTLLEKFPTFLYSSVKDLRKEKSYSLECAICLLEFDDD 116
+ +G + + +RGLD +++E FPTFLYS VK + K LECAICL EF+DD
Sbjct: 82 --SPAGGARSRATVNAAARGLDVSVVETFPTFLYSDVKTQKLGKG-ELECAICLNEFEDD 138
Query: 117 SMLRLLTICCHVFHQECIDLWLESHKTCPVCRTDLDSPPNQMSKHGEGNHNNNNNNLNVQ 176
LRLL C HVFH CID WLE+H TCPVCR +L + GE +V+
Sbjct: 139 ETLRLLPKCDHVFHPHCIDAWLEAHVTCPVCRANL----AEQVAEGE----------SVE 184
Query: 177 EGMTSLPCDDIHIDVRGEESDNVGEITRAQVHEGDQHDHHVGMSMQQQEDHRFSRSHSTG 236
G T P ++ V E + V G+ + + +FSRSH+TG
Sbjct: 185 PGGTE-PDLELQQVVVNPEPVVTAPVPEQLVTSEVDSRRLPGVPVDLKR-VKFSRSHTTG 242
Query: 237 HSIVTIRGEEKDHEKYTLRLPEHV 260
HS+V + E++TLRLPE V
Sbjct: 243 HSVVQ---PGECTERFTLRLPEDV 263
>At4g09120 putative protein
Length = 345
Score = 114 bits (285), Expect = 7e-26
Identities = 81/235 (34%), Positives = 111/235 (46%), Gaps = 37/235 (15%)
Query: 29 VLILCFVAFSVVYVCKYCFAGFFHTWALQRTTSGSLVRLSPDRSPSRGLDNTLLEKFPTF 88
V++ F++ +V C +C A + S R RGL+ ++E FPTF
Sbjct: 55 VVLAIFISLGMVSCCLHCIFYREEIGAAGQDVLHSRAR--------RGLEKEVIESFPTF 106
Query: 89 LYSSVKDLRKEKSYSLECAICLLEFDDDSMLRLLTICCHVFHQECIDLWLESHKTCPVCR 148
LYS VK L+ K +ECAICL EF+D LR + C H FH CID+WL S TCPVCR
Sbjct: 107 LYSEVKGLKIGKG-GVECAICLSEFEDQETLRWMPPCSHTFHANCIDVWLSSWSTCPVCR 165
Query: 149 TDLDSPPNQMSKHGEGNHNNNNNNLNVQE-GMTSLPCDDIHIDVRGEESDNVGEITRAQV 207
+L P + + N++V+ G+ LP E S +T
Sbjct: 166 ANLSLKPGESYPY---------LNMDVETGGVQKLP---------NERSLTGNSVTTRSR 207
Query: 208 HEGDQHDHHVGMSMQQQEDHRFSRSHSTGHSIVTIRGEEKDHEKYTLRLPEHVIR 262
G +S + + RSHSTGHS+V GE D ++TL+LPE V R
Sbjct: 208 STG-------LLSSWRMAEIFVPRSHSTGHSLVQQLGENLD--RFTLQLPEEVQR 253
>At1g72200 RING-H2 zinc finger protein ATL3, putative
Length = 404
Score = 114 bits (284), Expect = 9e-26
Identities = 64/143 (44%), Positives = 82/143 (56%), Gaps = 16/143 (11%)
Query: 25 FTLTVLILCFVAFSVVYVCKYCFAGFFHTW---ALQRTTSGSLVR-------LSPDRSPS 74
F T+ IL V SV + F GFF + L+R L+ +R +
Sbjct: 59 FDPTMAILMIVLVSVFF-----FLGFFSVYIRRCLERVMGMDYGNPNDAGNWLATNRQQA 113
Query: 75 RGLDNTLLEKFPTFLYSSVKDLRKEKSYSLECAICLLEFDDDSMLRLLTICCHVFHQECI 134
RGLD +++E FPTF YS+VK LR K +LEC++CL EF+DD LRL+ CCHVFH CI
Sbjct: 114 RGLDASIIETFPTFQYSTVKTLRIGKE-ALECSVCLNEFEDDETLRLIPKCCHVFHPGCI 172
Query: 135 DLWLESHKTCPVCRTDLDSPPNQ 157
D WL SH TCP+CR DL P +
Sbjct: 173 DAWLRSHTTCPLCRADLIPVPGE 195
>At5g27420 RING-H2 zinc finger protein-like
Length = 368
Score = 112 bits (280), Expect = 3e-25
Identities = 82/245 (33%), Positives = 114/245 (46%), Gaps = 56/245 (22%)
Query: 18 TPPALVAFTLTVLILCFVAFSVVYVCKYCFAGFFHTWALQR--TTSGSLVRLSPDRSPSR 75
+P V + + L F+ F VY+ ++C T A+ T +G R + + +R
Sbjct: 42 SPAMAVVVVVVIAALFFMGFFTVYI-RHC------TGAVDGSVTPAGGARRRVTNATVAR 94
Query: 76 GLDNTLLEKFPTFLYSSVKDLRKEKSYSLECAICLLEFDDDSMLRLLTICCHVFHQECID 135
GLD +E FPTF+YS VK + K +LECAICL EF+DD LRLL C HVFH CI
Sbjct: 95 GLDAETIETFPTFVYSEVKTQKIGKG-ALECAICLNEFEDDETLRLLPKCDHVFHPHCIG 153
Query: 136 LWLESHKTCPVCRTDLDSPPNQMSKHGEGNHNNNNNNLNVQEGMTSLPCDDIHIDVRGEE 195
WL+ H TCPVCRT+L T P + D+
Sbjct: 154 AWLQGHVTCPVCRTNLAE-------------------------QTPEPEVVVETDLEA-- 186
Query: 196 SDNVGEITRAQVHEGDQHDHHVGMSMQQQEDHRFSRSHSTGHSIVTIRGEEKDHEKYTLR 255
Q V + + + +F RSH+TGHS+V + GE D ++TLR
Sbjct: 187 ----------------QQQSAVPVPVVELPRVKFPRSHTTGHSVV-LPGESTD--RFTLR 227
Query: 256 LPEHV 260
+PE +
Sbjct: 228 VPEEL 232
>At4g40070 unknown protein
Length = 322
Score = 111 bits (277), Expect = 6e-25
Identities = 83/255 (32%), Positives = 115/255 (44%), Gaps = 50/255 (19%)
Query: 11 PPAPTYKTPPALVAFTLTVLILCFVAFSVVYVCKYCFAGFFHTWALQRTTSGSLVRLSPD 70
P T TP F + V + VY+ ++C R+ S R +
Sbjct: 35 PDLQTGHTPSKTTVFAVLVTLFFLTGLLSVYI-RHC----------ARSNPDSSTRYFRN 83
Query: 71 R-----SPSRGLDNTLLEKFPTFLYSSVKDLRKEKSYSLECAICLLEFDDDSMLRLLTIC 125
R S GLDN ++E FP F YSSVK+ K S LECAICL E +D +RLL IC
Sbjct: 84 RANDGSSRRGGLDNAVVESFPVFAYSSVKE-SKIGSKDLECAICLNELEDHETVRLLPIC 142
Query: 126 CHVFHQECIDLWLESHKTCPVCRTDLDSPPNQMSKHGEGNHNNNNNNLNVQEGMTSLPCD 185
H+FH +CID WL SH TCPVCR++L + N+ +G
Sbjct: 143 NHLFHIDCIDTWLYSHATCPVCRSNLTAKSNKPGDEDDG--------------------- 181
Query: 186 DIHIDVRGEESDNVGEITRAQVHEGDQHDHHVGMSMQQQEDHRFSRSHSTGHSIVTIRGE 245
+ + V +I +V + HH +S + +F RS+STGHS+
Sbjct: 182 ---VPLAAMRDHVVVDIETVEVAK----SHHRRLS--SEISGKFPRSNSTGHSMDRF--- 229
Query: 246 EKDHEKYTLRLPEHV 260
E++TLRLP+ V
Sbjct: 230 SDGTERFTLRLPDDV 244
>At4g09130 putative protein
Length = 357
Score = 105 bits (262), Expect = 3e-23
Identities = 80/236 (33%), Positives = 104/236 (43%), Gaps = 28/236 (11%)
Query: 29 VLILCFVAFSVVYVCKY--CFAGFFHTWALQRTTSGSLVRLSPDRSPSRGLDNTLLEKFP 86
VL+ F+ VVY Y C + T G V S R RG+D ++E FP
Sbjct: 53 VLLSLFLLLLVVYCLNYGCCIE--------ENETGGHEVLHSRVR---RGIDKDVIESFP 101
Query: 87 TFLYSSVKDLRKEKSYSLECAICLLEFDDDSMLRLLTICCHVFHQECIDLWLESHKTCPV 146
FLYS VK K + +ECAICL EF+D+ LR + C H FH CID WL S TCPV
Sbjct: 102 AFLYSEVKAF-KIGNGGVECAICLCEFEDEEPLRWMPPCSHTFHANCIDEWLSSRSTCPV 160
Query: 147 CRTDLDSPPNQMSKHGEGNHNNNNNNLNVQEGMTSLPCDDIHIDVRGEESDNVGEITRAQ 206
CR +L H + N VQE + ++N T
Sbjct: 161 CRANLSLKSGDSFPHPSMDVETGNAQRGVQESPDERSLTGSSVTC----NNNANYTTPRS 216
Query: 207 VHEGDQHDHHVGMSMQQQEDHRFSRSHSTGHSIVTIRGEEKDHEKYTLRLPEHVIR 262
G HV + RSHSTGHS+V ++ +++TL+LPE V R
Sbjct: 217 RSTGLLSSWHV-------PELFLPRSHSTGHSLVQ---PCQNIDRFTLQLPEEVQR 262
>At1g22500 Similar to zinc finger protein
Length = 381
Score = 104 bits (259), Expect = 7e-23
Identities = 68/187 (36%), Positives = 96/187 (50%), Gaps = 20/187 (10%)
Query: 76 GLDNTLLEKFPTFLYSSVKDLRKEKSYSLECAICLLEFDDDSMLRLLTICCHVFHQECID 135
GLD +++E FPTF YS+VK LR K +LEC +CL EF+DD LRL+ CCHVFH CID
Sbjct: 89 GLDASVIETFPTFPYSTVKTLRIGKE-ALECPVCLNEFEDDETLRLIPQCCHVFHPGCID 147
Query: 136 LWLESHKTCPVCRTDLDSPPNQMSKHGEGNHNNNNNNLNVQEGMTSL--PCDDIHIDVRG 193
WL S TCP+CR +L P + + ++ L + G SL P DD V
Sbjct: 148 AWLRSQTTCPLCRANLVPVPGE-------SVSSEIPGLARETGQNSLRTPIDDNRKRVLT 200
Query: 194 EESDNVGEITRAQVHEGDQHDHHVGMSMQQQEDHRFSRSHSTGHSIVTIRGEEKDHEKYT 253
+ + + G+Q MS + +S + S G E++ ++YT
Sbjct: 201 SPDE---RLIDSVAWTGNQSMPRKSMSTGWKLAELYSPASSPGQ-------PEENLDRYT 250
Query: 254 LRLPEHV 260
LRLP+ +
Sbjct: 251 LRLPQEI 257
>At4g09110 putative protein
Length = 302
Score = 91.7 bits (226), Expect = 5e-19
Identities = 43/83 (51%), Positives = 51/83 (60%), Gaps = 1/83 (1%)
Query: 75 RGLDNTLLEKFPTFLYSSVKDLRKEKSYSLECAICLLEFDDDSMLRLLTICCHVFHQECI 134
RGL+ L+E FP FLYS VK L+ K +ECAICL EF D LR + C H FH CI
Sbjct: 93 RGLEKELVESFPIFLYSEVKGLKIGKG-GVECAICLSEFVDKETLRWMPPCSHTFHANCI 151
Query: 135 DLWLESHKTCPVCRTDLDSPPNQ 157
D+WL S TCP CR +L P +
Sbjct: 152 DVWLSSQSTCPACRANLSLKPGE 174
>At3g60220 RING-H2 zinc finger protein ATL4
Length = 334
Score = 91.7 bits (226), Expect = 5e-19
Identities = 51/174 (29%), Positives = 90/174 (51%), Gaps = 9/174 (5%)
Query: 8 YDTPPAPTYKTPPALVAFTLTVLILCFVAFSVVYVCKYCFAGFFHTWALQRTTSGSLVRL 67
YD+ + P+++ L +L+ ++ S+ ++ + C H L ++S S+ +
Sbjct: 14 YDSHSSSLDSLKPSVLVIILILLMTLLISVSICFLLR-CLNRCSHRSVLPLSSSSSVATV 72
Query: 68 SPDRSPSRGLD-------NTLLEKFPTFLYSSVKDLRKEKSYSLECAICLLEFDDDSMLR 120
+ D G +++L+ P F +SSV R S +CA+CL +F+ + LR
Sbjct: 73 TSDSRRFSGHRVSPETERSSVLDSLPIFKFSSVTR-RSSSMNSGDCAVCLSKFEPEDQLR 131
Query: 121 LLTICCHVFHQECIDLWLESHKTCPVCRTDLDSPPNQMSKHGEGNHNNNNNNLN 174
LL +CCH FH +CID+WL S++TCP+CR+ L + + + K +NN N
Sbjct: 132 LLPLCCHAFHADCIDIWLVSNQTCPLCRSPLFASESDLMKSLAVVGSNNGGGEN 185
>At4g09100 putative protein
Length = 132
Score = 89.7 bits (221), Expect = 2e-18
Identities = 43/119 (36%), Positives = 65/119 (54%), Gaps = 4/119 (3%)
Query: 32 LCFVAFSVVYVCKYCFAGFFHTWALQRTTSGSL--VRLSPDRSPSRGLDNTLLEKFPTFL 89
+ F S+ ++ Y + + +G + R+SP R P RGLD ++ FP+F+
Sbjct: 11 IVFAFASIGFIAFYIINYYIRRCRNRAAAAGDIEEARMSP-RRPPRGLDAEAIKSFPSFV 69
Query: 90 YSSVKDLRKEKSYSLECAICLLEFDDDSMLRLLTICCHVFHQECIDLWLESHKTCPVCR 148
Y+ + + LEC +CL EF DD LRL+ C HVFH +C+D+WL TCP+CR
Sbjct: 70 YTEARGIEPGIG-ELECVVCLNEFKDDETLRLVPPCVHVFHADCVDIWLSHSSTCPICR 127
>At1g04360 unknown protein (At1g04360)
Length = 381
Score = 87.4 bits (215), Expect = 9e-18
Identities = 65/234 (27%), Positives = 103/234 (43%), Gaps = 47/234 (20%)
Query: 6 PKYDTPPAPTYKTP------------PALVAFTLTVLILCFVAFSV-VYVCKYCFAGFFH 52
P PP+ T+ +P P L + +L F+ S ++V K C +H
Sbjct: 16 PPPPPPPSTTFTSPIFPRTSSSGTNFPILAIAVIGILATAFLLVSYYIFVIKCCLN--WH 73
Query: 53 TWALQRTTSGSLVRLSPDRSP---------SRGLDNTLLEKFPTFLYSS---VKDLRKEK 100
+ R R S D++P +RGLD + + P F + V +
Sbjct: 74 QIDIFRRR-----RRSSDQNPLMIYSPHEVNRGLDESAIRAIPVFKFKKRDVVAGEEDQS 128
Query: 101 SYSLECAICLLEFDDDSMLRLLTICCHVFHQECIDLWLESHKTCPVCRT----------D 150
S EC++CL EF +D LR++ CCHVFH +CID+WL+ + CP+CRT D
Sbjct: 129 KNSQECSVCLNEFQEDEKLRIIPNCCHVFHIDCIDIWLQGNANCPLCRTSVSCEASFTLD 188
Query: 151 LDSPPNQMSKHGEGNHNNNNNNLNVQEGMTSLPCDDIHIDVRGEESDNVGEITR 204
L S P+ + ++ ++ N N++ G+ DD + G + N E R
Sbjct: 189 LISAPSSPRE-----NSPHSRNRNLEPGLVLGGDDDFVVIELGASNGNNRESVR 237
>At5g43420 unknown protein
Length = 375
Score = 85.9 bits (211), Expect = 3e-17
Identities = 55/178 (30%), Positives = 84/178 (46%), Gaps = 30/178 (16%)
Query: 11 PPAPTYKTP-------PALVAFTLTVLILCFVAFSV-VYVCKYCF-------AGFFHTWA 55
PP P + P L + +L F+ S V+V K C G F
Sbjct: 19 PPPPIFHRASSTGTSFPILAVAVIGILATAFLLVSYYVFVIKCCLNWHRIDILGRFSLSR 78
Query: 56 LQRTTSGSLVRLSPDRSPSRGLDNTLLEKFPTFLYSSVKDLR----------KEKSYSLE 105
+R L+ SP+ SRGLD +++ P F + D +E+ S E
Sbjct: 79 RRRNDQDPLMVYSPELR-SRGLDESVIRAIPIFKFKKRYDQNDGVFTGEGEEEEEKRSQE 137
Query: 106 CAICLLEFDDDSMLRLLTICCHVFHQECIDLWLESHKTCPVCRT----DLDSPPNQMS 159
C++CL EF D+ LR++ C H+FH +CID+WL+++ CP+CRT D PP+++S
Sbjct: 138 CSVCLSEFQDEEKLRIIPNCSHLFHIDCIDVWLQNNANCPLCRTRVSCDTSFPPDRVS 195
>At4g30400 unknown protein
Length = 472
Score = 85.5 bits (210), Expect = 3e-17
Identities = 36/76 (47%), Positives = 49/76 (64%), Gaps = 3/76 (3%)
Query: 76 GLDNTLLEKFPTFLYSSVKDLRKEKSYSLECAICLLEFDDDSMLRLLTICCHVFHQECID 135
G+D + ++ P F Y S+ L K+Y +CA+CL EF+ + LRLL C H FH +CID
Sbjct: 107 GVDQSFIDTLPVFHYKSIIGL---KNYPFDCAVCLCEFETEDKLRLLPKCSHAFHMDCID 163
Query: 136 LWLESHKTCPVCRTDL 151
WL SH TCP+CR+ L
Sbjct: 164 TWLLSHSTCPLCRSSL 179
>At2g20030 putative RING zinc finger protein
Length = 390
Score = 85.1 bits (209), Expect = 4e-17
Identities = 53/149 (35%), Positives = 76/149 (50%), Gaps = 16/149 (10%)
Query: 10 TPPAPTYKTPPALVAFTLTVLILCF-VAFSVVYVCKYCFAGFFHTWALQRTTSGSLVRLS 68
+PP P L +L ++ F + F++ +V +A FH L+ T R+
Sbjct: 24 SPPPPNLYATSDLFKPSLAIITGVFSIVFTLTFVL-LVYAKCFHN-DLRSETDSDGERIR 81
Query: 69 PDR---------SPSRGLDNTLLEKFPTFLYSSVKDLRKEKSYSLECAICLLEFDDDSML 119
DR S GLD +E P F +S++K L++ LEC++CL +F+D +L
Sbjct: 82 HDRLWQGLFNRSSRFSGLDKKAIESLPFFRFSALKGLKQ----GLECSVCLSKFEDVEIL 137
Query: 120 RLLTICCHVFHQECIDLWLESHKTCPVCR 148
RLL C H FH CID WLE H TCP+CR
Sbjct: 138 RLLPKCRHAFHIGCIDQWLEQHATCPLCR 166
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.319 0.134 0.420
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 7,887,275
Number of Sequences: 26719
Number of extensions: 359985
Number of successful extensions: 2275
Number of sequences better than 10.0: 302
Number of HSP's better than 10.0 without gapping: 234
Number of HSP's successfully gapped in prelim test: 69
Number of HSP's that attempted gapping in prelim test: 1886
Number of HSP's gapped (non-prelim): 344
length of query: 306
length of database: 11,318,596
effective HSP length: 99
effective length of query: 207
effective length of database: 8,673,415
effective search space: 1795396905
effective search space used: 1795396905
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.8 bits)
S2: 60 (27.7 bits)
Medicago: description of AC126787.10


