

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC126786.6 - phase: 0
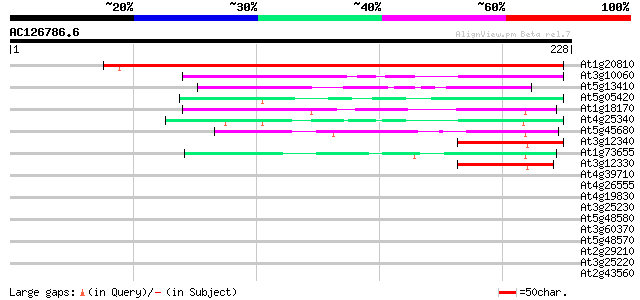
(228 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
At1g20810 immunophilin / FKBP-type peptidyl-prolyl cis-trans iso... 280 3e-76
At3g10060 unknown protein 106 1e-23
At5g13410 unknown protein 49 2e-06
At5g05420 putative protein 49 2e-06
At1g18170 unknown protein 48 5e-06
At4g25340 unknown protein 46 2e-05
At5g45680 unknown protein 44 5e-05
At3g12340 hypothetical protein 44 7e-05
At1g73655 peptidylprolyl isomerase like protein 43 1e-04
At3g12330 hypothetical protein 43 2e-04
At4g39710 unknown protein 40 0.001
At4g26555 unknown protein 40 0.001
At4g19830 unknown protein 40 0.001
At3g25230 rotamase FKBP (ROF1) 37 0.009
At5g48580 peptidyl-prolyl cis-trans isomerase-like protein 35 0.025
At3g60370 hypothetical protein 35 0.033
At5g48570 peptidylprolyl isomerase 33 0.12
At2g29210 proline-rich protein like 32 0.36
At3g25220 immunophilin (FKBP15-1) 31 0.47
At2g43560 putative FKBP type peptidyl-prolyl cis-trans isomerase 31 0.47
>At1g20810 immunophilin / FKBP-type peptidyl-prolyl cis-trans
isomerase
Length = 232
Score = 280 bits (717), Expect = 3e-76
Identities = 138/190 (72%), Positives = 159/190 (83%), Gaps = 3/190 (1%)
Query: 39 VVALS---QSRRSTAILISSLPFTFVFLSPPPPAEARERRKKKNIPIDDYITSPDGLKYY 95
VVA S R ++ IL+SS+P T F+ P +EARERR +K IP+++Y T P+GLK+Y
Sbjct: 36 VVAFSVPISRRDASIILLSSIPLTSFFVLTPSSSEARERRSRKVIPLEEYSTGPEGLKFY 95
Query: 96 DFLEGKGPIAEKGSTVQVHFDCLYRGITAVSSRESKLLAGNRVIAQPYEFKVGAPPGKER 155
D EGKGP+A +GST QVHFDC YR ITA+S+RESKLLAGNR IAQPYEFKVG+ PGKER
Sbjct: 96 DIEEGKGPVATEGSTAQVHFDCRYRSITAISTRESKLLAGNRSIAQPYEFKVGSTPGKER 155
Query: 156 KREFVDNPNGLFSAQAAPKPPQAMYTIVEGMRVGGKRTVIVPPEKGYGKKGMNEIPPGAT 215
KREFVDNPNGLFSAQAAPKPP AMY I EGM+VGGKRTVIVPPE GYG+KGMNEIPPGAT
Sbjct: 156 KREFVDNPNGLFSAQAAPKPPPAMYFITEGMKVGGKRTVIVPPEAGYGQKGMNEIPPGAT 215
Query: 216 FELNIELLQL 225
FELNIELL++
Sbjct: 216 FELNIELLRV 225
>At3g10060 unknown protein
Length = 230
Score = 106 bits (264), Expect = 1e-23
Identities = 66/155 (42%), Positives = 82/155 (52%), Gaps = 24/155 (15%)
Query: 71 ARERRKKKNIPIDDYITSPDGLKYYDFLEGKGPIAEKGSTVQVHFDCLYRGITAVSSRES 130
+R + +P D+ T P+GLKYYD G G A KGS V VH+ ++GIT ++SR+
Sbjct: 86 SRRALRASKLPESDFTTLPNGLKYYDIKVGNGAEAVKGSRVAVHYVAKWKGITFMTSRQG 145
Query: 131 KLLAGNRVIAQPYEFKVGAPPGKERKREFVDNPNGLFSAQAAPKPPQAMYTIVEGMRVGG 190
+ G PY F VG ER G VEGMRVGG
Sbjct: 146 LGVGGGT----PYGFDVGQ---SERGNVLKGLDLG-----------------VEGMRVGG 181
Query: 191 KRTVIVPPEKGYGKKGMNEIPPGATFELNIELLQL 225
+R VIVPPE YGKKG+ EIPP AT EL+IELL +
Sbjct: 182 QRLVIVPPELAYGKKGVQEIPPNATIELDIELLSI 216
>At5g13410 unknown protein
Length = 256
Score = 49.3 bits (116), Expect = 2e-06
Identities = 43/136 (31%), Positives = 57/136 (41%), Gaps = 20/136 (14%)
Query: 77 KKNIPIDDYITSPDGLKYYDFLEGKGPIAEKGSTVQVHFDCLYRGITAVSSRESKLLAGN 136
K + DY + GL+Y D G GPIA+KG V V +D G
Sbjct: 104 KTKMKYPDYTETQSGLQYKDLRVGTGPIAKKGDKVVVDWDGYTIGYY------------G 151
Query: 137 RVIAQPYEFKVGAPPGKERKREFVDNPNGLFSAQAAPKPPQAMYTIVEGMRVGGKRTVIV 196
R+ + K G+ G + +EF G S + P A V GM +GG R +IV
Sbjct: 152 RIFEARNKTKGGSFEGDD--KEFFKFTLG--SNEVIP----AFEEAVSGMALGGIRRIIV 203
Query: 197 PPEKGYGKKGMNEIPP 212
PPE GY N+ P
Sbjct: 204 PPELGYPDNDYNKSGP 219
>At5g05420 putative protein
Length = 143
Score = 48.9 bits (115), Expect = 2e-06
Identities = 43/158 (27%), Positives = 63/158 (39%), Gaps = 33/158 (20%)
Query: 70 EARERRKKKNIPIDDYITSPDGLKYYDFLEGK--GPIAEKGSTVQVHFDCLYRGITAVSS 127
EA K +I ++ DGL + G G AE G V VH+
Sbjct: 16 EATVESKAFSISVEKQTPDLDGLIVEELCMGNPNGKKAEPGKRVSVHYT----------- 64
Query: 128 RESKLLAGNRVIAQPYEFKVGAPPGKERKREFVDNPNGLFSAQAAPKPPQAMYTIVEGMR 187
KL ++ + GK R + +D A K + + + GM
Sbjct: 65 --GKLQGNGKIF--------DSTVGKSRYKFRLD----------AGKVIKGLDVGLNGML 104
Query: 188 VGGKRTVIVPPEKGYGKKGMNEIPPGATFELNIELLQL 225
VGGKR + +PPE GYG +G IPP + ++ELL +
Sbjct: 105 VGGKRKLTIPPEMGYGAEGAGSIPPDSWLVFDVELLNV 142
>At1g18170 unknown protein
Length = 247
Score = 47.8 bits (112), Expect = 5e-06
Identities = 38/162 (23%), Positives = 66/162 (40%), Gaps = 36/162 (22%)
Query: 71 ARERRKKKNIPIDDYITSPDGLKYYDFLEGKGPIAEKGSTVQVHFDCLYRG-----ITAV 125
++E +++ + I P+G++YYD G G G V + +G +
Sbjct: 104 SQEVANTRDVEEEKEIVLPNGIRYYDQRVGGGATPRAGDLVVIDLKGQVQGTGQVFVDTF 163
Query: 126 SSRESKLLAGNRVIAQPYEFKVGAPPGKERKREFVDNPNGLFSAQAAPKPPQAMYTIVEG 185
+++ K + +P VG+ P + E +D ++
Sbjct: 164 GTKDKKKM-------KPLALVVGSKPYSKGLCEGIDY-------------------VLRS 197
Query: 186 MRVGGKRTVIVPPEKGYGKKGMN-----EIPPGATFELNIEL 222
M+ GGKR VIVPP G+G G +IPP A+ E +E+
Sbjct: 198 MKAGGKRRVIVPPSLGFGVDGAELESGLQIPPNASLEYIVEI 239
>At4g25340 unknown protein
Length = 477
Score = 45.8 bits (107), Expect = 2e-05
Identities = 48/185 (25%), Positives = 71/185 (37%), Gaps = 54/185 (29%)
Query: 64 SPPPPAEARERRKKKNIPIDDYI--------------------TSPDGLKYYDFLEGK-- 101
+P AE + + KKK P D+ T P+GL + GK
Sbjct: 323 TPDKSAEKKTKNKKKKKPSDEAAEISGTVEKQTPADSKSSQVRTYPNGLIVEELSMGKPN 382
Query: 102 GPIAEKGSTVQVHFDCLYRGITAVSSRESKLLAGNRVIAQPYEFKVGAPPGKERKREFVD 161
G A+ G TV V + + K+ N + P++F++G G K V
Sbjct: 383 GKRADPGKTVSVRY-------IGKLQKNGKIFDSN-IGKSPFKFRLGI--GSVIKGWDVG 432
Query: 162 NPNGLFSAQAAPKPPQAMYTIVEGMRVGGKRTVIVPPEKGYGKKGM-NEIPPGATFELNI 220
V GMRVG KR + +PP GYG KG +IPP + ++
Sbjct: 433 ---------------------VNGMRVGDKRKLTIPPSMGYGVKGAGGQIPPNSWLTFDV 471
Query: 221 ELLQL 225
EL+ +
Sbjct: 472 ELINV 476
>At5g45680 unknown protein
Length = 208
Score = 44.3 bits (103), Expect = 5e-05
Identities = 38/147 (25%), Positives = 62/147 (41%), Gaps = 33/147 (22%)
Query: 84 DYITSPDGLKYYDFLEGKGPIAEKGSTVQVHFDCLYRGITAVSSRES-KLLAGNRVIAQP 142
++ SP GL + D + G GP A KG ++ H+ V E+ K+ + +P
Sbjct: 85 EFSVSPSGLAFCDKVVGYGPEAVKGQLIKAHY---------VGKLENGKVFDSSYNRGKP 135
Query: 143 YEFKVGAPPGKERKREFVDNPNGLFSAQAAPKPPQAMYTIVEGMRVGGKRTVIVPPEKGY 202
F++G + + + +G+ PP M GGKRT+ +PPE Y
Sbjct: 136 LTFRIGVGEVIKGWDQGILGSDGI--------PP---------MLTGGKRTLRIPPELAY 178
Query: 203 GKKGMN------EIPPGATFELNIELL 223
G +G IPP + +IE +
Sbjct: 179 GDRGAGCKGGSCLIPPASVLLFDIEYI 205
>At3g12340 hypothetical protein
Length = 647
Score = 43.9 bits (102), Expect = 7e-05
Identities = 20/44 (45%), Positives = 29/44 (65%), Gaps = 1/44 (2%)
Query: 183 VEGMRVGGKRTVIVPPEKGYGKKGMNE-IPPGATFELNIELLQL 225
VEGMRVG KR +I+PP GY K+G+ E +P A +E +++
Sbjct: 603 VEGMRVGDKRRLIIPPALGYSKRGLKEKVPKSAWLVYEVEAVKI 646
>At1g73655 peptidylprolyl isomerase like protein
Length = 234
Score = 43.1 bits (100), Expect = 1e-04
Identities = 39/158 (24%), Positives = 61/158 (37%), Gaps = 34/158 (21%)
Query: 72 RERRKKKNIPIDDYITSPDGLKYYDFLEGKGPIAEKGSTVQVHFDCLYRGITAVSSRESK 131
+E + + + I P+G++YYD G G G V V + +
Sbjct: 94 QEEDNTRGLEKQEEIILPNGIRYYDLQVGSGATPSSGYLV-------------VFDVKGQ 140
Query: 132 LLAGNRVIAQPYEFKVGAPPGKERKREFVDNP--NGLFSAQAAPKPPQAMYTIVEGMRVG 189
+ +V + K GK P GL Q + ++ M+ G
Sbjct: 141 VHGTEQVFVDTFGGK-----GKSLAMVMDSRPYSKGLC---------QGIEHVLRSMKAG 186
Query: 190 GKRTVIVPPEKGYGKKGMN-----EIPPGATFELNIEL 222
GKR VI+PP G+G + + EIPP AT + IE+
Sbjct: 187 GKRKVIIPPSLGFGDRNVEFGQGLEIPPSATLDYIIEV 224
>At3g12330 hypothetical protein
Length = 248
Score = 42.7 bits (99), Expect = 2e-04
Identities = 20/40 (50%), Positives = 26/40 (65%), Gaps = 1/40 (2%)
Query: 183 VEGMRVGGKRTVIVPPEKGYGKKGMNE-IPPGATFELNIE 221
VEGMRVG KR +I+PP GY K+G+ E +P A +E
Sbjct: 144 VEGMRVGDKRRLIIPPALGYSKRGLKEKVPKSAWLVYEVE 183
>At4g39710 unknown protein
Length = 217
Score = 39.7 bits (91), Expect = 0.001
Identities = 37/151 (24%), Positives = 60/151 (39%), Gaps = 31/151 (20%)
Query: 81 PIDDYITSPDGLKYYDFLEGKGPIAEKGSTVQVHFDCLYRGITAVSSRESKLLAGNRVIA 140
P+ +Y + GL + D G G A +G V +H+ + T S + A
Sbjct: 85 PLCEYSYAKSGLGFCDLDVGFGDEAPRGVLVNIHYTARFADGTLFDSSYKR--------A 136
Query: 141 QPYEFKVGAPPGKERKREFVDNPNGLFSAQAAPKPPQAMYTIVEGMRVGGKRTVIVPPEK 200
+P ++G GK + G+ + P MRVGGKR + +PP+
Sbjct: 137 RPLTMRIGV--GKVIR----GLDQGILGGEGVPP-----------MRVGGKRKLQIPPKL 179
Query: 201 GYGKK------GMNEIPPGATFELNIELLQL 225
YG + G IP AT +I +++
Sbjct: 180 AYGPEPAGCFSGDCNIPGNATLLYDINFVEI 210
>At4g26555 unknown protein
Length = 207
Score = 39.7 bits (91), Expect = 0.001
Identities = 28/123 (22%), Positives = 54/123 (43%), Gaps = 27/123 (21%)
Query: 89 PDGLKYYDFLEGKGPIAEKGSTVQVHFDCLYRGITAVSSRESKLLAGNRVIAQPYEFKVG 148
P G++Y + +EG+G A +G V++++ C + G V + +F
Sbjct: 85 PSGVRYQEIIEGEGREAHEGDLVELNYVC-------------RRANGYFVHSTVDQFSGE 131
Query: 149 APPGKERKREFVDNPNGLFSAQAAPKPPQAMYTIVEGMRVGGKRTVIVPPEKGYGKKGMN 208
+ P K + + N + + + ++ GM+ GGKR ++PP GY + +
Sbjct: 132 SSPVK-----LILDENDVI---------EGLKEVLVGMKAGGKRRALIPPSVGYINETLK 177
Query: 209 EIP 211
IP
Sbjct: 178 PIP 180
>At4g19830 unknown protein
Length = 229
Score = 39.7 bits (91), Expect = 0.001
Identities = 44/177 (24%), Positives = 71/177 (39%), Gaps = 50/177 (28%)
Query: 45 SRRSTAILI----SSLPFTFVFLSPPPPAEARERRKKKNIPIDDYITSPD--GLKYYDFL 98
SRRS ++ I SS+ +F F SP + D+ P+ G+K D
Sbjct: 51 SRRSISLSIIAVTSSVVSSFCFSSPA---------------LADFSEIPNSGGVKALDLR 95
Query: 99 EGKGPIAEKGSTVQVHFD---CLYRGITAVSSRESKLLAGNRVIAQPYEFKVGAPPGKER 155
G G + +G +++H+ +G S+ + K G V P+ F +G
Sbjct: 96 IGDGDVPIEGDQIEIHYYGRLAAKQGWRFDSTYDHKDSNGEAV---PFTFVLG------- 145
Query: 156 KREFVDNPNGLFSAQAAPKPPQAMYTIVEGMRVGGKRTVIVPPEKGYGKKGMNEIPP 212
S++ P + T V M+VGG R V++PP +GY +PP
Sbjct: 146 ------------SSKVIP----GIETAVRSMKVGGIRRVVIPPSQGYQNTSQEPLPP 186
>At3g25230 rotamase FKBP (ROF1)
Length = 551
Score = 37.0 bits (84), Expect = 0.009
Identities = 19/52 (36%), Positives = 30/52 (57%), Gaps = 6/52 (11%)
Query: 178 AMYTIVEGMRVGGKRTVIVPPEKGYGKKGM------NEIPPGATFELNIELL 223
A+ V+ M+ G K + V P+ G+G+KG +PP AT E+N+EL+
Sbjct: 208 ALTKAVKTMKKGEKVLLTVKPQYGFGEKGKPASAGEGAVPPNATLEINLELV 259
>At5g48580 peptidyl-prolyl cis-trans isomerase-like protein
Length = 163
Score = 35.4 bits (80), Expect = 0.025
Identities = 37/121 (30%), Positives = 53/121 (43%), Gaps = 34/121 (28%)
Query: 105 AEKGSTVQVHFDCLYRG-ITAVSSRESKLLAGNRVIAQPYEFKVGAPPGKERKREFVDNP 163
A KG T++VH YRG +T + +S G+ P+EFK+G
Sbjct: 49 AHKGDTIKVH----YRGKLTDGTVFDSSFERGD-----PFEFKLG--------------- 84
Query: 164 NGLFSAQAAPKPPQAMYTIVEGMRVGGKRTVIVPPEKGYGKKGM-NEIPPGATFELNIEL 222
S Q Q + G VG KR + +P + GYG++G IP GAT + EL
Sbjct: 85 ----SGQVIKGWDQGLL----GACVGEKRKLKIPAKLGYGEQGSPPTIPGGATLIFDTEL 136
Query: 223 L 223
+
Sbjct: 137 I 137
>At3g60370 hypothetical protein
Length = 242
Score = 35.0 bits (79), Expect = 0.033
Identities = 33/138 (23%), Positives = 51/138 (36%), Gaps = 39/138 (28%)
Query: 68 PAEARERRKKKNIPIDDYITSPDGLKYYDFLEGKGPIAEKGSTVQVHFDCLYRGITAVSS 127
P RE + K + D+YI + GL Y DF G+G + G V H+ +
Sbjct: 98 PNFVREGFEVKVLASDNYIKADSGLIYRDFNVGQGDFPKDGQQVTFHY---------IGY 148
Query: 128 RES-KLLAGNRVIAQPYEFKVGA---PPGKERKREFVDNPNGLFSAQAAPKPPQAMYTIV 183
ES + + + P ++G PG E +
Sbjct: 149 NESGRRIDSTYIQGSPARIRMGTNALVPGFEMG--------------------------I 182
Query: 184 EGMRVGGKRTVIVPPEKG 201
M+ GG+R +I+PPE G
Sbjct: 183 RDMKPGGRRRIIIPPELG 200
>At5g48570 peptidylprolyl isomerase
Length = 578
Score = 33.1 bits (74), Expect = 0.12
Identities = 15/42 (35%), Positives = 24/42 (56%), Gaps = 1/42 (2%)
Query: 183 VEGMRVGGKRTVIVPPEKGYGKKGM-NEIPPGATFELNIELL 223
++ M+ G +PPE YG+ G IPP AT + ++EL+
Sbjct: 109 IKTMKKGENAIFTIPPELAYGETGSPPTIPPNATLQFDVELI 150
Score = 30.0 bits (66), Expect = 1.1
Identities = 17/53 (32%), Positives = 29/53 (54%), Gaps = 7/53 (13%)
Query: 178 AMYTIVEGMRVGGKRTVIVPPEKGYGKKG-------MNEIPPGATFELNIELL 223
A+ V+ M+ G K + V P+ G+G+ G IPP AT ++++EL+
Sbjct: 216 ALSKAVKTMKRGEKVLLTVKPQYGFGEFGRPASDGLQAAIPPNATLQIDLELV 268
>At2g29210 proline-rich protein like
Length = 878
Score = 31.6 bits (70), Expect = 0.36
Identities = 37/136 (27%), Positives = 55/136 (40%), Gaps = 18/136 (13%)
Query: 64 SPPPPAEARE------RRKKKNIPIDDYITSPDGLKYYDFLEGKGPIAEKGSTVQVHFDC 117
SP PPA R RR++ P+ SP L Y + P+A++G + D
Sbjct: 371 SPSPPARRRRSPSPPARRRRSPSPLYRRNRSPSPL--YRRNRSRSPLAKRGRS-----DS 423
Query: 118 LYRGITAVSSRESKLLAGNRVIAQPYEFKVGAPPGKERKREFVDNPNGLFS---AQAAPK 174
R + V+ G R+ + E ++ +PP +R GL S AQ P
Sbjct: 424 PGRSPSPVARLRDP--TGARLPSPSIEQRLPSPPVAQRLPSPPPRRAGLPSPPPAQRLPS 481
Query: 175 PPQAMYTIVEGMRVGG 190
PP + MR+GG
Sbjct: 482 PPPRRAGLPSPMRIGG 497
>At3g25220 immunophilin (FKBP15-1)
Length = 153
Score = 31.2 bits (69), Expect = 0.47
Identities = 35/121 (28%), Positives = 51/121 (41%), Gaps = 34/121 (28%)
Query: 105 AEKGSTVQVHFDCLYRG-ITAVSSRESKLLAGNRVIAQPYEFKVGAPPGKERKREFVDNP 163
A KG ++VH YRG +T + +S G+ P EF++G
Sbjct: 49 AHKGDKIKVH----YRGKLTDGTVFDSSFERGD-----PIEFELG--------------- 84
Query: 164 NGLFSAQAAPKPPQAMYTIVEGMRVGGKRTVIVPPEKGYGKKGM-NEIPPGATFELNIEL 222
+ Q P Q + G VG KR + +P + GYG G +IP GAT + EL
Sbjct: 85 ----TGQVIPGWDQGLL----GACVGEKRKLKIPSKLGYGDNGSPPKIPGGATLIFDTEL 136
Query: 223 L 223
+
Sbjct: 137 V 137
>At2g43560 putative FKBP type peptidyl-prolyl cis-trans isomerase
Length = 167
Score = 31.2 bits (69), Expect = 0.47
Identities = 31/106 (29%), Positives = 47/106 (44%), Gaps = 16/106 (15%)
Query: 48 STAILISSLPFTFVFLSP---PPPAEARERRKKKNIPIDDYITSPDGLKYYDFLEGKGPI 104
S + +SSL L P P EA ++ +N+P+ +T+ GL+Y D G+GP
Sbjct: 8 SGGLSMSSLAAYAAGLPPEDKPRLCEAECEKELENVPM---VTTESGLQYKDIKVGRGPS 64
Query: 105 AEKGSTVQVHFDCLY-RGITAVSSRESKLLAGNRVIAQPYEFKVGA 149
G V ++ + G SS E L PY F+VG+
Sbjct: 65 PPVGFQVAANYVAMVPSGQIFDSSLEKGL---------PYLFRVGS 101
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.317 0.136 0.398
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 5,592,738
Number of Sequences: 26719
Number of extensions: 250175
Number of successful extensions: 1055
Number of sequences better than 10.0: 34
Number of HSP's better than 10.0 without gapping: 23
Number of HSP's successfully gapped in prelim test: 12
Number of HSP's that attempted gapping in prelim test: 892
Number of HSP's gapped (non-prelim): 166
length of query: 228
length of database: 11,318,596
effective HSP length: 96
effective length of query: 132
effective length of database: 8,753,572
effective search space: 1155471504
effective search space used: 1155471504
T: 11
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.6 bits)
S2: 58 (26.9 bits)
Medicago: description of AC126786.6


