

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC126786.2 - phase: 0
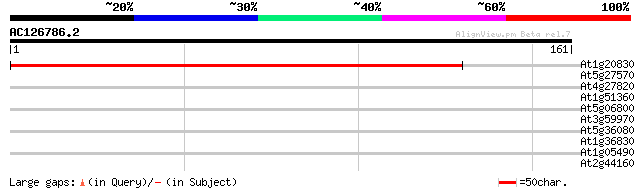
(161 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
At1g20830 unknown protein 221 2e-58
At5g27570 cdc20-like protein 29 1.3
At4g27820 putative beta-glucosidase 28 2.2
At1g51360 unknown protein 28 2.2
At5g06800 unknown protein 27 3.7
At3g59970 methylenetetrahydrofolate reductase MTHFR1 27 3.7
At5g36080 putative protein 27 4.8
At1g36830 hypothetical protein 27 4.8
At1g05490 hypothetical protein 27 4.8
At2g44160 methylenetetrahydrofolate reductase (MTHFR2) 26 8.2
>At1g20830 unknown protein
Length = 349
Score = 221 bits (562), Expect = 2e-58
Identities = 99/130 (76%), Positives = 117/130 (89%)
Query: 1 ISRNLKYEDPFDNPLVKVSKSKSSVEMCGKVYRLAPVTLTQEEQAVHQRRRSRAYQWKRP 60
IS++L YEDPF+NP VK+ K S+VEMCGKVYRLAPVTLT++EQ +HQ+RRSRAYQWKRP
Sbjct: 168 ISKSLNYEDPFNNPFVKLDKGSSTVEMCGKVYRLAPVTLTEKEQTIHQKRRSRAYQWKRP 227
Query: 61 TVFLKEGESVPPDVDPDTIRWIPANHPFATTSTDIGEDFAHKNVSQKHGVPFRIQAEHEA 120
T+FLKEG+S+PPDVDPDT+RWIPANHPFATT +DI +D A NV QK GVPFRI+AEHEA
Sbjct: 228 TIFLKEGDSIPPDVDPDTVRWIPANHPFATTVSDIDQDLAQNNVYQKQGVPFRIRAEHEA 287
Query: 121 LQRKLEALQN 130
+Q+KLEALQN
Sbjct: 288 MQKKLEALQN 297
>At5g27570 cdc20-like protein
Length = 411
Score = 28.9 bits (63), Expect = 1.3
Identities = 18/64 (28%), Positives = 28/64 (43%), Gaps = 3/64 (4%)
Query: 39 LTQEEQAVHQRRRSRAYQWKRPTVFLKEGESVPPDVDPDTI---RWIPANHPFATTSTDI 95
+TQ +A++Q R +P L S PP P ++ R+IP N + I
Sbjct: 30 MTQLAEAMNQNRTRILAFRNKPKALLSSNHSDPPHQQPISVKPRRYIPQNSERVLDAPGI 89
Query: 96 GEDF 99
+DF
Sbjct: 90 ADDF 93
>At4g27820 putative beta-glucosidase
Length = 498
Score = 28.1 bits (61), Expect = 2.2
Identities = 14/46 (30%), Positives = 26/46 (56%), Gaps = 1/46 (2%)
Query: 19 SKSKSSVEMCGKVYRLAPVTLTQEEQAVHQRRRSRAYQWK-RPTVF 63
SK K S+ + + L+P T +++++ QR ++ Y W +P VF
Sbjct: 245 SKQKGSIGLSIFAFGLSPYTNSKDDEIATQRAKTFLYGWMLKPLVF 290
>At1g51360 unknown protein
Length = 210
Score = 28.1 bits (61), Expect = 2.2
Identities = 21/67 (31%), Positives = 30/67 (44%), Gaps = 7/67 (10%)
Query: 28 CGKVYRLAPVTLTQEEQAVHQRRRSR----AYQWKRPTV-FLKEGESVPPDVDPDTIRWI 82
CG ++RL T + +H R RS+ AY V +KE ES+ D+ + WI
Sbjct: 44 CGSIHRLITTTASDFTHVLHSRYRSKEDLNAYAIHPDHVRVVKESESIREDI--MAVDWI 101
Query: 83 PANHPFA 89
P A
Sbjct: 102 AEQAPEA 108
>At5g06800 unknown protein
Length = 375
Score = 27.3 bits (59), Expect = 3.7
Identities = 16/61 (26%), Positives = 25/61 (40%)
Query: 26 EMCGKVYRLAPVTLTQEEQAVHQRRRSRAYQWKRPTVFLKEGESVPPDVDPDTIRWIPAN 85
E+C + Y + VT + HQ ++S P+ + G P V+ IRW
Sbjct: 142 ELCRRTYSNSNVTHLNFTSSQHQPKQSHPRFSSPPSFSIHGGSMAPNCVNKTRIRWTQDL 201
Query: 86 H 86
H
Sbjct: 202 H 202
>At3g59970 methylenetetrahydrofolate reductase MTHFR1
Length = 592
Score = 27.3 bits (59), Expect = 3.7
Identities = 20/80 (25%), Positives = 33/80 (41%), Gaps = 9/80 (11%)
Query: 46 VHQRRRSRAYQWKRPTVFLKEGESVPPDVDPDTIRWIPANHPFATTSTDIG-EDFAHKNV 104
+ + + SR+ W+RP + E V P I W AN P + S G DF H
Sbjct: 303 IDESKISRSLPWRRPANVFRTKEDVRP------IFW--ANRPKSYISRTKGWNDFPHGRW 354
Query: 105 SQKHGVPFRIQAEHEALQRK 124
H + ++++ + K
Sbjct: 355 GDSHSAAYSTLSDYQFARPK 374
>At5g36080 putative protein
Length = 153
Score = 26.9 bits (58), Expect = 4.8
Identities = 15/60 (25%), Positives = 30/60 (50%), Gaps = 4/60 (6%)
Query: 95 IGEDFAHKNVSQKHGVPFRIQAEHEAL--QRKLEALQNVSLSRIRNFNSPMFDTVLEVVV 152
+G + HK++ + H F ++ HE + L+ L L I NFN +F+ + +++
Sbjct: 79 VGVEVVHKDLGEAH--KFLLEVVHEEIVEDNLLKQLYKTLLLVIENFNDKVFNNFVPMLL 136
>At1g36830 hypothetical protein
Length = 243
Score = 26.9 bits (58), Expect = 4.8
Identities = 15/50 (30%), Positives = 26/50 (52%), Gaps = 3/50 (6%)
Query: 80 RWIPANHPFATTSTDIGEDFAHKNVSQKHGVPFRIQAEH--EALQRKLEA 127
+WI A T + EDF +N+ +HG+P+ I ++ RK++A
Sbjct: 28 KWIEAKAFQQVTEKQV-EDFLWENIVCQHGIPYEIITDNGTNLTSRKIKA 76
>At1g05490 hypothetical protein
Length = 731
Score = 26.9 bits (58), Expect = 4.8
Identities = 19/68 (27%), Positives = 31/68 (44%), Gaps = 5/68 (7%)
Query: 88 FATTSTDIGEDFAHKNVSQKHGVPFRIQAEHEALQRKLEALQNVSLSRIRNFNSPMFDTV 147
FA+T+ ED N + G P R + E+ ++L + S+SR +F T
Sbjct: 424 FASTNVSKYEDSVSINSGKTTGAPSRPEVENPETGKELNTPEKPSISR-----PEIFTTE 478
Query: 148 LEVVVQMP 155
+ VQ+P
Sbjct: 479 KAIDVQVP 486
>At2g44160 methylenetetrahydrofolate reductase (MTHFR2)
Length = 594
Score = 26.2 bits (56), Expect = 8.2
Identities = 29/109 (26%), Positives = 41/109 (37%), Gaps = 23/109 (21%)
Query: 26 EMCGK-----VYRLAPVTLTQEEQA---------VHQRRRSRAYQWKRPTVFLKEGESVP 71
EMC K V L TL E+ A + + + SR+ W+RP + E V
Sbjct: 269 EMCKKMLAHGVKSLHLYTLNMEKSALAILMNLGMIDESKISRSLPWRRPANVFRTKEDVR 328
Query: 72 PDVDPDTIRWIPANHPFATTSTDIG-EDFAHKNVSQKHGVPFRIQAEHE 119
P I W AN P + S G EDF + ++H+
Sbjct: 329 P------IFW--ANRPKSYISRTKGWEDFPQGRWGDSRSASYGALSDHQ 369
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.318 0.133 0.398
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 3,546,810
Number of Sequences: 26719
Number of extensions: 132213
Number of successful extensions: 382
Number of sequences better than 10.0: 10
Number of HSP's better than 10.0 without gapping: 4
Number of HSP's successfully gapped in prelim test: 6
Number of HSP's that attempted gapping in prelim test: 378
Number of HSP's gapped (non-prelim): 10
length of query: 161
length of database: 11,318,596
effective HSP length: 91
effective length of query: 70
effective length of database: 8,887,167
effective search space: 622101690
effective search space used: 622101690
T: 11
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 56 (26.2 bits)
Medicago: description of AC126786.2


