

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC126786.12 + phase: 0
(451 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
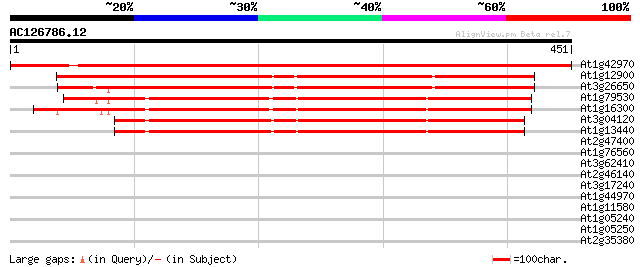
Score E
Sequences producing significant alignments: (bits) Value
At1g42970 putative glyceraldehyde-3-phosphate dehydrogenase (At1... 763 0.0
At1g12900 glyceraldehyde 3-phosphate dehydrogenase A, cholorplas... 571 e-163
At3g26650 glyceraldehyde 3-phosphate dehydrogenase A subunit (GapA) 563 e-161
At1g79530 glyceraldehyde-3-phosphate dehydrogenase (EC 1.2.1.12)... 330 1e-90
At1g16300 glyceraldehyde-3-phosphate dehydrogenase like protein 318 3e-87
At3g04120 glyceraldehyde-3-phosphate dehydrogenase C subunit (GapC) 298 5e-81
At1g13440 putative protein 296 2e-80
At2g47400 putative chloroplast protein CP12 40 0.003
At1g76560 unknown protein 39 0.006
At3g62410 CP12 protein precursor-like protein 35 0.065
At2g46140 putative desiccation related protein 30 2.1
At3g17240 lipoamide dehydrogenase precursor 30 3.5
At1g44970 peroxidase like protein 29 4.6
At1g11580 unknown protein 29 4.6
At1g05240 putative peroxidase ATP12a 29 4.6
At1g05250 putative peroxidase ATP12a 29 4.6
At2g35380 putative peroxidase 28 7.9
>At1g42970 putative glyceraldehyde-3-phosphate dehydrogenase
(At1g42970)
Length = 447
Score = 763 bits (1969), Expect = 0.0
Identities = 382/453 (84%), Positives = 416/453 (91%), Gaps = 8/453 (1%)
Query: 1 MATHAALASTRIPTSTRFPSKAS-HSFPTQCASKRLEVAEFSGLKSTSCITYASNARESS 59
MATHAALA +RIP + R SK++ HSFP QC+SKRLEVAEFSGL+ +S E+S
Sbjct: 1 MATHAALAVSRIPVTQRLQSKSAIHSFPAQCSSKRLEVAEFSGLRMSSI------GGEAS 54
Query: 60 FSDVVAAQLATKAIG-STAVRGETVAKLKVAINGFGRIGRNFLRCWHGRKDSPLEVIVVN 118
F D VAAQ+ KA+ ST VRGETVAKLKVAINGFGRIGRNFLRCWHGRKDSPLEV+V+N
Sbjct: 55 FFDAVAAQIIPKAVTTSTPVRGETVAKLKVAINGFGRIGRNFLRCWHGRKDSPLEVVVLN 114
Query: 119 DSGGVKNASHLLKYDSMLGTFKADVKILNNETITVDGKPILVVSSRDPLKLPWAELGIDI 178
DSGGVKNASHLLKYDSMLGTFKA+VKI++NETI+VDGK I VVS+RDPLKLPWAELGIDI
Sbjct: 115 DSGGVKNASHLLKYDSMLGTFKAEVKIVDNETISVDGKLIKVVSNRDPLKLPWAELGIDI 174
Query: 179 VIEGTGVFVDGPGAGKHIQAGAKKVIITAPAKGADIPTYVVGVNEQDYGHEVADIISNAS 238
VIEGTGVFVDGPGAGKHIQAGA KVIITAPAKGADIPTYV+GVNEQDYGH+VA+IISNAS
Sbjct: 175 VIEGTGVFVDGPGAGKHIQAGASKVIITAPAKGADIPTYVMGVNEQDYGHDVANIISNAS 234
Query: 239 CTTNCLAPFVKILDEEFGIVKGTMTTTHSYTGDQRLLDASHRDLRRARAAALNIVPTSTG 298
CTTNCLAPF K+LDEEFGIVKGTMTTTHSYTGDQRLLDASHRDLRRARAAALNIVPTSTG
Sbjct: 235 CTTNCLAPFAKVLDEEFGIVKGTMTTTHSYTGDQRLLDASHRDLRRARAAALNIVPTSTG 294
Query: 299 AAKAVSLVLPQLKGKLNGIALRVPTPNVSVVDLVVNVAKKGISAEDVNAAFRKAAEGPLK 358
AAKAVSLVLPQLKGKLNGIALRVPTPNVSVVDLV+NV KKG++AEDVN AFRKAA GP+K
Sbjct: 295 AAKAVSLVLPQLKGKLNGIALRVPTPNVSVVDLVINVEKKGLTAEDVNEAFRKAANGPMK 354
Query: 359 GVLDVCDVPLVSVDFRCTDVSSTIDSSLTMVMGDDMVKVVAWYDNEWGYSQRVVDLAHLV 418
G+LDVCD PLVSVDFRC+DVS+TIDSSLTMVMGDDMVKVVAWYDNEWGYSQRVVDLAHLV
Sbjct: 355 GILDVCDAPLVSVDFRCSDVSTTIDSSLTMVMGDDMVKVVAWYDNEWGYSQRVVDLAHLV 414
Query: 419 ANKWPGTPQAGSGDPLEDFCETNPSTDECKVFE 451
A+KWPG GSGDPLEDFC+TNP+ +ECKV++
Sbjct: 415 ASKWPGAEAVGSGDPLEDFCKTNPADEECKVYD 447
>At1g12900 glyceraldehyde 3-phosphate dehydrogenase A, cholorplast
precursor, putative
Length = 399
Score = 571 bits (1472), Expect = e-163
Identities = 290/385 (75%), Positives = 325/385 (84%), Gaps = 3/385 (0%)
Query: 38 AEFSGLKSTSCITYASNARESSFSDVVAAQLATKAIGSTAVRGETVAKLKVAINGFGRIG 97
+EFSGL+++S + +A + F V+ Q + +G T AK+KVAINGFGRIG
Sbjct: 17 SEFSGLRNSSALPFAKRSSSDEFVSFVSFQTSAMRSNGGYRKGVTEAKIKVAINGFGRIG 76
Query: 98 RNFLRCWHGRKDSPLEVIVVNDSGGVKNASHLLKYDSMLGTFKADVKILNNETITVDGKP 157
RNFLRCWHGRKDSPL+V+V+ND+GGVK ASHLLKYDS LG F ADVK + ++VDGK
Sbjct: 77 RNFLRCWHGRKDSPLDVVVINDTGGVKQASHLLKYDSTLGIFDADVKPSGDSALSVDGKI 136
Query: 158 ILVVSSRDPLKLPWAELGIDIVIEGTGVFVDGPGAGKHIQAGAKKVIITAPAKGADIPTY 217
I +VS R+P LPW ELGID+VIEGTGVFVD GAGKH+QAGAKKV+ITAP KG DIPTY
Sbjct: 137 IKIVSDRNPSNLPWGELGIDLVIEGTGVFVDRDGAGKHLQAGAKKVLITAPGKG-DIPTY 195
Query: 218 VVGVNEQDYGHEVADIISNASCTTNCLAPFVKILDEEFGIVKGTMTTTHSYTGDQRLLDA 277
VVGVN + Y HE IISNASCTTNCLAPFVK+LD++FGI+KGTMTTTHSYTGDQRLLDA
Sbjct: 196 VVGVNAELYSHE-DTIISNASCTTNCLAPFVKVLDQKFGIIKGTMTTTHSYTGDQRLLDA 254
Query: 278 SHRDLRRARAAALNIVPTSTGAAKAVSLVLPQLKGKLNGIALRVPTPNVSVVDLVVNVAK 337
SHRDLRRARAAALNIVPTSTGAAKAV+LVLP LKGKLNGIALRVPTPNVSVVDLVV V+K
Sbjct: 255 SHRDLRRARAAALNIVPTSTGAAKAVALVLPNLKGKLNGIALRVPTPNVSVVDLVVQVSK 314
Query: 338 KGISAEDVNAAFRKAAEGPLKGVLDVCDVPLVSVDFRCTDVSSTIDSSLTMVMGDDMVKV 397
K AE+VNAAFR AAE LKG+LDVCD PLVSVDFRC+DVSSTIDSSLTMVMGDDMVKV
Sbjct: 315 KTF-AEEVNAAFRDAAEKELKGILDVCDEPLVSVDFRCSDVSSTIDSSLTMVMGDDMVKV 373
Query: 398 VAWYDNEWGYSQRVVDLAHLVANKW 422
+AWYDNEWGYSQRVVDLA +VAN W
Sbjct: 374 IAWYDNEWGYSQRVVDLADIVANNW 398
>At3g26650 glyceraldehyde 3-phosphate dehydrogenase A subunit (GapA)
Length = 396
Score = 563 bits (1450), Expect = e-161
Identities = 294/387 (75%), Positives = 325/387 (83%), Gaps = 8/387 (2%)
Query: 39 EFSGLKSTSC-ITYASNARESSFSDVVAAQLATKAIGSTAV--RGETVAKLKVAINGFGR 95
EFSGL+S+S + + F +V+ Q T A+GS+ +G T AKLKVAINGFGR
Sbjct: 14 EFSGLRSSSASLPFGKKLSSDEFVSIVSFQ--TSAMGSSGGYRKGVTEAKLKVAINGFGR 71
Query: 96 IGRNFLRCWHGRKDSPLEVIVVNDSGGVKNASHLLKYDSMLGTFKADVKILNNETITVDG 155
IGRNFLRCWHGRKDSPL++I +ND+GGVK ASHLLKYDS LG F ADVK I+VDG
Sbjct: 72 IGRNFLRCWHGRKDSPLDIIAINDTGGVKQASHLLKYDSTLGIFDADVKPSGETAISVDG 131
Query: 156 KPILVVSSRDPLKLPWAELGIDIVIEGTGVFVDGPGAGKHIQAGAKKVIITAPAKGADIP 215
K I VVS+R+P LPW ELGIDIVIEGTGVFVD GAGKHI+AGAKKVIITAP KG DIP
Sbjct: 132 KIIQVVSNRNPSLLPWKELGIDIVIEGTGVFVDREGAGKHIEAGAKKVIITAPGKG-DIP 190
Query: 216 TYVVGVNEQDYGHEVADIISNASCTTNCLAPFVKILDEEFGIVKGTMTTTHSYTGDQRLL 275
TYVVGVN Y H+ IISNASCTTNCLAPFVK+LD++FGI+KGTMTTTHSYTGDQRLL
Sbjct: 191 TYVVGVNADAYSHD-EPIISNASCTTNCLAPFVKVLDQKFGIIKGTMTTTHSYTGDQRLL 249
Query: 276 DASHRDLRRARAAALNIVPTSTGAAKAVSLVLPQLKGKLNGIALRVPTPNVSVVDLVVNV 335
DASHRDLRRARAAALNIVPTSTGAAKAV+LVLP LKGKLNGIALRVPTPNVSVVDLVV V
Sbjct: 250 DASHRDLRRARAAALNIVPTSTGAAKAVALVLPNLKGKLNGIALRVPTPNVSVVDLVVQV 309
Query: 336 AKKGISAEDVNAAFRKAAEGPLKGVLDVCDVPLVSVDFRCTDVSSTIDSSLTMVMGDDMV 395
+KK AE+VNAAFR +AE LKG+LDVCD PLVSVDFRC+D S+TIDSSLTMVMGDDMV
Sbjct: 310 SKKTF-AEEVNAAFRDSAEKELKGILDVCDEPLVSVDFRCSDFSTTIDSSLTMVMGDDMV 368
Query: 396 KVVAWYDNEWGYSQRVVDLAHLVANKW 422
KV+AWYDNEWGYSQRVVDLA +VAN W
Sbjct: 369 KVIAWYDNEWGYSQRVVDLADIVANNW 395
>At1g79530 glyceraldehyde-3-phosphate dehydrogenase (EC 1.2.1.12)
precursor like protein
Length = 422
Score = 330 bits (845), Expect = 1e-90
Identities = 187/391 (47%), Positives = 251/391 (63%), Gaps = 21/391 (5%)
Query: 44 KSTSCITYASNARESSFSDVVAAQL------ATKAIGSTAV-------RGETVAKLKVAI 90
K S + ++ N + S FS +++ L + + I +TA R + K KV I
Sbjct: 32 KVLSSLGFSRNLKPSRFSSGISSSLQNGNARSVQPIKATATEVPSAVRRSSSSGKTKVGI 91
Query: 91 NGFGRIGRNFLRCWHGRKDSPLEVIVVNDSG-GVKNASHLLKYDSMLGTFKADVKILNNE 149
NGFGRIGR LR R D +EV+ VND K +++LKYDS G FK + ++++
Sbjct: 92 NGFGRIGRLVLRIATSRDD--IEVVAVNDPFIDAKYMAYMLKYDSTHGNFKGSINVIDDS 149
Query: 150 TITVDGKPILVVSSRDPLKLPWAELGIDIVIEGTGVFVDGPGAGKHIQAGAKKVIITAPA 209
T+ ++GK + VVS RDP ++PWA+LG D V+E +GVF A H++ GAKKVII+AP+
Sbjct: 150 TLEINGKKVNVVSKRDPSEIPWADLGADYVVESSGVFTTLSKAASHLKGGAKKVIISAPS 209
Query: 210 KGADIPTYVVGVNEQDYGHEVADIISNASCTTNCLAPFVKILDEEFGIVKGTMTTTHSYT 269
AD P +VVGVNE Y + DI+SNASCTTNCLAP K++ EEFGI++G MTT H+ T
Sbjct: 210 --ADAPMFVVGVNEHTYQPNM-DIVSNASCTTNCLAPLAKVVHEEFGILEGLMTTVHATT 266
Query: 270 GDQRLLDA-SHRDLRRARAAALNIVPTSTGAAKAVSLVLPQLKGKLNGIALRVPTPNVSV 328
Q+ +D S +D R R A+ NI+P+STGAAKAV VLP+L GKL G+A RVPT NVSV
Sbjct: 267 ATQKTVDGPSMKDWRGGRGASQNIIPSSTGAAKAVGKVLPELNGKLTGMAFRVPTSNVSV 326
Query: 329 VDLVVNVAKKGISAEDVNAAFRKAAEGPLKGVLDVCDVPLVSVDFRCTDVSSTIDSSLTM 388
VDL + +KG S EDV AA + A+EGPLKG+L D +VS DF SS D++ +
Sbjct: 327 VDLTCRL-EKGASYEDVKAAIKHASEGPLKGILGYTDEDVVSNDFVGDSRSSIFDANAGI 385
Query: 389 VMGDDMVKVVAWYDNEWGYSQRVVDLAHLVA 419
+ VK+V+WYDNEWGYS RV+DL +A
Sbjct: 386 GLSKSFVKLVSWYDNEWGYSNRVLDLIEHMA 416
>At1g16300 glyceraldehyde-3-phosphate dehydrogenase like protein
Length = 420
Score = 318 bits (816), Expect = 3e-87
Identities = 186/417 (44%), Positives = 259/417 (61%), Gaps = 23/417 (5%)
Query: 20 SKASHSFPTQCASKRLEV---AEFSGLKSTSCITYASNARESSFSDVVAAQLATKA---- 72
S S T A+ R+E+ + ++ + TS + ++ + S FS + A
Sbjct: 4 SSLLRSAATSAAAPRVELYPSSSYNHSQVTSSLGFSHSLTSSRFSGAAVSTGKYNAKRVQ 63
Query: 73 -IGSTAV-------RGETVAKLKVAINGFGRIGRNFLRCWHGRKDSPLEVIVVNDSG-GV 123
I +TA R + K KV INGFGRIGR LR R D +EV+ VND
Sbjct: 64 PIKATATEAPPAVHRSRSSGKTKVGINGFGRIGRLVLRIATFRDD--IEVVAVNDPFIDA 121
Query: 124 KNASHLLKYDSMLGTFKADVKILNNETITVDGKPILVVSSRDPLKLPWAELGIDIVIEGT 183
K +++ KYDS G +K + ++++ T+ ++GK + VVS RDP ++PWA+LG + V+E +
Sbjct: 122 KYMAYMFKYDSTHGNYKGTINVIDDSTLEINGKQVKVVSKRDPAEIPWADLGAEYVVESS 181
Query: 184 GVFVDGPGAGKHIQAGAKKVIITAPAKGADIPTYVVGVNEQDYGHEVADIISNASCTTNC 243
GVF A H++ GAKKVII+AP+ AD P +VVGVNE+ Y + DI+SNASCTTNC
Sbjct: 182 GVFTTVGQASSHLKGGAKKVIISAPS--ADAPMFVVGVNEKTYLPNM-DIVSNASCTTNC 238
Query: 244 LAPFVKILDEEFGIVKGTMTTTHSYTGDQRLLDA-SHRDLRRARAAALNIVPTSTGAAKA 302
LAP K++ EEFGI++G MTT H+ T Q+ +D S +D R R A+ NI+P+STGAAKA
Sbjct: 239 LAPLAKVVHEEFGILEGLMTTVHATTATQKTVDGPSMKDWRGGRGASQNIIPSSTGAAKA 298
Query: 303 VSLVLPQLKGKLNGIALRVPTPNVSVVDLVVNVAKKGISAEDVNAAFRKAAEGPLKGVLD 362
V VLP+L GKL G+A RVPTPNVSVVDL + +K S EDV AA + A+EGPL+G+L
Sbjct: 299 VGKVLPELNGKLTGMAFRVPTPNVSVVDLTCRL-EKDASYEDVKAAIKFASEGPLRGILG 357
Query: 363 VCDVPLVSVDFRCTDVSSTIDSSLTMVMGDDMVKVVAWYDNEWGYSQRVVDLAHLVA 419
+ +VS DF SS D++ + + +K+V+WYDNEWGYS RV+DL +A
Sbjct: 358 YTEEDVVSNDFLGDSRSSIFDANAGIGLSKSFMKLVSWYDNEWGYSNRVLDLIEHMA 414
>At3g04120 glyceraldehyde-3-phosphate dehydrogenase C subunit (GapC)
Length = 338
Score = 298 bits (762), Expect = 5e-81
Identities = 161/333 (48%), Positives = 224/333 (66%), Gaps = 9/333 (2%)
Query: 85 KLKVAINGFGRIGRNFLRCWHGRKDSPLEVIVVNDSG-GVKNASHLLKYDSMLGTFKA-D 142
K+++ INGFGRIGR R R D +E++ VND + +++ KYDS+ G +K +
Sbjct: 5 KIRIGINGFGRIGRLVARVVLQRDD--VELVAVNDPFITTEYMTYMFKYDSVHGQWKHNE 62
Query: 143 VKILNNETITVDGKPILVVSSRDPLKLPWAELGIDIVIEGTGVFVDGPGAGKHIQAGAKK 202
+KI + +T+ KP+ V R+P +PWAE G D V+E TGVF D A H++ GAKK
Sbjct: 63 LKIKDEKTLLFGEKPVTVFGIRNPEDIPWAEAGADYVVESTGVFTDKDKAAAHLKGGAKK 122
Query: 203 VIITAPAKGADIPTYVVGVNEQDYGHEVADIISNASCTTNCLAPFVKILDEEFGIVKGTM 262
V+I+AP+K D P +VVGVNE +Y ++ DI+SNASCTTNCLAP K++++ FGIV+G M
Sbjct: 123 VVISAPSK--DAPMFVVGVNEHEYKSDL-DIVSNASCTTNCLAPLAKVINDRFGIVEGLM 179
Query: 263 TTTHSYTGDQRLLDA-SHRDLRRARAAALNIVPTSTGAAKAVSLVLPQLKGKLNGIALRV 321
TT HS T Q+ +D S +D R RAA+ NI+P+STGAAKAV VLP L GKL G++ RV
Sbjct: 180 TTVHSITATQKTVDGPSMKDWRGGRAASFNIIPSSTGAAKAVGKVLPALNGKLTGMSFRV 239
Query: 322 PTPNVSVVDLVVNVAKKGISAEDVNAAFRKAAEGPLKGVLDVCDVPLVSVDFRCTDVSST 381
PT +VSVVDL V + +K + +++ A ++ +EG LKG+L + +VS DF + SS
Sbjct: 240 PTVDVSVVDLTVRL-EKAATYDEIKKAIKEESEGKLKGILGYTEDDVVSTDFVGDNRSSI 298
Query: 382 IDSSLTMVMGDDMVKVVAWYDNEWGYSQRVVDL 414
D+ + + D VK+V+WYDNEWGYS RVVDL
Sbjct: 299 FDAKAGIALSDKFVKLVSWYDNEWGYSSRVVDL 331
>At1g13440 putative protein
Length = 338
Score = 296 bits (758), Expect = 2e-80
Identities = 158/333 (47%), Positives = 224/333 (66%), Gaps = 9/333 (2%)
Query: 85 KLKVAINGFGRIGRNFLRCWHGRKDSPLEVIVVNDSG-GVKNASHLLKYDSMLGTFKA-D 142
K+++ INGFGRIGR R R D +E++ VND + +++ KYDS+ G +K +
Sbjct: 5 KIRIGINGFGRIGRLVARVVLQRDD--VELVAVNDPFITTEYMTYMFKYDSVHGQWKHHE 62
Query: 143 VKILNNETITVDGKPILVVSSRDPLKLPWAELGIDIVIEGTGVFVDGPGAGKHIQAGAKK 202
+K+ +++T+ KP+ V R+P +PW E G D V+E TGVF D A H++ GAKK
Sbjct: 63 LKVKDDKTLLFGEKPVTVFGIRNPEDIPWGEAGADFVVESTGVFTDKDKAAAHLKGGAKK 122
Query: 203 VIITAPAKGADIPTYVVGVNEQDYGHEVADIISNASCTTNCLAPFVKILDEEFGIVKGTM 262
V+I+AP+K D P +VVGVNE +Y ++ DI+SNASCTTNCLAP K++++ FGIV+G M
Sbjct: 123 VVISAPSK--DAPMFVVGVNEHEYKSDL-DIVSNASCTTNCLAPLAKVINDRFGIVEGLM 179
Query: 263 TTTHSYTGDQRLLDA-SHRDLRRARAAALNIVPTSTGAAKAVSLVLPQLKGKLNGIALRV 321
TT HS T Q+ +D S +D R RAA+ NI+P+STGAAKAV VLP L GKL G++ RV
Sbjct: 180 TTVHSITATQKTVDGPSMKDWRGGRAASFNIIPSSTGAAKAVGKVLPSLNGKLTGMSFRV 239
Query: 322 PTPNVSVVDLVVNVAKKGISAEDVNAAFRKAAEGPLKGVLDVCDVPLVSVDFRCTDVSST 381
PT +VSVVDL V + +K + +++ A ++ +EG +KG+L + +VS DF + SS
Sbjct: 240 PTVDVSVVDLTVRL-EKAATYDEIKKAIKEESEGKMKGILGYTEDDVVSTDFVGDNRSSI 298
Query: 382 IDSSLTMVMGDDMVKVVAWYDNEWGYSQRVVDL 414
D+ + + D VK+V+WYDNEWGYS RVVDL
Sbjct: 299 FDAKAGIALSDKFVKLVSWYDNEWGYSSRVVDL 331
>At2g47400 putative chloroplast protein CP12
Length = 124
Score = 39.7 bits (91), Expect = 0.003
Identities = 13/25 (52%), Positives = 19/25 (76%)
Query: 427 QAGSGDPLEDFCETNPSTDECKVFE 451
+AG DPLE++C NP TDEC+ ++
Sbjct: 99 KAGGSDPLEEYCNDNPETDECRTYD 123
>At1g76560 unknown protein
Length = 134
Score = 38.9 bits (89), Expect = 0.006
Identities = 13/20 (65%), Positives = 17/20 (85%)
Query: 432 DPLEDFCETNPSTDECKVFE 451
DPLE FC+ NP TDEC+++E
Sbjct: 114 DPLESFCQENPETDECRIYE 133
>At3g62410 CP12 protein precursor-like protein
Length = 131
Score = 35.4 bits (80), Expect = 0.065
Identities = 11/25 (44%), Positives = 19/25 (76%)
Query: 427 QAGSGDPLEDFCETNPSTDECKVFE 451
+A DPLE++C+ NP T+EC+ ++
Sbjct: 106 KADGSDPLEEYCKDNPETNECRTYD 130
>At2g46140 putative desiccation related protein
Length = 166
Score = 30.4 bits (67), Expect = 2.1
Identities = 21/90 (23%), Positives = 39/90 (43%), Gaps = 4/90 (4%)
Query: 300 AKAVSLVLPQLKGKLNGIALRVPTPNVSVVDLVVNVAKKGISAEDVNAAFRKAAEGPLKG 359
A +S +L + KG +PTP +V D V KG++ + V+ + + + P
Sbjct: 13 ASVISSLLDKAKGFFAEKLANIPTPEATVDD----VDFKGVTRDGVDYHAKVSVKNPYSQ 68
Query: 360 VLDVCDVPLVSVDFRCTDVSSTIDSSLTMV 389
+ +C + + T S TI ++V
Sbjct: 69 SIPICQISYILKSATRTIASGTIPDPGSLV 98
>At3g17240 lipoamide dehydrogenase precursor
Length = 507
Score = 29.6 bits (65), Expect = 3.5
Identities = 17/62 (27%), Positives = 28/62 (44%), Gaps = 1/62 (1%)
Query: 124 KNASHLLK-YDSMLGTFKADVKILNNETITVDGKPILVVSSRDPLKLPWAELGIDIVIEG 182
KN + +K Y L + V ++ E + V GK I+V + D LP + ++
Sbjct: 145 KNKVNYVKGYGKFLSPSEVSVDTIDGENVVVKGKHIIVATGSDVKSLPGITIDEKKIVSS 204
Query: 183 TG 184
TG
Sbjct: 205 TG 206
>At1g44970 peroxidase like protein
Length = 346
Score = 29.3 bits (64), Expect = 4.6
Identities = 11/32 (34%), Positives = 20/32 (62%)
Query: 314 LNGIALRVPTPNVSVVDLVVNVAKKGISAEDV 345
LNG +P PN ++ +L+ +KG++ ED+
Sbjct: 174 LNGANTNIPAPNSTIQNLLTMFQRKGLNEEDL 205
>At1g11580 unknown protein
Length = 557
Score = 29.3 bits (64), Expect = 4.6
Identities = 40/180 (22%), Positives = 69/180 (38%), Gaps = 24/180 (13%)
Query: 229 EVADIISNASCTTNCLAPFVKILD--EEFGIVKGTMTTTHSYTGDQRLLDASHRDLRRAR 286
++ D++S A LA FV +L ++ ++ + D++LL++S + L+
Sbjct: 189 QLEDLVSRARVA---LAIFVSVLPARDDLKMIISNRFPSWLTALDRKLLESSPKTLK--- 242
Query: 287 AAALNIVPTSTGAAKAVSLVLPQLKGKLNGIALRVPTPNVSVVDLVVNVAKKGISAEDVN 346
N+V G K K +A N V V KKG+ E ++
Sbjct: 243 -VTANVVVAKDGTGK--------FKTVNEAVAAAPENSNTRYVIYV----KKGVYKETID 289
Query: 347 AAFRKAAEGPLKGVLDVCDVPLVSVDFRCTDVSSTIDSSLTMVMGDDMVKVVAWYDNEWG 406
+K L V D D +++ D S+T S+ GD + W+ N G
Sbjct: 290 IGKKKKN---LMLVGDGKDATIITGSLNVIDGSTTFRSATVAANGDGFMAQDIWFQNTAG 346
>At1g05240 putative peroxidase ATP12a
Length = 325
Score = 29.3 bits (64), Expect = 4.6
Identities = 13/33 (39%), Positives = 20/33 (60%)
Query: 313 KLNGIALRVPTPNVSVVDLVVNVAKKGISAEDV 345
KLN L +P+P + L N A KG++A+D+
Sbjct: 152 KLNDALLNLPSPFADIKTLKKNFANKGLNAKDL 184
>At1g05250 putative peroxidase ATP12a
Length = 325
Score = 29.3 bits (64), Expect = 4.6
Identities = 13/33 (39%), Positives = 20/33 (60%)
Query: 313 KLNGIALRVPTPNVSVVDLVVNVAKKGISAEDV 345
KLN L +P+P + L N A KG++A+D+
Sbjct: 152 KLNDALLNLPSPFADIKTLKKNFANKGLNAKDL 184
>At2g35380 putative peroxidase
Length = 336
Score = 28.5 bits (62), Expect = 7.9
Identities = 12/38 (31%), Positives = 22/38 (57%)
Query: 310 LKGKLNGIALRVPTPNVSVVDLVVNVAKKGISAEDVNA 347
LK G +P PN S+ L++N ++G++ +D+ A
Sbjct: 155 LKASFAGANQFIPAPNSSLDSLIINFKQQGLNIQDLIA 192
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.316 0.133 0.386
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 9,682,903
Number of Sequences: 26719
Number of extensions: 408614
Number of successful extensions: 975
Number of sequences better than 10.0: 17
Number of HSP's better than 10.0 without gapping: 15
Number of HSP's successfully gapped in prelim test: 2
Number of HSP's that attempted gapping in prelim test: 931
Number of HSP's gapped (non-prelim): 17
length of query: 451
length of database: 11,318,596
effective HSP length: 103
effective length of query: 348
effective length of database: 8,566,539
effective search space: 2981155572
effective search space used: 2981155572
T: 11
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.6 bits)
S2: 62 (28.5 bits)
Medicago: description of AC126786.12


