

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC126782.8 + phase: 0 /pseudo
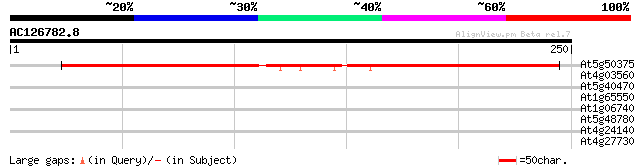
(250 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
At5g50375 cyclopropyl isomerase (CPI1) 267 3e-72
At4g03560 two-pore calcium channel (AtTPC1) 30 1.6
At5g40470 unknown protein 29 2.7
At1g65550 hypothetical protein 29 2.7
At1g06740 mudrA-like protein 29 2.7
At5g48780 putative protein 28 6.0
At4g24140 unknown protein 28 6.0
At4g27730 unknown protein 27 7.9
>At5g50375 cyclopropyl isomerase (CPI1)
Length = 280
Score = 267 bits (683), Expect = 3e-72
Identities = 150/283 (53%), Positives = 175/283 (61%), Gaps = 66/283 (23%)
Query: 24 SASPTPSLWFAPNLSKRWGELFFLLYTPFWLSLSLGIVVPYKLYENFTELEYLLLGLISA 83
S S +PSLW APN SKRWGELFFL YTPFWL+L LGIVVPYKLYE FTELEYLLL L+SA
Sbjct: 2 SGSSSPSLWLAPNPSKRWGELFFLFYTPFWLTLCLGIVVPYKLYETFTELEYLLLALVSA 61
Query: 84 VPAFLIPMLFVGKADKGISWQDRYWVKSMDNNLQLC---------WELFLDSLL------ 128
VPAF+IPML VGKAD+ + W+DRYWVK+ NL + W + +L
Sbjct: 62 VPAFVIPMLLVGKADRSLCWKDRYWVKA---NLWIIVFSYVGNYFWTHYFFKVLGASYTF 118
Query: 129 -------------FHSSGCILYLSFMEN---------------------EQSYLETLAIS 154
F + C L+ N E +++ LA+S
Sbjct: 119 PSWKMNNVPHTTFFLTHVCFLFYHVASNITLRRLRHSTADLPDSLKWCFEAAWI--LALS 176
Query: 155 NFPYY------------EFVDRESMYKVGSLFYAIYFIVSFPMFLRIDEKPGDKWDLPRV 202
F Y EFVDR +MY+VG LFYAIYFIVSFPMF R+DEK D+WDL RV
Sbjct: 177 YFIAYLETIAIANFPYYEFVDRSAMYRVGCLFYAIYFIVSFPMFFRMDEKSTDEWDLSRV 236
Query: 203 AVDALGAAMLVTIILDLWRIFLGPIVPVSEAKQCSPTGLPWFS 245
AVDALGAAMLVTIILDLWR+FLGPIVP+ E + C +GLPWFS
Sbjct: 237 AVDALGAAMLVTIILDLWRLFLGPIVPLPEGQNCLQSGLPWFS 279
>At4g03560 two-pore calcium channel (AtTPC1)
Length = 733
Score = 29.6 bits (65), Expect = 1.6
Identities = 13/49 (26%), Positives = 24/49 (48%), Gaps = 1/49 (2%)
Query: 24 SASPTPSLWFAPNLSKRWGELFFLLYTPFWLSLSLGIVVPYKLYENFTE 72
+ S P +W S RW +FF+LY + +++ +Y++F E
Sbjct: 263 TTSNNPDVWIPAYKSSRWSSVFFVLYVLIGVYFVTNLILAV-VYDSFKE 310
>At5g40470 unknown protein
Length = 496
Score = 28.9 bits (63), Expect = 2.7
Identities = 17/38 (44%), Positives = 20/38 (51%)
Query: 20 SSSSSASPTPSLWFAPNLSKRWGELFFLLYTPFWLSLS 57
SSSSS+S PS P +SKRW L+ T L S
Sbjct: 23 SSSSSSSSLPSFESVPLVSKRWLRLYRASKTSMSLQFS 60
>At1g65550 hypothetical protein
Length = 515
Score = 28.9 bits (63), Expect = 2.7
Identities = 41/171 (23%), Positives = 71/171 (40%), Gaps = 43/171 (25%)
Query: 75 YLL-LGLISAVPAFLIPMLFVGKADKGISWQDRYWVKSMDNNLQLCWELFLDSLLFHSSG 133
YLL LG+ +P+ L+P++ G A+K VK + +LLF S
Sbjct: 51 YLLSLGITVLIPSVLVPLMGGGYAEK---------VK------------VIQTLLFVSGL 89
Query: 134 CILYLSFMENEQSYLETLAISNFPYYEFVDRESMYKVGSLFYA---IYFIVSFPMFLRID 190
L+ SF L +A++++ Y + + S+ Y+ Y+ F F+R
Sbjct: 90 TTLFQSFFGTR---LPVIAVASYAY--------IIPITSIIYSTRFTYYTDPFERFVRTM 138
Query: 191 EKPGDKWDLPRVAVDALGAAMLVTIILDLWRIFLGPIVPVSEAKQCSPTGL 241
+ A+ G ++ IL +WR + + P+S A + TGL
Sbjct: 139 RS-------IQGALIITGCFQVLICILGVWRNIVRFLSPLSIAPLATFTGL 182
>At1g06740 mudrA-like protein
Length = 726
Score = 28.9 bits (63), Expect = 2.7
Identities = 15/45 (33%), Positives = 20/45 (44%), Gaps = 4/45 (8%)
Query: 6 LFSKSLFCVSVLEISSS----SSASPTPSLWFAPNLSKRWGELFF 46
LF ++ C++VLE S SP SLW RW +F
Sbjct: 463 LFWEAAHCLTVLEFKSKINKIEQISPEASLWIQNKSPARWASSYF 507
>At5g48780 putative protein
Length = 669
Score = 27.7 bits (60), Expect = 6.0
Identities = 28/109 (25%), Positives = 47/109 (42%), Gaps = 16/109 (14%)
Query: 118 LCWELFLDSLLFHSSGCILYLS-FMENEQSYLETLAISNFPYYEFVDRESMYKVGSLFYA 176
LC + L L FH SG ++ + F E + + + + +RE+ KV + A
Sbjct: 82 LCLDSLLKILKFHQSGSLVLIPIFYEVDPMDVRKQIGKLYEAFSLHERENPEKVQTWRQA 141
Query: 177 IYFIVSFPMFLRIDEKPGDKWDLPRVAVDALGAAMLV-TIILDLWRIFL 224
+ +VS P + + WD G A L+ I +D+W IF+
Sbjct: 142 LSQLVSIP-----GGQYSEIWD---------GDAELIHQITVDIWNIFV 176
>At4g24140 unknown protein
Length = 498
Score = 27.7 bits (60), Expect = 6.0
Identities = 19/75 (25%), Positives = 36/75 (47%), Gaps = 2/75 (2%)
Query: 87 FLIPMLFVGKADKGIS--WQDRYWVKSMDNNLQLCWELFLDSLLFHSSGCILYLSFMENE 144
F + +L GK+ K + R V+ ++ ++ + + ++ HS GCIL LS
Sbjct: 251 FAVDLLGFGKSPKPADSLYTLREHVEMIEKSVLHKYNVKSFHIVAHSLGCILALSLAARH 310
Query: 145 QSYLETLAISNFPYY 159
+++L + PYY
Sbjct: 311 GGLIKSLTLLAPPYY 325
>At4g27730 unknown protein
Length = 736
Score = 27.3 bits (59), Expect = 7.9
Identities = 16/46 (34%), Positives = 25/46 (53%), Gaps = 2/46 (4%)
Query: 70 FTELEYLLLGLISAVPAFLIPMLFVGKADKGISWQDRYWVKSMDNN 115
+T+ Y+L G + A AF+ +LF+ KGI Q +W S D +
Sbjct: 672 WTKYNYVLSGGLDAGSAFMTILLFLALGRKGIEVQ--WWGNSGDRD 715
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.326 0.141 0.449
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 5,819,725
Number of Sequences: 26719
Number of extensions: 248787
Number of successful extensions: 788
Number of sequences better than 10.0: 8
Number of HSP's better than 10.0 without gapping: 3
Number of HSP's successfully gapped in prelim test: 5
Number of HSP's that attempted gapping in prelim test: 782
Number of HSP's gapped (non-prelim): 10
length of query: 250
length of database: 11,318,596
effective HSP length: 97
effective length of query: 153
effective length of database: 8,726,853
effective search space: 1335208509
effective search space used: 1335208509
T: 11
A: 40
X1: 15 ( 7.0 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 40 (21.6 bits)
S2: 59 (27.3 bits)
Medicago: description of AC126782.8


