

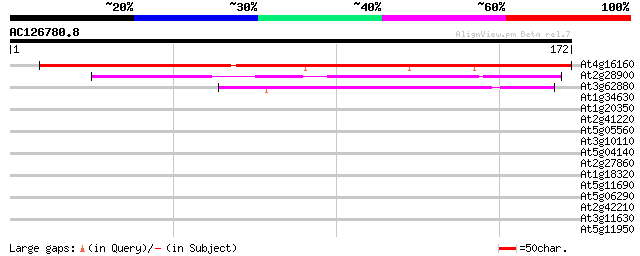
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC126780.8 - phase: 0 /pseudo
(172 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
At4g16160 pore protein homolog 202 7e-53
At2g28900 putative membrane channel protein 74 5e-14
At3g62880 unknown protein 49 1e-06
At1g34630 unknown protein 34 0.045
At1g20350 unknown protein 30 0.49
At2g41220 ferredoxin-dependent glutamate synthase (GLU2) 30 0.84
At5g05560 meiotic check point regulator-like protein 29 1.1
At3g10110 hypothetical protein 29 1.4
At5g04140 ferredoxin-dependent glutamate synthase 28 1.9
At2g27860 putative dTDP-glucose 4-6-dehydratase 28 2.5
At1g18320 hypothetical protein 28 3.2
At5g11690 membrane translocase - like protein 27 4.2
At5g06290 2-cys peroxiredoxin-like protein 27 4.2
At2g42210 unknown protein 27 4.2
At3g11630 putative 2-cys peroxiredoxin 27 7.1
At5g11950 lysine decarboxylase - like protein 26 9.3
>At4g16160 pore protein homolog
Length = 176
Score = 202 bits (514), Expect = 7e-53
Identities = 107/170 (62%), Positives = 130/170 (75%), Gaps = 8/170 (4%)
Query: 10 RTLLDELSDFNKGGLFDFGHPLVNRIAESFVKAAGIGAVQAVSREAYFTVIEGTGIDNAG 69
R ++DE+ F K LFD GHPL+NRIA+SFVKAAG+GA+QAVSREAYFTV++G +N
Sbjct: 7 RIVMDEIRSFEKAHLFDLGHPLLNRIADSFVKAAGVGALQAVSREAYFTVVDGFDSNNV- 65
Query: 70 GMPPEISGAKKNRFHGLRDQ-----Q*IY*GNGKESFQWGLAAGLYSGLTYGMKEAR-GT 123
G P EI+G KK+RF LR + + GKES QWGLAAGLYSG+TYGM E R G
Sbjct: 66 GPPSEITGNKKHRFPNLRGESSKSLDALVKNTGKESLQWGLAAGLYSGITYGMTEVRGGA 125
Query: 124 HDWKNSAVAGAITGAALA-CTSDNTSHEQIAQCAITGAAISTAANLLTGI 172
HDW+NSAVAGA+TGAA+A TS+ TSHEQ+ Q A+TGAAISTAANLL+ +
Sbjct: 126 HDWRNSAVAGALTGAAMAMTTSERTSHEQVVQSALTGAAISTAANLLSSV 175
>At2g28900 putative membrane channel protein
Length = 148
Score = 73.6 bits (179), Expect = 5e-14
Identities = 43/144 (29%), Positives = 70/144 (47%), Gaps = 21/144 (14%)
Query: 26 DFGHPLVNRIAESFVKAAGIGAVQAVSREAYFTVIEGTGIDNAGGMPPEISGAKKNRFHG 85
D G+P +N ++F+K +G ++++ + Y + +G S +K H
Sbjct: 20 DMGNPFLNLTVDAFLKIGAVGVTKSLAEDTYKAIDKG-------------SLSKSTLEHA 66
Query: 86 LRDQQ*IY*GNGKESFQWGLAAGLYSGLTYGMKEARGTHDWKNSAVAGAITGAALACTSD 145
L+ KE WG A G+Y G YG++ RG+ DWKN+ +AGA TGA L+
Sbjct: 67 LKKLC-------KEGVYWGAAGGVYIGTEYGIERIRGSRDWKNAMLAGAATGAVLSAVG- 118
Query: 146 NTSHEQIAQCAITGAAISTAANLL 169
+ I AI G A++TA+ +
Sbjct: 119 KKGKDTIVIDAILGGALATASQFV 142
>At3g62880 unknown protein
Length = 136
Score = 49.3 bits (116), Expect = 1e-06
Identities = 33/106 (31%), Positives = 55/106 (51%), Gaps = 5/106 (4%)
Query: 65 IDNAGGMPPEISG---AKKNRFHGLRDQQ*IY*GNGKESFQWGLAAGLYSGLTYGMKEAR 121
+ AGG+ +G A+K G+ + G+ FQ GL +G+++ G++ R
Sbjct: 22 VATAGGLYGLCAGPRDARKIGLSGVSQASFVAKSIGRFGFQCGLVSGVFTMTHCGLQRYR 81
Query: 122 GTHDWKNSAVAGAITGAALACTSDNTSHEQIAQCAITGAAISTAAN 167
G +DW N+ V GA+ GAA+A ++ N + Q+ A +A S AN
Sbjct: 82 GKNDWVNALVGGAVAGAAVAISTRNWT--QVVGMAGLVSAFSVLAN 125
>At1g34630 unknown protein
Length = 481
Score = 33.9 bits (76), Expect = 0.045
Identities = 20/72 (27%), Positives = 35/72 (47%), Gaps = 8/72 (11%)
Query: 98 KESFQWGLAAGLYSGLTYGMKEA-------RGTHDWKNSAVAGAITGAALACTSDNTSHE 150
KE+ ++GL G ++G + EA + T W+ + AG + G ++ T NT H
Sbjct: 112 KETLRYGLFLGTFAGTFVSVDEAIAALAGDKRTAKWR-ALFAGLVAGPSMLLTGPNTQHT 170
Query: 151 QIAQCAITGAAI 162
+A + AA+
Sbjct: 171 SLAVYILMRAAV 182
>At1g20350 unknown protein
Length = 218
Score = 30.4 bits (67), Expect = 0.49
Identities = 34/112 (30%), Positives = 45/112 (39%), Gaps = 17/112 (15%)
Query: 30 PLVNRIAESFVKAAGIGAVQAVSREAYFTVIEGTGIDNAGGMPPEISGAKKNRFHGLRDQ 89
P +RI + G A+ AV AY + GI N+ G G + R G R
Sbjct: 10 PCPDRILDD---VGGAFAMGAVGGSAYHLI---RGIYNSPGGARLSGGVQALRMSGPR-- 61
Query: 90 Q*IY*GNGKESFQWGLAAGLYSGLTYGMKEARGTHDWKNSAVAGAITGAALA 141
+G WG GLYS + AR D NS ++GA TG L+
Sbjct: 62 ------SGGSFSVWG---GLYSTFDCALVYARQKEDPWNSILSGAATGGFLS 104
>At2g41220 ferredoxin-dependent glutamate synthase (GLU2)
Length = 1629
Score = 29.6 bits (65), Expect = 0.84
Identities = 31/118 (26%), Positives = 50/118 (42%), Gaps = 5/118 (4%)
Query: 34 RIAESFVKAAGIGAVQAVSREAYFTVIEGTGIDNAGGMPP--EISGAKKNRFHGLRDQQ* 91
+++ V GIG V + +A +I+ +G D G P I A GL + Q
Sbjct: 1132 KVSVKLVSETGIGTVASGVAKANADIIQISGYDGGTGASPISSIKHAGGPWELGLAETQK 1191
Query: 92 IY*GNG-KESFQWGLAAGLYSGLTYGMKEARGTHDWKNSAVAGAITGAALA--CTSDN 146
GNG +E + G SG+ + A G ++ +A TG +A C ++N
Sbjct: 1192 TLIGNGLRERVIIRVDGGFKSGVDVLIAAAMGADEYGFGTLAMIATGCIMARICHTNN 1249
>At5g05560 meiotic check point regulator-like protein
Length = 1678
Score = 29.3 bits (64), Expect = 1.1
Identities = 15/50 (30%), Positives = 28/50 (56%), Gaps = 1/50 (2%)
Query: 118 KEARGTHDWKNSAVAGAITGAALACTSDNTSHEQIAQCAITGAAISTAAN 167
+ A+ ++N + G ++ A L + + +H QI +CA+TG S+ AN
Sbjct: 412 ESAKTDSGFRNLKITG-LSDAVLGSINLSVNHSQIFRCALTGKPSSSLAN 460
>At3g10110 hypothetical protein
Length = 173
Score = 28.9 bits (63), Expect = 1.4
Identities = 10/25 (40%), Positives = 19/25 (76%)
Query: 117 MKEARGTHDWKNSAVAGAITGAALA 141
+++AR HD N+A+AG +TG +++
Sbjct: 119 VEKARAKHDTVNTAIAGCVTGGSMS 143
>At5g04140 ferredoxin-dependent glutamate synthase
Length = 1648
Score = 28.5 bits (62), Expect = 1.9
Identities = 31/118 (26%), Positives = 50/118 (42%), Gaps = 5/118 (4%)
Query: 34 RIAESFVKAAGIGAVQAVSREAYFTVIEGTGIDNAGGMPP--EISGAKKNRFHGLRDQQ* 91
+++ V AGIG V + + +I+ +G D G P I A GL +
Sbjct: 1156 KVSVKLVAEAGIGTVASGVAKGNADIIQISGHDGGTGASPISSIKHAGGPWELGLTETHQ 1215
Query: 92 IY*GNG-KESFQWGLAAGLYSGLTYGMKEARGTHDWKNSAVAGAITGAALA--CTSDN 146
NG +E + GL SG+ M A G ++ ++A TG +A C ++N
Sbjct: 1216 TLIANGLRERVILRVDGGLKSGVDVLMAAAMGADEYGFGSLAMIATGCVMARICHTNN 1273
>At2g27860 putative dTDP-glucose 4-6-dehydratase
Length = 389
Score = 28.1 bits (61), Expect = 2.5
Identities = 18/81 (22%), Positives = 37/81 (45%), Gaps = 8/81 (9%)
Query: 4 NSNLETRTLLDELSDFNKGGLFDFGHP--------LVNRIAESFVKAAGIGAVQAVSREA 55
N +E L+ E + G +F+ G+P L + E + K +G GA+++ + +
Sbjct: 266 NDAIEAVLLMIENPERANGHIFNVGNPNNEVTVRQLAEMMTEVYAKVSGEGAIESPTVDV 325
Query: 56 YFTVIEGTGIDNAGGMPPEIS 76
G G D++ P+++
Sbjct: 326 SSKEFYGEGYDDSDKRIPDMT 346
>At1g18320 hypothetical protein
Length = 156
Score = 27.7 bits (60), Expect = 3.2
Identities = 10/22 (45%), Positives = 16/22 (72%)
Query: 120 ARGTHDWKNSAVAGAITGAALA 141
AR HD N+A+AG +TG +++
Sbjct: 105 ARAKHDTVNTAIAGCVTGGSMS 126
>At5g11690 membrane translocase - like protein
Length = 133
Score = 27.3 bits (59), Expect = 4.2
Identities = 15/34 (44%), Positives = 17/34 (49%)
Query: 108 GLYSGLTYGMKEARGTHDWKNSAVAGAITGAALA 141
GL S Y + R D NS VAGA TG L+
Sbjct: 71 GLLSTFDYALVRIRKKEDPWNSIVAGAATGGVLS 104
>At5g06290 2-cys peroxiredoxin-like protein
Length = 273
Score = 27.3 bits (59), Expect = 4.2
Identities = 12/29 (41%), Positives = 18/29 (61%)
Query: 17 SDFNKGGLFDFGHPLVNRIAESFVKAAGI 45
+D GGL D +PLV+ I +S K+ G+
Sbjct: 164 TDRKSGGLGDLNYPLVSDITKSISKSFGV 192
>At2g42210 unknown protein
Length = 159
Score = 27.3 bits (59), Expect = 4.2
Identities = 18/71 (25%), Positives = 31/71 (43%), Gaps = 3/71 (4%)
Query: 97 GKESFQWGLAAGLYSGLTYGMKEARGTHDWKNSAVAGAITGAAL---ACTSDNTSHEQIA 153
G + G+Y G+ ++ R D+ N A+ G + GA++ S T+ A
Sbjct: 63 GTHGLTFAAIGGVYIGVEQLVQNFRSKRDFYNGAIGGFVAGASVLGYRARSIPTAIAAGA 122
Query: 154 QCAITGAAIST 164
A+T A I +
Sbjct: 123 TLAVTSALIDS 133
>At3g11630 putative 2-cys peroxiredoxin
Length = 266
Score = 26.6 bits (57), Expect = 7.1
Identities = 10/29 (34%), Positives = 18/29 (61%)
Query: 17 SDFNKGGLFDFGHPLVNRIAESFVKAAGI 45
+D GGL D +PL++ + +S K+ G+
Sbjct: 157 TDRKSGGLGDLNYPLISDVTKSISKSFGV 185
>At5g11950 lysine decarboxylase - like protein
Length = 216
Score = 26.2 bits (56), Expect = 9.3
Identities = 16/52 (30%), Positives = 23/52 (43%)
Query: 26 DFGHPLVNRIAESFVKAAGIGAVQAVSREAYFTVIEGTGIDNAGGMPPEISG 77
+ G+ LV R + +G + +SR Y + GI MP EISG
Sbjct: 32 ELGNELVKRKIDLVYGGGSVGLMGLISRRVYEGGLHVLGIIPKALMPIEISG 83
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.322 0.137 0.407
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 3,596,074
Number of Sequences: 26719
Number of extensions: 136232
Number of successful extensions: 280
Number of sequences better than 10.0: 16
Number of HSP's better than 10.0 without gapping: 12
Number of HSP's successfully gapped in prelim test: 5
Number of HSP's that attempted gapping in prelim test: 262
Number of HSP's gapped (non-prelim): 17
length of query: 172
length of database: 11,318,596
effective HSP length: 92
effective length of query: 80
effective length of database: 8,860,448
effective search space: 708835840
effective search space used: 708835840
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.9 bits)
S2: 56 (26.2 bits)
Medicago: description of AC126780.8


