

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC126019.3 + phase: 0
(219 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
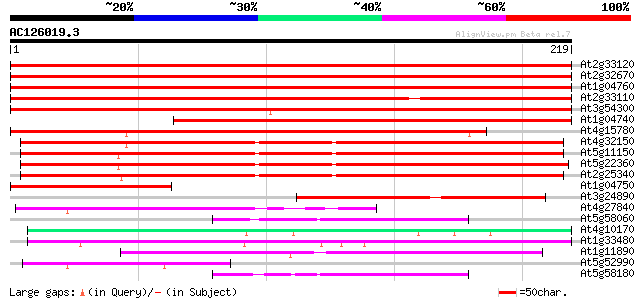
Score E
Sequences producing significant alignments: (bits) Value
At2g33120 putative synaptobrevin 382 e-106
At2g32670 synaptobrevin like protein 380 e-106
At1g04760 synaptobrevin 7B, putative 379 e-106
At2g33110 synaptobrevin like protein 302 8e-83
At3g54300 synaptobrevin -like protein 281 2e-76
At1g04740 putative vesicle-associated membrane protein, synaptob... 269 7e-73
At4g15780 SYBL1 like protein 265 2e-71
At4g32150 vesicle-associated membrane protein 7C (At VAMP7C) 164 4e-41
At5g11150 unknown protein (At5g11150) 160 4e-40
At5g22360 synaptobrevin-like protein 158 2e-39
At2g25340 putative synaptobrevin 153 6e-38
At1g04750 synaptobrevin 7B, putative, 3' partial 112 2e-25
At3g24890 synaptobrevin, putative 100 5e-22
At4g27840 unknown protein 54 8e-08
At5g58060 ATGP1 53 1e-07
At4g10170 unknown protein 52 3e-07
At1g33480 unknown protein 52 3e-07
At1g11890 putative vesicle transport protein 49 3e-06
At5g52990 unknown protein 45 3e-05
At5g58180 ATGP1-like protein 44 5e-05
>At2g33120 putative synaptobrevin
Length = 221
Score = 382 bits (980), Expect = e-106
Identities = 182/219 (83%), Positives = 207/219 (94%)
Query: 1 MGQQSLIYSFVARGTVILAEYSDFTGNFTTIALQCLQKLPASNNRFTYNCDGHTFSFLVD 60
M QQSLIYSFVARGTVIL E++DF GNFT+IA QCLQKLP+SNN+FTYNCDGHTF++LV+
Sbjct: 1 MAQQSLIYSFVARGTVILVEFTDFKGNFTSIAAQCLQKLPSSNNKFTYNCDGHTFNYLVE 60
Query: 61 NGFTYCVVAVESVGRQIPIAFLERIKDDFNKRYGGGRATTATAKSLNKEFGPKLKEQMQY 120
NGFTYCVVAV+S GRQIP+AFLER+K+DFNKRYGGG+A TA A SLNKEFG KLKE MQY
Sbjct: 61 NGFTYCVVAVDSAGRQIPMAFLERVKEDFNKRYGGGKAATAQANSLNKEFGSKLKEHMQY 120
Query: 121 CVEHPEEVSKLAKVKAQVSEVKGVMMENIDKVIDRGEKIEVLVDKTENLRSQAQDFRQQG 180
C++HP+E+SKLAKVKAQVSEVKGVMMENI+KV+DRGEKIE+LVDKTENLRSQAQDFR QG
Sbjct: 121 CMDHPDEISKLAKVKAQVSEVKGVMMENIEKVLDRGEKIELLVDKTENLRSQAQDFRTQG 180
Query: 181 TQLRRKMWYQNMKIKLIVLAIIIALILIIVLSVCHGFSC 219
TQ+RRKMW+QNMKIKLIVLAIIIALILII+LS+C GF+C
Sbjct: 181 TQMRRKMWFQNMKIKLIVLAIIIALILIIILSICGGFNC 219
>At2g32670 synaptobrevin like protein
Length = 285
Score = 380 bits (975), Expect = e-106
Identities = 180/219 (82%), Positives = 206/219 (93%)
Query: 1 MGQQSLIYSFVARGTVILAEYSDFTGNFTTIALQCLQKLPASNNRFTYNCDGHTFSFLVD 60
MGQQ+LIYSFVARGTVIL EY++F GNFT +A QCLQKLP+SNN+FTYNCDGHTF++LV+
Sbjct: 66 MGQQNLIYSFVARGTVILVEYTEFKGNFTAVAAQCLQKLPSSNNKFTYNCDGHTFNYLVE 125
Query: 61 NGFTYCVVAVESVGRQIPIAFLERIKDDFNKRYGGGRATTATAKSLNKEFGPKLKEQMQY 120
NGFTYCVVAVESVGRQIP+AFLER+K+DFNKRYGGG+ATTA A SLN+EFG KLKE MQY
Sbjct: 126 NGFTYCVVAVESVGRQIPMAFLERVKEDFNKRYGGGKATTAQANSLNREFGSKLKEHMQY 185
Query: 121 CVEHPEEVSKLAKVKAQVSEVKGVMMENIDKVIDRGEKIEVLVDKTENLRSQAQDFRQQG 180
CV+HP+E+SKLAKVKAQV+EVKGVMMENI+KV+DRGEKIE+LVDKTENLRSQAQDFR QG
Sbjct: 186 CVDHPDEISKLAKVKAQVTEVKGVMMENIEKVLDRGEKIELLVDKTENLRSQAQDFRTQG 245
Query: 181 TQLRRKMWYQNMKIKLIVLAIIIALILIIVLSVCHGFSC 219
T++RRKMW++NMKIKLIVL III LILII+LSVC GF C
Sbjct: 246 TKIRRKMWFENMKIKLIVLGIIITLILIIILSVCGGFKC 284
>At1g04760 synaptobrevin 7B, putative
Length = 220
Score = 379 bits (972), Expect = e-106
Identities = 176/219 (80%), Positives = 205/219 (93%)
Query: 1 MGQQSLIYSFVARGTVILAEYSDFTGNFTTIALQCLQKLPASNNRFTYNCDGHTFSFLVD 60
MGQQSLIYSFVARGTVILAEY++F GNFT++A QCLQKLP+SNN+FTYNCDGHTF++L D
Sbjct: 1 MGQQSLIYSFVARGTVILAEYTEFKGNFTSVAAQCLQKLPSSNNKFTYNCDGHTFNYLAD 60
Query: 61 NGFTYCVVAVESVGRQIPIAFLERIKDDFNKRYGGGRATTATAKSLNKEFGPKLKEQMQY 120
NGFTYCVV +ES GRQIP+AFLER+K+DFNKRYGGG+A+TA A SLNKEFG KLKE MQY
Sbjct: 61 NGFTYCVVVIESAGRQIPMAFLERVKEDFNKRYGGGKASTAKANSLNKEFGSKLKEHMQY 120
Query: 121 CVEHPEEVSKLAKVKAQVSEVKGVMMENIDKVIDRGEKIEVLVDKTENLRSQAQDFRQQG 180
C +HPEE+SKL+KVKAQV+EVKGVMMENI+KV+DRGEKIE+LVDKTENLRSQAQDFR QG
Sbjct: 121 CADHPEEISKLSKVKAQVTEVKGVMMENIEKVLDRGEKIELLVDKTENLRSQAQDFRTQG 180
Query: 181 TQLRRKMWYQNMKIKLIVLAIIIALILIIVLSVCHGFSC 219
T+++RK+W++NMKIKLIV II+ALILII+LSVCHGF C
Sbjct: 181 TKMKRKLWFENMKIKLIVFGIIVALILIIILSVCHGFKC 219
>At2g33110 synaptobrevin like protein
Length = 217
Score = 302 bits (774), Expect = 8e-83
Identities = 148/219 (67%), Positives = 180/219 (81%), Gaps = 4/219 (1%)
Query: 1 MGQQSLIYSFVARGTVILAEYSDFTGNFTTIALQCLQKLPASNNRFTYNCDGHTFSFLVD 60
M QQSL YSF+ARGTVIL E++DF GNFT++A Q L+ LP+SNN+FTYNCDGHTF+ LV+
Sbjct: 1 MAQQSLFYSFIARGTVILVEFTDFKGNFTSVAAQYLENLPSSNNKFTYNCDGHTFNDLVE 60
Query: 61 NGFTYCVVAVESVGRQIPIAFLERIKDDFNKRYGGGRATTATAKSLNKEFGPKLKEQMQY 120
NGFTYCVVAV+S GR+IP+AFLER+K+DF KRYGG +A T A SLNKEFG LKE MQY
Sbjct: 61 NGFTYCVVAVDSAGREIPMAFLERVKEDFYKRYGGEKAATDQANSLNKEFGSNLKEHMQY 120
Query: 121 CVEHPEEVSKLAKVKAQVSEVKGVMMENIDKVIDRGEKIEVLVDKTENLRSQAQDFRQQG 180
C++HP+E+S LAK KAQVSEVK +MMENI+KV+ RG V+ + + SQ Q F +
Sbjct: 121 CMDHPDEISNLAKAKAQVSEVKSLMMENIEKVLARG----VICEMLGSSESQPQAFYIKR 176
Query: 181 TQLRRKMWYQNMKIKLIVLAIIIALILIIVLSVCHGFSC 219
TQ++RK W+QNMKIKLIVLAIIIALILII+LSVC GF+C
Sbjct: 177 TQMKRKKWFQNMKIKLIVLAIIIALILIIILSVCGGFNC 215
>At3g54300 synaptobrevin -like protein
Length = 240
Score = 281 bits (719), Expect = 2e-76
Identities = 133/238 (55%), Positives = 180/238 (74%), Gaps = 19/238 (7%)
Query: 1 MGQQSLIYSFVARGTVILAEYSDFTGNFTTIALQCLQKLPASNNRFTYNCDGHTFSFLVD 60
M Q+ LIYSFVA+GTV+LAE++ ++GNF+TIA+QCLQKLP +++++TY+CDGHTF+FLVD
Sbjct: 1 MSQKGLIYSFVAKGTVVLAEHTPYSGNFSTIAVQCLQKLPTNSSKYTYSCDGHTFNFLVD 60
Query: 61 NGFTYCVVAVESVGRQIPIAFLERIKDDFNKRYGGGRATT-------------------A 101
NGF + VVA ES GR +P FLER+K+DF KRY +
Sbjct: 61 NGFVFLVVADESTGRSVPFVFLERVKEDFKKRYEASIKNDERHPLADEDEDDDLFGDRFS 120
Query: 102 TAKSLNKEFGPKLKEQMQYCVEHPEEVSKLAKVKAQVSEVKGVMMENIDKVIDRGEKIEV 161
A +L++EFGP LKE MQYC+ HPEE+SKL+K+KAQ++EVKG+MM+NI+KV+DRGEKIE+
Sbjct: 121 VAYNLDREFGPILKEHMQYCMSHPEEMSKLSKLKAQITEVKGIMMDNIEKVLDRGEKIEL 180
Query: 162 LVDKTENLRSQAQDFRQQGTQLRRKMWYQNMKIKLIVLAIIIALILIIVLSVCHGFSC 219
LVDKTENL+ QA F++QG QLRRKMW Q++++KL+V + + ILI+ + C GF C
Sbjct: 181 LVDKTENLQFQADSFQRQGRQLRRKMWLQSLQMKLMVAGAVFSFILIVWVVACGGFKC 238
>At1g04740 putative vesicle-associated membrane protein,
synaptobrevin 7B, 5' partial
Length = 166
Score = 269 bits (688), Expect = 7e-73
Identities = 131/155 (84%), Positives = 145/155 (93%)
Query: 65 YCVVAVESVGRQIPIAFLERIKDDFNKRYGGGRATTATAKSLNKEFGPKLKEQMQYCVEH 124
YCVVAV+S GRQIP++FLER+K+DFNKRYGGG+A TA A SLNKEFG KLKE MQYC++H
Sbjct: 12 YCVVAVDSAGRQIPMSFLERVKEDFNKRYGGGKAATAQANSLNKEFGSKLKEHMQYCMDH 71
Query: 125 PEEVSKLAKVKAQVSEVKGVMMENIDKVIDRGEKIEVLVDKTENLRSQAQDFRQQGTQLR 184
P+E+SKLAKVKAQVSEVKGVMMENI+KV+DRGEKIE+LVDKTENLRSQAQDFR GTQ+R
Sbjct: 72 PDEISKLAKVKAQVSEVKGVMMENIEKVLDRGEKIELLVDKTENLRSQAQDFRTTGTQMR 131
Query: 185 RKMWYQNMKIKLIVLAIIIALILIIVLSVCHGFSC 219
RKMW QNMKIKLIVLAIIIALILIIVLSVCHGF C
Sbjct: 132 RKMWLQNMKIKLIVLAIIIALILIIVLSVCHGFKC 166
>At4g15780 SYBL1 like protein
Length = 194
Score = 265 bits (676), Expect = 2e-71
Identities = 129/188 (68%), Positives = 160/188 (84%), Gaps = 2/188 (1%)
Query: 1 MGQQSLIYSFVARGTVILAEYSDFTGNFTTIALQCLQKLPASNN-RFTYNCDGHTFSFLV 59
MGQ+S IYSFVARGT+ILAEY++FTGNF +IA QCLQKLP+S+N +FTYNCD HTF+FLV
Sbjct: 1 MGQESFIYSFVARGTMILAEYTEFTGNFPSIAAQCLQKLPSSSNSKFTYNCDHHTFNFLV 60
Query: 60 DNGFTYCVVAVESVGRQIPIAFLERIKDDFNKRYGGGRATTATAKSLNKEFGPKLKEQMQ 119
++ + YCVVA +S+ +QI IAFLER+K DF KRYGGG+A+TA AKSLNKEFGP +KE M
Sbjct: 61 EDAYAYCVVAKDSLSKQISIAFLERVKADFKKRYGGGKASTAIAKSLNKEFGPVMKEHMN 120
Query: 120 YCVEHPEEVSKLAKVKAQVSEVKGVMMENIDKVIDRGEKIEVLVDKTENLRSQAQDFRQ- 178
Y V+H EE+ KL KVKAQVSEVK +M+ENIDK IDRGE + VL DKTENLRSQA+++++
Sbjct: 121 YIVDHAEEIEKLIKVKAQVSEVKSIMLENIDKAIDRGENLTVLTDKTENLRSQAREYKKT 180
Query: 179 QGTQLRRK 186
+G + RRK
Sbjct: 181 KGHRWRRK 188
>At4g32150 vesicle-associated membrane protein 7C (At VAMP7C)
Length = 219
Score = 164 bits (414), Expect = 4e-41
Identities = 82/213 (38%), Positives = 139/213 (64%), Gaps = 3/213 (1%)
Query: 5 SLIYSFVARGTVILAEYSDFTGNFTTIALQCLQKLPASNN-RFTYNCDGHTFSFLVDNGF 63
+++Y+ VARGTV+L+E++ + N +TIA Q L+K+P N+ +Y+ D + F +G
Sbjct: 2 AILYALVARGTVVLSEFTATSTNASTIAKQILEKVPGDNDSNVSYSQDRYVFHVKRTDGL 61
Query: 64 TYCVVAVESVGRQIPIAFLERIKDDFNKRYGGGRATTATAKSLNKEFGPKLKEQMQYCVE 123
T +A E+ GR+IP AFLE I F + YG TA A ++N+EF L +Q+ Y
Sbjct: 62 TVLCMAEETAGRRIPFAFLEDIHQRFVRTYGRA-VHTALAYAMNEEFSRVLSQQIDYYSN 120
Query: 124 HPEEVSKLAKVKAQVSEVKGVMMENIDKVIDRGEKIEVLVDKTENLRSQAQDFRQQGTQL 183
P ++ ++K ++++V+GVM+ENIDKV+DRGE++E+LVDKT N++ FR+Q +
Sbjct: 121 DPN-ADRINRIKGEMNQVRGVMIENIDKVLDRGERLELLVDKTANMQGNTFRFRKQARRF 179
Query: 184 RRKMWYQNMKIKLIVLAIIIALILIIVLSVCHG 216
R +W++N K+ ++++ +++ +I I V +CHG
Sbjct: 180 RSNVWWRNCKLTVLLILLLLVIIYIAVAFLCHG 212
>At5g11150 unknown protein (At5g11150)
Length = 221
Score = 160 bits (406), Expect = 4e-40
Identities = 83/214 (38%), Positives = 139/214 (64%), Gaps = 4/214 (1%)
Query: 5 SLIYSFVARGTVILAEYSDFTGNFTTIALQCLQKLPA--SNNRFTYNCDGHTFSFLVDNG 62
++I++ VARGTV+L+E+S + N ++I+ Q L+KLP S++ +Y+ D + F +G
Sbjct: 2 AIIFALVARGTVVLSEFSATSTNASSISKQILEKLPGNDSDSHMSYSQDRYIFHVKRTDG 61
Query: 63 FTYCVVAVESVGRQIPIAFLERIKDDFNKRYGGGRATTATAKSLNKEFGPKLKEQMQYCV 122
T +A E+ GR IP AFL+ I F K YG +A A S+N EF L +QM++
Sbjct: 62 LTVLCMADETAGRNIPFAFLDDIHQRFVKTYGRA-IHSAQAYSMNDEFSRVLSQQMEFYS 120
Query: 123 EHPEEVSKLAKVKAQVSEVKGVMMENIDKVIDRGEKIEVLVDKTENLRSQAQDFRQQGTQ 182
P +++++K ++S+V+ VM+ENIDKV+DRGE++E+LVDKTEN++ FR+Q +
Sbjct: 121 NDPN-ADRMSRIKGEMSQVRNVMIENIDKVLDRGERLELLVDKTENMQGNTFRFRKQARR 179
Query: 183 LRRKMWYQNMKIKLIVLAIIIALILIIVLSVCHG 216
R MW++N+K+ + ++ ++ ++ I + VCHG
Sbjct: 180 YRTIMWWRNVKLTIALILVLALVVYIAMAFVCHG 213
>At5g22360 synaptobrevin-like protein
Length = 221
Score = 158 bits (399), Expect = 2e-39
Identities = 82/216 (37%), Positives = 139/216 (63%), Gaps = 4/216 (1%)
Query: 5 SLIYSFVARGTVILAEYSDFTGNFTTIALQCLQKLPA--SNNRFTYNCDGHTFSFLVDNG 62
+++Y+ VARGTV+LAE+S TGN + + L+KL S+ R ++ D + F L +G
Sbjct: 2 AIVYAVVARGTVVLAEFSAVTGNTGAVVRRILEKLSPEISDERLCFSQDRYIFHILRSDG 61
Query: 63 FTYCVVAVESVGRQIPIAFLERIKDDFNKRYGGGRATTATAKSLNKEFGPKLKEQMQYCV 122
T+ +A ++ GR++P ++LE I F K YG A A A ++N EF L +QM++
Sbjct: 62 LTFLCMANDTFGRRVPFSYLEEIHMRFMKNYGKV-AHNAPAYAMNDEFSRVLHQQMEFFS 120
Query: 123 EHPEEVSKLAKVKAQVSEVKGVMMENIDKVIDRGEKIEVLVDKTENLRSQAQDFRQQGTQ 182
+P V L +V+ +VSE++ VM+ENI+K+++RG++IE+LVDKT ++ + FR+Q +
Sbjct: 121 SNPS-VDTLNRVRGEVSEIRSVMVENIEKIMERGDRIELLVDKTATMQDSSFHFRKQSKR 179
Query: 183 LRRKMWYQNMKIKLIVLAIIIALILIIVLSVCHGFS 218
LRR +W +N K+ +++ +I+ L+ II+ S C G +
Sbjct: 180 LRRALWMKNAKLLVLLTCLIVFLLYIIIASFCGGIT 215
>At2g25340 putative synaptobrevin
Length = 219
Score = 153 bits (387), Expect = 6e-38
Identities = 79/213 (37%), Positives = 134/213 (62%), Gaps = 3/213 (1%)
Query: 5 SLIYSFVARGTVILAEYSDFTGNFTTIALQCLQKLPAS-NNRFTYNCDGHTFSFLVDNGF 63
S++Y+ VARGTV+LAE S + N +TIA Q L+K+P + ++ +Y+ D + F +G
Sbjct: 2 SILYALVARGTVVLAELSTTSTNASTIAKQILEKIPGNGDSHVSYSQDRYVFHVKRTDGL 61
Query: 64 TYCVVAVESVGRQIPIAFLERIKDDFNKRYGGGRATTATAKSLNKEFGPKLKEQMQYCVE 123
T +A E GR+IP +FLE I F + YG +A A ++N EF L +Q++Y
Sbjct: 62 TVLCMADEDAGRRIPFSFLEDIHQRFVRTYGRA-IHSAQAYAMNDEFSRVLNQQIEYYSN 120
Query: 124 HPEEVSKLAKVKAQVSEVKGVMMENIDKVIDRGEKIEVLVDKTENLRSQAQDFRQQGTQL 183
P ++++K ++++V+ VM+ENID ++DRGE++E+LVDKT N++ FR+Q +
Sbjct: 121 DPN-ADTISRIKGEMNQVRDVMIENIDNILDRGERLELLVDKTANMQGNTFRFRKQTRRF 179
Query: 184 RRKMWYQNMKIKLIVLAIIIALILIIVLSVCHG 216
+W++N K+ L+++ +++ +I I V CHG
Sbjct: 180 NNTVWWRNCKLTLLLILVLLVIIYIGVAFACHG 212
>At1g04750 synaptobrevin 7B, putative, 3' partial
Length = 63
Score = 112 bits (280), Expect = 2e-25
Identities = 50/63 (79%), Positives = 59/63 (93%)
Query: 1 MGQQSLIYSFVARGTVILAEYSDFTGNFTTIALQCLQKLPASNNRFTYNCDGHTFSFLVD 60
M QQSLIYSFVARGTVIL E++DF GNFT+IA QCLQKLP+SNN+FTYNCDGHTF++LV+
Sbjct: 1 MAQQSLIYSFVARGTVILVEFTDFKGNFTSIAAQCLQKLPSSNNKFTYNCDGHTFNYLVE 60
Query: 61 NGF 63
+GF
Sbjct: 61 DGF 63
>At3g24890 synaptobrevin, putative
Length = 114
Score = 100 bits (250), Expect = 5e-22
Identities = 51/97 (52%), Positives = 73/97 (74%), Gaps = 4/97 (4%)
Query: 113 KLKEQMQYCVEHPEEVSKLAKVKAQVSEVKGVMMENIDKVIDRGEKIEVLVDKTENLRSQ 172
+L++++ + HPEE+SKLAKVKA V+++KGVMMENI+K +DR EKI++LVD LRS+
Sbjct: 14 QLEQRVLVLLAHPEEISKLAKVKALVTKMKGVMMENIEKALDRSEKIKILVD----LRSK 69
Query: 173 AQDFRQQGTQLRRKMWYQNMKIKLIVLAIIIALILII 209
+D GT++ RKMW+QNMK KLIVL + ++I
Sbjct: 70 YKDIITPGTKITRKMWFQNMKFKLIVLGTSSSRFVLI 106
>At4g27840 unknown protein
Length = 260
Score = 53.5 bits (127), Expect = 8e-08
Identities = 38/142 (26%), Positives = 69/142 (47%), Gaps = 18/142 (12%)
Query: 3 QQSLIYSFVARGTVILAEY-SDFTGNFTTIALQCLQKLPASNNRFTYNCDGHTFSFLVDN 61
Q+ L YS +A+GTVILAE+ S +AL+C++ +P ++ ++ T++ ++D
Sbjct: 5 QRMLFYSCIAKGTVILAEFASKEEPGIEDLALRCIENVPPHHSMISHTVHKRTYALIIDG 64
Query: 62 GFTYCVVAVESVGRQIPIAFLERIKDDFNKRYGGGRATTATAKSLNKEFGPKLKEQMQYC 121
F+Y + E V + + R+K G +TA SL+ Q+C
Sbjct: 65 LFSYFAILDEVVAKSESVWLFNRLKSATESLMEDG----STADSLD--------NPTQHC 112
Query: 122 VEHPEEVSKLAKVKAQVSEVKG 143
++ SKL V A+++ + G
Sbjct: 113 LQ-----SKLDPVFAEIAAIGG 129
>At5g58060 ATGP1
Length = 199
Score = 52.8 bits (125), Expect = 1e-07
Identities = 30/100 (30%), Positives = 57/100 (57%), Gaps = 4/100 (4%)
Query: 80 AFLERIKDDFNKRYGGGRATTATAKSLNKEFGPKLKEQMQYCVEHPEEVSKLAKVKAQVS 139
+ L ++ D++ K +G + +AK + + P L E + + P E KL K++ ++
Sbjct: 93 SLLNQVLDEYQKSFG---ESWRSAKEDSNQPWPYLTEALNK-FQDPAEADKLLKIQRELD 148
Query: 140 EVKGVMMENIDKVIDRGEKIEVLVDKTENLRSQAQDFRQQ 179
E K ++ + ID V+ RGEK++ LV+K+ +L +Q F +Q
Sbjct: 149 ETKIILHKTIDSVLARGEKLDSLVEKSSDLSMASQMFYKQ 188
>At4g10170 unknown protein
Length = 254
Score = 51.6 bits (122), Expect = 3e-07
Identities = 55/242 (22%), Positives = 93/242 (37%), Gaps = 30/242 (12%)
Query: 8 YSFVARGTVILAEYSDFTGNFTTIALQCLQKLPASNNRFTYNCDGHTFSFLVDNGFTYCV 67
Y V+R IL Y+ ++A CL+K P + + F FL+ +GF Y
Sbjct: 10 YCCVSRDNQILYSYNGGDQTNESLAALCLEKSPPFHTWYFETIGKRRFGFLIGDGFVYFA 69
Query: 68 VAVESVGRQIPIAFLERIKDDFNK--RYGGGRATTATAKSLNKE---------FGPKLKE 116
+ E + R + FLE ++D+F K R + TA S+N E L+
Sbjct: 70 IVDEVLKRSSVLKFLEHLRDEFKKAARENSRGSFTAMIGSINVEDQLVPVVTRLIASLER 129
Query: 117 QMQYCVEHPEEVSKLAKVKAQVSEVKGVMMENIDKVIDRGEK---IEVLVDKTENLRSQ- 172
+ + + S L + + K ++ + K + K IE+ + N R
Sbjct: 130 VAESSSNNELKSSNLGEQSEGSNSTKAPLLGRLSKQEKKKGKDHVIELEEHRKSNDRGNI 189
Query: 173 AQDFRQQGTQLRRK---------------MWYQNMKIKLIVLAIIIALILIIVLSVCHGF 217
D GT L ++ W + ++I L + A I + I L++C G
Sbjct: 190 TDDSAGAGTSLEKECVSSSGRSVTQSFEWKWRRLVQIVLAIDAAICLTLFGIWLAICRGI 249
Query: 218 SC 219
C
Sbjct: 250 EC 251
>At1g33480 unknown protein
Length = 508
Score = 51.6 bits (122), Expect = 3e-07
Identities = 50/243 (20%), Positives = 100/243 (40%), Gaps = 31/243 (12%)
Query: 8 YSFVARGTVILAEYSDFTG--NFTTIALQCLQKLPASNNRFTYNCDGHTFSFLVDNGFTY 65
Y V+R I+ Y++ N ++A CL+K P + + TF FL+ + F Y
Sbjct: 263 YCCVSRDNQIMYAYNNAGDHRNNESLAALCLEKTPPFHKWYFETRGKKTFGFLMKDDFVY 322
Query: 66 CVVAVESVGRQIPIAFLERIKDDFN----KRYGGGRATTATAKSLNKEFGPKLKEQMQY- 120
+ + + + FLE+++D+ K G + + + ++ + +L +++
Sbjct: 323 FAIVDDVFKKSSVLDFLEKLRDELKEANKKNSRGSFSGSISFSNVQDQIVRRLIASLEFD 382
Query: 121 ---------CVEHPEEV-------------SKLAKVKAQ--VSEVKGVMMENIDKVIDRG 156
++ E+ +K K K + ++G+ +E K DRG
Sbjct: 383 HTCLPLSSPSIDGAEQSYASNSKAPLLGRSNKQDKKKGRDHAHSLRGIEIEEHRKSNDRG 442
Query: 157 EKIEVLVDKTENLRSQAQDFRQQGTQLRRKMWYQNMKIKLIVLAIIIALILIIVLSVCHG 216
E +E+ + R G+Q + W + +KI L + I +L + L++CHG
Sbjct: 443 NVTECSNASSESATYVPRRGRSGGSQSIERKWRRQVKIVLAIDIAICLTLLGVWLAICHG 502
Query: 217 FSC 219
C
Sbjct: 503 IEC 505
>At1g11890 putative vesicle transport protein
Length = 218
Score = 48.5 bits (114), Expect = 3e-06
Identities = 33/167 (19%), Positives = 74/167 (43%), Gaps = 6/167 (3%)
Query: 44 NRFTYNCDGHTFSFLVDNGFTYCVVAVESVGRQIPIAFLERIKDDFNKRYGGGRATTATA 103
+R + + F ++++ Y + S +++ +LE +K++F + G T A
Sbjct: 51 SRMSVETGPYVFHYIIEGRVCYLTMCDRSYPKKLAFQYLEDLKNEFERVNGPNIETAARP 110
Query: 104 KSLNK--EFGPKLKEQMQYCVEHPEEVSKLAKVKAQVSEVKGVMMENIDKVIDRGEKIEV 161
+ K F K K+ Q +AK+ ++ EV +M N+ +V+ GEK++
Sbjct: 111 YAFIKFDTFIQKTKKLYQ----DTRTQRNIAKLNDELYEVHQIMTRNVQEVLGVGEKLDQ 166
Query: 162 LVDKTENLRSQAQDFRQQGTQLRRKMWYQNMKIKLIVLAIIIALILI 208
+ + + L S+++ + + L R+ + IV ++ L +
Sbjct: 167 VSEMSSRLTSESRIYADKAKDLNRQALIRKWAPVAIVFGVVFLLFWV 213
>At5g52990 unknown protein
Length = 272
Score = 45.1 bits (105), Expect = 3e-05
Identities = 25/83 (30%), Positives = 47/83 (56%), Gaps = 2/83 (2%)
Query: 6 LIYSFVARGTVILAEY-SDFTGNFTTIALQCLQKLPASNNRFTYNCDGHTFSFLV-DNGF 63
L Y+ +A+GTV+LAE+ S IAL+C++ P ++ F++ T++F + D+ F
Sbjct: 8 LSYTCIAKGTVVLAEFVSRQEPGIEAIALRCIENTPPHHSMFSHTVHKKTYTFAIDDDSF 67
Query: 64 TYCVVAVESVGRQIPIAFLERIK 86
Y ++ ES+ + L R++
Sbjct: 68 VYFSISDESMEKPESFWVLNRLR 90
>At5g58180 ATGP1-like protein
Length = 199
Score = 44.3 bits (103), Expect = 5e-05
Identities = 30/100 (30%), Positives = 54/100 (54%), Gaps = 6/100 (6%)
Query: 80 AFLERIKDDFNKRYGGGRATTATAKSLNKEFGPKLKEQMQYCVEHPEEVSKLAKVKAQVS 139
+ L ++ D + K YG T ++ ++ + P LKE P E KL K++ ++
Sbjct: 95 SLLNQVLDVYQKDYGD----TWRFENSSQPW-PYLKEASDK-FRDPAEADKLLKIQRELD 148
Query: 140 EVKGVMMENIDKVIDRGEKIEVLVDKTENLRSQAQDFRQQ 179
E K ++ + ID V+ RGEK++ LV+K+ L ++ F +Q
Sbjct: 149 ETKIILHKTIDGVLARGEKLDSLVEKSSELSLASKMFYKQ 188
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.323 0.137 0.395
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 4,470,995
Number of Sequences: 26719
Number of extensions: 177654
Number of successful extensions: 696
Number of sequences better than 10.0: 51
Number of HSP's better than 10.0 without gapping: 33
Number of HSP's successfully gapped in prelim test: 18
Number of HSP's that attempted gapping in prelim test: 639
Number of HSP's gapped (non-prelim): 58
length of query: 219
length of database: 11,318,596
effective HSP length: 95
effective length of query: 124
effective length of database: 8,780,291
effective search space: 1088756084
effective search space used: 1088756084
T: 11
A: 40
X1: 16 ( 7.5 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (22.0 bits)
S2: 58 (26.9 bits)
Medicago: description of AC126019.3


