

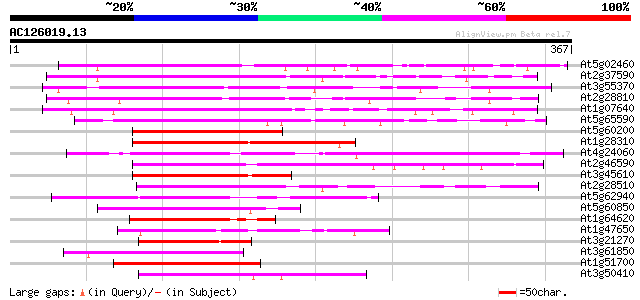
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC126019.13 - phase: 0
(367 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
At5g02460 putative zinc finger protein 223 1e-58
At2g37590 putative DOF zinc finger protein 208 4e-54
At3g55370 zinc finger protein OBP3 182 3e-46
At2g28810 putative DOF zinc finger protein 161 6e-40
At1g07640 zinc finger protein OBP2 like 156 1e-38
At5g65590 DOF zinc finger protein-like 138 4e-33
At5g60200 zinc finger protein - like 134 6e-32
At1g28310 zinc finger like protein OBP3 130 1e-30
At4g24060 unknown protein 129 2e-30
At2g46590 DOF zinc finger like protein 128 6e-30
At3g45610 dof6 zinc finger protein 127 1e-29
At2g28510 DOF zinc finger like protein 126 2e-29
At5g62940 Dof zinc finger protein - like 126 2e-29
At5g60850 zinc finger protein OBP4 - like 125 5e-29
At1g64620 zinc finger protein, putative 123 2e-28
At1g47650 hypothetical protein 119 2e-27
At3g21270 Dof zinc finger protein 119 3e-27
At3g61850 transcription factor BBFa 118 4e-27
At1g51700 unknown protein 118 4e-27
At3g50410 DNA binding protein 116 2e-26
>At5g02460 putative zinc finger protein
Length = 399
Score = 223 bits (568), Expect = 1e-58
Identities = 155/390 (39%), Positives = 204/390 (51%), Gaps = 79/390 (20%)
Query: 33 GGSVSQQLLQTQTPPPQQSQSQPH------------GGGSTGSIRPGSMSDRARMANMPM 80
G +++QQ L PPQQ Q+ P GG G IRPGSM++RAR+AN+P+
Sbjct: 31 GNNINQQFLPHHPLPPQQQQTPPQLHHNNGNGGVAVPGGPGGLIRPGSMAERARLANIPL 90
Query: 81 PETALKCPRCESTNTKFCYFNNYSLSQPRHFCKTCRRYWTRGGALRNVPVGGGFRRNKRS 140
PETALKCPRC+STNTKFCYFNNYSL+QPRHFCK CRRYWTRGGALR+VPVGGG RRNKR+
Sbjct: 91 PETALKCPRCDSTNTKFCYFNNYSLTQPRHFCKACRRYWTRGGALRSVPVGGGCRRNKRT 150
Query: 141 SKSSNNGNSTKSPASSDRQTGSASSTNSITSNNNIHSSA---DILGLTPQMSSLRF---- 193
SS G + S +G++ S +S TSN+ H A + +G SSL
Sbjct: 151 KNSSGGGGGSTS-------SGNSKSQDSATSNDQYHHRAMANNQMGPPSSSSSLSSLLSS 203
Query: 194 ----MTPLHHNFSNPNDFVGGG------------DLGLNYGFSYMGGVG----DLGSALG 233
+ P H + SN N+ +G G D N+ Y G V +G
Sbjct: 204 YNAGLIPGHDHNSNNNNILGLGSSLPPLKLMPPLDFTDNFTLQY-GAVSAPSYHIGGGSS 262
Query: 234 GNSILSSSGLEQWRMPMSMTHQQHQFPFLANLEGTNSNLYPFEGNVQNHEMITSGGYVRP 293
G + +G +QWR P + +Q P L L+ + + E + ++T G RP
Sbjct: 263 GGAAALLNGFDQWRFPAT-----NQLP-LGGLDPFDQQ-HQMEQQNPGYGLVTGSGQYRP 315
Query: 294 KVI---------STSGIM-----TQLASVKMEDSINRGFLGINTSNNNSNQGSEQFWN-- 337
K I S S M +QLASVKMEDS N+ +N S +Q WN
Sbjct: 316 KNIFHNLISSSSSASSAMVTATASQLASVKMEDSNNQ----LNLSRQLFGD-EQQLWNIH 370
Query: 338 --SAVVTGNSAANWTDHVSGFSSSSNTTSN 365
+A T + ++W++ + FSSSS TSN
Sbjct: 371 GAAAASTAAATSSWSEVSNNFSSSS--TSN 398
>At2g37590 putative DOF zinc finger protein
Length = 330
Score = 208 bits (529), Expect = 4e-54
Identities = 136/335 (40%), Positives = 189/335 (55%), Gaps = 40/335 (11%)
Query: 25 NHHQQPGNGGSVSQ-QLLQTQTPPPQQSQSQPH-------GGGSTGSIRPGSMSDRARMA 76
+++ G G S + +L+ Q P QQ Q QPH GGG GSIR GSM DRAR A
Sbjct: 21 SNYNHDGTGASANGGHVLRPQLQPQQQPQQQPHPNGSGGGGGGGGGSIRAGSMVDRARQA 80
Query: 77 NMPMPETALKCPRCESTNTKFCYFNNYSLSQPRHFCKTCRRYWTRGGALRNVPVGGGFRR 136
N+ +PE ALKCPRCESTNTKFCYFNNYSL+QPRHFCKTCRRYWTRGGALRNVPVGGG RR
Sbjct: 81 NVALPEAALKCPRCESTNTKFCYFNNYSLTQPRHFCKTCRRYWTRGGALRNVPVGGGCRR 140
Query: 137 NKRS-SKSSNNGNSTKSPASSDRQTGSASSTNSITSNNNIHSSADILGLTPQMSSLRFMT 195
N+R+ S S+NN NST + ++ +G+AS+ ++I S++ + I L+ +S R M
Sbjct: 141 NRRTKSNSNNNNNSTATSNNTSFSSGNASTISTILSSHYGGNQESI--LSQILSPARLMN 198
Query: 196 PLHHNFSN--PNDFVGGGDLGLNYGFSYMGGVGDLGSALGGNSILSSSGLEQWRMPMSMT 253
P +++ + N LNYG + + G S++S +++WR S +
Sbjct: 199 PTYNHLGDLTSNTKTDNNMSLLNYG-GLSQDLRSIHMGASGGSLMSC--VDEWR---SAS 252
Query: 254 HQQHQFPFLANLEGTNSNLYPFEGNVQNHEMITSGGYVRPKVIS---TSGIMTQLASVKM 310
+ Q NLE +SN P + E P++ S +S + +Q +SVK+
Sbjct: 253 YHQQSSMGGGNLE-DSSNPNPSANGFYSFE--------SPRITSASISSALASQFSSVKV 303
Query: 311 EDSINRGFLGINTSNNNSNQGSEQFWNSAVVTGNS 345
ED+ + +N + N S+ WN G+S
Sbjct: 304 EDN---PYKWVNVNGNCSS------WNDLSAFGSS 329
>At3g55370 zinc finger protein OBP3
Length = 323
Score = 182 bits (462), Expect = 3e-46
Identities = 132/358 (36%), Positives = 175/358 (48%), Gaps = 78/358 (21%)
Query: 22 QQQNHHQQP--GNGGSVSQQLLQTQTPPPQQSQSQPHGGGSTGSIRPGSMSDRARMANMP 79
QQ N HQ + + L Q +PP Q + R SM +RAR+A +P
Sbjct: 18 QQGNQHQLECVTTDQNPNNYLRQLSSPPTSQV-------AGSSQARVNSMVERARIAKVP 70
Query: 80 MPETALKCPRCESTNTKFCYFNNYSLSQPRHFCKTCRRYWTRGGALRNVPVGGGFRRNKR 139
+PE AL CPRC+STNTKFCYFNNYSL+QPRHFCKTCRRYWTRGG+LRNVPVGGGFRRNKR
Sbjct: 71 LPEAALNCPRCDSTNTKFCYFNNYSLTQPRHFCKTCRRYWTRGGSLRNVPVGGGFRRNKR 130
Query: 140 SSKSSNNGNSTKSPASSDRQTGSASSTNSITSNNNIHSSADILGLTPQM-SSLRFMTPLH 198
S S + ++ + T S +S S ++ + HS G P+ S+L + PL
Sbjct: 131 S--KSRSKSTVVVSTDNTTSTSSLTSRPSYSNPSKFHS----YGQIPEFNSNLPILPPLQ 184
Query: 199 ----HNFSNPN-DFVGGGDLGLNYGFSYMGGVGDLGSALGGNSILSSSGLEQWRMPMSMT 253
+N SN DF G + G S GG+ L+ WR+P S
Sbjct: 185 SLGDYNSSNTGLDFGGTQISNMISGMSSSGGI-----------------LDAWRIPPS-- 225
Query: 254 HQQHQFPFLANLEG---TNSNLYP-FEGNVQNHEMITSGGYVRPKVISTSGIMTQLASVK 309
Q QFPFL N G +++ LYP EG V TQ +VK
Sbjct: 226 QQAQQFPFLINTTGLVQSSNALYPLLEGGVS---------------------ATQTRNVK 264
Query: 310 MED-------------SINRGFLGINTSNNNSNQGSEQFWNSAVVTGNSAANWTDHVS 354
E+ +++R FLG N+ N+ + ++ TG ++ N T H+S
Sbjct: 265 AEENDQDRGRDGDGVNNLSRNFLGNININSGRNEEYTSWGGNSSWTGFTSNNSTGHLS 322
>At2g28810 putative DOF zinc finger protein
Length = 320
Score = 161 bits (407), Expect = 6e-40
Identities = 117/336 (34%), Positives = 165/336 (48%), Gaps = 65/336 (19%)
Query: 25 NHHQQPGNGGSVS--QQLLQTQTPPPQQSQSQPHGGGSTGSIRPGSMS----DRARMA-N 77
+HHQ NG VS Q+L P + ++ P S++ +RAR+A N
Sbjct: 27 HHHQLQENGSLVSGHHQVLSHHFPQNPNPNHHHVETAAATTVDPSSLNGQAAERARLAKN 86
Query: 78 MPMPETALKCPRCESTNTKFCYFNNYSLSQPRHFCKTCRRYWTRGGALRNVPVGGGFRRN 137
PE ALKCPRC+S NTKFCYFNNY+L+QPRHFCK CRRYWTRGGALRNVPVGGG RRN
Sbjct: 87 SQPPEGALKCPRCDSANTKFCYFNNYNLTQPRHFCKACRRYWTRGGALRNVPVGGGCRRN 146
Query: 138 KRSSKSSNNGNSTKSPASSDRQTGSASSTNSITSNNNIHSSADILGLTPQMSSLRFMTPL 197
K+ +GNS S +S ++Q+ S + S T+ +N+ + P + +L+ +T L
Sbjct: 147 KK----GKSGNSKSSSSSQNKQSTSMVNATSPTNTSNVQLQTN--SQFPFLPTLQNLTQL 200
Query: 198 HHNFSNPNDFVGGGDLGLNYGFSYMGGVGDLGSALGG---NSILSSSGLEQWRMPMSMTH 254
G +GLN + G G G +G N++++S G T
Sbjct: 201 -------------GGIGLNLA-AINGNNGGNGPVMGNNNENNLMTSLGSSSHFALFDRTM 246
Query: 255 QQHQFPFLANLEGTNSNLYPFEGNVQNHEMITSGGYVRPKVISTSGIMTQLASVKMED-- 312
+ FP N+ ++S G R ++Q A VKMED
Sbjct: 247 GLYNFPNEVNMG------------------LSSIGATR---------VSQTAQVKMEDNH 279
Query: 313 --SINRGFLGINTSNNNSNQGSEQFWNSAVVTGNSA 346
+I+R G+ + N SN Q+W + G+S+
Sbjct: 280 LGNISRPVSGLTSPGNQSN----QYWTGQGLPGSSS 311
>At1g07640 zinc finger protein OBP2 like
Length = 331
Score = 156 bits (395), Expect = 1e-38
Identities = 118/345 (34%), Positives = 169/345 (48%), Gaps = 46/345 (13%)
Query: 22 QQQNHHQQPGNGGSVSQQ--LLQTQTPPPQQSQSQPHGGGSTGSIRP----GSMSDRARM 75
Q HQ NG +S +L Q PP Q + + H +T + P GSM++RAR
Sbjct: 8 QHHLQHQLNENGSIISGHGLVLSHQLPPLQANPNPNHHHVATSAGLPSRMGGSMAERARQ 67
Query: 76 ANMPMPETALKCPRCESTNTKFCYFNNYSLSQPRHFCKTCRRYWTRGGALRNVPVGGGFR 135
AN+P LKCPRC+S+NTKFCY+NNY+L+QPRHFCK CRRYWT+GGALRNVPVGGG R
Sbjct: 68 ANIPPLAGPLKCPRCDSSNTKFCYYNNYNLTQPRHFCKGCRRYWTQGGALRNVPVGGGCR 127
Query: 136 RNKRSSKSSNNGNSTKSPASSDRQTGSASSTNSITSNNNIHSSADILGLTPQMSSLRFMT 195
RN + K+ N +S+ S S + S+ + +N+ S + LT Q+ +
Sbjct: 128 RNNKKGKNGNLKSSSSSSKQSSSVNAQSPSSGQLRTNHQFPFSPTLYNLT-QLGGIGL-- 184
Query: 196 PLHHNFSNPNDFVGGGDLGLNYGFSYMGGVGDLGSALGGNSILSSSGLEQWRMPMSMTHQ 255
N + N G + G S M + DLG G N+ +G + + +
Sbjct: 185 ----NLAATN----GNNQAHQIGSSLM--MSDLGFLHGRNTSTPMTG----NIHENNNNN 230
Query: 256 QHQFPFLAN--------LEGTNSNLYPF--EGNVQNHEMITSGGYVRPKVISTSGI---M 302
++ +A+ L + LY F +GN+ N+ I+ STS + +
Sbjct: 231 NNENNLMASVGSLSPFALFDPTTGLYAFQNDGNIGNNVGISGS--------STSMVDSRV 282
Query: 303 TQLASVKMEDSINRGFLGINTSNNNS--NQGSEQFWNSAVVTGNS 345
Q VKME+ N L S S NQ ++ FW + +G S
Sbjct: 283 YQTPPVKMEEQPNLANLSRPVSGLTSPGNQTNQYFWPGSDFSGPS 327
>At5g65590 DOF zinc finger protein-like
Length = 316
Score = 138 bits (348), Expect = 4e-33
Identities = 109/341 (31%), Positives = 156/341 (44%), Gaps = 69/341 (20%)
Query: 43 TQTPPPQQSQSQPHGGGSTGSIRPGSMSDRARMANMPMPETALKCPRCESTNTKFCYFNN 102
T P P+ H T S + + +++ + LKCPRC S NTKFCY+NN
Sbjct: 5 TNLPSPKPVPKPDHRISGT------SQTKKPPSSSVAQDQQNLKCPRCNSPNTKFCYYNN 58
Query: 103 YSLSQPRHFCKTCRRYWTRGGALRNVPVGGGFRRNKRSSKSSNNGNSTKSPASSDRQTGS 162
YSLSQPRHFCK+CRRYWTRGGALRNVP+GGG R+ K+S K +++ N+ S +SS R S
Sbjct: 59 YSLSQPRHFCKSCRRYWTRGGALRNVPIGGGCRKTKKSIKPNSSMNTLPSSSSSQRFFSS 118
Query: 163 ASSTN-----------------SITSNNNIH--SSADILGLTPQMSSLRFMTPLHHNFSN 203
+ S+ N++ ++ ++L L P MSS R TP+ + +
Sbjct: 119 IMEDSSKFFPPPTTMDFQLAGLSLNKMNDLQLLNNQEVLDLRPMMSSGRENTPV--DVGS 176
Query: 204 PNDFVGGGDLGLNY---GFSYMGGV-GDLGSALGGNSILSSS---GLEQWRMPMSMTHQQ 256
+G GD N+ GF+ G G+L S++ S L+ L+Q RM M + +
Sbjct: 177 GLSLMGFGDFNNNHSPTGFTTAGASDGNLASSIETLSCLNQDLHWRLQQQRMAMLFGNSK 236
Query: 257 HQFPFLANLEGTNSNLYPFEGNVQNHEMITSGGYVRPKVISTSGIMTQLASVKMEDSINR 316
+ +E LY +N E++ S P K D+
Sbjct: 237 EE---TVVVERPQPILY------RNLEIVNSSSPSSP--------------TKKGDNQTE 273
Query: 317 GFLGINT------SNNNSNQGSEQFWNSAVVTGNSAANWTD 351
+ G N+ SNN + G WN N WTD
Sbjct: 274 WYFGNNSDNEGVISNNANTGGGGSEWN------NGIQAWTD 308
>At5g60200 zinc finger protein - like
Length = 257
Score = 134 bits (338), Expect = 6e-32
Identities = 60/99 (60%), Positives = 77/99 (77%), Gaps = 1/99 (1%)
Query: 81 PETALKCPRCESTNTKFCYFNNYSLSQPRHFCKTCRRYWTRGGALRNVPVGGGFRRNKRS 140
PE AL+CPRC+STNTKFCY+NNYSL+QPR+FCK+CRRYWT+GG LRN+PVGGG R+NKRS
Sbjct: 51 PELALRCPRCDSTNTKFCYYNNYSLTQPRYFCKSCRRYWTKGGTLRNIPVGGGCRKNKRS 110
Query: 141 SKSSNNG-NSTKSPASSDRQTGSASSTNSITSNNNIHSS 178
+ S+ +T PAS D + SA+ N ++N +I S
Sbjct: 111 TSSAARSLRTTPEPASHDGKVFSAAGFNGYSNNEHIDLS 149
>At1g28310 zinc finger like protein OBP3
Length = 311
Score = 130 bits (326), Expect = 1e-30
Identities = 67/150 (44%), Positives = 93/150 (61%), Gaps = 5/150 (3%)
Query: 81 PETALKCPRCESTNTKFCYFNNYSLSQPRHFCKTCRRYWTRGGALRNVPVGGGFRRNKRS 140
P+ LKCPRC+S+NTKFCY+NNYSLSQPRHFCK C+RYWTRGG LRNVPVGG +R+NKR
Sbjct: 23 PQPQLKCPRCDSSNTKFCYYNNYSLSQPRHFCKACKRYWTRGGTLRNVPVGGSYRKNKRV 82
Query: 141 SKSSNNGNSTKSPASSDRQTGSASSTNSITSNNNIHSSADILGLTPQMSSLRFMTP-LHH 199
+ S +T S S+ + S ++ + I+ ++++ GL+ MSS P +
Sbjct: 83 KRPSTATTTTASTVST-TNSSSPNNPHQISHFSSMNHHPLFYGLSDHMSSCNNNLPMIPS 141
Query: 200 NFSNPNDFVGGGDLG---LNYGFSYMGGVG 226
FS+ + L L+ GFS + +G
Sbjct: 142 RFSDSSKTCSSSGLESEFLSSGFSSLSALG 171
>At4g24060 unknown protein
Length = 342
Score = 129 bits (325), Expect = 2e-30
Identities = 105/331 (31%), Positives = 143/331 (42%), Gaps = 45/331 (13%)
Query: 38 QQLLQTQTPPPQQSQSQPHGGGSTGSIRPGSMSDRARMANMPMPETALKCPRCESTNTKF 97
++++ P PQ QP S G R +AR P + A+ CPRC STNTKF
Sbjct: 16 EEIVTNTCPKPQPQPLQPQQPPSVGGER------KAR----PEKDQAVNCPRCNSTNTKF 65
Query: 98 CYFNNYSLSQPRHFCKTCRRYWTRGGALRNVPVGGGFRRNKRSSKSSNNGNSTKSPASSD 157
CY+NNYSL+QPR+FCK CRRYWT GG+LRN+PVGGG R+NKRS SS S S++
Sbjct: 66 CYYNNYSLTQPRYFCKGCRRYWTEGGSLRNIPVGGGSRKNKRSHSSS-------SDISNN 118
Query: 158 RQTGSASSTNSITSNNNIH-SSADILGLTPQMSSLRFMTPLHHNFSNPNDFVGGGDLGLN 216
+ +T S+++ H S GLT Q NP F+ LN
Sbjct: 119 HSDSTQPATKKHLSDHHHHLMSMSQQGLTGQ---------------NPK-FLETTQQDLN 162
Query: 217 YGFSYMGGV----GDLGSALGGNSILSSSGLEQWRMPMSMTHQQHQFPFLANLEGT-NSN 271
GFS G + DL +G N+ S++ + + N NS+
Sbjct: 163 LGFSPHGMIRTNFTDLIHNIGNNTNKSNNNNNPLIVSSCSAMATSSLDLIRNNSNNGNSS 222
Query: 272 LYPFEGNVQNHEMITSGGYVRPKVISTSGIMTQLASVKMEDSINRGFLGINTSNNNSNQG 331
F G +++ SGG+ T L ++ N GF G +G
Sbjct: 223 NSSFMGFPVHNQDPASGGFSMQDHYKPCNTNTTLLGFSLDHHHNNGFHGGFQGGEEGGEG 282
Query: 332 SEQFWNSAVVTGNSAANWTDHVSGFSSSSNT 362
+ V G + D SSSS T
Sbjct: 283 GDD------VNGRHLFPFEDLKLPVSSSSAT 307
>At2g46590 DOF zinc finger like protein
Length = 357
Score = 128 bits (321), Expect = 6e-30
Identities = 93/301 (30%), Positives = 143/301 (46%), Gaps = 39/301 (12%)
Query: 81 PETALKCPRCESTNTKFCYFNNYSLSQPRHFCKTCRRYWTRGGALRNVPVGGGFRRNKRS 140
P+ L CPRC STNTKFCY+NNYSL+QPR+FCK CRRYWT GG+LRNVPVGG R+NKRS
Sbjct: 64 PQEKLNCPRCNSTNTKFCYYNNYSLTQPRYFCKGCRRYWTEGGSLRNVPVGGSSRKNKRS 123
Query: 141 SKSSNNGNSTKSPASSDRQTGSASSTNSITSNNNIHSSADILGLTPQMSSLRFMTPLHHN 200
S SS++ P+S I +N IH+ + + S M HH+
Sbjct: 124 SSSSSSNILQTIPSS------LPDLNPPILFSNQIHNKSKGSSQDLNLLSFPVMQDQHHH 177
Query: 201 FSNPNDFVGGGDLGLNYGFSYMGGVGDLGSALGGNS--------------ILSSSGLEQW 246
+ + F+ + N ++ S G +S + S SG+
Sbjct: 178 HVHMSQFLQMPKMEGNGNITHQQQPSSSSSVYGSSSSPVSALELLRTGVNVSSRSGINSS 237
Query: 247 RMPM-------SMTHQQHQFPFLANLEGTN---SNLYPFEGNVQNH--EMITSGGYVRPK 294
MP ++ + FP + + + +N S + G+ N+ E + S + + +
Sbjct: 238 FMPSGSMMDSNTVLYTSSGFPTMVDYKPSNLSFSTDHQGLGHNSNNRSEALHSDHHQQGR 297
Query: 295 VISTSG-IMTQLAS-----VKMEDSINRGFLGINTSNNNSNQGSEQFWNSAVVTGNSAAN 348
V+ G M +L+S V +D+ + G N +NNNS+ + +W+ T ++
Sbjct: 298 VLFPFGDQMKELSSSITQEVDHDDNQQQKSHGNNNNNNNSSP-NNGYWSGMFSTTGGGSS 356
Query: 349 W 349
W
Sbjct: 357 W 357
>At3g45610 dof6 zinc finger protein
Length = 245
Score = 127 bits (319), Expect = 1e-29
Identities = 58/105 (55%), Positives = 78/105 (74%), Gaps = 3/105 (2%)
Query: 81 PETALKCPRCESTNTKFCYFNNYSLSQPRHFCKTCRRYWTRGGALRNVPVGGGFRRNKRS 140
PE +L+CPRC+STNTKFCY+NNYSLSQPR+FCK+CRRYWT+GG LRN+P+GG +R++KRS
Sbjct: 36 PEQSLRCPRCDSTNTKFCYYNNYSLSQPRYFCKSCRRYWTKGGILRNIPIGGAYRKHKRS 95
Query: 141 SKSSNNGNSTKSPASSDRQTGSASSTNSI-TSNNNIHSSADILGL 184
S ++ + +T P + G + T S +NNNI + LGL
Sbjct: 96 SSATKSLRTTPEPTMT--HDGKSFPTASFGYNNNNISNEQMELGL 138
>At2g28510 DOF zinc finger like protein
Length = 288
Score = 126 bits (317), Expect = 2e-29
Identities = 97/269 (36%), Positives = 124/269 (46%), Gaps = 63/269 (23%)
Query: 84 ALKCPRCESTNTKFCYFNNYSLSQPRHFCKTCRRYWTRGGALRNVPVGGGFRRNKRSSKS 143
A CPRCES NTKFCY+NNYSLSQPR+FCK+CRRYWT+GG LRNVPVGGG RRNKRSS S
Sbjct: 47 AQNCPRCESPNTKFCYYNNYSLSQPRYFCKSCRRYWTKGGTLRNVPVGGGCRRNKRSSSS 106
Query: 144 S-NNGNSTKSPASSDRQTGSASSTNSITSNNNIHSSADILGLTPQMSSLRFMTPLHHNFS 202
+ + N+ KS + T S+ S D+ +L F T H+ S
Sbjct: 107 AFSKNNNNKSINFHTDPLQNPLITGMPPSSFGYDHSIDL--------NLAFATLQKHHLS 158
Query: 203 N---PNDFVGGGDLGLNYGFSYMGGVGDLGSALGGNSILSSSGLEQWRMPMSMTHQQHQF 259
+ F GGDL + Y D+G G N +
Sbjct: 159 SQATTPSFGFGGDLSI-----YGNSTNDVGIFGGQNGTYN-------------------- 193
Query: 260 PFLANLEGTNSNLYPF-EGNVQNHEMITSGGYVRPKVISTSGIMTQLASVKMEDSINRGF 318
NS Y F GN N++ K+ ST G+ + K E+
Sbjct: 194 ---------NSLCYGFMSGNGNNNQN-------EIKMASTLGMSLEGNERKQEN------ 231
Query: 319 LGINTSNNNSNQGSEQFWNSA-VVTGNSA 346
+N +NNNS S+ FW +TG+SA
Sbjct: 232 --VNNNNNNSENPSKVFWGFPWQMTGDSA 258
>At5g62940 Dof zinc finger protein - like
Length = 372
Score = 126 bits (316), Expect = 2e-29
Identities = 80/215 (37%), Positives = 108/215 (50%), Gaps = 29/215 (13%)
Query: 28 QQPGNGGSVSQQLLQTQTPPPQQSQSQPHGGGSTGSIRPGSMSDRARMANMPMPETALKC 87
+ +GGS+ + + + S ++ ++ D R P KC
Sbjct: 17 ESESSGGSMLDSSTNSPSAADILAACSTRPQASAVAVAAAALMDGGRRLRPPHDHPQ-KC 75
Query: 88 PRCESTNTKFCYFNNYSLSQPRHFCKTCRRYWTRGGALRNVPVGGGFRRNKRSSKSSNNG 147
PRCEST+TKFCY+NNYSLSQPR+FCKTCRRYWT+GG LRN+PVGGG R+NK+ S S+
Sbjct: 76 PRCESTHTKFCYYNNYSLSQPRYFCKTCRRYWTKGGTLRNIPVGGGCRKNKKPSSSN--- 132
Query: 148 NSTKSPASSDRQTGSASSTNSITSNNNIHSSADILGLTPQMSSLRFMTPLHHNFS-NPND 206
S+SST+S +NI +T S L + H N+ +P
Sbjct: 133 --------------SSSSTSSGKKPSNI--------VTANTSDLMALAHSHQNYQHSPLG 170
Query: 207 FVGGGDLGLNYGFSYMGGVGDLGSALGGNSILSSS 241
F G + +Y G VG L S GG +LS S
Sbjct: 171 FSHFGGMMGSYSTPEHGNVGFLESKYGG--LLSQS 203
>At5g60850 zinc finger protein OBP4 - like
Length = 307
Score = 125 bits (313), Expect = 5e-29
Identities = 67/136 (49%), Positives = 81/136 (59%), Gaps = 10/136 (7%)
Query: 58 GGSTGSIRPGSMSDRARMANMPMPETALKCPRCESTNTKFCYFNNYSLSQPRHFCKTCRR 117
GG G G RA N+ +LKCPRC S NTKFCY+NNY+LSQPRHFCK CRR
Sbjct: 24 GGGIGGGGGGDRRMRAHQNNILNHHQSLKCPRCNSLNTKFCYYNNYNLSQPRHFCKNCRR 83
Query: 118 YWTRGGALRNVPVGGGFRRNKRS-SKSSNNGNSTKSPASS--DRQTGSASSTNSITSNNN 174
YWT+GG LRNVPVGGG R+ KRS +K + +S P ++ D SST S
Sbjct: 84 YWTKGGVLRNVPVGGGCRKAKRSKTKQVPSSSSADKPTTTQDDHHVEEKSSTGS------ 137
Query: 175 IHSSADILGLTPQMSS 190
HSS++ LT S+
Sbjct: 138 -HSSSESSSLTASNST 152
>At1g64620 zinc finger protein, putative
Length = 352
Score = 123 bits (308), Expect = 2e-28
Identities = 58/96 (60%), Positives = 71/96 (73%), Gaps = 10/96 (10%)
Query: 79 PMPETALKCPRCESTNTKFCYFNNYSLSQPRHFCKTCRRYWTRGGALRNVPVGGGFRRNK 138
P + AL CPRC S NTKFCY+NNYSL+QPR+FCK CRRYWT GG+LRN+PVGGG R+NK
Sbjct: 43 PEKDQALNCPRCNSLNTKFCYYNNYSLTQPRYFCKDCRRYWTAGGSLRNIPVGGGVRKNK 102
Query: 139 RSSKSSNNGNSTKSPASSDRQTGSASSTNSITSNNN 174
RSS +S S+ SP+S S+SS + +NNN
Sbjct: 103 RSSSNS----SSSSPSS------SSSSKKPLFANNN 128
>At1g47650 hypothetical protein
Length = 1409
Score = 119 bits (299), Expect = 2e-27
Identities = 75/186 (40%), Positives = 107/186 (57%), Gaps = 24/186 (12%)
Query: 71 DRARMANMPMPETA--LKCPRCESTNTKFCYFNNYSLSQPRHFCKTCRRYWTRGGALRNV 128
D+ A P P A L CPRC ST TKFCY+NNY+L+QPR++CK+CRRYWT+GG LR+V
Sbjct: 1213 DQPSTAAYPPPNLAEPLPCPRCNSTTTKFCYYNNYNLAQPRYYCKSCRRYWTQGGTLRDV 1272
Query: 129 PVGGGFRRNKRSSKSSNNGNSTKSPASSDRQTGSASSTNSITSNNNIHSSADILGLTPQM 188
PVGGG R RSS + ST + +SS S+SS + T+ + A +T +
Sbjct: 1273 PVGGGTR---RSSSKRHRSFSTTATSSS-----SSSSVITTTTQEPATTEASQTKVTNLI 1324
Query: 189 SSLRFMTPLHHNFSNPNDFVGGGDLGLNYGFSYMGG-----VGDLG-SALGGNSILSSSG 242
S H +F++ +G G+ GL+YGF Y G +G LG S++G ++ G
Sbjct: 1325 SG-------HGSFASLLG-LGSGNGGLDYGFGYGYGLEEMSIGYLGDSSVGEIPVVDGCG 1376
Query: 243 LEQWRM 248
+ W++
Sbjct: 1377 GDTWQI 1382
>At3g21270 Dof zinc finger protein
Length = 204
Score = 119 bits (298), Expect = 3e-27
Identities = 50/74 (67%), Positives = 65/74 (87%), Gaps = 1/74 (1%)
Query: 85 LKCPRCESTNTKFCYFNNYSLSQPRHFCKTCRRYWTRGGALRNVPVGGGFRRNKRSSKSS 144
LKCPRC+S NTKFCY+NNY+LSQPRHFCK+CRRYWT+GGALRNVPVGGG R+N + +S+
Sbjct: 29 LKCPRCDSPNTKFCYYNNYNLSQPRHFCKSCRRYWTKGGALRNVPVGGGSRKN-ATKRST 87
Query: 145 NNGNSTKSPASSDR 158
++ +S SP++S +
Sbjct: 88 SSSSSASSPSNSSQ 101
>At3g61850 transcription factor BBFa
Length = 284
Score = 118 bits (296), Expect = 4e-27
Identities = 56/124 (45%), Positives = 74/124 (59%), Gaps = 6/124 (4%)
Query: 36 VSQQLLQTQTPPPQQ------SQSQPHGGGSTGSIRPGSMSDRARMANMPMPETALKCPR 89
+ Q + T PQQ + ++P+ S G + + P+ + CPR
Sbjct: 7 MEQMISSTNNNTPQQQPTFIATNTRPNATASNGGSGGNTNNTATMETRKARPQEKVNCPR 66
Query: 90 CESTNTKFCYFNNYSLSQPRHFCKTCRRYWTRGGALRNVPVGGGFRRNKRSSKSSNNGNS 149
C STNTKFCY+NNYSL+QPR+FCK CRRYWT GG+LRNVPVGG R+NKRSS + ++
Sbjct: 67 CNSTNTKFCYYNNYSLTQPRYFCKGCRRYWTEGGSLRNVPVGGSSRKNKRSSTPLASPSN 126
Query: 150 TKSP 153
K P
Sbjct: 127 PKLP 130
>At1g51700 unknown protein
Length = 194
Score = 118 bits (296), Expect = 4e-27
Identities = 56/98 (57%), Positives = 68/98 (69%), Gaps = 2/98 (2%)
Query: 69 MSDRARMANMPMPETA-LKCPRCESTNTKFCYFNNYSLSQPRHFCKTCRRYWTRGGALRN 127
M A+ +PE LKCPRC+S NTKFCY+NNY+LSQPRHFCK CRRYWT+GGALRN
Sbjct: 16 MMTTAKQNQPELPEQEQLKCPRCDSPNTKFCYYNNYNLSQPRHFCKNCRRYWTKGGALRN 75
Query: 128 VPVGGGFRR-NKRSSKSSNNGNSTKSPASSDRQTGSAS 164
+PVGGG R+ NKRS S ++ ++ A GS S
Sbjct: 76 IPVGGGTRKSNKRSGSSPSSNLKNQTVAEKPDHHGSGS 113
>At3g50410 DNA binding protein
Length = 253
Score = 116 bits (290), Expect = 2e-26
Identities = 67/158 (42%), Positives = 87/158 (54%), Gaps = 9/158 (5%)
Query: 85 LKCPRCESTNTKFCYFNNYSLSQPRHFCKTCRRYWTRGGALRNVPVGGGFRRN-KRSSKS 143
L CPRC+S+NTKFCY+NNY+ SQPRHFCK CRRYWT GG LR+VPVGGG R++ KRS
Sbjct: 30 LPCPRCDSSNTKFCYYNNYNFSQPRHFCKACRRYWTHGGTLRDVPVGGGTRKSAKRSRTC 89
Query: 144 SNNGNSTKSPASSDR-----QTGSASSTNSITSNNNIH--SSADILGLTPQMSSLRFMTP 196
SN+ +S+ S S+ QT S SN H + +D G + T
Sbjct: 90 SNSSSSSVSGVVSNSNGVPLQTTPVLFPQSSISNGVTHTVTESDGKGSALSLCGSFTSTL 149
Query: 197 LHHNFSNPNDFVGGGDLGL-NYGFSYMGGVGDLGSALG 233
L+HN + G +G+ +G G D+ LG
Sbjct: 150 LNHNAAATATHGSGSVIGIGGFGIGLGSGFDDVSFGLG 187
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.313 0.128 0.386
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 8,945,272
Number of Sequences: 26719
Number of extensions: 409645
Number of successful extensions: 1820
Number of sequences better than 10.0: 100
Number of HSP's better than 10.0 without gapping: 51
Number of HSP's successfully gapped in prelim test: 49
Number of HSP's that attempted gapping in prelim test: 1682
Number of HSP's gapped (non-prelim): 159
length of query: 367
length of database: 11,318,596
effective HSP length: 101
effective length of query: 266
effective length of database: 8,619,977
effective search space: 2292913882
effective search space used: 2292913882
T: 11
A: 40
X1: 16 ( 7.2 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 42 (22.0 bits)
S2: 61 (28.1 bits)
Medicago: description of AC126019.13


