

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC126019.10 + phase: 0
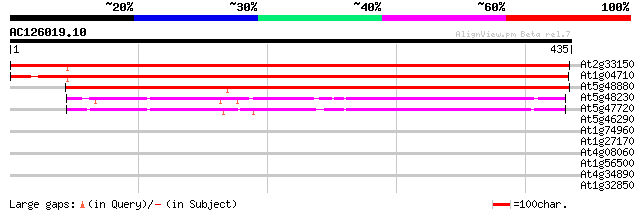
(435 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
At2g33150 3-ketoacyl-CoA thiolase 669 0.0
At1g04710 acetyl-CoA acyltransferase like protein 649 0.0
At5g48880 peroxisomal-3-keto-acyl-CoA thiolase 1 (PKT1) 617 e-177
At5g48230 acetoacetyl-CoA thiolase (AAT1) 228 5e-60
At5g47720 acetoacyl-CoA-thiolase 216 3e-56
At5g46290 3-oxoacyl-[acyl-carrier-protein] synthase I precursor ... 31 1.5
At1g74960 unknown protein 30 2.0
At1g27170 disease resistance protein, putative 30 2.6
At4g08060 putative protein 29 5.8
At1g56500 unknown protein 29 5.8
At4g34890 xanthine dehydrogenase - like protein 28 7.6
At1g32850 hypothetical protein 28 7.6
>At2g33150 3-ketoacyl-CoA thiolase
Length = 462
Score = 669 bits (1725), Expect = 0.0
Identities = 337/440 (76%), Positives = 383/440 (86%), Gaps = 6/440 (1%)
Query: 1 MEKAVERQRVLLQHLNPNSVTSSHHSTHLSASLCSAGQTGGSE------NDVVIVAAYRT 54
MEKA+ERQRVLL+HL P+S +S ++ LSAS C AG + + +DVVIVAA+RT
Sbjct: 1 MEKAIERQRVLLEHLRPSSSSSHNYEASLSASACLAGDSAAYQRTSLYGDDVVIVAAHRT 60
Query: 55 AICKAKRGGFKDTLPDDLLASVLKAVIEKTNVEPSEVGDIVVGTVLGPGSERAIECRMAA 114
+CK+KRG FKDT PDDLLA VL+A+IEKTN+ PSEVGDIVVGTVL PGS+RA ECRMAA
Sbjct: 61 PLCKSKRGNFKDTYPDDLLAPVLRALIEKTNLNPSEVGDIVVGTVLAPGSQRASECRMAA 120
Query: 115 FYAGFPDTVPLRTVNRQCSSGLQAVADVAAYIKAGFYDIGIGAGLECMTQDTISGVRKTN 174
FYAGFP+TV +RTVNRQCSSGLQAVADVAA IKAGFYDIGIGAGLE MT + ++ N
Sbjct: 121 FYAGFPETVAVRTVNRQCSSGLQAVADVAAAIKAGFYDIGIGAGLESMTTNPMAWEGSVN 180
Query: 175 PKVEIFAQARDCLLSMGITSENVAQRYGVTRQEQDQAAVESHRRAAAATASGKFKDEIIP 234
P V+ FAQA++CLL MG+TSENVAQR+GV+RQEQDQAAV+SHR+AAAATA+GKFKDEIIP
Sbjct: 181 PAVKKFAQAQNCLLPMGVTSENVAQRFGVSRQEQDQAAVDSHRKAAAATAAGKFKDEIIP 240
Query: 235 VSTKFVDPKTGEEKQIIVSVDDGIRPNANLVDLAKLKPAFKKDGTTTAGNASQVSDGAAA 294
V TK VDPKTG+EK I VSVDDGIRP L L KLKP FKKDGTTTAGN+SQVSDGA A
Sbjct: 241 VKTKLVDPKTGDEKPITVSVDDGIRPTTTLASLGKLKPVFKKDGTTTAGNSSQVSDGAGA 300
Query: 295 VLLMKRSVAVQKGLPILGIFRSFSAVGVDPAVMGVGPAFAIPAAVKSAGLELSNIDLYEI 354
VLLMKRSVA+QKGLP+LG+FR+F+AVGVDPA+MG+GPA AIPAAVK+AGLEL +IDL+EI
Sbjct: 301 VLLMKRSVAMQKGLPVLGVFRTFAAVGVDPAIMGIGPAVAIPAAVKAAGLELDDIDLFEI 360
Query: 355 NEAFASQFVYSCKKLGLDPTKVNVNGGAMAFGHPLGATGARCVATLLNEMKRRGKDCRFG 414
NEAFASQFVY KLGLDP K+NVNGGAMA GHPLGATGARCVATLL+EMKRRGKDCRFG
Sbjct: 361 NEAFASQFVYCRNKLGLDPEKINVNGGAMAIGHPLGATGARCVATLLHEMKRRGKDCRFG 420
Query: 415 VISMCIGSGMGAAAVFERGD 434
V+SMCIG+GMGAAAVFERGD
Sbjct: 421 VVSMCIGTGMGAAAVFERGD 440
>At1g04710 acetyl-CoA acyltransferase like protein
Length = 443
Score = 649 bits (1675), Expect = 0.0
Identities = 333/435 (76%), Positives = 374/435 (85%), Gaps = 6/435 (1%)
Query: 1 MEKAVERQRVLLQHLNPNSVTSSHHSTHLSASLCSAGQTGGSE--NDVVIVAAYRTAICK 58
MEKA ERQR+LL+HL P SS LSAS C + + + +DVVIVAA RTA+CK
Sbjct: 1 MEKATERQRILLRHLQP----SSSSDASLSASACLSKDSAAYQYGDDVVIVAAQRTALCK 56
Query: 59 AKRGGFKDTLPDDLLASVLKAVIEKTNVEPSEVGDIVVGTVLGPGSERAIECRMAAFYAG 118
AKRG FKDT PD+LLASVL+A+IEKTNV PSEVGDIVVGTVLGPGS+RA ECRMAAFYAG
Sbjct: 57 AKRGSFKDTFPDELLASVLRALIEKTNVNPSEVGDIVVGTVLGPGSQRASECRMAAFYAG 116
Query: 119 FPDTVPLRTVNRQCSSGLQAVADVAAYIKAGFYDIGIGAGLECMTQDTISGVRKTNPKVE 178
FP+TVP+RTVNRQCSSGLQAVADVAA IKAGFYDIGIGAGLE MT + NP V+
Sbjct: 117 FPETVPIRTVNRQCSSGLQAVADVAAAIKAGFYDIGIGAGLESMTTNPRGWKGSVNPNVK 176
Query: 179 IFAQARDCLLSMGITSENVAQRYGVTRQEQDQAAVESHRRAAAATASGKFKDEIIPVSTK 238
F QA +CLL MGITSENVA R+ V+R+EQDQAAV+SHR+AA+ATASGKFKDEI PV TK
Sbjct: 177 KFEQAHNCLLPMGITSENVAHRFNVSREEQDQAAVDSHRKAASATASGKFKDEITPVKTK 236
Query: 239 FVDPKTGEEKQIIVSVDDGIRPNANLVDLAKLKPAFKKDGTTTAGNASQVSDGAAAVLLM 298
VDPKTG+EK I VSVDDGIRPN L LAKLKP FK+DGTTTAGN+SQ+SDGA AVLLM
Sbjct: 237 IVDPKTGDEKPITVSVDDGIRPNTTLSGLAKLKPVFKEDGTTTAGNSSQLSDGAGAVLLM 296
Query: 299 KRSVAVQKGLPILGIFRSFSAVGVDPAVMGVGPAFAIPAAVKSAGLELSNIDLYEINEAF 358
+R+VA+QKGLPILG+FR+FSAVGVDPA+MGVGPA AIPAAVK+AGLEL+++DL+EINEAF
Sbjct: 297 RRNVAMQKGLPILGVFRTFSAVGVDPAIMGVGPAVAIPAAVKAAGLELNDVDLFEINEAF 356
Query: 359 ASQFVYSCKKLGLDPTKVNVNGGAMAFGHPLGATGARCVATLLNEMKRRGKDCRFGVISM 418
ASQFVY KLGLD K+NVNGGA+A GHPLGATGARCVATLL+EMKRRGKDCRFGV+SM
Sbjct: 357 ASQFVYCRNKLGLDAEKINVNGGAIAIGHPLGATGARCVATLLHEMKRRGKDCRFGVVSM 416
Query: 419 CIGSGMGAAAVFERG 433
CIGSGMGAAAVFERG
Sbjct: 417 CIGSGMGAAAVFERG 431
>At5g48880 peroxisomal-3-keto-acyl-CoA thiolase 1 (PKT1)
Length = 414
Score = 617 bits (1590), Expect = e-177
Identities = 304/393 (77%), Positives = 357/393 (90%), Gaps = 2/393 (0%)
Query: 44 NDVVIVAAYRTAICKAKRGGFKDTLPDDLLASVLKAVIEKTNVEPSEVGDIVVGTVLGPG 103
+D+VIVAAYRTAICKA+RGGFKDTLPDDLLASVLKAV+E+T+++PSEVGDIVVGTV+ PG
Sbjct: 6 DDIVIVAAYRTAICKARRGGFKDTLPDDLLASVLKAVVERTSLDPSEVGDIVVGTVIAPG 65
Query: 104 SERAIECRMAAFYAGFPDTVPLRTVNRQCSSGLQAVADVAAYIKAGFYDIGIGAGLECMT 163
S+RA+ECR+AA++AGFPD+VP+RTVNRQCSSGLQAVADVAA I+AG+YDIGIGAG+E M+
Sbjct: 66 SQRAMECRVAAYFAGFPDSVPVRTVNRQCSSGLQAVADVAASIRAGYYDIGIGAGVESMS 125
Query: 164 QDTI--SGVRKTNPKVEIFAQARDCLLSMGITSENVAQRYGVTRQEQDQAAVESHRRAAA 221
D I G +NP+ + F +ARDCLL MGITSENVA+R+GVTR+EQD AAVESH+RAAA
Sbjct: 126 TDHIPGGGFHGSNPRAQDFPKARDCLLPMGITSENVAERFGVTREEQDMAAVESHKRAAA 185
Query: 222 ATASGKFKDEIIPVSTKFVDPKTGEEKQIIVSVDDGIRPNANLVDLAKLKPAFKKDGTTT 281
A ASGK KDEIIPV+TK VDP+T EK I+VSVDDG+RPN+N+ DLAKLK FK++G+TT
Sbjct: 186 AIASGKLKDEIIPVATKIVDPETKAEKAIVVSVDDGVRPNSNMADLAKLKTVFKQNGSTT 245
Query: 282 AGNASQVSDGAAAVLLMKRSVAVQKGLPILGIFRSFSAVGVDPAVMGVGPAFAIPAAVKS 341
AGNASQ+SDGA AVLLMKRS+A++KGLPILG+FRSF+ GV+P+VMG+GPA AIPAA K
Sbjct: 246 AGNASQISDGAGAVLLMKRSLAMKKGLPILGVFRSFAVTGVEPSVMGIGPAVAIPAATKL 305
Query: 342 AGLELSNIDLYEINEAFASQFVYSCKKLGLDPTKVNVNGGAMAFGHPLGATGARCVATLL 401
AGL +S+IDL+EINEAFASQ+VYSCKKL LD KVNVNGGA+A GHPLGATGARCVATLL
Sbjct: 306 AGLNVSDIDLFEINEAFASQYVYSCKKLELDMEKVNVNGGAIAIGHPLGATGARCVATLL 365
Query: 402 NEMKRRGKDCRFGVISMCIGSGMGAAAVFERGD 434
+EMKRRGKDCRFGVISMCIG+GMGAAAVFERGD
Sbjct: 366 HEMKRRGKDCRFGVISMCIGTGMGAAAVFERGD 398
>At5g48230 acetoacetyl-CoA thiolase (AAT1)
Length = 403
Score = 228 bits (581), Expect = 5e-60
Identities = 157/405 (38%), Positives = 226/405 (55%), Gaps = 33/405 (8%)
Query: 45 DVVIVAAYRTAICKAKRGGFK---DTLPDDLLASV-LKAVIEKTNVEPSEVGDIVVGTVL 100
DV IV RT + GGF +LP L S+ + A +++ NV+P+ V ++V G VL
Sbjct: 12 DVCIVGVARTPM-----GGFLGSLSSLPATKLGSLAIAAALKRANVDPALVQEVVFGNVL 66
Query: 101 GPGSERAIECRMAAFYAGFPDTVPLRTVNRQCSSGLQAVADVAAYIKAGFYDIGIGAGLE 160
+A R AA AG P++V TVN+ C+SG++AV A I+ G D+ + G+E
Sbjct: 67 SANLGQA-PARQAALGAGIPNSVICTTVNKVCASGMKAVMIAAQSIQLGINDVVVAGGME 125
Query: 161 CM--TQDTISGVRKTNP-----------KVEIFAQARDCLLSMGITSENVAQRYGVTRQE 207
M T ++ RK + K ++ DC MG +E A+++ +TR++
Sbjct: 126 SMSNTPKYLAEARKGSRFGHDSLVDGMLKDGLWDVYNDC--GMGSCAELCAEKFQITREQ 183
Query: 208 QDQAAVESHRRAAAATASGKFKDEIIPVSTKFVDPKTGEEKQIIVSVDDGIRPNANLVDL 267
QD AV+S R AA +G F EI+PV V G I V D+G+ + L
Sbjct: 184 QDDYAVQSFERGIAAQEAGAFTWEIVPVE---VSGGRGRPSTI-VDKDEGLG-KFDAAKL 238
Query: 268 AKLKPAFKKDG-TTTAGNASQVSDGAAAVLLMKRSVAVQKGLPILGIFRSFSAVGVDPAV 326
KL+P+FK++G T TAGNAS +SDGAAA++L+ A+Q GL +L + + +P
Sbjct: 239 RKLRPSFKENGGTVTAGNASSISDGAAALVLVSGEKALQLGLLVLAKIKGYGDAAQEPEF 298
Query: 327 MGVGPAFAIPAAVKSAGLELSNIDLYEINEAFASQFVYSCKKLGLDPTKVNVNGGAMAFG 386
PA AIP A+ AGLE S +D YEINEAFA + + K LG+ P KVNVNGGA++ G
Sbjct: 299 FTTAPALAIPKAIAHAGLESSQVDYYEINEAFAVVALANQKLLGIAPEKVNVNGGAVSLG 358
Query: 387 HPLGATGARCVATLLNEMKRRGKDCRFGVISMCIGSGMGAAAVFE 431
HPLG +GAR + TLL +K+R + ++GV +C G G +A V E
Sbjct: 359 HPLGCSGARILITLLGILKKR--NGKYGVGGVCNGGGGASALVLE 401
>At5g47720 acetoacyl-CoA-thiolase
Length = 415
Score = 216 bits (549), Expect = 3e-56
Identities = 144/402 (35%), Positives = 222/402 (54%), Gaps = 27/402 (6%)
Query: 45 DVVIVAAYRTAICKAKRGGFKDTLPDDLLASV-LKAVIEKTNVEPSEVGDIVVGTVLGPG 103
DV +V RT I G +L L S+ ++A +++ +V+P+ V ++ G VL
Sbjct: 14 DVCVVGVARTPIGDFL--GSLSSLTATRLGSIAIQAALKRAHVDPALVEEVFFGNVLTAN 71
Query: 104 SERAIECRMAAFYAGFPDTVPLRTVNRQCSSGLQAVADVAAYIKAGFYDIGIGAGLECMT 163
+A R AA AG P +V T+N+ C++G+++V + I+ G DI + G+E M+
Sbjct: 72 LGQA-PARQAALGAGIPYSVICTTINKVCAAGMKSVMLASQSIQLGLNDIVVAGGMESMS 130
Query: 164 Q------DTISGVRKTNPKVEIFAQARDCL------LSMGITSENVAQRYGVTRQEQDQA 211
D G R + V + +D L MG+ E A +Y +TR+EQD
Sbjct: 131 NVPKYLPDARRGSRLGHDTV-VDGMMKDGLWDVYNDFGMGVCGEICADQYRITREEQDAY 189
Query: 212 AVESHRRAAAATASGKFKDEIIPVSTKFVDPKTGEEK-QIIVSVDDGIRPNANLVDLAKL 270
A++S R AA + F EI+PV TG + +++ D+G+ + L KL
Sbjct: 190 AIQSFERGIAAQNTQLFAWEIVPVEVS-----TGRGRPSVVIDKDEGLG-KFDAAKLKKL 243
Query: 271 KPAFKKDG-TTTAGNASQVSDGAAAVLLMKRSVAVQKGLPILGIFRSFSAVGVDPAVMGV 329
+P+FK+DG + TAGNAS +SDGAAA++L+ A++ GL ++ R ++ P +
Sbjct: 244 RPSFKEDGGSVTAGNASSISDGAAALVLVSGEKALELGLHVIAKIRGYADAAQAPELFTT 303
Query: 330 GPAFAIPAAVKSAGLELSNIDLYEINEAFASQFVYSCKKLGLDPTKVNVNGGAMAFGHPL 389
PA AIP A+K AGL+ S +D YEINEAF+ + + K LGLDP ++N +GGA++ GHPL
Sbjct: 304 TPALAIPKAIKRAGLDASQVDYYEINEAFSVVALANQKLLGLDPERLNAHGGAVSLGHPL 363
Query: 390 GATGARCVATLLNEMKRRGKDCRFGVISMCIGSGMGAAAVFE 431
G +GAR + TLL + R K ++GV S+C G G +A V E
Sbjct: 364 GCSGARILVTLLGVL--RAKKGKYGVASICNGGGGASALVLE 403
>At5g46290 3-oxoacyl-[acyl-carrier-protein] synthase I precursor
(beta-ketoacyl-acp synthase I) (KAS I) (sp|P52410)
Length = 473
Score = 30.8 bits (68), Expect = 1.5
Identities = 29/126 (23%), Positives = 54/126 (42%), Gaps = 16/126 (12%)
Query: 288 VSDGAAAVLLMKRSVAVQKGLPILGIFRSFSAVG------VDPAVMGVGPAFAIPAAVKS 341
+ +GA +++ A+++G PI+ + AV DP G+G + I ++
Sbjct: 292 MGEGAGVLVMESLEHAMKRGAPIVAEYLG-GAVNCDAHHMTDPRADGLGVSSCIERCLED 350
Query: 342 AGLELSNIDLYEINEAFASQF------VYSCKKLGLDPTKVNVNGGAMAFGHPLGATGA- 394
AG+ ++ IN S + + KK+ + + +N GH LGA G
Sbjct: 351 AGVSPEEVNY--INAHATSTLAGDLAEINAIKKVFKSTSGIKINATKSMIGHCLGAAGGL 408
Query: 395 RCVATL 400
+AT+
Sbjct: 409 EAIATV 414
>At1g74960 unknown protein
Length = 541
Score = 30.4 bits (67), Expect = 2.0
Identities = 37/126 (29%), Positives = 53/126 (41%), Gaps = 16/126 (12%)
Query: 288 VSDGAAAVLLMKRSVAVQKGLPILGIFR--SFSAVG---VDPAVMGVGPAFAIPAAVKSA 342
+ +GA +LL + A ++G I F SF+ +P G G I A+ SA
Sbjct: 360 MGEGAGVLLLEELEHAKKRGATIYAEFLGGSFTCDAYHMTEPHPDGAGVILCIERALASA 419
Query: 343 GLELSNIDLYEINEAFASQFVYSCKK-------LGLDPTKVNVNGGAMAFGHPLGATGA- 394
G+ I+ IN S K+ G +P ++ VN GH LGA GA
Sbjct: 420 GISKEQINY--INAHATSTHAGDIKEYQALAHCFGQNP-ELKVNSTKSMIGHLLGAAGAV 476
Query: 395 RCVATL 400
VAT+
Sbjct: 477 EAVATV 482
>At1g27170 disease resistance protein, putative
Length = 1560
Score = 30.0 bits (66), Expect = 2.6
Identities = 12/33 (36%), Positives = 22/33 (66%)
Query: 65 KDTLPDDLLASVLKAVIEKTNVEPSEVGDIVVG 97
KD+ DD++ V+K V+ + + P +VG+ +VG
Sbjct: 306 KDSKDDDMIELVVKRVLAELSNTPEKVGEFIVG 338
>At4g08060 putative protein
Length = 756
Score = 28.9 bits (63), Expect = 5.8
Identities = 19/64 (29%), Positives = 31/64 (47%), Gaps = 11/64 (17%)
Query: 237 TKFVDPKTGEEKQIIVSVDDGIRPNANLVDLAKLK----PAFKKDGTTTAGNASQVSDGA 292
TKF + KT +++ +RP N ++ +LK KKDGT NA++ A
Sbjct: 261 TKFREAKTNQQRM-------NLRPKTNPTEVTRLKVWVKSRTKKDGTPVNTNAAEKIKKA 313
Query: 293 AAVL 296
A ++
Sbjct: 314 AEIV 317
>At1g56500 unknown protein
Length = 1055
Score = 28.9 bits (63), Expect = 5.8
Identities = 26/91 (28%), Positives = 41/91 (44%), Gaps = 8/91 (8%)
Query: 321 GVDPAVMGVGPAFAIPAAVKSAGLELSNIDLYEINEAFASQ------FVYSCKKLGLDPT 374
G+ AV + A +K+AGL L+ D +AF + F+ + K LG+ +
Sbjct: 176 GLKVAVASSADRIKVDANLKAAGLSLTMFDAIVSADAFENLKPAPDIFLAAAKILGVPTS 235
Query: 375 KVNVNGGAMAFGHPLGATGARCVA--TLLNE 403
+ V A+A A RC+A T L+E
Sbjct: 236 ECVVIEDALAGVQAAQAANMRCIAVKTTLSE 266
>At4g34890 xanthine dehydrogenase - like protein
Length = 1359
Score = 28.5 bits (62), Expect = 7.6
Identities = 17/54 (31%), Positives = 26/54 (47%)
Query: 301 SVAVQKGLPILGIFRSFSAVGVDPAVMGVGPAFAIPAAVKSAGLELSNIDLYEI 354
+V++ KG P S AVG P + FAI A+K+A E+ D + +
Sbjct: 1274 NVSLLKGNPNTKAIHSSKAVGEPPFFLASSVFFAIKEAIKAARTEVGLTDWFPL 1327
>At1g32850 hypothetical protein
Length = 887
Score = 28.5 bits (62), Expect = 7.6
Identities = 27/108 (25%), Positives = 48/108 (44%), Gaps = 19/108 (17%)
Query: 293 AAVLLMKRSVAVQKGLPILGIFRSFSAVGVDPAVMGVGPAFAIPAAVKSAGLELSNIDLY 352
A++ + V KG I F +V +DP+ +V+ AGL+ + L
Sbjct: 169 ASIGQLYEMVCAGKGARIWDYFEKKKSVLLDPSS---------EQSVEEAGLQFNQDILL 219
Query: 353 EINEAFASQFVYSCKK-----LGLDPTKVNV-----NGGAMAFGHPLG 390
E++ + +SQFV S + + L+P + + GG ++ GH G
Sbjct: 220 EVDGSASSQFVMSLAENELAMVPLEPMRSDAMDIVRGGGTLSNGHSNG 267
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.318 0.133 0.378
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 9,195,740
Number of Sequences: 26719
Number of extensions: 375423
Number of successful extensions: 944
Number of sequences better than 10.0: 12
Number of HSP's better than 10.0 without gapping: 7
Number of HSP's successfully gapped in prelim test: 5
Number of HSP's that attempted gapping in prelim test: 926
Number of HSP's gapped (non-prelim): 13
length of query: 435
length of database: 11,318,596
effective HSP length: 102
effective length of query: 333
effective length of database: 8,593,258
effective search space: 2861554914
effective search space used: 2861554914
T: 11
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 61 (28.1 bits)
Medicago: description of AC126019.10


