

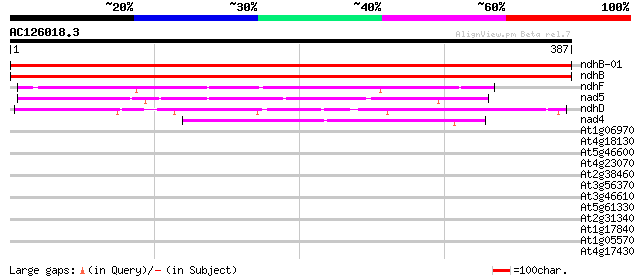
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC126018.3 + phase: 0
(387 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
ndhB-01 -chloroplast genome- NADH dehydrogenase ND2 735 0.0
ndhB -chloroplast genome- NADH dehydrogenase ND2 735 0.0
ndhF -chloroplast genome- NADH dehydrogenase ND5 103 1e-22
nad5 -mitochondrial genome- NADH dehydrogenase subunit 5 79 3e-15
ndhD -chloroplast genome- NADH dehydrogenase ND4 66 4e-11
nad4 -mitochondrial genome- NADH dehydrogenase subunit 4 48 1e-05
At1g06970 hypothetical protein 33 0.35
At4g18130 phytochrome E 30 2.2
At5g46600 putative protein 30 2.9
At4g23070 putative membrane protein 30 2.9
At2g38460 unknown protein 29 3.8
At3g56370 putative protein 29 5.0
At3g46610 unknown protein 29 5.0
At5g61330 unknown protein 28 6.5
At2g31340 unknown protein 28 6.5
At1g17840 putative ABC transporter (At1g17840) 28 6.5
At1g05570 putative glucan synthase 28 6.5
At4g17430 unknown protein 28 8.5
>ndhB-01 -chloroplast genome- NADH dehydrogenase ND2
Length = 389
Score = 735 bits (1898), Expect = 0.0
Identities = 369/387 (95%), Positives = 379/387 (97%)
Query: 1 MAITEFLLFILTATLGGMFLCSANDLITIFVALECFSLCSYLLSGYTKKDVRSNEATMKY 60
MAITEFLLFILTATLGGMFLC ANDLITIFVA ECFSLCSYLLSGYTKKD+RSNEATMKY
Sbjct: 1 MAITEFLLFILTATLGGMFLCGANDLITIFVAPECFSLCSYLLSGYTKKDIRSNEATMKY 60
Query: 61 LLMGGASSSILVHGFSWLYGLSGGEIELQEIVNGLINTQMYNSPGISIALIFITVGIGFK 120
LLMGGASSSILVHGFSWLYG SGGEIELQEIVNGLINTQMYNSPGISIALIFITVGIGFK
Sbjct: 61 LLMGGASSSILVHGFSWLYGSSGGEIELQEIVNGLINTQMYNSPGISIALIFITVGIGFK 120
Query: 121 LSLAPSHQWTPDVYEGSPTPVVAFLSVTSKVAASASATRIFDILFYFSSNEWHLLLEILA 180
LSLAPSHQWTPDVYEGSPTPVVAFLSVTSKVAASASATRIFDI FYFSSNEWHLLLEILA
Sbjct: 121 LSLAPSHQWTPDVYEGSPTPVVAFLSVTSKVAASASATRIFDIPFYFSSNEWHLLLEILA 180
Query: 181 ILSMLLGNLIAITQTSMKRMLAYSSIGQIGYVIIGIIVGDSNDGYASMITYMLFYISMNL 240
ILSM+ GNLIAITQTSMKRMLAYSSIGQIGYVIIGIIVGDSN GYASMITYMLFYI+MNL
Sbjct: 181 ILSMIFGNLIAITQTSMKRMLAYSSIGQIGYVIIGIIVGDSNGGYASMITYMLFYIAMNL 240
Query: 241 GTFACIVLFGLRTGTDNIRDYAGLYTKDPFLALSLALCLLSLGGLPPLAGFFGKLHLFWC 300
GTFACI+LFGLRTGTDNIRDYAGLYTKDPFLALSLALCLLSLGGLPPLAGFFGKLHLFWC
Sbjct: 241 GTFACIILFGLRTGTDNIRDYAGLYTKDPFLALSLALCLLSLGGLPPLAGFFGKLHLFWC 300
Query: 301 GWQAGLYFLVSIGLITSVVSIYYYLKIIKLLMTGRNQEITPHVRNYRRSPVRSNNSIELS 360
GWQAGLYFLVSIGL+TSV+SIYYYLKIIKLLMTGRNQEITPH+RNYR SP+RSNNSIELS
Sbjct: 301 GWQAGLYFLVSIGLLTSVLSIYYYLKIIKLLMTGRNQEITPHMRNYRISPLRSNNSIELS 360
Query: 361 MIICVIASTVPGISMNPIIEIAQDTLF 387
MI+CVIAST+PGISMNPII IAQDTLF
Sbjct: 361 MIVCVIASTIPGISMNPIIAIAQDTLF 387
>ndhB -chloroplast genome- NADH dehydrogenase ND2
Length = 389
Score = 735 bits (1898), Expect = 0.0
Identities = 369/387 (95%), Positives = 379/387 (97%)
Query: 1 MAITEFLLFILTATLGGMFLCSANDLITIFVALECFSLCSYLLSGYTKKDVRSNEATMKY 60
MAITEFLLFILTATLGGMFLC ANDLITIFVA ECFSLCSYLLSGYTKKD+RSNEATMKY
Sbjct: 1 MAITEFLLFILTATLGGMFLCGANDLITIFVAPECFSLCSYLLSGYTKKDIRSNEATMKY 60
Query: 61 LLMGGASSSILVHGFSWLYGLSGGEIELQEIVNGLINTQMYNSPGISIALIFITVGIGFK 120
LLMGGASSSILVHGFSWLYG SGGEIELQEIVNGLINTQMYNSPGISIALIFITVGIGFK
Sbjct: 61 LLMGGASSSILVHGFSWLYGSSGGEIELQEIVNGLINTQMYNSPGISIALIFITVGIGFK 120
Query: 121 LSLAPSHQWTPDVYEGSPTPVVAFLSVTSKVAASASATRIFDILFYFSSNEWHLLLEILA 180
LSLAPSHQWTPDVYEGSPTPVVAFLSVTSKVAASASATRIFDI FYFSSNEWHLLLEILA
Sbjct: 121 LSLAPSHQWTPDVYEGSPTPVVAFLSVTSKVAASASATRIFDIPFYFSSNEWHLLLEILA 180
Query: 181 ILSMLLGNLIAITQTSMKRMLAYSSIGQIGYVIIGIIVGDSNDGYASMITYMLFYISMNL 240
ILSM+ GNLIAITQTSMKRMLAYSSIGQIGYVIIGIIVGDSN GYASMITYMLFYI+MNL
Sbjct: 181 ILSMIFGNLIAITQTSMKRMLAYSSIGQIGYVIIGIIVGDSNGGYASMITYMLFYIAMNL 240
Query: 241 GTFACIVLFGLRTGTDNIRDYAGLYTKDPFLALSLALCLLSLGGLPPLAGFFGKLHLFWC 300
GTFACI+LFGLRTGTDNIRDYAGLYTKDPFLALSLALCLLSLGGLPPLAGFFGKLHLFWC
Sbjct: 241 GTFACIILFGLRTGTDNIRDYAGLYTKDPFLALSLALCLLSLGGLPPLAGFFGKLHLFWC 300
Query: 301 GWQAGLYFLVSIGLITSVVSIYYYLKIIKLLMTGRNQEITPHVRNYRRSPVRSNNSIELS 360
GWQAGLYFLVSIGL+TSV+SIYYYLKIIKLLMTGRNQEITPH+RNYR SP+RSNNSIELS
Sbjct: 301 GWQAGLYFLVSIGLLTSVLSIYYYLKIIKLLMTGRNQEITPHMRNYRISPLRSNNSIELS 360
Query: 361 MIICVIASTVPGISMNPIIEIAQDTLF 387
MI+CVIAST+PGISMNPII IAQDTLF
Sbjct: 361 MIVCVIASTIPGISMNPIIAIAQDTLF 387
>ndhF -chloroplast genome- NADH dehydrogenase ND5
Length = 746
Score = 103 bits (258), Expect = 1e-22
Identities = 90/337 (26%), Positives = 154/337 (44%), Gaps = 15/337 (4%)
Query: 6 FLLFILTATLGGMFLCSANDLITIFVALECFSLCSYLLSGYTKKDVRSNEATMKYLLMGG 65
++ F T+ LG L ++++LI ++ E +CSYLL G+ + A K +
Sbjct: 127 YMGFFNTSMLG---LVTSSNLIQVYFFWELVGMCSYLLIGFWFTRPIAANACQKAFVTNR 183
Query: 66 ASSSILVHGFSWLYGLSGGEI--ELQEIVNGLINTQMYNSPGISIALIFITVGIGFKLSL 123
L+ G LY ++G +L EI N LI N +++ + VG K +
Sbjct: 184 VGDFGLLLGILGLYWITGSFEFQDLFEIFNNLILNNRVNLLFLTLCAFLLFVGPIAKSAQ 243
Query: 124 APSHQWTPDVYEGSPTPVVAFLSVTSKVAASAS-ATRIFDILFYFSSNEWHLLLEILAIL 182
P H W PD EG PTP+ A + + VAA R+ + S + ++ ++ I+
Sbjct: 244 FPLHVWLPDAMEG-PTPISALIHAATMVAAGIFLVARLLPLFIVIPSIMY--IISLIGII 300
Query: 183 SMLLGNLIAITQTSMKRMLAYSSIGQIGYVIIGIIVGDSNDGYASMITYMLFYISMNLGT 242
++LLG +A+ Q +KR LAYS++ Q+GY+++ + +G +IT+ + LG+
Sbjct: 301 TVLLGATLALAQKDIKRGLAYSTMSQLGYMMLALGMGSYRSALFHLITHAYSKALLFLGS 360
Query: 243 FACIVLFGLRTG-----TDNIRDYAGLYTKDPFLALSLALCLLSLGGLPPLAGFFGKLHL 297
+ I G + N+ GL P + + LSL G+PPLA F+ K +
Sbjct: 361 GSIIHSMEAIVGYSPDKSQNMILMGGLTKHVPITKTAFLIGTLSLCGIPPLACFWSKDEI 420
Query: 298 FWCGWQAGLYFLVSIGLITSVVSIYYYLKIIKLLMTG 334
F + I T+ ++ +Y +I L G
Sbjct: 421 LNDSLLFSPIFAI-IACSTAGLTAFYMFRIYLLTFEG 456
>nad5 -mitochondrial genome- NADH dehydrogenase subunit 5
Length = 669
Score = 79.3 bits (194), Expect = 3e-15
Identities = 79/335 (23%), Positives = 142/335 (41%), Gaps = 17/335 (5%)
Query: 6 FLLFILTATLGGMFLCSANDLITIFVALECFSLCSYLLSGYTKKDVRSNEATMKYLLMGG 65
F+ ++ T L + ++ + +F+ E L SYLL + +++++A K +L+
Sbjct: 113 FMCYLSILTFFMPMLVTGDNSLQLFLGWEGVGLASYLLIHFWFTRLQADKAATKAMLVNR 172
Query: 66 ASSSILVHGFSWLYGLSGGEIELQEIV-------NGLINTQMYNSPGISIALIFITVGIG 118
L G S + L ++ I N I+ M + IS+ I + +G
Sbjct: 173 VGDFGLALGISGRFTLFQ-TVDFSTIFARASAPRNSWISCNMRLN-AISLICILLLIGAV 230
Query: 119 FKLSLAPSHQWTPDVYEGSPTPVVAFLSVTSKVAASASATRIFDILFYFSSNEWHLLLEI 178
K + SH W+PD EG PTPV A + + V A LF + ++
Sbjct: 231 GKSAQIGSHTWSPDAMEG-PTPVSASIHAATMVTAGVFMIARCSPLFEYPPTALIVITSA 289
Query: 179 LAILSMLLGNLIAITQTSMKRMLAYSSIGQIGYVIIGIIVGDSNDGYASMITYMLFYISM 238
A S L I Q +KR++AYS+ Q+GY+I + + + ++ + F +
Sbjct: 290 GATTSFLAATT-GILQNDLKRVIAYSTCSQLGYMIFACGISNYSVSVFHLMNHAFFKALL 348
Query: 239 NLGTFACIVLFGLRTGTDNIRDYAGLYTKDPFLALSLALCLLSLGGLPPLAGFFGK---L 295
L + I + ++R GL + P + + LSL G P L GF+ K L
Sbjct: 349 FLSAGSVI---HAMSDEQDMRKMGGLASSFPLTYAMMLIGSLSLIGFPFLTGFYSKDVIL 405
Query: 296 HLFWCGWQAGLYFLVSIGLITSVVSIYYYLKIIKL 330
L + + F +G ++ + + YY +++ L
Sbjct: 406 ELAYTKYTISGNFAFWLGSVSVLFTSYYSFRLLFL 440
>ndhD -chloroplast genome- NADH dehydrogenase ND4
Length = 506
Score = 65.9 bits (159), Expect = 4e-11
Identities = 88/397 (22%), Positives = 164/397 (41%), Gaps = 35/397 (8%)
Query: 4 TEFLLFILTATLGGMF-LCSANDLITIFVALECFSLCSYLLSGYTKKDVRSNEATMKYLL 62
+ F F++ A G S+ DL+ F+ E + YLL R AT L
Sbjct: 117 SRFFHFLMLAMYSGQIGSFSSRDLLLFFIMWELELIPVYLLLSMWGGKKRLYSATKFILY 176
Query: 63 MGGASSSILVH--GFSWLYGLSGGEIELQEIVNGLINTQMYNSPGISIALIF---ITVGI 117
G+S +L+ G S LYG + + L+ + N S +++ ++F +
Sbjct: 177 TAGSSIFLLIGVLGIS-LYGSNEPTLNLELLAN--------KSYPVTLEILFYIGFLIAF 227
Query: 118 GFKLSLAPSHQWTPDVY-EGSPTPVVAFLSVTSKVAASASATRIFDILFYFSS--NEWHL 174
K + P H W PD + E + + + K+ A ++L + S + W L
Sbjct: 228 AVKSPIIPLHTWLPDTHGEAHYSTCMLLAGILLKMGAYGLVRINMELLPHAHSMFSPWLL 287
Query: 175 LLEILAILSMLLGNLIAITQTSMKRMLAYSSIGQIGYVIIGIIVGDSNDGYASMITYMLF 234
++ + ++ + Q ++K+ +AYSS+ +G++IIG I ++ G I ++
Sbjct: 288 ---VVGTIQIIYAASTSPGQRNLKKRIAYSSVSHMGFIIIG-ISSITDPGLNGAILQIIS 343
Query: 235 YISMNLGTFACIVLFGLRTGTDNIR-----DYAGLYTKDPFLALSLALCLLSLGGLPPLA 289
+ G + F T D IR + G+ P + + ++ LP ++
Sbjct: 344 H-----GFIGAALFFLAGTSYDRIRLVYLDEMGGMAISIPKIFTMFTILSMASLALPGMS 398
Query: 290 GFFGKLHLFWCGWQAGLYFLVSIGLITSVVSIYYYLKIIKLLMTGRNQEITPHVRNYRRS 349
GF + +F+ + YFL+S I V++I L I LL R + N +
Sbjct: 399 GFIAEFIVFFGIITSQKYFLISKIFIIFVMAIGMILTPIYLLSMLRQMFYGYKLINIKNF 458
Query: 350 PVRSNNSIELSMIICVIASTVPGISMNP--IIEIAQD 384
+ EL + I ++ + GI + P ++ +A D
Sbjct: 459 SFFDSGPRELFLSISILLPII-GIGIYPDFVLSLASD 494
>nad4 -mitochondrial genome- NADH dehydrogenase subunit 4
Length = 495
Score = 47.8 bits (112), Expect = 1e-05
Identities = 45/212 (21%), Positives = 92/212 (43%), Gaps = 4/212 (1%)
Query: 120 KLSLAPSHQWTPDVYEGSPTPVVAFLSVTSKVAASASATRIFDILFYFSSNEWHLLLEIL 179
K+ + P H W P+ + +PT L+ + R +F ++ + L
Sbjct: 227 KVPMVPVHIWLPEAHVEAPTAGSVILAGIPLKFGTHGFLRFSIPMFPEATLCSTPFIYTL 286
Query: 180 AILSMLLGNLIAITQTSMKRMLAYSSIGQIGYVIIGIIVGDSNDGYASMITYMLFYISMN 239
+ ++++ +L Q +K+++AYSS+ + V IG+ + G I ML + +
Sbjct: 287 SAIAIIYTSLTTSRQIDLKKIIAYSSVAHMNLVTIGMF-SPNIQGIGGSILPMLSHGLVP 345
Query: 240 LGTFACIVLFGLRTGTDNIRDYAGLYTKDPFLALSLALCLLSLGGLPPLAGFFGKLHLFW 299
F C+ + R T +R Y GL + P L+ L+ P + F G+ +
Sbjct: 346 SALFLCVGVLYDRHKTRLVRYYGGLVSTMPNLSTIFFSFTLANMSSPGTSSFIGEFLILV 405
Query: 300 CGWQAG--LYFLVSIGLIT-SVVSIYYYLKII 328
+Q + L ++G+I + S++ Y +++
Sbjct: 406 GAFQRNSLVATLAALGMILGAAYSLWLYNRVV 437
>At1g06970 hypothetical protein
Length = 829
Score = 32.7 bits (73), Expect = 0.35
Identities = 30/87 (34%), Positives = 39/87 (44%), Gaps = 9/87 (10%)
Query: 182 LSMLLGNLIAITQTSMKRMLAYSSIGQIG-YVIIGIIVGDSNDGYASMITYMLFYIS--- 237
L +L ++I IT + R+L G I V+ GII+G S G +S M IS
Sbjct: 52 LMLLQMSVIIITSRLLYRLLKPLKQGMISAQVLAGIILGPSLFGQSSAYMQMFLPISGKI 111
Query: 238 -----MNLGTFACIVLFGLRTGTDNIR 259
NLG F + L GLR IR
Sbjct: 112 TLQTLSNLGFFIHLFLLGLRIDASIIR 138
>At4g18130 phytochrome E
Length = 1112
Score = 30.0 bits (66), Expect = 2.2
Identities = 18/70 (25%), Positives = 33/70 (46%), Gaps = 2/70 (2%)
Query: 38 LCSYLLSGYTKKDVRSNEATMKYLLMGGASSSILVHGFSWLYGLSGGEIELQEIVNGLIN 97
LC LL V + M + GA+ + G WL G++ E +++++VN L+
Sbjct: 405 LCDMLLRDTVSAIVTQSPGIMDLVKCDGAA--LYYKGKCWLVGVTPNESQVKDLVNWLVE 462
Query: 98 TQMYNSPGIS 107
+S G++
Sbjct: 463 NHGDDSTGLT 472
>At5g46600 putative protein
Length = 539
Score = 29.6 bits (65), Expect = 2.9
Identities = 33/104 (31%), Positives = 44/104 (41%), Gaps = 18/104 (17%)
Query: 186 LGNLIAITQTSMKRMLAYSSIGQIGYVIIGIIVGDSNDGYASMITYMLF--YISMN---- 239
LG LIA + +A S G ++ GI +G S SMITYM F YI N
Sbjct: 108 LGTLIAGSLAFFIEWVAIHS----GKILGGIFIGTSVFTIGSMITYMRFIPYIKKNYDYG 163
Query: 240 ----LGTFACIVLFGLRTGTDNIRDYAGLYTKDPFLALSLALCL 279
L TF I + R T + LYT + + + +CL
Sbjct: 164 MLVFLLTFNLITVSSYRVDTVIKIAHERLYT----IGMGIGICL 203
>At4g23070 putative membrane protein
Length = 313
Score = 29.6 bits (65), Expect = 2.9
Identities = 19/57 (33%), Positives = 30/57 (52%), Gaps = 5/57 (8%)
Query: 69 SILVHGFSWLYGLSGGEIELQEIVNGLINTQMYNSPGISIALIFITVGIGFKLSLAP 125
+I V S L+GL G L E+ LIN Y++ G++I ++ + VG+ L P
Sbjct: 165 AISVGASSALFGLLGAM--LSEL---LINWTTYDNKGVAIVMLLVIVGVNLGLGTLP 216
>At2g38460 unknown protein
Length = 524
Score = 29.3 bits (64), Expect = 3.8
Identities = 19/54 (35%), Positives = 30/54 (55%), Gaps = 9/54 (16%)
Query: 104 PGISIALIFITVGIGFKLSLAPSHQWTPDVYEGSPTPVVAF---LSVTSKVAAS 154
PG+S+AL+F TV + F + + QW EG PT ++ +S T +AA+
Sbjct: 317 PGVSLALLFFTV-LSFGTLMTATLQW-----EGIPTYIIGIGRGISATVGLAAT 364
>At3g56370 putative protein
Length = 964
Score = 28.9 bits (63), Expect = 5.0
Identities = 25/98 (25%), Positives = 44/98 (44%), Gaps = 5/98 (5%)
Query: 72 VHGFSWLYGLSGGEIELQEIVNGLINTQMYNSPGISIALIFITVGIGFKLSLAPSHQWTP 131
+H F+ + GE+ I NGL + + +PGI A++ + + + T
Sbjct: 522 LHTFNISHNHLFGELPAGGIFNGLSPSSVSGNPGICGAVVNKSCPAISPKPIVLNPNATF 581
Query: 132 DVYEGSPTPVVA-----FLSVTSKVAASASATRIFDIL 164
D Y G P A LS++S +A SA+A + ++
Sbjct: 582 DPYNGEIVPPGAGHKRILLSISSLIAISAAAAIVVGVI 619
>At3g46610 unknown protein
Length = 665
Score = 28.9 bits (63), Expect = 5.0
Identities = 20/102 (19%), Positives = 40/102 (38%), Gaps = 2/102 (1%)
Query: 26 LITIFVALECFSLCSYLLSGYTKKDVRSNEATMKYLLMGGASSSILVHGFSWLYGLSGGE 85
+ ++ + F+L LL K + + T ++ G A + + + W + +
Sbjct: 534 MASVLTGQQKFNLLDTLLKEMASKGIEPSVVTFNAVISGCARNGLSGVAYEWFHRMKSEN 593
Query: 86 IELQEIVNGLINTQMYN--SPGISIALIFITVGIGFKLSLAP 125
+E EI ++ + N P ++ L G KLS P
Sbjct: 594 VEPNEITYEMLIEALANDAKPRLAYELHVKAQNEGLKLSSKP 635
>At5g61330 unknown protein
Length = 436
Score = 28.5 bits (62), Expect = 6.5
Identities = 15/49 (30%), Positives = 23/49 (46%)
Query: 336 NQEITPHVRNYRRSPVRSNNSIELSMIICVIASTVPGISMNPIIEIAQD 384
NQ ++ V +Y R P R ++ S + TVP +M P E Q+
Sbjct: 289 NQNVSEQVASYMRDPSRMIKQMQQSRSTVAVFGTVPQEAMEPNPEEKQE 337
>At2g31340 unknown protein
Length = 258
Score = 28.5 bits (62), Expect = 6.5
Identities = 14/42 (33%), Positives = 24/42 (56%)
Query: 166 YFSSNEWHLLLEILAILSMLLGNLIAITQTSMKRMLAYSSIG 207
YFS++EW ++ ++L S L L A+ Q++ + L S G
Sbjct: 5 YFSASEWTIIADVLGANSRHLFELYALKQSNHYQSLMGSKAG 46
>At1g17840 putative ABC transporter (At1g17840)
Length = 703
Score = 28.5 bits (62), Expect = 6.5
Identities = 13/37 (35%), Positives = 24/37 (64%), Gaps = 3/37 (8%)
Query: 310 VSIGLITSVVSIYYYLKIIKLLMTGRNQEITPHVRNY 346
+++ +I S++ IY +II +M N+++TP VR Y
Sbjct: 628 INLSVILSMIIIY---RIIFFIMIKTNEDVTPWVRGY 661
>At1g05570 putative glucan synthase
Length = 1858
Score = 28.5 bits (62), Expect = 6.5
Identities = 42/205 (20%), Positives = 79/205 (38%), Gaps = 14/205 (6%)
Query: 26 LITIFVALECFSLCSYLLS---GYTKKDVRSNEATMKYLL-MGGASSSILVHGFSWLYGL 81
++++F+ L +L + + S ++Y++ +G A+ ++V ++ Y
Sbjct: 516 VLSVFITAAILKLAQAVLDIALSWKARHSMSLYVKLRYVMKVGAAAVWVVVMAVTYAYSW 575
Query: 82 SGGEIELQEIVNGLINTQMYNSPGISIALIFITVGIGFKLSLAPSHQWTPDVYEGSPTPV 141
Q I N +NSP + I I I + +L + E S +
Sbjct: 576 KNASGFSQTIKNWF-GGHSHNSPSLFIVAILIYLSPNMLSALLFLFPFIRRYLERSDYKI 634
Query: 142 VAFLSVTSKVAASASATRIFDIL-FYFSSNEWHLLLEILAILSMLLGNLIAITQTSMKRM 200
+ + S++ T+ DI+ + S WH E LG +IA+ +
Sbjct: 635 MMLMMWWSQIKPLVGPTK--DIMRIHISVYSWH---EFFPHAKNNLGVVIALWSPVI--- 686
Query: 201 LAYSSIGQIGYVIIGIIVGDSNDGY 225
L Y QI Y I+ +VG N +
Sbjct: 687 LVYFMDTQIWYAIVSTLVGGLNGAF 711
>At4g17430 unknown protein
Length = 507
Score = 28.1 bits (61), Expect = 8.5
Identities = 47/209 (22%), Positives = 79/209 (37%), Gaps = 25/209 (11%)
Query: 18 MFLCSANDLITIFVALECFSLCSYLLSGYTKKDVRSNEATMKYLLMGGASSSILVHGFSW 77
+FLCS + L+ F+ + F S + S + ++L F W
Sbjct: 27 IFLCSVSILVVFFIVV-FFITYSEMPKSLFSISAFSGSVQFPQCRSEILTRTLLGQKFLW 85
Query: 78 LYGLSGGEIELQEIVNGLINTQMYNSPGISIALI-FITVGIG----FKLSLAPSH----Q 128
SG +L E N L+ + N I ++ V +G F++ L+PS
Sbjct: 86 YAPHSGFSNQLSEFKNALLMAGILNRTLIIPPILDHHAVALGSCPKFRV-LSPSEIRISV 144
Query: 129 WTPDVYEGSPTPVVAFLSVTS-KVAASASATRIFDILFYFSSNEWHLLLEILAILSMLLG 187
W + V+ + S+SA R+ D YF+S + + LE L
Sbjct: 145 WNHSIELLKTDRYVSMADIVDISSLVSSSAVRVIDFR-YFASLQCGVDLETLC------- 196
Query: 188 NLIAITQTSMKRMLAYSSIGQIGYVIIGI 216
T ++ AY S+ Q GY++ G+
Sbjct: 197 -----TDDLAEQSQAYESLKQCGYLLSGV 220
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.326 0.142 0.421
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 8,025,611
Number of Sequences: 26719
Number of extensions: 327929
Number of successful extensions: 1004
Number of sequences better than 10.0: 18
Number of HSP's better than 10.0 without gapping: 7
Number of HSP's successfully gapped in prelim test: 11
Number of HSP's that attempted gapping in prelim test: 986
Number of HSP's gapped (non-prelim): 18
length of query: 387
length of database: 11,318,596
effective HSP length: 101
effective length of query: 286
effective length of database: 8,619,977
effective search space: 2465313422
effective search space used: 2465313422
T: 11
A: 40
X1: 15 ( 7.1 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 40 (21.6 bits)
S2: 61 (28.1 bits)
Medicago: description of AC126018.3


