

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC126018.10 + phase: 0
(72 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
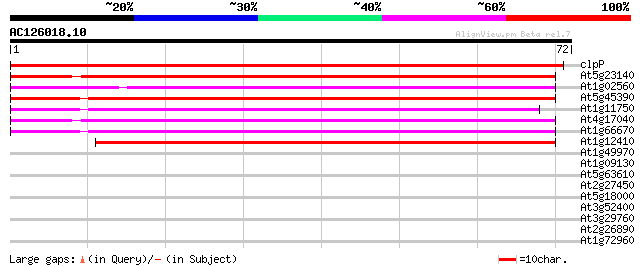
Score E
Sequences producing significant alignments: (bits) Value
clpP -chloroplast genome- ATP-dependent protease subunit 105 3e-24
At5g23140 ATP-dependent protease proteolytic subunit ClpP-like p... 52 7e-08
At1g02560 ATP-dependent clp protease proteolytic subunit (nClpP1) 50 2e-07
At5g45390 ATP-dependent Clp protease-like protein 47 1e-06
At1g11750 putative ATP-dependent Clp protease proteolytic subunit 45 7e-06
At4g17040 ClpP protease complex subunit ClpR4 43 3e-05
At1g66670 nClpP3 protein 43 3e-05
At1g12410 ClpP protease complex subunit ClpR2 42 7e-05
At1g49970 ClpP protease complex subunit ClpR1 35 0.005
At1g09130 ClpP protease complex subunit ClpR3 32 0.075
At5g63610 cdc2-like protein kinase-like protein 25 5.4
At2g27450 nitrilase like protein 25 5.4
At5g18000 putative protein 25 7.1
At3g52400 syntaxin-like protein synt4 25 7.1
At3g29760 unknown protein 25 7.1
At2g26890 unknown protein 25 7.1
At1g72960 hypothetical protein 25 7.1
>clpP -chloroplast genome- ATP-dependent protease subunit
Length = 196
Score = 105 bits (263), Expect = 3e-24
Identities = 51/71 (71%), Positives = 58/71 (80%)
Query: 1 MIHQPATSLYEGQVGECMLEAEELLKMRKTITNIYAQRTGKASWQIYRDMERDLFMSAEE 60
MIHQPA+S YE Q GE +LEAEELLK+R+TIT +Y QRTGK W I DMERD+FMSA E
Sbjct: 124 MIHQPASSFYEAQTGEFILEAEELLKLRETITRVYVQRTGKPIWVISEDMERDVFMSATE 183
Query: 61 AEAHGIVDTVA 71
A+AHGIVD VA
Sbjct: 184 AQAHGIVDLVA 194
>At5g23140 ATP-dependent protease proteolytic subunit ClpP-like
protein
Length = 241
Score = 51.6 bits (122), Expect = 7e-08
Identities = 24/70 (34%), Positives = 43/70 (61%), Gaps = 1/70 (1%)
Query: 1 MIHQPATSLYEGQVGECMLEAEELLKMRKTITNIYAQRTGKASWQIYRDMERDLFMSAEE 60
MIHQP+ Y GQ + + ++++++ + +Y + TG+ + +M+RD FM+ EE
Sbjct: 150 MIHQPSGG-YSGQAKDITIHTKQIVRVWDALNELYVKHTGQPLDVVANNMDRDHFMTPEE 208
Query: 61 AEAHGIVDTV 70
A+A GI+D V
Sbjct: 209 AKAFGIIDEV 218
>At1g02560 ATP-dependent clp protease proteolytic subunit (nClpP1)
Length = 298
Score = 50.4 bits (119), Expect = 2e-07
Identities = 26/70 (37%), Positives = 42/70 (59%), Gaps = 1/70 (1%)
Query: 1 MIHQPATSLYEGQVGECMLEAEELLKMRKTITNIYAQRTGKASWQIYRDMERDLFMSAEE 60
MIHQP GQ + ++A E+L + + A TG++ +I +D +RD FMSA+E
Sbjct: 216 MIHQPLGGAQGGQT-DIDIQANEMLHHKANLNGYLAYHTGQSLEKINQDTDRDFFMSAKE 274
Query: 61 AEAHGIVDTV 70
A+ +G++D V
Sbjct: 275 AKEYGLIDGV 284
>At5g45390 ATP-dependent Clp protease-like protein
Length = 292
Score = 47.4 bits (111), Expect = 1e-06
Identities = 24/70 (34%), Positives = 43/70 (61%), Gaps = 1/70 (1%)
Query: 1 MIHQPATSLYEGQVGECMLEAEELLKMRKTITNIYAQRTGKASWQIYRDMERDLFMSAEE 60
MIHQP GQ + ++A+E++ + +T+I A T ++ Q+ +D++RD +MS E
Sbjct: 181 MIHQPLGGA-SGQAIDVEIQAKEVMHNKNNVTSIIAGCTSRSFEQVLKDIDRDRYMSPIE 239
Query: 61 AEAHGIVDTV 70
A +G++D V
Sbjct: 240 AVEYGLIDGV 249
>At1g11750 putative ATP-dependent Clp protease proteolytic subunit
Length = 271
Score = 45.1 bits (105), Expect = 7e-06
Identities = 25/68 (36%), Positives = 38/68 (55%), Gaps = 1/68 (1%)
Query: 1 MIHQPATSLYEGQVGECMLEAEELLKMRKTITNIYAQRTGKASWQIYRDMERDLFMSAEE 60
MIHQP T G V + + E ++ R+ I +YA TG+ ++ + ERD F+SA E
Sbjct: 198 MIHQPQTGC-GGHVEDVRRQVNEAIEARQKIDRMYAAFTGQPLEKVQQYTERDRFLSASE 256
Query: 61 AEAHGIVD 68
A G++D
Sbjct: 257 ALEFGLID 264
>At4g17040 ClpP protease complex subunit ClpR4
Length = 305
Score = 42.7 bits (99), Expect = 3e-05
Identities = 22/70 (31%), Positives = 39/70 (55%), Gaps = 1/70 (1%)
Query: 1 MIHQPATSLYEGQVGECMLEAEELLKMRKTITNIYAQRTGKASWQIYRDMERDLFMSAEE 60
MI QP ++GQ + + +E+ ++ + +Y++ GK+ QI DM+R + S E
Sbjct: 215 MIKQPIAR-FQGQATDVEIARKEIKHIKTEMVKLYSKHIGKSPEQIEADMKRPKYFSPTE 273
Query: 61 AEAHGIVDTV 70
A +GI+D V
Sbjct: 274 AVEYGIIDKV 283
>At1g66670 nClpP3 protein
Length = 309
Score = 42.7 bits (99), Expect = 3e-05
Identities = 22/70 (31%), Positives = 40/70 (56%), Gaps = 1/70 (1%)
Query: 1 MIHQPATSLYEGQVGECMLEAEELLKMRKTITNIYAQRTGKASWQIYRDMERDLFMSAEE 60
MIHQP + G+ E + E++ + + I+++ TGK +I D +RD F++ E
Sbjct: 187 MIHQPLGTA-GGKATEMSIRIREMMYHKIKLNKIFSRITGKPESEIESDTDRDNFLNPWE 245
Query: 61 AEAHGIVDTV 70
A+ +G++D V
Sbjct: 246 AKEYGLIDAV 255
>At1g12410 ClpP protease complex subunit ClpR2
Length = 279
Score = 41.6 bits (96), Expect = 7e-05
Identities = 19/59 (32%), Positives = 37/59 (62%)
Query: 12 GQVGECMLEAEELLKMRKTITNIYAQRTGKASWQIYRDMERDLFMSAEEAEAHGIVDTV 70
GQ + EA+EL ++R + N A+ TG+ + ++++D+ R +AEEA +G++D +
Sbjct: 201 GQADDIQNEAKELSRIRDYLFNELAKNTGQPAERVFKDLSRVKRFNAEEAIEYGLIDKI 259
>At1g49970 ClpP protease complex subunit ClpR1
Length = 387
Score = 35.4 bits (80), Expect = 0.005
Identities = 19/70 (27%), Positives = 37/70 (52%)
Query: 2 IHQPATSLYEGQVGECMLEAEELLKMRKTITNIYAQRTGKASWQIYRDMERDLFMSAEEA 61
++ P + G + ++A+EL + + A+ TGK+ QI D++R ++ A+ A
Sbjct: 283 LYLPKVNRSSGAAIDMWIKAKELDANTEYYIELLAKGTGKSKEQINEDIKRPKYLQAQAA 342
Query: 62 EAHGIVDTVA 71
+GI D +A
Sbjct: 343 IDYGIADKIA 352
>At1g09130 ClpP protease complex subunit ClpR3
Length = 330
Score = 31.6 bits (70), Expect = 0.075
Identities = 12/55 (21%), Positives = 30/55 (53%)
Query: 16 ECMLEAEELLKMRKTITNIYAQRTGKASWQIYRDMERDLFMSAEEAEAHGIVDTV 70
+ ++ A+E++ R + + ++ TG + + M R +M A +A+ G++D +
Sbjct: 248 DVLIRAKEVITNRDILVELLSKHTGNSVETVANVMRRPYYMDAPKAKEFGVIDRI 302
>At5g63610 cdc2-like protein kinase-like protein
Length = 470
Score = 25.4 bits (54), Expect = 5.4
Identities = 10/26 (38%), Positives = 17/26 (64%)
Query: 42 ASWQIYRDMERDLFMSAEEAEAHGIV 67
++W I+RD++ + +AE HGIV
Sbjct: 147 SNWIIHRDLKPSNILVMGDAEEHGIV 172
>At2g27450 nitrilase like protein
Length = 326
Score = 25.4 bits (54), Expect = 5.4
Identities = 16/55 (29%), Positives = 27/55 (49%), Gaps = 11/55 (20%)
Query: 7 TSLYEGQVGECMLEAEELLKMRKTITNIYAQ------RTGKASWQIYRDMERDLF 55
TS G GE + EA++ K+ + AQ ++ + SW ++RD DL+
Sbjct: 267 TSFIAGPTGEIVAEADD-----KSEAVLVAQFDLDMIKSKRQSWGVFRDRRPDLY 316
>At5g18000 putative protein
Length = 307
Score = 25.0 bits (53), Expect = 7.1
Identities = 11/45 (24%), Positives = 25/45 (55%)
Query: 20 EAEELLKMRKTITNIYAQRTGKASWQIYRDMERDLFMSAEEAEAH 64
+ E+L+ RKTIT+ + + IY+D++++ + + +H
Sbjct: 115 DGTEMLRPRKTITSSSGRNKREERKSIYKDVKKEEEIESWSESSH 159
>At3g52400 syntaxin-like protein synt4
Length = 341
Score = 25.0 bits (53), Expect = 7.1
Identities = 12/40 (30%), Positives = 22/40 (55%)
Query: 7 TSLYEGQVGECMLEAEELLKMRKTITNIYAQRTGKASWQI 46
TS+ G + E E+ ++R+TITN Y + G+ + +
Sbjct: 135 TSVVNGLRKKLKDEMEKFSRVRETITNEYKETVGRMCFTV 174
>At3g29760 unknown protein
Length = 533
Score = 25.0 bits (53), Expect = 7.1
Identities = 18/72 (25%), Positives = 30/72 (41%), Gaps = 7/72 (9%)
Query: 3 HQPATSLYEGQVGECMLEAEELLKMRKTITNIYAQRTGKA-------SWQIYRDMERDLF 55
HQ T G G+ L E+L++ I G+ SW+ YR++ R L
Sbjct: 461 HQNKTDTSLGNGGDLRLHLEKLVEAENVQDFIKKNPFGQEAITEVSESWEFYREIIRMLR 520
Query: 56 MSAEEAEAHGIV 67
+S++ I+
Sbjct: 521 ISSQTYRLRNII 532
>At2g26890 unknown protein
Length = 2535
Score = 25.0 bits (53), Expect = 7.1
Identities = 8/23 (34%), Positives = 13/23 (55%)
Query: 44 WQIYRDMERDLFMSAEEAEAHGI 66
W Y+D + DLF+ + A G+
Sbjct: 2489 WSAYKDQKHDLFLPSNTQSAAGV 2511
>At1g72960 hypothetical protein
Length = 748
Score = 25.0 bits (53), Expect = 7.1
Identities = 23/79 (29%), Positives = 37/79 (46%), Gaps = 14/79 (17%)
Query: 7 TSLYEGQVGECMLEAEELL------KMRKTITNIYAQRT-----GKASWQIYRDME---R 52
TSLYE +V E + E E L + T+ ++ + T G +S DME R
Sbjct: 412 TSLYESKVHEALSEPVEALLDGANDETWSTVKKLHRRETESAVSGLSSALAGFDMEEETR 471
Query: 53 DLFMSAEEAEAHGIVDTVA 71
D + + + A G+++T A
Sbjct: 472 DRMVKSLQDYARGVIETKA 490
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.317 0.129 0.363
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 1,393,377
Number of Sequences: 26719
Number of extensions: 39870
Number of successful extensions: 94
Number of sequences better than 10.0: 17
Number of HSP's better than 10.0 without gapping: 14
Number of HSP's successfully gapped in prelim test: 3
Number of HSP's that attempted gapping in prelim test: 80
Number of HSP's gapped (non-prelim): 17
length of query: 72
length of database: 11,318,596
effective HSP length: 48
effective length of query: 24
effective length of database: 10,036,084
effective search space: 240866016
effective search space used: 240866016
T: 11
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 52 (24.6 bits)
Medicago: description of AC126018.10


