

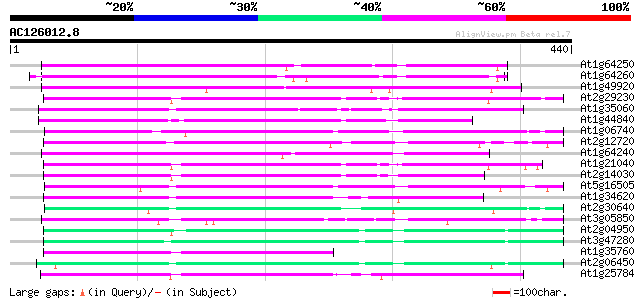
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC126012.8 + phase: 0
(440 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
At1g64250 hypothetical protein 167 8e-42
At1g64260 hypothetical protein 163 2e-40
At1g49920 hypothetical protein 162 4e-40
At2g29230 Mutator-like transposase 154 9e-38
At1g35060 hypothetical protein 153 2e-37
At1g44840 hypothetical protein 152 5e-37
At1g06740 mudrA-like protein 152 5e-37
At2g12720 putative protein 149 3e-36
At1g64240 hypothetical protein 148 7e-36
At1g21040 hypothetical protein 143 2e-34
At2g14030 Mutator-like transposase 142 5e-34
At5g16505 mutator-like transposase-like protein (MQK4.25) 137 9e-33
At1g34620 Mutator-like protein 133 2e-31
At2g30640 Mutator-like transposase 133 2e-31
At3g05850 unknown protein 132 3e-31
At2g04950 putative transposase of transposon FARE2.9 (cds1) 130 1e-30
At3g47280 putative protein 130 2e-30
At1g35760 hypothetical protein 126 3e-29
At2g06450 putative transposase of FARE2.11 124 1e-28
At1g25784 mutator-like transposase, putative 123 2e-28
>At1g64250 hypothetical protein
Length = 790
Score = 167 bits (424), Expect = 8e-42
Identities = 106/377 (28%), Positives = 171/377 (45%), Gaps = 25/377 (6%)
Query: 26 AQFHRLFWAFYPCINGFKYCKPVVHVDGTWLYDKYKGTLLLAVAQDGNNKTIPIAFALVE 85
A F +FWAF I GF++C+P++ VD L KY L++A + ++ P+AFA
Sbjct: 384 ASFRSVFWAFPQSIEGFQHCRPLIVVDTKDLKGKYPMKLMIASGVEADDCYFPLAFAFTT 443
Query: 86 GETKEGWSFFLKNLRRHVTKGISVCMVSDRHESIKSAFNDPRNGWQATGSAHVYCIRHIK 145
+ + W +FL +R VT+ +C++S H I N+P + WQ + H++C+ I
Sbjct: 444 EVSSDTWRWFLSGIREKVTQRKDICLISRPHPDILDVINEPGSQWQEPWAYHMFCLDDIC 503
Query: 146 QNFMRTIKDGDLKDVVNNMGYALNVPLFNYYRGVIQEANQRALDWVDNIPKEKWTQAYDE 205
F +D LK++V G F+ Y I++ N A W+D P+ +W QA+D
Sbjct: 504 TQFHYVFQDDYLKNLVYEAGSTSEKEEFDSYMNEIEKKNSEARKWLDQFPQYQWAQAHDS 563
Query: 206 GRRWGHMTSN------IVESWNSVFKGTRNLPVTPIVQSTYYRLACLFADRAQKAFARVG 259
GRR+ MT + ES+ S+ LPVT + + F + RV
Sbjct: 564 GRRYRVMTIDAENLFAFCESFQSL-----GLPVTATALLLFDEMRFFFYSGLCDSSGRVN 618
Query: 260 SGDLFSEYCQNAIQDDIVKSNTHHVEQFDRERYTFSVRETVNYREGRPMGTFKVDLRAGW 319
GD++++ + ++ + S H V E+ F V E + E + V L
Sbjct: 619 RGDMYTKPVMDQLEKLMTDSIPHVV--MPLEKGLFQVTEPLQEDE------WIVQLSEWS 670
Query: 320 CDCGKFQALHLPCSHVIAACSSFRHDYTTLIPHVFKNESVYSIYNTAFKVVHDKSYWLPY 379
C CG+FQ PC HV+A C + + + + + +Y Y F V + + W
Sbjct: 671 CTCGEFQLKKFPCLHVLAVCEKLKINPLQYVDDCYSLDRLYKTYAATFSPVPEVAAWPEA 730
Query: 380 DG------PVICHNPNM 390
G PVI PN+
Sbjct: 731 SGVPTLFPPVILPPPNV 747
>At1g64260 hypothetical protein
Length = 1490
Score = 163 bits (413), Expect = 2e-40
Identities = 102/381 (26%), Positives = 174/381 (44%), Gaps = 28/381 (7%)
Query: 16 FKNADWREWRAQFHRLFWAFYPCINGFKYCKPVVHVDGTWLYDKYKGTLLLAVAQDGNNK 75
F N D+ A F +FW+F I GF++C+P++ VD L KY+ L++A D NK
Sbjct: 1121 FPNPDF----ASFRGVFWSFSQSIEGFQHCRPLIVVDTKSLNGKYQLKLMIASGVDAANK 1176
Query: 76 TIPIAFALVEGETKEGWSFFLKNLRRHVTKGISVCMVSDRHESIKSAFNDPRNGWQATGS 135
P+AFA+ + + + W +F +R VT+ +C++S I + N+P + WQ +
Sbjct: 1177 FFPLAFAVTKEVSTDSWRWFFTKIREKVTQRKDLCLISSPLRDIVAVVNEPGSLWQEPWA 1236
Query: 136 AHVYCIRHIKQNFMRTIKDGDLKDVVNNMGYALNVPLFNYYRGVIQEANQRALDWVDNIP 195
H +C+ H++ F+ +D +L+ +V G F+ Y I+E N A W+D IP
Sbjct: 1237 HHKFCLNHLRSQFLGVFRDYNLESLVEQAGSTNQKEEFDSYMNDIKEKNPEAWKWLDQIP 1296
Query: 196 KEKWTQAYDEGRRWGHMTSNIVE-SWNSVFKGTRNLP-----VTPIVQSTYYRLACLFAD 249
+ KW A+D G R+G I+E ++F R P +T V + L F
Sbjct: 1297 RHKWALAHDSGLRYG-----IIEIDREALFAVCRGFPYCTVAMTGGVMLMFDELRSSFDK 1351
Query: 250 RAQKAFARVGSGDLFSEYCQNAIQDDIVKSNTHHVEQFDRERYTFSVRETVNYREGRPMG 309
++ + G +++E + +++ + S + + Q +R+ +F V E+ E
Sbjct: 1352 SLSSIYSSLNRGVVYTEPFMDKLEEFMTDSIPYVITQLERD--SFKVSESSEKEE----- 1404
Query: 310 TFKVDLRAGWCDCGKFQALHLPCSHVIAACSSFRHDYTTLIPHVFKNESVYSIYNTAFKV 369
+ V L C C KFQ+ PC H +A + + + + E Y F
Sbjct: 1405 -WIVQLNVSTCTCRKFQSYKFPCLHALAVFEKLKINPLQYVDECYTVEQYCKTYAATFSP 1463
Query: 370 VHDKSYWLPYDGPVICHNPNM 390
V D + W P C P +
Sbjct: 1464 VPDVAAW-----PEDCRVPTL 1479
Score = 141 bits (356), Expect = 6e-34
Identities = 98/371 (26%), Positives = 160/371 (42%), Gaps = 15/371 (4%)
Query: 26 AQFHRLFWAFYPCINGFKYCKPVVHVDGTWLYDKYKGTLLLAVAQDGNNKTIPIAFALVE 85
A F +FWAF I GF++C+P++ VD L +Y+ L++A D NK P+AFA+ +
Sbjct: 363 ASFCGVFWAFPQSIEGFQHCRPLIVVDTKNLNCEYQLKLMIASGVDAANKYFPLAFAVTK 422
Query: 86 GETKEGWSFFLKNLRRHVTKGISVCMVSDRHESIKSAFNDPRNGWQATGSAHVYCIRHIK 145
+ + W +FL +R VT+ +C++S H I + N+ + WQ + H + + H
Sbjct: 423 EVSTDIWRWFLTGIREKVTQRKGLCLISSPHPDIIAVVNESGSQWQEPWAYHRFSLNHFY 482
Query: 146 QNFMRTIKDGDLKDVVNNMGYALNVPLFNYYRGVIQEANQRALDWVDNIPKEKWTQAYDE 205
F R L + G F Y I+E N A W+D P+ +W A+D
Sbjct: 483 SQFSRVFPSFCLGARIRRAGSTSQKDEFVSYMNDIKEKNPEARKWLDQFPQNRWALAHDN 542
Query: 206 GRRWGHMTSNIVESWN--SVFKGTRNLPVTPIVQSTYYRLACLFADRAQKAFARVGSGDL 263
GRR+G M N + + F+ ++ VT V + L F + + + GD+
Sbjct: 543 GRRYGIMEINTKALFAVCNAFEQAGHV-VTGSVLLLFDELRSKFDKSFSCSRSSLNCGDV 601
Query: 264 FSEYCQNAIQDDIVKSNTHHVEQFDRERYTFSVRETVNYREGRPMGTFKVDLRAGWCDCG 323
++E + +++ T+ + F V ++ G V L C CG
Sbjct: 602 YTEPVMDKLEEFRTTFVTYSYIVTPLDNNAFQVATALD------KGECIVQLSDCSCTCG 655
Query: 324 KFQALHLPCSHVIAACSSFRHDYTTLIPHVFKNESVYSIYNTAFKVVHDKSYWLPYDG-- 381
FQ PC H +A C + + + + E + Y T F V + S W G
Sbjct: 656 DFQRYKFPCLHALAVCKKLKFNPLQYVDDCYTLERLKRTYATIFSHVPEMSAWPEASGVP 715
Query: 382 ----PVICHNP 388
PVI +P
Sbjct: 716 RLLPPVIPPSP 726
>At1g49920 hypothetical protein
Length = 785
Score = 162 bits (409), Expect = 4e-40
Identities = 103/388 (26%), Positives = 182/388 (46%), Gaps = 13/388 (3%)
Query: 26 AQFHRLFWAFYPCINGFKYCKPVVHVDGTWLYDKYKGTLLLAVAQDGNNKTIPIAFALVE 85
A F LFWAF I GF++C+P++ VD L KYK L++A A D N+ P+AFA+ +
Sbjct: 359 ASFRGLFWAFSQSIQGFQHCRPLIVVDTKNLGGKYKMKLMIASAFDATNQYFPLAFAVTK 418
Query: 86 GETKEGWSFFLKNLRRHVTKGISVCMVSDRHESIKSAFNDPRNGWQATGSAHVYCIRHIK 145
+ + W +FL +R VT+ +C++S I + N+P + W+ + H +C+ H+
Sbjct: 419 EVSVDSWRWFLTRIREKVTQRQGICLISSPDPDILAVINEPGSQWKEPWAYHRFCLYHLC 478
Query: 146 QNFMRTIK--DGDLKDVVNNMGYALNVPLFNYYRGVIQEANQRALDWVDNIPKEKWTQAY 203
D ++ +V+ G + F+ Y I+E N A W+D P +W A+
Sbjct: 479 SKLCSVSPGFDYNMHFLVDEAGSSSQKEEFDSYMKEIKERNPEAWKWLDQFPPHQWALAH 538
Query: 204 DEGRRWGHMTSNIVESWNSVFKGTRNLPVTPIVQSTYYRLACLFADRAQKAFARVGSGDL 263
D+GRR+G M + E+ +V K R + + V + +L FA+ + + + GD+
Sbjct: 539 DDGRRYGIMRID-TEALFAVCKRFRKVAMAGGVMLLFGQLKDAFAESFKLSRGSLKHGDV 597
Query: 264 FSEYCQNAIQDDIVKSNTH--HVEQFDRERYTFSV---RETVNYREGRPMGTFKVDLRAG 318
++E+ +++ S+T + +R+ Y S+ ++T + + V L
Sbjct: 598 YTEHVMEKLEEFETDSDTWVITITPLERDAYQVSMAPKKKTRLMGQSNDSTSGIVQLNDT 657
Query: 319 WCDCGKFQALHLPCSHVIAACSSFRHDYTTLIPHVFKNESVYSIYNTAFKVVHDKSYW-- 376
C CG+FQ PC H +A C + + + + E + Y+ F V + S W
Sbjct: 658 TCTCGEFQKNKFPCLHALAVCDELKINPLQYVDDCYTVERYHKTYSAKFSPVPELSAWPE 717
Query: 377 ---LPYDGPVICHNPNMRRLKKGRPNST 401
+P P + P + KG+ T
Sbjct: 718 AYGVPTLIPPVIEPPPPKVSGKGKEKDT 745
>At2g29230 Mutator-like transposase
Length = 915
Score = 154 bits (389), Expect = 9e-38
Identities = 121/412 (29%), Positives = 177/412 (42%), Gaps = 25/412 (6%)
Query: 27 QFHRLFWAFYPCINGFKYCKPVVHVDGTWLYDKYKGTLLLAVAQDGNNKTIPIAFALVEG 86
+F +F AF I GF K V+ +DG L KY G LL A QD N + PIAF +V+
Sbjct: 521 RFKYMFLAFAASIQGFSCMKRVIVIDGAHLKGKYGGCLLTASGQDANFQVFPIAFGVVDS 580
Query: 87 ETKEGWSFFLKNLRRHVTKGISVCMVSDRHESIKSAFND--PRNGWQATGSAHVYCIRHI 144
E + W +F + L + G ++ VSDRH SI + P+ + H CI H+
Sbjct: 581 ENDDAWEWFFRVLSTAIPDGDNLTFVSDRHSSIYTGLRRVYPK-------AKHGACIVHL 633
Query: 145 KQNFMRTIKDGDLKDVVNNMGYALNVPLFNYYRGVIQEANQRALDWVDNIPKEKWTQAYD 204
++N + K L V+ A + F+ Y + + +++++ WT+AY
Sbjct: 634 QRNIATSYKKKHLLFHVSRAARAFRICEFHTYFNEVIRLDPACARYLESVGFCHWTRAYF 693
Query: 205 EGRRWGHMTSNIVESWNSVFKGTRNLPVTPIVQSTYYRLACLFADRAQKAFARVGSGDLF 264
G+R+ MTSN+ ES N+V K R LP+ +++ L FA R + A
Sbjct: 694 LGKRYNVMTSNVAESLNAVLKEARELPIISLLEFIRTTLISWFAMRREAARTEASP---L 750
Query: 265 SEYCQNAIQDDIVKSNTHHVEQFDRERYTFSVRETVNYREGRPMGTFKVDLRAGWCDCGK 324
+ + + KS V + D RY + +RE EG + V L C C
Sbjct: 751 PPKMREVVHRNFEKSVRFAVHRLD--RYDYEIRE-----EG--ASVYHVKLMERTCSCRA 801
Query: 325 FQALHLPCSHVIAACSSFRHDYTTLIPHVFKNESVYSIYNTAFKVVHDKS--YWLPYDGP 382
F LHLPC H IAA + L+ + ES Y K V + + LP
Sbjct: 802 FDLLHLPCPHAIAAAVAEGVPIQGLMAPEYSVESWRMSYLGTIKPVPEVGDVFALPEPIA 861
Query: 383 VICHNPNMRRLKKGRPNSTRIRTEMDEEVVERTPTPRRCGLCRHTGHIRRNC 434
+ P R GRP RI + + +R T RC C GH R C
Sbjct: 862 SLRLFPPATRRPSGRPKKKRITSRGEFTCPQRQVT--RCSRCTGAGHNRATC 911
>At1g35060 hypothetical protein
Length = 873
Score = 153 bits (386), Expect = 2e-37
Identities = 117/383 (30%), Positives = 175/383 (45%), Gaps = 20/383 (5%)
Query: 23 EWRAQFHRLFWAFYPCINGFKYCKPVVHVDGTWLYDKYKGTLLLAVAQDGNNKTIPIAFA 82
E + +F +F AF I GFK+ + V+ VDGT L KYKG LL A QD N + P+AFA
Sbjct: 500 ERKERFLYMFLAFGASIQGFKHLRRVLVVDGTHLKGKYKGVLLTASGQDANFQVYPLAFA 559
Query: 83 LVEGETKEGWSFFLKNLRRHVTKGISVCMVSDRHESIKSAFNDPRNGWQATGSAHVYCIR 142
+V+ E + W++F L R + ++ ++SDRHESIK + H CI
Sbjct: 560 VVDSENDDAWTWFFTKLERIIADNNTLTILSDRHESIKVGVKK-----VFPQAHHGACII 614
Query: 143 HIKQNFMRTIKDGDLKDVVNNMGYALNVPLFNYYRGVIQEANQRALDWVDNIPKEKWTQA 202
H+ +N K+ L +V N GY F I N + ++ ++ WT+
Sbjct: 615 HLCRNIQARFKNRGLTQLVKNAGYEFTSGKFKTLYNQINAINPLCIKYLHDVGMAHWTRL 674
Query: 203 YDEGRRWGHMTSNIVESWN-SVFKGTRNLPVTPIVQSTYYRLACLFADRAQKAFARVGSG 261
Y G+R+ MTSNI E+ N ++FKG R+ + +++ L F +K+ A SG
Sbjct: 675 YFPGQRFNLMTSNIAETLNKALFKG-RSSHIVELLRFIRSMLTRWFNAHRKKSQAH--SG 731
Query: 262 DLFSEYCQNAIQDDIVKSNTHHVEQFDRERYTFSVRETVNYREGRPMGTFKVDLRAGWCD 321
+ E I ++ S+ V + Y E V G+ G+ VDL C
Sbjct: 732 PVPPE-VDKQISKNLTTSSGSKVGRVTSWSY-----EVV----GKLGGSNVVDLEKKQCT 781
Query: 322 CGKFQALHLPCSHVIAACSSFRHDYTTLIPHVFKNESVYSIYNTA-FKVVHDKSYWLPYD 380
C ++ L +PC H + A +S Y L+ FK S + Y A F + K +P +
Sbjct: 782 CKRYDKLKIPCGHALVAANSINLSYKALVDDYFKPHSWVASYKGAVFPEANGKEEDIPEE 841
Query: 381 GPVICHNPNMRRLKKGRPNSTRI 403
P P R GRP RI
Sbjct: 842 LPHRSMLPPYTRRPSGRPKVARI 864
>At1g44840 hypothetical protein
Length = 926
Score = 152 bits (383), Expect = 5e-37
Identities = 103/341 (30%), Positives = 156/341 (45%), Gaps = 17/341 (4%)
Query: 23 EWRAQFHRLFWAFYPCINGFKYCKPVVHVDGTWLYDKYKGTLLLAVAQDGNNKTIPIAFA 82
E + +F +F AF I GF++ + ++ VDGT L KYKG LL + QD N + P+ FA
Sbjct: 549 ESKERFLYMFLAFGASIAGFRHLRRILVVDGTHLKGKYKGVLLTSSGQDANFQVYPLGFA 608
Query: 83 LVEGETKEGWSFFLKNLRRHVTKGISVCMVSDRHESIKSAFNDPRNGWQATGSAHVYCIR 142
+V+ E E W++F L R + ++ ++SDRH SI A R QA H CI
Sbjct: 609 VVDSENDESWTWFFTKLERIIADSKTLTILSDRHSSILVAVK--RVFPQAN---HGACII 663
Query: 143 HIKQNFMRTIKDGDLKDVVNNMGYALNVPLFNYYRGVIQEANQRALDWVDNIPKEKWTQA 202
H+ +N K+ L +V N GYA F + G I+ NQ ++ +I WT+
Sbjct: 664 HLCRNIQTKYKNKALTQLVKNAGYAFTGTKFKEFYGQIETTNQNCGKYLHDIGMANWTRH 723
Query: 203 YDEGRRWGHMTSNIVESWNSVFKGTRNLPVTPIVQSTYYRLACLFADRAQKAFARVGSGD 262
Y G+R+ MTSNI E+ N R+ + +++ L F R +K+ G
Sbjct: 724 YFRGQRFNLMTSNIAETLNKALNKGRSSHIVELIRFIRSMLTRWFNARRKKSLKHKGP-- 781
Query: 263 LFSEYCQNAIQDDIVKSNTHHVEQFDRERYTFSVRETVNYREGRPMGTFKVDLRAGWCDC 322
I ++ +N V + Y + G G VDL C C
Sbjct: 782 -VPPEVDKQITKTMLTTNGSKVGRITNWSYEIN---------GMLGGRNVVDLEKKQCTC 831
Query: 323 GKFQALHLPCSHVIAACSSFRHDYTTLIPHVFKNESVYSIY 363
++ L +PCSH + A +SF+ Y L+ FK + S Y
Sbjct: 832 KRYDKLKIPCSHALVAANSFKISYKALVDDCFKPHAWVSSY 872
>At1g06740 mudrA-like protein
Length = 726
Score = 152 bits (383), Expect = 5e-37
Identities = 109/412 (26%), Positives = 177/412 (42%), Gaps = 28/412 (6%)
Query: 28 FHRLFWAFYPCINGF-KYCKPVVHVDGTWLYDKYKGTLLLAVAQDGNNKTIPIAFALVEG 86
F LF +F I+GF C+P++ +D T L KY GTLLLA DG+ P+AFA+V
Sbjct: 334 FQHLFISFQASISGFLNACRPLIALDSTVLKSKYPGTLLLATGFDGDGAVFPLAFAIVNE 393
Query: 87 ETKEGWSFFLKNLRRHVTKGISVCMVSDRHESIKSAFNDPRNGWQATGSA--HVYCIRHI 144
E + W FL LR+ + + + + + S +G +A A H +C+ ++
Sbjct: 394 ENDDNWHRFLSELRKILDENMPKLTI------LSSGERPVVDGVEANFPAAFHGFCLHYL 447
Query: 145 KQNFMRTIKDGDLKDVVNNMGYALNVPLFNYYRGVIQEANQRALDWVDNIPKEKWTQAYD 204
+ F R + L D+ + L V F I++ + A W+ N +W +Y
Sbjct: 448 TERFQREFQSSVLVDLFWEAAHCLTVLEFKSKINKIEQISPEASLWIQNKSPARWASSYF 507
Query: 205 EGRRWGHMTSNIV-ESWNSVFKGTRNLPVTPIVQSTYYRLACLFADRAQKAFARVGSGDL 263
EG R+G +T+N++ ES ++ + T LP+ ++ + L + +R + + + ++
Sbjct: 508 EGTRFGQLTANVITESLSNWVEDTSGLPIIQTMECIHRHLINMLKERRETS---LHWSNV 564
Query: 264 FSEYCQNAIQDDIVKSNTHHVEQFDRERYTFSVRETVNYREGRPMGTFKVDLRAGWCDCG 323
+ + I +S H V + + + E G V++ C CG
Sbjct: 565 LVPSAEKQMLAAIEQSRAHRVYRANEAEFEVMTCE----------GNVVVNIENCSCLCG 614
Query: 324 KFQALHLPCSHVIAACSSFRHDYTTLIPHVFKNESVYSIYNTAFKVVHDKSYWLPYDGPV 383
++Q LPCSH + A S D F E+ Y + + DK W D
Sbjct: 615 RWQVYGLPCSHAVGALLSCEEDVYRYTESCFTVENYRRAYAETLEPISDKVQWKENDSER 674
Query: 384 ICHNP-NMRRLKKGRPNSTRIRTEMDEEVVERTPTPRRCGLCRHTGHIRRNC 434
N + KG P R+R E D + V R CG C TGH R C
Sbjct: 675 DSENVIKTPKAMKGAPRKRRVRAE-DRDRVRRVV---HCGRCNQTGHFRTTC 722
>At2g12720 putative protein
Length = 819
Score = 149 bits (376), Expect = 3e-36
Identities = 113/424 (26%), Positives = 185/424 (42%), Gaps = 50/424 (11%)
Query: 27 QFHRLFWAFYPCINGFKYCKPVVHVDGTWLYDKYKGTLLLAVAQDGNNKTIPIAFALVEG 86
+F+ +F F I GF Y + VV VDGT+L+ YKGTLL A+AQDGN + P+AF +V+
Sbjct: 428 RFNYMFIVFGASIAGFHYMRRVVVVDGTFLHGSYKGTLLTALAQDGNFQIFPLAFGVVDT 487
Query: 87 ETKEGWSFFLKNLRRHVTKGISVCMVSDRHESIKSAFNDPRNGWQATGSAHVYCIRHIKQ 146
E + W + L+ + + ++SDRH+SI A G +A C H+ +
Sbjct: 488 ENDDSWRWLFTQLKVVIPDATDLAIISDRHKSIGKAI-----GEVYPLAARGICTYHLYK 542
Query: 147 NFMRTIKDGDLKDVVNNMGYALNVPLFNYYRGVIQEANQRALDWVDNIPKEKWTQAYDEG 206
N + K DL +V + F I+E + ++ E W +A+ G
Sbjct: 543 NILVKFKRKDLFPLVKKAARCYRLNDFTNAFNEIEELDPLLHAYLQRAGVEMWARAHFPG 602
Query: 207 RRWGHMTSNIVESWNSVFKGTRNLPVTPIVQSTYYRLACLFADR---AQKAFARVGSGDL 263
R+ MT+NI ES N +NLP+ ++++ + FA+R A K ++ G
Sbjct: 603 DRYNLMTTNIAESMNRALSQAKNLPIVRMLEAIRQMMTRWFAERRDDASKQHTQLTPG-- 660
Query: 264 FSEYCQNAIQDDIVKSNTHHVEQFDRERYTFSVRETVNYREGRPMGTFKVDLRAGWCDCG 323
+ +Q + S V+ D R + +++ V++ C C
Sbjct: 661 ----VEKLLQTRVTSSRLLDVQTIDASRVQVAYEASLHV----------VNVDEKQCTCR 706
Query: 324 KFQALHLPCSHVIAACSSFRHDYTTLIPHVFKNESVYSIYNTAF----------KVVHDK 373
F LPC H IAA +L +K+ + ++Y A ++V D+
Sbjct: 707 LFNKEKLPCIHAIAAAEHMGVSRISLCSPYYKSSHLVNVYACAIMPSDTEVPVPQIVIDQ 766
Query: 374 SYWLPYDGPVICHNPNMRRLKKGRPNSTRIRTEMDEEVVERTPTPRR---CGLCRHTGHI 430
P P++ + P GRP R+++ + EV T PR+ C C+ TGH
Sbjct: 767 ----PCLPPIVANQP-------GRPKKLRMKSAL--EVAVETKRPRKEHACSRCKETGHN 813
Query: 431 RRNC 434
+ C
Sbjct: 814 VKTC 817
>At1g64240 hypothetical protein
Length = 938
Score = 148 bits (373), Expect = 7e-36
Identities = 92/354 (25%), Positives = 158/354 (43%), Gaps = 13/354 (3%)
Query: 26 AQFHRLFWAFYPCINGFKYCKPVVHVDGTWLYDKYKGTLLLAVAQDGNNKTIPIAFALVE 85
A F +FWAF I GF +C+P++ VD L KY+ L++A A D + +AFA
Sbjct: 553 ASFRGVFWAFSQSIEGFHHCRPLIIVDTKNLNCKYQWKLMIASAVDAADNFFLLAFAFTT 612
Query: 86 GETKEGWSFFLKNLRRHVTKGISVCMVSDRHESIKSAFNDPRNGWQATGSAHVYCIRHIK 145
+ + W +FL +R VT+ +C++S + + N+ + WQ + + +C+RH+
Sbjct: 613 ELSTDSWRWFLSGIRERVTQRKGLCLISSPDPDLLAVINESGSQWQEPWAYNRFCLRHLL 672
Query: 146 QNFMRTIKDGDLKDVVNNMGYALNVPLFNYYRGVIQEANQRALDWVDNIPKEKWTQAYDE 205
F +D L+D+V G F+ Y I++ N A W+D P+ +W A+D
Sbjct: 673 SQFSGIFRDYYLEDLVKRAGSTSQKEEFDSYMKDIEKKNSEARKWLDQFPQNQWALAHDN 732
Query: 206 GRRWGHM---TSNIVESWNSVFKGTRNLPVTPIVQSTYYRLACLFADRAQKAFARVGSGD 262
GRR+G M T+ + E +N N +T V + L F + + G+
Sbjct: 733 GRRYGIMEIETTTLFEDFN--VSHLDNHVLTGYVLRLFDELRHSFDEFFCFSRGSRKCGN 790
Query: 263 LFSEYCQNAIQDDIVKSNTHHVEQFDRERYTFSVRETVNYREGRPMGTFKVDLRAGWCDC 322
+++E + + S T+ V D + + + + + V L C C
Sbjct: 791 VYTEPVTEKLAESRKDSVTYDVMPLDNNAFQVTAPQEND--------EWTVQLSDCSCTC 842
Query: 323 GKFQALHLPCSHVIAACSSFRHDYTTLIPHVFKNESVYSIYNTAFKVVHDKSYW 376
G+FQ+ PC H +A C + + + + E +Y Y F V + S W
Sbjct: 843 GEFQSCKFPCLHALAVCKLLKINPLEYVDDCYTLERLYKTYTATFSPVPELSAW 896
>At1g21040 hypothetical protein
Length = 904
Score = 143 bits (360), Expect = 2e-34
Identities = 119/400 (29%), Positives = 172/400 (42%), Gaps = 27/400 (6%)
Query: 27 QFHRLFWAFYPCINGFKYCKPVVHVDGTWLYDKYKGTLLLAVAQDGNNKTIPIAFALVEG 86
+F +F AF I GF K V+ +DG L KY G LL A QD N + PIAF +V+
Sbjct: 521 RFKYMFLAFAASIQGFSCMKRVIVIDGAHLKGKYGGCLLTASGQDANFQVFPIAFGVVDS 580
Query: 87 ETKEGWSFFLKNLRRHVTKGISVCMVSDRHESIKSAFND--PRNGWQATGSAHVYCIRHI 144
E + W +F + L + G ++ VSDRH SI + P+ + H CI H+
Sbjct: 581 ENDDAWEWFFRVLSTAIPDGDNLTFVSDRHSSIYTGLRRVYPK-------AKHGACIVHL 633
Query: 145 KQNFMRTIKDGDLKDVVNNMGYALNVPLFNYYRGVIQEANQRALDWVDNIPKEKWTQAYD 204
++N + K L V+ A + F+ Y + + +++++ WT+AY
Sbjct: 634 QRNIATSYKKKHLLFHVSRAARAFRICEFHTYFNEVIRLDPACARYLESVGFCHWTRAYF 693
Query: 205 EGRRWGHMTSNIVESWNSVFKGTRNLPVTPIVQSTYYRLACLFADRAQKAFARVGSGDLF 264
G R+ MTSN+ ES N+V K R LP+ +++ L FA R + A
Sbjct: 694 LGERYNVMTSNVAESLNAVLKEARELPIISLLEFIRTTLISWFAMRREAARTEASP---L 750
Query: 265 SEYCQNAIQDDIVKSNTHHVEQFDRERYTFSVRETVNYREGRPMGTFKVDLRAGWCDCGK 324
+ + + KS V + D RY + +RE EG + V L C C
Sbjct: 751 PPKMREVVHRNFEKSVRFAVHRLD--RYDYEIRE-----EG--ASVYHVKLMERTCSCRA 801
Query: 325 FQALHLPCSHVIAACSSFRHDYTTLIPHVFKNESVYSIYNTAFKVVHDKS--YWLPYDGP 382
F LHLPC H IAA + L+ + ES Y K V + + LP
Sbjct: 802 FDLLHLPCPHAIAAAVAEGVPIQGLMAPEYSVESWRMSYLGTIKPVPEVGDVFALPEPIA 861
Query: 383 VICHNPNMRRLKKGRPNSTRI--RTEMDEEVV--ERTPTP 418
+ P R GRP RI R E V+ E T TP
Sbjct: 862 SLRLFPPATRRPSGRPKKKRITSRGEFTVNVIHLEFTVTP 901
>At2g14030 Mutator-like transposase
Length = 874
Score = 142 bits (357), Expect = 5e-34
Identities = 106/348 (30%), Positives = 152/348 (43%), Gaps = 21/348 (6%)
Query: 27 QFHRLFWAFYPCINGFKYCKPVVHVDGTWLYDKYKGTLLLAVAQDGNNKTIPIAFALVEG 86
+F LF AF I GF K V+ + G L KY G LL A AQD N + P+AF +V+
Sbjct: 506 RFKYLFLAFAASIQGFSCMKRVIVIGGAHLKGKYGGCLLTASAQDANFQVYPLAFGVVDS 565
Query: 87 ETKEGWSFFLKNLRRHVTKGISVCMVSDRHESIKSAFND--PRNGWQATGSAHVYCIRHI 144
+ + W +F + L + G + VSDRH SI + P+ + H CI H+
Sbjct: 566 KNDDAWEWFFRVLSTAIPDGEILTFVSDRHSSIYTGLRKVYPK-------ARHGACIVHL 618
Query: 145 KQNFMRTIKDGDLKDVVNNMGYALNVPLFNYYRGVIQEANQRALDWVDNIPKEKWTQAYD 204
++N + K L V+ A + F Y + +++ + WT+AY
Sbjct: 619 QRNIATSYKKKHLLFHVSRAARAYRICEFQTYFNEGIRLDPACARYLELVGFCHWTRAYF 678
Query: 205 EGRRWGHMTSNIVESWNSVFKGTRNLPVTPIVQSTYYRLACLFADRAQKAFARVGSGDLF 264
G R+ MTSN+VES N V K R LP+ +++ L FA R + A ++ +
Sbjct: 679 LGERYNVMTSNVVESLNPVLKEARELPIFSLLEFIRTTLISWFAMRRKAARSKTSA---L 735
Query: 265 SEYCQNAIQDDIVKSNTHHVEQFDRERYTFSVRETVNYREGRPMGTFKVDLRAGWCDCGK 324
+ + + KS T V + D RY + VR G F V L C C
Sbjct: 736 LPKMREVVHQNFEKSVTFAVHRID--RYDYKVR-------GEGSSVFHVKLMERTCSCRA 786
Query: 325 FQALHLPCSHVIAACSSFRHDYTTLIPHVFKNESVYSIYNTAFKVVHD 372
F LHLPC H IAA + L+ + ES Y K VHD
Sbjct: 787 FDLLHLPCPHAIAAAVAEGVPIQGLMAPEYSIESWRMSYQRTIKPVHD 834
>At5g16505 mutator-like transposase-like protein (MQK4.25)
Length = 597
Score = 137 bits (346), Expect = 9e-33
Identities = 103/421 (24%), Positives = 176/421 (41%), Gaps = 39/421 (9%)
Query: 28 FHRLFWAFYPCINGF-KYCKPVVHVDGTWLYDKYKGTLLLAVAQDGNNKTIPIAFALVEG 86
F RLF A+ CI+GF C+P++ +D L KY G +L A A D ++ P+A A+V+
Sbjct: 198 FQRLFIAYRACISGFFSSCRPLLELDRAHLKGKYLGAILCAAAVDADDGLFPLAIAIVDN 257
Query: 87 ETKEGWSFFLKNLRR----HVTKGISVCMVSDRHESIKSAFNDPRNGWQATGSAHVYCIR 142
E+ E WS+FL LR+ + + ++S+R ++ A + H +C+R
Sbjct: 258 ESDENWSWFLSELRKLLGMNTDSMPKLTILSERQSAVVEAVET-----HFPTAFHGFCLR 312
Query: 143 HIKQNFMRTIKDGDLKDVVNNMGYALNVPLFNYYRGVIQEANQRALDWVDNIPKEKWTQA 202
++ +NF T K+ L ++ + YAL F + E +Q + W + W A
Sbjct: 313 YVSENFRDTFKNTKLVNIFWSAVYALTPAEFETKSNEMIEISQDVVQWFELYLPHLWAVA 372
Query: 203 YDEGRRWGHMTSNIVESWNSVFKGTRNLPVTPIVQSTYYRLACLFADRAQKAFARVGSGD 262
Y +G R+GH I E + LP+ +++ ++++ F +R + + +G
Sbjct: 373 YFQGVRYGHFGLGITEVLYNWALECHELPIIQMMEHIRHQISSWFDNRRELS---MGWNS 429
Query: 263 LFSEYCQNAIQDDIVKSNTHHVEQFDRERYTFSVRETVNYREGRPMGTFKVDLRAGWCDC 322
+ + I + + + + V + + + E N VD+R C C
Sbjct: 430 ILVPSAERRITEAVADARCYQVLRANEVEFEIVSTERTNI----------VDIRTRDCSC 479
Query: 323 GKFQALHLPCSHVIAACSSFRHDYTTLIPHVFKNESVYSIYNTAFKVVHDKSYWLPYDGP 382
++Q LPC+H AA S + F S Y+ + D+S W
Sbjct: 480 RRWQIYGLPCAHAAAALISCGRNVHLFAEPCFTVSSYQQTYSQMIGEIPDRSLWKDEGEE 539
Query: 383 V------ICHNPNMRRLKKGRPNSTRIRTEMDEEVVERTPTPRR---CGLCRHTGHIRRN 433
+ P R GRP +R VE P+R CG C GH ++
Sbjct: 540 AGGGVESLRIRPPKTRRPPGRPKKKVVR-------VENLKRPKRIVQCGRCHLLGHSQKK 592
Query: 434 C 434
C
Sbjct: 593 C 593
>At1g34620 Mutator-like protein
Length = 1034
Score = 133 bits (335), Expect = 2e-31
Identities = 96/348 (27%), Positives = 154/348 (43%), Gaps = 23/348 (6%)
Query: 27 QFHRLFWAFYPCINGFKYCKPVVHVDGTWLYDKYKGTLLLAVAQDGNNKTIPIAFALVEG 86
+F F AF I G+ + + V+ VDGT + +Y G L+ A QDGN + P+AF +V
Sbjct: 542 RFKYQFIAFAASIKGYAFMRKVIVVDGTSMKGRYGGCLISACCQDGNFQIFPLAFGIVNS 601
Query: 87 ETKEGWSFFLKNLRRHVTKGISVCMVSDRHESIKSAFNDPRNGWQATGSAHVYCIRHIKQ 146
E + +F + L + +SDRHESI + + T + H C H+ +
Sbjct: 602 ENDSAYEWFFQRLSIIAPDNPDLMFISDRHESIYTGLSK-----VYTQANHAACTVHLWR 656
Query: 147 NFMRTIKDGDLKDVVNNMGYALNVPLFNYYRGVIQEANQRALDWVDNIPKEKWTQAYDEG 206
N K L +++ A +V FN IQ+ N ++ ++ +WT+ + +G
Sbjct: 657 NIRHLYKPKSLCRLMSEAAQAFHVTDFNRIFLKIQKLNPGCAAYLVDLGFSEWTRVHSKG 716
Query: 207 RRWGHMTSNIVESWNSVFKGTRNLPVTPIVQSTYYRLACLFADRAQKAFARVGSGDLFSE 266
RR+ M SNI ESWN+V + R P+ +++ L FA R +A E
Sbjct: 717 RRFNIMDSNICESWNNVIREAREYPLICMLEYIRTTLMDWFATRRAQA-----------E 765
Query: 267 YCQNAIQDDIVKSNTHHVEQFDRERYTFSVRETVNYR---EGRPMGTFKVDLRAGWCDCG 323
C + + VE+ + + SV+ N+ + R F V L C C
Sbjct: 766 DCPTTLAPRV----QERVEENYQSAMSMSVKPICNFEFQVQERTGECFIVKLDESTCSCL 821
Query: 324 KFQALHLPCSHVIAACSSFRHDYTTLIPHVFKNESVYSIYNTAFKVVH 371
+FQ L +PC+H IAA + +L + + NE V Y +H
Sbjct: 822 EFQGLGIPCAHAIAAAARIGVPTDSLAANGYFNELVKLSYEEKIYPIH 869
>At2g30640 Mutator-like transposase
Length = 754
Score = 133 bits (334), Expect = 2e-31
Identities = 110/422 (26%), Positives = 169/422 (39%), Gaps = 39/422 (9%)
Query: 28 FHRLFWAFYPCINGF-KYCKPVVHVDGTWLYDKYKGTLLLAVAQDGNNKTIPIAFALVEG 86
F + F +F I GF C+P++ +D T L KY GTLLLA DG P+AFA++
Sbjct: 353 FQQFFISFQASICGFLNACRPLIGLDRTVLKSKYVGTLLLATGFDGEGAVFPLAFAIISE 412
Query: 87 ETKEGWSFFLKNLRRHVTKGI----SVCMVSDRHESIKSAFNDPRNGWQATGSAHVYCIR 142
E W +FL LR+ + + ++S R +SI + + H C+
Sbjct: 413 ENDSSWQWFLSELRQLLEVNSENMPKLTILSSRDQSIVDGVDT-----NFPTAFHGLCVH 467
Query: 143 HIKQNFMRTIKDGDLKDVVNNMGYALNVPLFNYYRGVIQEANQRALDWVDNIPKEKWTQA 202
+ ++ + L ++V L F G I + + A W+ NI +W
Sbjct: 468 CLTESVRTQFNNSILVNLVWEAAKCLTDFEFEGKMGEIAQISPEAASWIRNIQHSQWATY 527
Query: 203 YDEGRRWGHMTSNIVESWNSVFKGTRNLPVTPIVQSTYYRLACLFADRAQKAFARVGSGD 262
EG R+GH+T+N+ ES NS + LP+ +++S +L LF +R + + G
Sbjct: 528 CFEGTRFGHLTANVSESLNSWVQDASGLPIIQMLESIRRQLMTLFNERRETSMQWSGM-- 585
Query: 263 LFSEYCQNAIQDDIVKSNTHHVEQFDRERYTFSVRET--VNYREGRPMGTFKVDLRAGWC 320
+V S HV + E + V + + G + VD+R C
Sbjct: 586 -------------LVPSAERHVLEAIEECRLYPVHKANEAQFEVMTSEGKWIVDIRCRTC 632
Query: 321 DCGKFQALHLPCSHVIAACSSFRHDYTTLIPHVFKNESVYSIYNTAFKVVHDKSYWLP-- 378
C ++ LPCSH +AA + R + F + Y V DK+ W
Sbjct: 633 YCRGWELYGLPCSHAVAALLACRQNVYRFTESYFTVANYRRTYAETIHPVPDKTEWKTTE 692
Query: 379 ------YDGPVICHNPNMRRLKKGRPNSTRIRTEMDEEVVERTPTPRRCGLCRHTGHIRR 432
DG I P + RP R + E D +R RC C GH R
Sbjct: 693 PAGESGEDGEEIVIRPPRDLRQPPRPKKRRSQGE-DRGRQKRVV---RCSRCNQAGHFRT 748
Query: 433 NC 434
C
Sbjct: 749 TC 750
>At3g05850 unknown protein
Length = 777
Score = 132 bits (333), Expect = 3e-31
Identities = 112/426 (26%), Positives = 189/426 (44%), Gaps = 46/426 (10%)
Query: 26 AQFHRLFWAFYPCINGF-KYCKPVVHVDGTWLYDKYKGTLLLAVAQDGNNKTIPIAFALV 84
+ FHR+F +F+ ++GF + C+P+V +D L KY+GTLL A + DG+++ P+AFA+V
Sbjct: 374 SSFHRVFVSFHASVHGFLEACRPLVFLDSMQLKSKYQGTLLAATSVDGDDEVFPLAFAVV 433
Query: 85 EGETKEGWSFFLKNLRRHVTKGISVCMVSDR----HESIKSAFNDPRNGWQATGSAHVYC 140
+ ET + W +FL LR ++ + V+DR ESI F S H YC
Sbjct: 434 DAETDDNWEWFLLQLRSLLSTPCYITFVADRQKNLQESIPKVFEK---------SFHAYC 484
Query: 141 IRHIKQNFMRTIK---DGDLK----DVVNNMGYALNVPLFNYYRGVIQEANQRALDW-VD 192
+R++ ++ +K ++K D + YA F + I+ + A DW V
Sbjct: 485 LRYLTDELIKDLKGPFSHEIKRLIVDDFYSAAYAPRADSFERHVENIKGLSPEAYDWIVQ 544
Query: 193 NIPKEKWTQAYDEGRRWGHMTSNIVESWNSVFKGTRNLPVTPIVQSTYYRLACLFADRAQ 252
+ W AY G R+ HMTS+ E + S +LP+T +V ++ L +
Sbjct: 545 KSQPDHWANAYFRGARYNHMTSHSGEPFFSWASDANDLPITQMVDVIRGKIMGLI--HVR 602
Query: 253 KAFARVGSGDLFSEYCQNAIQDDIVKSNTHHVEQFDRERYTFSVR-ETVNYREGRPMGTF 311
+ A +G+L + + ++ + +++ T HV + F VR ET
Sbjct: 603 RISANEANGNL-TPSMEVKLEKESLRAQTVHVAP-SADNNLFQVRGETYEL--------- 651
Query: 312 KVDLRAGWCDCGKFQALHLPCSHVIAACSSF---RHDYTTLIPHVFKNESVYSIYNTAFK 368
V++ C C +Q LPC H +A + + +DY + V S Y+
Sbjct: 652 -VNMAECDCSCKGWQLTGLPCHHAVAVINYYGRNPYDYCSKYFTVAYYRSTYAQSINPVP 710
Query: 369 VVHDKSYWLPYDGPVICHNPNMRRLKKGRPNSTRIRTEMDEEVVERTPTPRRCGLCRHTG 428
++ + G + P R GRP + EEV++R +C C+ G
Sbjct: 711 LLEGEMCRESSGGSAVTVTPPPTRRPPGRPPKKKTPA---EEVMKR---QLQCSRCKGLG 764
Query: 429 HIRRNC 434
H + C
Sbjct: 765 HNKSTC 770
>At2g04950 putative transposase of transposon FARE2.9 (cds1)
Length = 616
Score = 130 bits (327), Expect = 1e-30
Identities = 107/414 (25%), Positives = 166/414 (39%), Gaps = 28/414 (6%)
Query: 27 QFHRLFWAFYPCINGFKYCKPVVHVDGTWLYDKYKGTLLLAVAQDGNNKTIPIAFALVEG 86
+F F AF I G+K K V+ +DG L K+KGTLL A AQDGN PIAFA+V+
Sbjct: 221 KFKYAFLAFGASIRGYKLMKKVISIDGAHLISKFKGTLLGASAQDGNFNLYPIAFAIVDS 280
Query: 87 ETKEGWSFFLKNLRRHVTKGISVCMVSDRHESIKSAFND----PRNGWQATGSAHVYCIR 142
E W +FLK L + + VSDR SI S + NG C
Sbjct: 281 ENDASWDWFLKCLLNIIPDENDLVFVSDRAASIASGLSGNYPLAHNG---------LCTF 331
Query: 143 HIKQNFMRTIKDGDLKDVVNNMGYALNVPLFNYYRGVIQEANQRALDWVDNIPKEKWTQA 202
H+++N + L V F+ I ++++ ++ + KW++A
Sbjct: 332 HLQKNLETHFRGSSLIPVYYAASRVYTKTEFDSLFWEITNSDKKLAQYLWEVDVRKWSRA 391
Query: 203 YDEGRRWGHMTSNIVESWNSVFKGTRNLPVTPIVQSTYYRLACLFADRAQKAFARVGSGD 262
Y R+ MTSN+ ES N++ K R P+ + +S + F +R +++ +
Sbjct: 392 YSPSNRYNIMTSNLAESVNALLKQNREYPIVCLFESIRSIMTRWFNERREESSQHPSAVT 451
Query: 263 LFSEYCQNAIQDDIVKSNTHHVEQFDRERYTFSVRETVNYREGRPMGTFKVDLRAGWCDC 322
+ A D ++T +E + F V+ T V+L C C
Sbjct: 452 INVGKKMKASYD----TSTRWLEVCQVNQEEFEVKGDTK--------THLVNLDKRTCTC 499
Query: 323 GKFQALHLPCSHVIAACSSFRHDYTTLIPHVFKNESVYSIYNTAFKVVHDKSYW-LPYD- 380
F PC+H IA+ + + Y+ + D YW +P
Sbjct: 500 CMFDIDKFPCAHGIASAKHINLNENMFVDEFHSTYKWRQAYSKSIHPNGDMEYWEIPETI 559
Query: 381 GPVICHNPNMRRLKKGRPNSTRIRTEMDEEVVERTPTPRRCGLCRHTGHIRRNC 434
VIC P+ R+ GR RI + + + P +C C +GH C
Sbjct: 560 SEVICLPPS-TRVPSGRRKKKRIPSVWEHGRSQPKPKLHKCSRCGQSGHNNSTC 612
>At3g47280 putative protein
Length = 739
Score = 130 bits (326), Expect = 2e-30
Identities = 106/410 (25%), Positives = 167/410 (39%), Gaps = 20/410 (4%)
Query: 27 QFHRLFWAFYPCINGFKYCKPVVHVDGTWLYDKYKGTLLLAVAQDGNNKTIPIAFALVEG 86
+F F AF I G+K + V+ +DG L K+KGTLL A AQDGN PIAFA+V+
Sbjct: 344 KFKYAFLAFGASIRGYKLMRKVISIDGAHLTSKFKGTLLGASAQDGNFNLYPIAFAIVDS 403
Query: 87 ETKEGWSFFLKNLRRHVTKGISVCMVSDRHESIKSAFNDPRNGWQATGSAHVYCIRHIKQ 146
E W +FLK L + + VSDR SI S R + H C H+++
Sbjct: 404 ENDASWDWFLKCLLNIIPDENDLVFVSDRAASIAS-----RLSGNYPLAHHGLCTFHLQK 458
Query: 147 NFMRTIKDGDLKDVVNNMGYALNVPLFNYYRGVIQEANQRALDWVDNIPKEKWTQAYDEG 206
N + L V F+ I ++++ ++ + KW++AY
Sbjct: 459 NLETHFRGSSLIPVYYAASRIYTKTEFDSLFWEITNSDKKLAQYLCEVDVRKWSRAYSPS 518
Query: 207 RRWGHMTSNIVESWNSVFKGTRNLPVTPIVQSTYYRLACLFADRAQKAFARVGSGDLFSE 266
R+ MTSN+ ES N++ K R PV + +S + F +R +++ + +
Sbjct: 519 NRYNIMTSNLAESVNALLKQNREYPVVCLFESIRSIMTRWFNERREESSQHPSAVTINVG 578
Query: 267 YCQNAIQDDIVKSNTHHVEQFDRERYTFSVRETVNYREGRPMGTFKVDLRAGWCDCGKFQ 326
A D ++T +E + F V+ T V+L C C F
Sbjct: 579 KKMKASYD----TSTRWLEVCQVNQEEFEVKGDTK--------THLVNLDKRTCTCCMFD 626
Query: 327 ALHLPCSHVIAACSSFRHDYTTLIPHVFKNESVYSIYNTAFKVVHDKSYW-LPYD-GPVI 384
PC+H IA+ + + Y+ + D YW +P VI
Sbjct: 627 IDKFPCAHGIASAKHINLNENMFVDEFHSTYRWRQAYSESIHPNGDMEYWEIPETISEVI 686
Query: 385 CHNPNMRRLKKGRPNSTRIRTEMDEEVVERTPTPRRCGLCRHTGHIRRNC 434
C P+ R+ G+ RI + + + P +C C +GH + C
Sbjct: 687 CLPPS-TRVPTGKRKKKRIPSVWEHGRSQPKPKLHKCSRCGQSGHNKSTC 735
>At1g35760 hypothetical protein
Length = 937
Score = 126 bits (316), Expect = 3e-29
Identities = 74/228 (32%), Positives = 116/228 (50%), Gaps = 7/228 (3%)
Query: 27 QFHRLFWAFYPCINGFKYCKPVVHVDGTWLYDKYKGTLLLAVAQDGNNKTIPIAFALVEG 86
+F LF+A C+ G+ Y + V+ +DGT L +Y G L+ A AQD N + PIAF +V
Sbjct: 543 RFKYLFFALDACVQGYAYMRKVIVIDGTHLRGRYGGCLVAASAQDANFQVFPIAFGIVNS 602
Query: 87 ETKEGWSFFLKNLRRHVTKGISVCMVSDRHESIKSAFNDPRNGWQATGSAHVYCIRHIKQ 146
+ E W++F+ L + + SDRH SI A P S+H C+ H+K+
Sbjct: 603 KNDEAWTWFMTKLTEALPDDPELVFASDRHNSITPASVYPM-------SSHAACVVHLKR 655
Query: 147 NFMRTIKDGDLKDVVNNMGYALNVPLFNYYRGVIQEANQRALDWVDNIPKEKWTQAYDEG 206
N K + +V++ A + FN ++ + D+++ I E WT+++ G
Sbjct: 656 NIEAYFKYEQVGSLVSSAARAYRLTDFNRIFVEVRAKHGPCADYLEGIGFEHWTRSHFVG 715
Query: 207 RRWGHMTSNIVESWNSVFKGTRNLPVTPIVQSTYYRLACLFADRAQKA 254
+ MTSNIVES N++ R+ P+ I++S L LFA R Q A
Sbjct: 716 NMYNVMTSNIVESLNNMLTMARDYPIISILESIRTTLVTLFALRRQAA 763
>At2g06450 putative transposase of FARE2.11
Length = 580
Score = 124 bits (310), Expect = 1e-28
Identities = 104/422 (24%), Positives = 165/422 (38%), Gaps = 27/422 (6%)
Query: 22 REWRAQFHRLFWA-------FYPCINGFKYCKPVVHVDGTWLYDKYKGTLLLAVAQDGNN 74
+ WR Q H A F G+K + V+ +DG L K+KGTLL A AQDGN
Sbjct: 173 KAWRVQEHAAELARGLPDDSFEVLPRGYKLMRKVISIDGAHLTSKFKGTLLGASAQDGNF 232
Query: 75 KTIPIAFALVEGETKEGWSFFLKNLRRHVTKGISVCMVSDRHESIKSAFNDPRNGWQATG 134
PIAFA+V+ E W +FLK L + + VSDR I S +
Sbjct: 233 NLYPIAFAIVDSENDASWDWFLKCLLNIIPDENDLVFVSDRAAFIASGLSG-----NYPL 287
Query: 135 SAHVYCIRHIKQNFMRTIKDGDLKDVVNNMGYALNVPLFNYYRGVIQEANQRALDWVDNI 194
+ H C H+++N + L V F+ I ++++ ++ +
Sbjct: 288 AHHGLCTFHLQKNLETHFRGSSLIPVYYAASRVYTKTEFDSLFWEITNSDKKLAQYLWEV 347
Query: 195 PKEKWTQAYDEGRRWGHMTSNIVESWNSVFKGTRNLPVTPIVQSTYYRLACLFADRAQKA 254
KW++ Y R+ MTSN+ ES N++ K R P+ + +S + F +R +++
Sbjct: 348 DVRKWSRTYSPSNRYNIMTSNLAESVNALLKQNREYPIVCLFESIRSIMTRWFNERREES 407
Query: 255 FARVGSGDLFSEYCQNAIQDDIVKSNTHHVEQFDRERYTFSVRETVNYREGRPMGTFKVD 314
+ + A D ++T +E + F V+ T V+
Sbjct: 408 SQHPSAVTINVGKKMKASYD----TSTRWLEVCQVNQEEFEVKGDTK--------THLVN 455
Query: 315 LRAGWCDCGKFQALHLPCSHVIAACSSFRHDYTTLIPHVFKNESVYSIYNTAFKVVHDKS 374
L C C F PC+H IA+ + + Y+ + D
Sbjct: 456 LDKRTCTCCMFDIDKFPCAHGIASAKHINLNENMFVDEFHSTYRWRQAYSESIHPNGDME 515
Query: 375 YW--LPYDGPVICHNPNMRRLKKGRPNSTRIRTEMDEEVVERTPTPRRCGLCRHTGHIRR 432
YW L VIC P+ R+ GR RI + + + P +C C +GH +
Sbjct: 516 YWEILETISEVICLPPS-TRVPSGRREKKRIPSVWEHGRSQPKPKLHKCSRCGRSGHNKS 574
Query: 433 NC 434
C
Sbjct: 575 TC 576
>At1g25784 mutator-like transposase, putative
Length = 1127
Score = 123 bits (308), Expect = 2e-28
Identities = 105/388 (27%), Positives = 171/388 (44%), Gaps = 34/388 (8%)
Query: 25 RAQFHRLFWAFYPCINGFKYCKPVVHVDGTWLYDKYKGTLLLAVAQDGNNKTIPIAFALV 84
+ +F +F + I G Y + VV VDGT L YKG LL+A AQDGN + PIAF +V
Sbjct: 552 KQRFKYVFVSLGASIKGLIYMRKVVVVDGTQLVGPYKGCLLIACAQDGNFQIFPIAFGVV 611
Query: 85 EGETKEGWSFFLKNLRRHVTKGISVCMVSDRHESIKSAFN--DPRNGWQATGSAHVYCIR 142
+GET W++F + L V + +VSDRH SI + PR + H C
Sbjct: 612 DGETDASWAWFFEKLAEIVPDSDDLMIVSDRHSSIYKGLSVVYPR-------AHHGACAV 664
Query: 143 HIKQNFMRTIKDGDLKDVVNNMGYALNVPLFNYYRGVIQEANQRALDWVDNIPKEKWTQA 202
H+++N + + + A V F Y +++E + + ++++I E WT+A
Sbjct: 665 HLERNLSTYYGKFGVSALFFSAAKAYRVRDFEKYFELLREKSAKCAKYLEDIGFEHWTRA 724
Query: 203 YDEGRRWGHMTSNIVESWNSVFKGTRNLPVTPIVQSTYYRLACLFADRAQKAFARVGSGD 262
+ G R+ M+SN ES N V + P+ +++ L FA R +K AR
Sbjct: 725 HCRGERYNIMSSNNSESMNHVLTKAKTYPIVYMIEFIRDVLMRWFASRRKKV-AR----- 778
Query: 263 LFSEYCQNAIQDDIVKSNTHHVEQFDRE-----RYTFSVRETVNYREGRPMGT-FKVDLR 316
C++++ ++ E+F +E +Y + +Y+ G F V L
Sbjct: 779 -----CKSSVTPEV-------DERFLQELPASGKYAVKMSGPWSYQVTSKSGEHFHVVLD 826
Query: 317 AGWCDCGKFQALHLPCSHVIAACSSFRHDYTTLIPHVFKNESVYSIYNTAFKVVHD-KSY 375
C C ++ L +PC H +AA D + + + ++ + + D K
Sbjct: 827 QCTCTCLRYTKLRIPCEHALAAAIEHGIDPKSCVGWWYGLQTFSDSFQEPILPIADPKDV 886
Query: 376 WLPYDGPVICHNPNMRRLKKGRPNSTRI 403
+P + P R GRP S RI
Sbjct: 887 VIPQHIVDLILIPPYSRRPPGRPLSKRI 914
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.323 0.138 0.452
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 10,941,124
Number of Sequences: 26719
Number of extensions: 483806
Number of successful extensions: 1223
Number of sequences better than 10.0: 93
Number of HSP's better than 10.0 without gapping: 85
Number of HSP's successfully gapped in prelim test: 8
Number of HSP's that attempted gapping in prelim test: 992
Number of HSP's gapped (non-prelim): 131
length of query: 440
length of database: 11,318,596
effective HSP length: 102
effective length of query: 338
effective length of database: 8,593,258
effective search space: 2904521204
effective search space used: 2904521204
T: 11
A: 40
X1: 16 ( 7.5 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (22.0 bits)
S2: 62 (28.5 bits)
Medicago: description of AC126012.8


