

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC126012.10 + phase: 0 /pseudo
(498 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
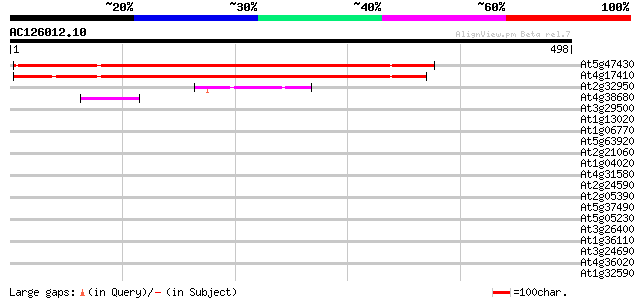
Score E
Sequences producing significant alignments: (bits) Value
At5g47430 DNA-binding protein-like 427 e-120
At4g17410 hypothetical protein 413 e-115
At2g32950 photomorphogenesis repressor (COP1) 54 2e-07
At4g38680 glycine-rich protein 2 (GRP2) 42 6e-04
At3g29500 hypothetical protein 41 0.001
At1g13020 eukaryotic initiation factor 4B (EIF4B5) 38 0.011
At1g06770 unknown protein 38 0.015
At5g63920 DNA topoisomerase III 37 0.019
At2g21060 glycine-rich protein (AtGRP2) 37 0.033
At1g04020 unknown protein 37 0.033
At4g31580 splicing factor 9G8-like SR protein / SRZ-22 36 0.043
At2g24590 RSZp22 splicing factor like protein 36 0.056
At2g05390 putative retroelement pol polyprotein 36 0.056
At5g37490 putative protein 35 0.073
At5g05230 unknown protein 35 0.073
At3g26400 eukaryotic initiation factor 4B (EIF4B7) 35 0.073
At1g36110 hypothetical protein 35 0.073
At3g24690 unknown protein 35 0.096
At4g36020 glycine-rich protein 35 0.13
At1g32590 hypothetical protein, 5' partial 35 0.13
>At5g47430 DNA-binding protein-like
Length = 889
Score = 427 bits (1098), Expect = e-120
Identities = 220/377 (58%), Positives = 271/377 (71%), Gaps = 7/377 (1%)
Query: 4 PENLDWDEFGNDLYSIPDQLPVQSSTMIPD-APLTSKADEDSKIKALIDTPALDWQQQGS 62
PE+ ++DEFG DLYSIPD Q P A K DE+SKI+ALIDTPALDWQQQG
Sbjct: 113 PED-EYDEFGTDLYSIPDTQDAQHIIPRPHLATADDKVDEESKIQALIDTPALDWQQQGQ 171
Query: 63 D-FGAGRGFRRGAVGGRIGGGRGFGLERKTPPEGYVCHRCKVSGHFIQHCPTNGDSTFDI 121
D FGAGRG+ RG G GRGFG+ERKTPP GYVCHRC + GHFIQHCPTNGD +D+
Sbjct: 172 DTFGAGRGYGRGMPGRM--NGRGFGMERKTPPPGYVCHRCNIPGHFIQHCPTNGDPNYDV 229
Query: 122 KKVRQPTGIPRSMLMVNPQGSYALQNGSVAVLKPNEAAFDKEMEGL-SSTRSVGDLPPEL 180
K+V+ PTGIP+SMLM P GSY+L +G+VAVLKPNE AF+KEMEGL S+TRSVG+LPPEL
Sbjct: 230 KRVKPPTGIPKSMLMATPDGSYSLPSGAVAVLKPNEDAFEKEMEGLPSTTRSVGELPPEL 289
Query: 181 HCPLCNNVMKDAVLTSKCCFKSFCDKCIRNYIMSKSACVCLATNILADDLLPNKTLRDAI 240
CPLC VMKDA LTSKCC+KSFCDKCIR++I+SKS CVC +++LADDLLPNKTLRD I
Sbjct: 290 KCPLCKEVMKDAALTSKCCYKSFCDKCIRDHIISKSMCVCGRSDVLADDLLPNKTLRDTI 349
Query: 241 HRILESGNSSTENAGSTYQVQDMLSSRCPQPKIPSPTSSATSKGEPKVSQVNEGMANIQE 300
+RILE+GN STEN GS + D+ S+RCP PK SPT+S SKGE K N A+ +
Sbjct: 350 NRILEAGNDSTENVGSVGHIPDLESARCPPPKALSPTTSVASKGEKKPVLSNNNDASTLK 409
Query: 301 IVAERKEVSATQQVSEQVKIPRAAVVSEVTHESMSVKEPEPASQGSAKLVEEEVQQKLVP 360
E E+++ + S +V + + E T S+ VKE S+ + + +EE+QQ++
Sbjct: 410 APMEVAEITSAPRASAEVNVEKPVDACESTQGSVIVKE-ATVSKLNTQAPKEEMQQQVAA 468
Query: 361 TDGGKKKKKKKVRMPAN 377
+ KKKKKK R+P N
Sbjct: 469 GEPAGKKKKKKPRVPGN 485
Score = 32.0 bits (71), Expect = 0.81
Identities = 21/47 (44%), Positives = 29/47 (61%), Gaps = 9/47 (19%)
Query: 459 NVPPPVMN-------MNREPVMNKNREDFEARNA-ILKKQENERRVE 497
N+PPP + MN + M R++ EARNA +L+K+ENERR E
Sbjct: 586 NIPPPHRDLAEMGNRMNLQRAM-MGRDEAEARNAEMLRKRENERRPE 631
>At4g17410 hypothetical protein
Length = 744
Score = 413 bits (1062), Expect = e-115
Identities = 215/369 (58%), Positives = 265/369 (71%), Gaps = 7/369 (1%)
Query: 4 PENLDWDEFGNDLYSIPDQLPVQSSTMIPDAPLTSKADEDSKIKALIDTPALDWQQQGSD 63
P ++DEFGNDLYSIPD V S+ + D+ DE++K+KALIDTPALDW QQG+D
Sbjct: 30 PVEDEFDEFGNDLYSIPDAPAVHSNNLCHDSAPAD--DEETKLKALIDTPALDWHQQGAD 87
Query: 64 -FGAGRGFRRGAVGGRIGGGRGFGLERKTPPEGYVCHRCKVSGHFIQHCPTNGDSTFDIK 122
FG GRG+ RG G GGRGFG+ER TPP GYVCHRC VSGHFIQHC TNG+ FD+K
Sbjct: 88 SFGPGRGYGRGMAGRM--GGRGFGMERTTPPPGYVCHRCNVSGHFIQHCSTNGNPNFDVK 145
Query: 123 KVRQPTGIPRSMLMVNPQGSYALQNGSVAVLKPNEAAFDKEMEGLSST-RSVGDLPPELH 181
+V+ PTGIP+SMLM P GSY+L +G+VAVLKPNE AF+KEMEGL+ST RSVG+ PPEL
Sbjct: 146 RVKPPTGIPKSMLMATPNGSYSLPSGAVAVLKPNEDAFEKEMEGLTSTTRSVGEFPPELK 205
Query: 182 CPLCNNVMKDAVLTSKCCFKSFCDKCIRNYIMSKSACVCLATNILADDLLPNKTLRDAIH 241
CPLC VM+DA L SKCC KS+CDKCIR++I++KS CVC AT++LADDLLPNKTLRD I+
Sbjct: 206 CPLCKEVMRDAALASKCCLKSYCDKCIRDHIIAKSMCVCGATHVLADDLLPNKTLRDTIN 265
Query: 242 RILESGNSSTENAGSTYQVQDMLSSRCPQPKIPSPTSSATSKGEPKVSQVNEGMANIQEI 301
RILESGNSS ENAGS QVQDM S RCP PK SPT+SA S GE K + N + +
Sbjct: 266 RILESGNSSAENAGSMCQVQDMESVRCPPPKALSPTTSAASGGEKKPAPSNNNETSTLKP 325
Query: 302 VAERKEVSATQQVSEQVKIPRAAVVSEVTHESMSVKEPEPASQGSAKLVEEEVQQKLVPT 361
E E+++ +E VK+ + S S + KE SQ + + +EE+ Q++
Sbjct: 326 SIEIAEITSAWASAEIVKVEKPVDASANIQGSSNGKE-AAVSQLNTQPPKEEMPQQVASG 384
Query: 362 DGGKKKKKK 370
+ GK+KKKK
Sbjct: 385 EQGKRKKKK 393
Score = 30.0 bits (66), Expect = 3.1
Identities = 19/49 (38%), Positives = 26/49 (52%), Gaps = 11/49 (22%)
Query: 448 FRDFAEFNIGMNVPPPVMNMNREPVMNKNREDFEARNAILK-KQENERR 495
+RD AE MN+ P+M RE+FEA+ +K K+ENE R
Sbjct: 489 YRDLAEMGNRMNLQHPIM----------GREEFEAKKTEMKRKRENEIR 527
>At2g32950 photomorphogenesis repressor (COP1)
Length = 675
Score = 53.9 bits (128), Expect = 2e-07
Identities = 33/106 (31%), Positives = 53/106 (49%), Gaps = 6/106 (5%)
Query: 165 EGLSSTRSVG--DLPPELHCPLCNNVMKDAVLTSKCCFKSFCDKCIRNYIMSKSACVCLA 222
+G S +G DL +L CP+C ++KDA LT+ C SFC CI ++ +KS C C +
Sbjct: 33 DGGSGGSEIGAPDLDKDLLCPICMQIIKDAFLTA--CGHSFCYMCIITHLRNKSDCPCCS 90
Query: 223 TNILADDLLPNKTLRDAIHRILESGNSSTENAGSTYQVQDMLSSRC 268
++ + L PN L + + S ++ A Q ++ L C
Sbjct: 91 QHLTNNQLYPNFLLDKLLKK--TSARHVSKTASPLDQFREALQRGC 134
>At4g38680 glycine-rich protein 2 (GRP2)
Length = 203
Score = 42.4 bits (98), Expect = 6e-04
Identities = 21/52 (40%), Positives = 26/52 (49%)
Query: 64 FGAGRGFRRGAVGGRIGGGRGFGLERKTPPEGYVCHRCKVSGHFIQHCPTNG 115
FG GRG RG+ GG GGG G+G G C++C GH + C G
Sbjct: 97 FGGGRGGGRGSGGGYGGGGGGYGGRGGGGRGGSDCYKCGEPGHMARDCSEGG 148
Score = 38.5 bits (88), Expect = 0.009
Identities = 22/55 (40%), Positives = 27/55 (49%), Gaps = 7/55 (12%)
Query: 61 GSDFGAGRGFRRGAVGGRIGGGRGFGLERKTPPEGYVCHRCKVSGHFIQHCPTNG 115
G +G G G+ G GG GGGRG G G C+ C SGHF + C + G
Sbjct: 155 GGGYGGGGGYGGGG-GGYGGGGRGGG------GGGGSCYSCGESGHFARDCTSGG 202
>At3g29500 hypothetical protein
Length = 341
Score = 41.2 bits (95), Expect = 0.001
Identities = 36/155 (23%), Positives = 66/155 (42%), Gaps = 21/155 (13%)
Query: 11 EFGNDLYSIPDQLPVQSSTMIPDAPLTSKADEDSKIKALIDTPALDWQQQGSDFGAGRGF 70
E N+L + L ST +P+A A ++ +A T ++ +G+ G GRG
Sbjct: 176 EKNNELLLMNSALRPPGSTAVPEANRAEMAKAPNEPQA---TKESNYVHRGNPHGRGRGR 232
Query: 71 RRGA------------VGGR------IGGGRGFGLERKTPPEGYVCHRCKVSGHFIQHCP 112
RG GGR G GRG G+ + VC+RC + H+ + C
Sbjct: 233 GRGGRGRGNFYGQGNHYGGRGRGNYGRGRGRGRGVNKPRGKAKSVCYRCGMDDHWAKTCR 292
Query: 113 TNGDSTFDIKKVRQPTGIPRSMLMVNPQGSYALQN 147
T+ +++ + G +++ ++ +G + +N
Sbjct: 293 TSKHLIEAYQEMIKQKGPEANLVHLDGEGDFDHEN 327
>At1g13020 eukaryotic initiation factor 4B (EIF4B5)
Length = 549
Score = 38.1 bits (87), Expect = 0.011
Identities = 25/63 (39%), Positives = 32/63 (50%), Gaps = 3/63 (4%)
Query: 27 SSTMIPDAPLTSKADEDSKI-KALIDTPALDWQQQGSDFGAGRGFRRGAVGGRIG--GGR 83
S++ + D P S+ADED K P+ D +QGS +G G G G GG G GG
Sbjct: 147 SNSRVSDFPQVSRADEDDDWGKGKKSLPSFDQGRQGSRYGGGGGSFGGGGGGGAGSYGGG 206
Query: 84 GFG 86
G G
Sbjct: 207 GAG 209
>At1g06770 unknown protein
Length = 421
Score = 37.7 bits (86), Expect = 0.015
Identities = 21/64 (32%), Positives = 31/64 (47%), Gaps = 6/64 (9%)
Query: 180 LHCPLCNNVMKDAVLTSKCCFKSFCDKCIRNYIMSKSACVCLATNI-----LADDLLPNK 234
L C +C+N+++DA S+ C +FC KCI I C NI + L P+
Sbjct: 14 LSCSICDNILRDATTISE-CLHTFCRKCIYEKITEDEIETCPVCNIDLGSTPLEKLRPDH 72
Query: 235 TLRD 238
L+D
Sbjct: 73 NLQD 76
>At5g63920 DNA topoisomerase III
Length = 926
Score = 37.4 bits (85), Expect = 0.019
Identities = 21/54 (38%), Positives = 23/54 (41%), Gaps = 1/54 (1%)
Query: 65 GAGRGFRRG-AVGGRIGGGRGFGLERKTPPEGYVCHRCKVSGHFIQHCPTNGDS 117
G GRG R G + GGR G G F P G C C HF CP +S
Sbjct: 868 GRGRGGRGGQSSGGRRGSGTSFVSATGEPVSGIRCFSCGDPSHFANACPNRNNS 921
>At2g21060 glycine-rich protein (AtGRP2)
Length = 201
Score = 36.6 bits (83), Expect = 0.033
Identities = 23/58 (39%), Positives = 27/58 (45%), Gaps = 11/58 (18%)
Query: 58 QQQGSDFGAGRGFRRGAVGGRIGGGRGFGLERKTPPEGYVCHRCKVSGHFIQHCPTNG 115
Q G G G G R G+ GG GGG G GL C+ C SGHF + C + G
Sbjct: 153 QGGGGYSGGGGGGRYGSGGG--GGGGGGGLS---------CYSCGESGHFARDCTSGG 199
Score = 32.0 bits (71), Expect = 0.81
Identities = 19/55 (34%), Positives = 22/55 (39%), Gaps = 6/55 (10%)
Query: 61 GSDFGAGRGFRRGAVGGRIGGGRGFGLERKTPPEGYVCHRCKVSGHFIQHCPTNG 115
G G G G G GGR GGRG G + C +C GH + C G
Sbjct: 107 GRGGGRGGGSYGGGYGGRGSGGRGGGGGDNS------CFKCGEPGHMARECSQGG 155
>At1g04020 unknown protein
Length = 714
Score = 36.6 bits (83), Expect = 0.033
Identities = 34/119 (28%), Positives = 50/119 (41%), Gaps = 15/119 (12%)
Query: 179 ELHCPLCNNVMKDAVLTSKCCFKSFCDKCIRNYIMSKSAC-VCLATNILADDLLPNKTLR 237
EL CPLC ++ VL C FCD C+ +S C VC + + P K R
Sbjct: 22 ELKCPLCLKLLNRPVLLP--CDHVFCDSCVHKSSQVESGCPVCKSKH-------PKKARR 72
Query: 238 DAIHRILESGNS---STENAGSTYQVQDMLSSRCPQPKIPSPTSSATSKGEPKVSQVNE 293
D R +ES S S A S + Q + + C S++ GE + S++ +
Sbjct: 73 DL--RFMESVISIYKSLNAAVSVHLPQLQIPNDCNYKNDALNNSNSPKHGESEDSEMTD 129
>At4g31580 splicing factor 9G8-like SR protein / SRZ-22
Length = 200
Score = 36.2 bits (82), Expect = 0.043
Identities = 23/69 (33%), Positives = 29/69 (41%), Gaps = 9/69 (13%)
Query: 60 QGSDFGAGRGFRRGAVGGRIGGGRGFGLERKTPPEGYVCHRCKVSGHFIQHCPTNGDSTF 119
+G G GRG RG GG GG G L+ C+ C +GHF + C G +
Sbjct: 72 RGERGGGGRGGDRGGGGGGRGGRGGSDLK---------CYECGETGHFARECRNRGGTGR 122
Query: 120 DIKKVRQPT 128
K R T
Sbjct: 123 RRSKSRSRT 131
>At2g24590 RSZp22 splicing factor like protein
Length = 196
Score = 35.8 bits (81), Expect = 0.056
Identities = 24/62 (38%), Positives = 30/62 (47%), Gaps = 9/62 (14%)
Query: 57 WQQQGSDFGAGRGFRRGAVGGRIGG-GRGFGLERKTPPEGYVCHRCKVSGHFIQHCPTNG 115
W+ + S G G R G GG GG GRG G + K C+ C SGHF + C + G
Sbjct: 64 WRVEQSHNRGGGGGRGGGRGGGDGGRGRG-GSDLK-------CYECGESGHFARECRSRG 115
Query: 116 DS 117
S
Sbjct: 116 GS 117
>At2g05390 putative retroelement pol polyprotein
Length = 1307
Score = 35.8 bits (81), Expect = 0.056
Identities = 18/49 (36%), Positives = 24/49 (48%), Gaps = 3/49 (6%)
Query: 63 DFGAGRGFRRGAVGGRIGGGRGFGLERKTPPEGYVCHRCKVSGHFIQHC 111
D GRG RG GR GG G+ K+ +C+RC +GH+ C
Sbjct: 183 DTRGGRGRGRGRSSGRGRGGYGYQQRDKSK---VICYRCDKTGHYASEC 228
>At5g37490 putative protein
Length = 435
Score = 35.4 bits (80), Expect = 0.073
Identities = 32/121 (26%), Positives = 48/121 (39%), Gaps = 26/121 (21%)
Query: 176 LPPELHCPLCNNVMKDAVLTSKCCFKSFCDKCIRNYIMS-KSACVCLATNILADDLLPNK 234
+PPE CP+ ++MKD V+ S ++ I +I S C T + D +PN
Sbjct: 31 IPPEFQCPISIDLMKDPVIISTGI--TYDRVSIETWINSGNKTCPVTNTVLTTFDQIPNH 88
Query: 235 TLRDAIHRILESGNSSTENAGSTYQVQDMLSSRCPQPKIP----------SPTSSATSKG 284
T+R I G + L R P P++P SSAT +G
Sbjct: 89 TIRKMIQ-------------GWCVEKGSPLIQRIPTPRVPLMPCEVYEISRKLSSATRRG 135
Query: 285 E 285
+
Sbjct: 136 D 136
>At5g05230 unknown protein
Length = 363
Score = 35.4 bits (80), Expect = 0.073
Identities = 28/115 (24%), Positives = 48/115 (41%), Gaps = 4/115 (3%)
Query: 140 QGSYALQNGSVAVLKPNEAAFDKEME-GLSSTRSVGDLPPELHCPLCNNVMKDAVLTSKC 198
+ S+ +N ++ N F + SS+ S+ L L P ++M DA++
Sbjct: 145 EASFVSRNEESSIAADNGCDFSGRRDLSSSSSNSIESLRTILSDPTTGSLMADAMILP-- 202
Query: 199 CFKSFCDKCIRNYIMSKSACVCLATNILADDLLPNKTLRDAIHRILESGNSSTEN 253
C +F I K+ C C + + D + PN TLR A+ NS + +
Sbjct: 203 CGHTFGAGGIEQVKQMKACCTC-SQPVSEDSITPNLTLRVAVQAFCREENSQSNH 256
>At3g26400 eukaryotic initiation factor 4B (EIF4B7)
Length = 532
Score = 35.4 bits (80), Expect = 0.073
Identities = 26/65 (40%), Positives = 32/65 (49%), Gaps = 2/65 (3%)
Query: 27 SSTMIPDAPLTSKADE-DSKIKALIDTPALDWQQQGSDFGAGRGFRRGAVG-GRIGGGRG 84
+ + + D P S+ADE D K P+ D +QG G G GF G G G GGG G
Sbjct: 155 NQSRVSDFPQPSRADEVDDWGKEKKPLPSFDQGRQGRYSGDGGGFGGGGSGFGGGGGGGG 214
Query: 85 FGLER 89
GL R
Sbjct: 215 GGLSR 219
>At1g36110 hypothetical protein
Length = 745
Score = 35.4 bits (80), Expect = 0.073
Identities = 31/111 (27%), Positives = 46/111 (40%), Gaps = 18/111 (16%)
Query: 66 AGRGFRRGAVGGRIGGGRGFGLERKTPPEGYVCHRCKVSGHFIQHCPTN-GDSTFDIKKV 124
A RG RG GG G GRG G + + C+RC GH+ +CP + S F+ +
Sbjct: 219 ASRG--RGQGGGFNGRGRGRGRGSRDTSK-VTCYRCDKLGHYASNCPDSVKPSVFETE-- 273
Query: 125 RQPTGIPRSMLMVNPQGSYALQNGSVAVLKPNEAAFDKEMEGLSSTRSVGD 175
++ + L NG+ + N A F K E ++ GD
Sbjct: 274 ------------LDGDNVWYLDNGASNHMTGNRAYFSKIDESITGKVRFGD 312
>At3g24690 unknown protein
Length = 158
Score = 35.0 bits (79), Expect = 0.096
Identities = 29/151 (19%), Positives = 65/151 (42%), Gaps = 19/151 (12%)
Query: 234 KTLRDAIHRILESGNSSTENAGSTYQVQDMLSSR---------CPQPKIPSPTSSATSKG 284
+TLR +RI + S ++ + +D L S+ + +IP P+++ + +
Sbjct: 17 RTLRKKYNRITDMEESISQGKTLNKEQEDTLRSKPIVSALIDELVKLRIPPPSAAISEET 76
Query: 285 EPKVSQVNEGMANIQEIVAERKEVSATQQVSEQVKIPRAAVVSEVTHESMSVKEPEPASQ 344
P + Q++ RKEV+ + V+ + I + + + +S + P+S+
Sbjct: 77 SPPAKNKKQ-----QKLSHARKEVAEEENVTAKTDIEDCKISDDGINSEVSKDDTSPSSE 131
Query: 345 GSAKLVEEEVQQKLVPTDGGKKKKKKKVRMP 375
S + + +K +K+ KK ++P
Sbjct: 132 SSQGVTPSPLSRK-----ARRKRNAKKPQIP 157
>At4g36020 glycine-rich protein
Length = 299
Score = 34.7 bits (78), Expect = 0.13
Identities = 27/89 (30%), Positives = 35/89 (38%), Gaps = 17/89 (19%)
Query: 43 DSKIKALIDTPALDWQQQGSDFGAGRGFRRGAVGGR---------------IGGGRGFGL 87
D K KA+ T + + G G RRG G IGGG G G
Sbjct: 66 DGKTKAVNVTAPGGGSLKKENNSRGNGARRGGGGSGCYNCGELGHISKDCGIGGGGGGGE 125
Query: 88 ERKTPPEGYVCHRCKVSGHFIQHCPTNGD 116
R EG C+ C +GHF + C + G+
Sbjct: 126 RRSRGGEG--CYNCGDTGHFARDCTSAGN 152
>At1g32590 hypothetical protein, 5' partial
Length = 1263
Score = 34.7 bits (78), Expect = 0.13
Identities = 26/89 (29%), Positives = 38/89 (42%), Gaps = 10/89 (11%)
Query: 25 VQSSTMIPDAPLTSKADEDSKIKALIDTPALDWQQQGSDFGAGRGFRRGAVGGRIGGGRG 84
+QSS M+ + L+ E+ +KA W+ G G R GG G GRG
Sbjct: 149 LQSSLMVHEQNLSRHDVEERVLKA-----ETQWRPDGGRGRGGSPSRGRGRGGYQGRGRG 203
Query: 85 FGLERKTPPEGYVCHRCKVSGHFIQHCPT 113
+ + R T C +C GH+ CP+
Sbjct: 204 Y-VNRDTVE----CFKCHKMGHYKAECPS 227
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.320 0.135 0.416
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 11,741,329
Number of Sequences: 26719
Number of extensions: 522829
Number of successful extensions: 2852
Number of sequences better than 10.0: 96
Number of HSP's better than 10.0 without gapping: 27
Number of HSP's successfully gapped in prelim test: 69
Number of HSP's that attempted gapping in prelim test: 2483
Number of HSP's gapped (non-prelim): 340
length of query: 498
length of database: 11,318,596
effective HSP length: 103
effective length of query: 395
effective length of database: 8,566,539
effective search space: 3383782905
effective search space used: 3383782905
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.8 bits)
S2: 62 (28.5 bits)
Medicago: description of AC126012.10


