

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC126012.1 + phase: 0
(220 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
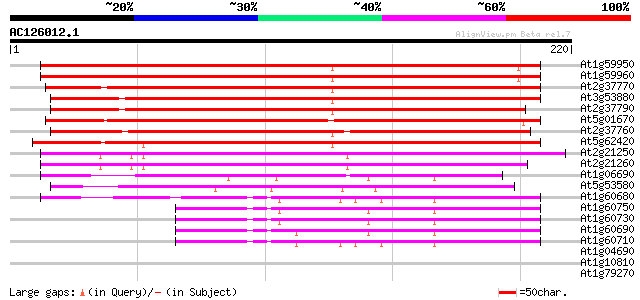
Score E
Sequences producing significant alignments: (bits) Value
At1g59950 hypothetical protein 239 1e-63
At1g59960 putative protein 233 8e-62
At2g37770 putative aldo/keto reductase 164 3e-41
At3g53880 putative alcohol dehydrogenase 161 3e-40
At2g37790 putative alcohol dehydrogenase 160 5e-40
At5g01670 aldose reductase-like protein 153 8e-38
At2g37760 putative alcohol dehydrogenase 147 3e-36
At5g62420 aldose reductase-like protein 145 2e-35
At2g21250 putative NADPH dependent mannose 6-phosphate reductase 124 3e-29
At2g21260 putative NADPH dependent mannose 6-phosphate reductase 124 4e-29
At1g06690 unknown protein 68 4e-12
At5g53580 aldo/keto reductase-like protein 62 2e-10
At1g60680 unknown protein 48 4e-06
At1g60750 47 1e-05
At1g60730 unknown protein 44 7e-05
At1g60690 43 1e-04
At1g60710 unknown protein 41 4e-04
At1g04690 putative K+ channel, beta subunit 37 0.008
At1g10810 putative auxin-induced protein 33 0.090
At1g79270 unknown protein 31 0.45
>At1g59950 hypothetical protein
Length = 320
Score = 239 bits (609), Expect = 1e-63
Identities = 115/198 (58%), Positives = 157/198 (79%), Gaps = 2/198 (1%)
Query: 13 SCRTLQLDYLDLYLIHWPLSSQPGKFTFPIDVADLLPFDVKGVWESMEEGLKLGLTKAIG 72
S TL+LDYLDLYLIHWP+SS+PGK+ FPI+ D LP D + VW MEE +LG+ K IG
Sbjct: 102 SLETLKLDYLDLYLIHWPVSSKPGKYKFPIEEDDFLPMDYETVWSEMEECQRLGVAKCIG 161
Query: 73 VSNFSVKKLENLLSVATILPAVNQVEMNLAWQQKKLREFCNANGIVLTAFSPL-RKGASR 131
VSNFS KKL+++LS+A I P+VNQVEM+ WQQ+KLRE C + GIV+TA+S L +GA
Sbjct: 162 VSNFSCKKLQHILSIAKIPPSVNQVEMSPVWQQRKLRELCKSKGIVVTAYSVLGSRGAFW 221
Query: 132 GPNEVMENDMLKEIADAHGKSVAQISLRWLYEQGVTFVPKSYDKERMNQNLCIFDWSLTK 191
G +++ME+D+LKEIA+A GK+VAQ+S+RW YE+GV+ V KS+ K+R+ +NL IFDWSLT+
Sbjct: 222 GTHKIMESDVLKEIAEAKGKTVAQVSMRWAYEEGVSMVVKSFRKDRLEENLKIFDWSLTE 281
Query: 192 EDHEKID-QIKQNRLIPG 208
E+ ++I +I Q+R++ G
Sbjct: 282 EEKQRISTEISQSRIVDG 299
>At1g59960 putative protein
Length = 326
Score = 233 bits (593), Expect = 8e-62
Identities = 115/198 (58%), Positives = 154/198 (77%), Gaps = 2/198 (1%)
Query: 13 SCRTLQLDYLDLYLIHWPLSSQPGKFTFPIDVADLLPFDVKGVWESMEEGLKLGLTKAIG 72
S + L+LDYLDLY+IHWP+SS+PGK+ FPID D +P D + VW MEE +LGL K IG
Sbjct: 108 SLKNLKLDYLDLYIIHWPVSSKPGKYKFPIDEDDFMPMDFEVVWSEMEECQRLGLAKCIG 167
Query: 73 VSNFSVKKLENLLSVATILPAVNQVEMNLAWQQKKLREFCNANGIVLTAFSPL-RKGASR 131
VSNFS KKL+++LS+ATI P+VNQVEM+ WQQ+KLRE C +N IV+TA+S L +GA
Sbjct: 168 VSNFSCKKLQHILSIATIPPSVNQVEMSPIWQQRKLRELCRSNDIVVTAYSVLGSRGAFW 227
Query: 132 GPNEVMENDMLKEIADAHGKSVAQISLRWLYEQGVTFVPKSYDKERMNQNLCIFDWSLTK 191
G ++ME+D+LKEIA+A K+VAQ+S+RW YEQGV+ V KS+ KER+ +NL IFDWSLT+
Sbjct: 228 GTPKIMESDVLKEIAEAKEKTVAQVSMRWAYEQGVSMVVKSFTKERLEENLKIFDWSLTE 287
Query: 192 EDHEKID-QIKQNRLIPG 208
++ ++I +I Q R + G
Sbjct: 288 DETQRISTEIPQFRNVHG 305
>At2g37770 putative aldo/keto reductase
Length = 315
Score = 164 bits (415), Expect = 3e-41
Identities = 85/195 (43%), Positives = 128/195 (65%), Gaps = 3/195 (1%)
Query: 15 RTLQLDYLDLYLIHWPLSSQPGKFTFPIDVADLLPFDVKGVWESMEEGLKLGLTKAIGVS 74
+ LQL+Y+DLYLIHWP + G + I +LLP D+ W++ME G +AIGVS
Sbjct: 101 KDLQLEYVDLYLIHWPARIKKG--SVGIKPENLLPVDIPSTWKAMEALYDSGKARAIGVS 158
Query: 75 NFSVKKLENLLSVATILPAVNQVEMNLAWQQKKLREFCNANGIVLTAFSPL-RKGASRGP 133
NFS KKL +LL +A + PAVNQVE + +W+Q KL+EFC + G+ L+A+SPL G +
Sbjct: 159 NFSTKKLADLLELARVPPAVNQVECHPSWRQTKLQEFCKSKGVHLSAYSPLGSPGTTWLK 218
Query: 134 NEVMENDMLKEIADAHGKSVAQISLRWLYEQGVTFVPKSYDKERMNQNLCIFDWSLTKED 193
++V++N +L +A+ GKS AQ++LRW + G + +PKS ++ R+ +N +FDWS+
Sbjct: 219 SDVLKNPILNMVAEKLGKSPAQVALRWGLQMGHSVLPKSTNEGRIKENFNVFDWSIPDYM 278
Query: 194 HEKIDQIKQNRLIPG 208
K +I+Q RL+ G
Sbjct: 279 FAKFAEIEQARLVTG 293
>At3g53880 putative alcohol dehydrogenase
Length = 315
Score = 161 bits (407), Expect = 3e-40
Identities = 82/193 (42%), Positives = 125/193 (64%), Gaps = 3/193 (1%)
Query: 17 LQLDYLDLYLIHWPLSSQPGKFTFPIDVADLLPFDVKGVWESMEEGLKLGLTKAIGVSNF 76
LQLDY+DLYL+HWP+ + G F + +++P D+ W++ME + G +AIGVSNF
Sbjct: 103 LQLDYVDLYLMHWPVRLKKGTVDFKPE--NIMPIDIPSTWKAMEALVDSGKARAIGVSNF 160
Query: 77 SVKKLENLLSVATILPAVNQVEMNLAWQQKKLREFCNANGIVLTAFSPL-RKGASRGPNE 135
S KKL +L+ A + PAVNQVE + +WQQ KL EFC + GI L+ +SPL G + +
Sbjct: 161 STKKLSDLVEAARVPPAVNQVECHPSWQQHKLHEFCKSKGIHLSGYSPLGSPGTTWVKAD 220
Query: 136 VMENDMLKEIADAHGKSVAQISLRWLYEQGVTFVPKSYDKERMNQNLCIFDWSLTKEDHE 195
V+++ +++ IA GKS AQ +LRW + G + +PKS ++ R+ +N + WS+ KE +
Sbjct: 221 VLKSPVIEMIAKEIGKSPAQTALRWGLQMGHSILPKSTNEGRIRENFDVLGWSIPKEMFD 280
Query: 196 KIDQIKQNRLIPG 208
K +I+Q RL+ G
Sbjct: 281 KFSKIEQARLVQG 293
>At2g37790 putative alcohol dehydrogenase
Length = 350
Score = 160 bits (405), Expect = 5e-40
Identities = 82/187 (43%), Positives = 124/187 (65%), Gaps = 3/187 (1%)
Query: 17 LQLDYLDLYLIHWPLSSQPGKFTFPIDVADLLPFDVKGVWESMEEGLKLGLTKAIGVSNF 76
LQLDY+DLYLIHWP+S + G F + ++LP D+ W++ME G +AIGVSNF
Sbjct: 103 LQLDYVDLYLIHWPVSLKKGSTGFKPE--NILPTDIPSTWKAMESLFDSGKARAIGVSNF 160
Query: 77 SVKKLENLLSVATILPAVNQVEMNLAWQQKKLREFCNANGIVLTAFSPL-RKGASRGPNE 135
S KKL +LL VA + PAVNQVE + +WQQ LR+FC + G+ L+ +SPL G + ++
Sbjct: 161 SSKKLADLLVVARVPPAVNQVECHPSWQQNVLRDFCKSKGVHLSGYSPLGSPGTTWLTSD 220
Query: 136 VMENDMLKEIADAHGKSVAQISLRWLYEQGVTFVPKSYDKERMNQNLCIFDWSLTKEDHE 195
V++N +L +A+ GK+ AQ++LRW + G + +PKS ++R+ QN +F+WS+ ++
Sbjct: 221 VLKNPILGGVAEKLGKTPAQVALRWGLQMGQSVLPKSTHEDRIKQNFDVFNWSIPEDMLS 280
Query: 196 KIDQIKQ 202
K +I Q
Sbjct: 281 KFSEIGQ 287
>At5g01670 aldose reductase-like protein
Length = 322
Score = 153 bits (386), Expect = 8e-38
Identities = 78/195 (40%), Positives = 122/195 (62%), Gaps = 4/195 (2%)
Query: 15 RTLQLDYLDLYLIHWPLSSQPGKFTFPIDVADLLPFDVKGVWESMEEGLKLGLTKAIGVS 74
+ LQL+YLDLYLIHWP+ + G + P D+L FD++GVW ME K L + IGV
Sbjct: 108 KELQLEYLDLYLIHWPIRLREGA-SKPPKAGDVLDFDMEGVWREMENLSKDSLVRNIGVC 166
Query: 75 NFSVKKLENLLSVATILPAVNQVEMNLAWQQKKLREFCNANGIVLTAFSPLRKGASRGPN 134
NF+V KL LL A ++PAV Q+EM+ W+ ++ EFC N I +TA+SPL G+ G
Sbjct: 167 NFTVTKLNKLLGFAELIPAVCQMEMHPGWRNDRILEFCKKNEIHVTAYSPL--GSQEGGR 224
Query: 135 EVMENDMLKEIADAHGKSVAQISLRWLYEQGVTFVPKSYDKERMNQNLCIFDWSLTKEDH 194
+++ + + IA K+ QI ++W ++G + +PKS + ER+ +N+ +FDW + ++D
Sbjct: 225 DLIHDQTVDRIAKKLNKTPGQILVKWGLQRGTSVIPKSLNPERIKENIKVFDWVIPEQDF 284
Query: 195 EKIDQI-KQNRLIPG 208
+ ++ I Q R+I G
Sbjct: 285 QALNSITDQKRVIDG 299
>At2g37760 putative alcohol dehydrogenase
Length = 311
Score = 147 bits (372), Expect = 3e-36
Identities = 80/191 (41%), Positives = 118/191 (60%), Gaps = 7/191 (3%)
Query: 17 LQLDYLDLYLIHWPLSSQPGKFTFPIDVADLLPFDVKGVWESMEEGLKLGLTKAIGVSNF 76
LQ+DY+DLYLIHWP S + ++ L D+ W++ME G +AIGVSNF
Sbjct: 99 LQIDYVDLYLIHWPASLKKESLMPTPEM--LTKPDITSTWKAMEALYDSGKARAIGVSNF 156
Query: 77 SVKKLENLLSVATILPAVNQVEMNLAWQQKKLREFCNANGIVLTAFSPL---RKGASRGP 133
S KKL +LL+VA + PAVNQVE + WQQ+ L E C + G+ L+ +SPL KG R
Sbjct: 157 SSKKLTDLLNVARVTPAVNQVECHPVWQQQGLHELCKSKGVHLSGYSPLGSQSKGEVR-- 214
Query: 134 NEVMENDMLKEIADAHGKSVAQISLRWLYEQGVTFVPKSYDKERMNQNLCIFDWSLTKED 193
+V++N ++ E+A+ GK+ AQ++LRW + G + +PKS R+ +NL +FDWS+ ++
Sbjct: 215 LKVLQNPIVTEVAEKLGKTTAQVALRWGLQTGHSVLPKSSSGARLKENLDVFDWSIPEDL 274
Query: 194 HEKIDQIKQNR 204
K I Q +
Sbjct: 275 FTKFSNIPQEK 285
>At5g62420 aldose reductase-like protein
Length = 316
Score = 145 bits (365), Expect = 2e-35
Identities = 78/202 (38%), Positives = 126/202 (61%), Gaps = 4/202 (1%)
Query: 10 LLISCRTLQLDYLDLYLIHWPLSSQPGKFTFPIDVADLLPFD--VKGVWESMEEGLKLGL 67
L+ + +T+ LDYLD YL+HWP+ +PG + PI D D ++ W+ ME L++GL
Sbjct: 94 LIQTLKTMGLDYLDNYLVHWPIKLKPG-VSEPIPKEDEFEKDLGIEETWQGMERCLEMGL 152
Query: 68 TKAIGVSNFSVKKLENLLSVATILPAVNQVEMNLAWQQKKLREFCNANGIVLTAFSPL-R 126
++IGVSNFS KK+ +LL A++ P+VNQVEM+ W+Q+KLR+ C N I ++ +SPL
Sbjct: 153 CRSIGVSNFSSKKIFDLLDFASVSPSVNQVEMHPLWRQRKLRKVCEENNIHVSGYSPLGG 212
Query: 127 KGASRGPNEVMENDMLKEIADAHGKSVAQISLRWLYEQGVTFVPKSYDKERMNQNLCIFD 186
G G V+E+ ++K IA H + AQ++LRW +G + + KS++ RM +N +
Sbjct: 213 PGNCWGSTAVIEHPIIKSIALKHNATPAQVALRWGMSKGASVIVKSFNGARMIENKRALE 272
Query: 187 WSLTKEDHEKIDQIKQNRLIPG 208
L +D ID +++ +++ G
Sbjct: 273 IKLDDQDLSLIDHLEEWKIMRG 294
>At2g21250 putative NADPH dependent mannose 6-phosphate reductase
Length = 309
Score = 124 bits (312), Expect = 3e-29
Identities = 73/217 (33%), Positives = 126/217 (57%), Gaps = 11/217 (5%)
Query: 13 SCRTLQLDYLDLYLIHWPLSSQ-PGKFTFPIDVAD--LLPFD----VKGVWESMEEGLKL 65
S + LQLDYLDL+L+H+P++++ G T + D +L D ++ W ME+ + +
Sbjct: 92 SLKKLQLDYLDLFLVHFPVATKHTGVGTTDSALGDDGVLDIDTTISLETTWHDMEKLVSM 151
Query: 66 GLTKAIGVSNFSVKKLENLLSVATILPAVNQVEMNLAWQQKKLREFCNANGIVLTAFSPL 125
GL ++IG+SN+ V + L+ + I PAVNQ+E + +Q+ L +FC +GI +TA +PL
Sbjct: 152 GLVRSIGISNYDVFLTRDCLAYSKIKPAVNQIETHPYFQRDSLVKFCQKHGICVTAHTPL 211
Query: 126 RKGASR----GPNEVMENDMLKEIADAHGKSVAQISLRWLYEQGVTFVPKSYDKERMNQN 181
+ G +++ +LK++A+ + K+VAQ+ LRW ++ +PK+ R+ +N
Sbjct: 212 GGATANAEWFGTVSCLDDPVLKDVAEKYKKTVAQVVLRWGIQRKTVVIPKTSKPARLEEN 271
Query: 182 LCIFDWSLTKEDHEKIDQIKQNRLIPGPTKPGINDLY 218
+FD+ L+KED E I +++ P K DLY
Sbjct: 272 FQVFDFELSKEDMEVIKSMERKYRTNQPAKFWNIDLY 308
>At2g21260 putative NADPH dependent mannose 6-phosphate reductase
Length = 309
Score = 124 bits (311), Expect = 4e-29
Identities = 70/202 (34%), Positives = 122/202 (59%), Gaps = 11/202 (5%)
Query: 13 SCRTLQLDYLDLYLIHWPLSSQ-PGKFTFPIDVAD--LLPFD----VKGVWESMEEGLKL 65
S + LQLDYLDL+L+H P++++ G T + D +L D ++ W ME+ + +
Sbjct: 92 SLKKLQLDYLDLFLVHIPIATKHTGIGTTDSALGDDGVLDIDTTISLETTWHDMEKLVSM 151
Query: 66 GLTKAIGVSNFSVKKLENLLSVATILPAVNQVEMNLAWQQKKLREFCNANGIVLTAFSPL 125
GL ++IG+SN+ V + L+ + I PAVNQ+E + +Q+ L +FC +GI +TA +PL
Sbjct: 152 GLVRSIGISNYDVFLTRDCLAYSKIKPAVNQIETHPYFQRDSLVKFCQKHGICVTAHTPL 211
Query: 126 RKGASR----GPNEVMENDMLKEIADAHGKSVAQISLRWLYEQGVTFVPKSYDKERMNQN 181
+ G +++ +LK++A+ + ++VAQI LRW ++ +PK+ ER+ +N
Sbjct: 212 GGATANAEWFGTVSCLDDPVLKDVAEKYKQTVAQIVLRWGIQRNTVVIPKTSKPERLEEN 271
Query: 182 LCIFDWSLTKEDHEKIDQIKQN 203
+FD+ L+KED E I +++N
Sbjct: 272 FQVFDFQLSKEDMEVIKSMERN 293
>At1g06690 unknown protein
Length = 377
Score = 67.8 bits (164), Expect = 4e-12
Identities = 57/210 (27%), Positives = 94/210 (44%), Gaps = 47/210 (22%)
Query: 13 SCRTLQLDYLDLYLIHWPLSSQPGKFTFPIDVADLLPFDVKGVWESMEEGLKLGLTKAIG 72
S L+L +DLY +HWP + +G + + + ++ GL KA+G
Sbjct: 164 SLSRLELSSVDLYQLHWPGL-----------------WGNEGYLDGLGDAVEQGLVKAVG 206
Query: 73 VSNFSVKKLENL---LSVATILPAVNQVEMNLAW---QQKKLREFCNANGIVLTAFSPLR 126
VSN+S K+L + L I A NQV +L + +Q ++ C+ G+ L A+SP+
Sbjct: 207 VSNYSEKRLRDAYERLKKRGIPLASNQVNYSLIYRAPEQTGVKAACDELGVTLIAYSPIA 266
Query: 127 KGASRGPNEVMEN----------------------DMLKEIADAHGKSVAQISLRWLYEQ 164
+GA G EN + +K+I + + K+ QI+L WL Q
Sbjct: 267 QGALTG-KYTPENPPSGPRGRIYTREFLTKLQPLLNRIKQIGENYSKTPTQIALNWLVAQ 325
Query: 165 G-VTFVPKSYDKERMNQNLCIFDWSLTKED 193
G V +P + + E+ + WSLT +
Sbjct: 326 GNVIPIPGAKNAEQAKEFAGAIGWSLTDNE 355
>At5g53580 aldo/keto reductase-like protein
Length = 365
Score = 62.0 bits (149), Expect = 2e-10
Identities = 47/208 (22%), Positives = 87/208 (41%), Gaps = 39/208 (18%)
Query: 17 LQLDYLDLYLIHWPLSSQPGKFTFPIDVADLLPFDVKGVWESMEEGLKLGLTKAIGVSNF 76
LQ+D L + +HW A P +W+ + + + GL +A+GVSN+
Sbjct: 153 LQIDQLGIGQLHW-------------STASYAPLQELVLWDGLVQMYEKGLVRAVGVSNY 199
Query: 77 SVK---KLENLLSVATILPAVNQVEMNL---AWQQKKLREFCNANGIVLTAFSPLRKGA- 129
+ K+ + L + QV+ +L +Q +++ C+ GI L ++SPL G
Sbjct: 200 GPQQLVKIHDYLKTRGVPLCSAQVQFSLLSMGKEQLEIKSICDELGIRLISYSPLGLGML 259
Query: 130 ---------SRGPNEVMENDML----------KEIADAHGKSVAQISLRWLYEQGVTFVP 170
GP ++ +L EIA GK++ Q+++ W +G +P
Sbjct: 260 TGKYSSSKLPTGPRSLLFRQILPGLEPLLLALSEIAKKRGKTMPQVAINWCICKGTVPIP 319
Query: 171 KSYDKERMNQNLCIFDWSLTKEDHEKID 198
+ NL W LT ++ +++
Sbjct: 320 GIKSVRHVEDNLGALGWKLTNDEQLQLE 347
>At1g60680 unknown protein
Length = 346
Score = 48.1 bits (113), Expect = 4e-06
Identities = 55/226 (24%), Positives = 96/226 (42%), Gaps = 49/226 (21%)
Query: 13 SCRTLQLDYLDLYLIHWPLSSQPGKFTFPIDVADLLPFDVKGVWESMEEGLKLGLTKAIG 72
S + L + +DLY H ID + ++ + + +EEG K IG
Sbjct: 116 SLKRLDIACIDLYYQH------------RIDTRVPIEITMRELKKLVEEGK----IKYIG 159
Query: 73 VSNFSVKKLENLLSVATILPAVNQVEMNLAWQ---QKKLREFCNANGIVLTAFSPLRKG- 128
+S S + +V I Q+E +L W ++ + C GI + A+SPL +G
Sbjct: 160 LSEASASTIRRAHAVHPITAV--QIEWSL-WSRDAEEDIIPICRELGIGIVAYSPLGRGF 216
Query: 129 ASRGPN--EVMENDMLKE----------------------IADAHGKSVAQISLRWLYEQ 164
+ GP E +END ++ +A+ G + AQ++L W++ Q
Sbjct: 217 LAAGPKLAENLENDDFRKTLPRFQQENVDHNKILFEKVSAMAEKKGCTPAQLALAWVHHQ 276
Query: 165 G--VTFVPKSYDKERMNQNLCIFDWSLTKEDHEKIDQIKQNRLIPG 208
G V +P + E +NQN+ LT E+ ++D + + + G
Sbjct: 277 GDDVCPIPGTTKIENLNQNIRALSVKLTPEEISELDSLAKPESVKG 322
>At1g60750
Length = 330
Score = 46.6 bits (109), Expect = 1e-05
Identities = 41/156 (26%), Positives = 71/156 (45%), Gaps = 16/156 (10%)
Query: 66 GLTKAIGVSNFSVKKLENLLSVATILPAVNQVEMNLAWQ---QKKLREFCNANGIVLTAF 122
G K IG+S S + +V I Q+E +L W ++ + C GI + A+
Sbjct: 154 GKIKYIGLSEASASTIRRAHAVHPITAV--QIEWSL-WSRDVEEDIIPTCRELGIGIVAY 210
Query: 123 SPLRKGASRGPNEVMEN--------DMLKEIADAHGKSVAQISLRWLYEQG--VTFVPKS 172
SPL +G P EN + ++ +A + AQ++L W++ QG V +P +
Sbjct: 211 SPLGRGFLGLPRFQQENLENNKILYEKVQAMATKKSCTPAQLALAWVHHQGDDVCPIPGT 270
Query: 173 YDKERMNQNLCIFDWSLTKEDHEKIDQIKQNRLIPG 208
+ +NQN+ LT E+ +++ I Q + G
Sbjct: 271 SKIQNLNQNIGALSVKLTPEEMVELEAIAQPDFVKG 306
>At1g60730 unknown protein
Length = 345
Score = 43.9 bits (102), Expect = 7e-05
Identities = 40/173 (23%), Positives = 74/173 (42%), Gaps = 33/173 (19%)
Query: 66 GLTKAIGVSNFSVKKLENLLSVATILPAVNQVEMNLAWQ---QKKLREFCNANGIVLTAF 122
G K IG+S S + +V I Q+E +L W ++ + C GI + A+
Sbjct: 152 GKIKYIGLSEASASTIRRAHAVHPITAL--QIEWSL-WSRDVEEDIIPTCRELGIGIVAY 208
Query: 123 SPLRKGASRGPNEVMEN-------------------------DMLKEIADAHGKSVAQIS 157
SPL +G +++EN + + +++ G + AQ++
Sbjct: 209 SPLGRGFFASGPKLVENLDNNDVRKTLPRFQQENLDHNKILFEKVSAMSEKKGCTPAQLA 268
Query: 158 LRWLYEQG--VTFVPKSYDKERMNQNLCIFDWSLTKEDHEKIDQIKQNRLIPG 208
L W++ QG V +P + E +NQN+ LT E+ +++ + Q + G
Sbjct: 269 LAWVHHQGDDVCPIPGTTKIENLNQNIGALSVKLTPEEMSELESLAQPGFVKG 321
>At1g60690
Length = 345
Score = 43.1 bits (100), Expect = 1e-04
Identities = 41/173 (23%), Positives = 73/173 (41%), Gaps = 33/173 (19%)
Query: 66 GLTKAIGVSNFSVKKLENLLSVATILPAVNQVEMNLAWQQKKLREF---CNANGIVLTAF 122
G K IG+S S + +V I Q+E +L W + E C GI + ++
Sbjct: 152 GKIKYIGLSEASASTIRRAHTVHPITAV--QLEWSL-WTRDVEEEIVPTCRELGIGIVSY 208
Query: 123 SPLRKGASRGPNEVMEN-------------------------DMLKEIADAHGKSVAQIS 157
SPL +G +++EN + + +++ G + AQ++
Sbjct: 209 SPLGRGFFASGPKLVENLDNNDFRKALPRFQQENLDHNKILYEKVSAMSEKKGCTPAQLA 268
Query: 158 LRWLYEQG--VTFVPKSYDKERMNQNLCIFDWSLTKEDHEKIDQIKQNRLIPG 208
L W++ QG V +P + E +NQN+ LT E+ +++ I Q + G
Sbjct: 269 LAWVHHQGDDVCPIPGTTKIENLNQNIRALSVKLTPEEMSELETIAQPESVKG 321
>At1g60710 unknown protein
Length = 345
Score = 41.2 bits (95), Expect = 4e-04
Identities = 44/173 (25%), Positives = 73/173 (41%), Gaps = 33/173 (19%)
Query: 66 GLTKAIGVSNFSVKKLENLLSVATILPAVNQVEMNLAWQQKKLREF---CNANGIVLTAF 122
G K IG+S S + +V I Q+E +L W + E C GI + A+
Sbjct: 152 GKIKYIGLSEASASTIRRAHAVHPITAV--QIEWSL-WTRDVEEEIIPTCRELGIGIVAY 208
Query: 123 SPLRKG-ASRGPN--EVMENDMLKE----------------------IADAHGKSVAQIS 157
SPL +G + GP E +E D ++ I++ G + Q++
Sbjct: 209 SPLGRGFFASGPKLVENLEKDDFRKALPRFQEENLDHNKIVYEKVCAISEKKGCTPGQLA 268
Query: 158 LRWLYEQG--VTFVPKSYDKERMNQNLCIFDWSLTKEDHEKIDQIKQNRLIPG 208
L W++ QG V +P + E + QN+ LT E+ +++ I Q + G
Sbjct: 269 LAWVHHQGDDVCPIPGTTKIENLKQNIGALSVKLTPEEMTELEAIAQPGFVKG 321
>At1g04690 putative K+ channel, beta subunit
Length = 328
Score = 37.0 bits (84), Expect = 0.008
Identities = 44/182 (24%), Positives = 69/182 (37%), Gaps = 50/182 (27%)
Query: 13 SCRTLQLDYLDLYLIHWPLSSQPGKFTFPIDVADLLPFDVKGVWESMEEGLKLGLTKAIG 72
S + L +DY+D+ H P +S P ++ +M + G G
Sbjct: 107 SLKRLDMDYVDVLYCHRPDASTP----------------IEETVRAMNYVIDKGWAFYWG 150
Query: 73 VSNFSVKKLENLLSVATIL----PAVNQVEMNLAWQQKKLREFC---NANGIVLTAFSPL 125
S +S +++ A L P V Q E N+ + K EF +GI LT +SPL
Sbjct: 151 TSEWSAQQITEAWGAADRLDLVGPIVEQPEYNMFARHKVETEFLPLYTNHGIGLTTWSPL 210
Query: 126 RKGASRGP---------------------NEVMENDML------KEIADAHGKSVAQISL 158
G G N + +D+L K IAD G ++AQ+++
Sbjct: 211 ASGVLTGKYNKGAIPSDSRFALENYKNLANRSLVDDVLRKVSGLKPIADELGVTLAQLAI 270
Query: 159 RW 160
W
Sbjct: 271 AW 272
>At1g10810 putative auxin-induced protein
Length = 344
Score = 33.5 bits (75), Expect = 0.090
Identities = 50/216 (23%), Positives = 90/216 (41%), Gaps = 49/216 (22%)
Query: 13 SCRTLQLDYLDLYLIHWPLSSQPGKFTFPIDVADLLPFDVKGVWESMEEGLKLGLTKAIG 72
S R L + +DLY H + T PI+V + + + +EEG K IG
Sbjct: 115 SLRRLGVSCIDLYYQHRIDT------TVPIEVT------IGELKKLVEEGK----IKYIG 158
Query: 73 VSNFSVKKLENLLSVATILPAVNQVEMNLAWQ---QKKLREFCNANGIVLTAFSPL---- 125
+S + +V + Q+E +L W ++ + C GI + A+SPL
Sbjct: 159 LSEACASTIRRAHAVHPLTAV--QLEWSL-WSRDVEEDIIPTCRELGIGIVAYSPLGLGF 215
Query: 126 ----------------RKGASRGPNEVMENDML-----KEIADAHGKSVAQISLRWLYEQ 164
RKG R E ++++ + +A+ + AQ++L W++ Q
Sbjct: 216 FAAGPKFIESMDNGDYRKGLPRFQQENLDHNKILYEKVNAMAEKKSCTPAQLALAWVHHQ 275
Query: 165 G--VTFVPKSYDKERMNQNLCIFDWSLTKEDHEKID 198
G V +P + + +NQN+ L+ E+ ++D
Sbjct: 276 GNDVCPIPGTSKIKNLNQNIGALSVKLSIEEMAELD 311
>At1g79270 unknown protein
Length = 528
Score = 31.2 bits (69), Expect = 0.45
Identities = 16/57 (28%), Positives = 29/57 (50%)
Query: 157 SLRWLYEQGVTFVPKSYDKERMNQNLCIFDWSLTKEDHEKIDQIKQNRLIPGPTKPG 213
S + YE+ + FV KSY ++ +++++ WS T ++K+D Q K G
Sbjct: 313 SFQTKYEEAIFFVIKSYSEDDIHKSIKYNVWSSTLNGNKKLDSAYQESQKKAADKSG 369
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.319 0.138 0.414
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 4,994,255
Number of Sequences: 26719
Number of extensions: 204727
Number of successful extensions: 569
Number of sequences better than 10.0: 38
Number of HSP's better than 10.0 without gapping: 21
Number of HSP's successfully gapped in prelim test: 17
Number of HSP's that attempted gapping in prelim test: 520
Number of HSP's gapped (non-prelim): 38
length of query: 220
length of database: 11,318,596
effective HSP length: 95
effective length of query: 125
effective length of database: 8,780,291
effective search space: 1097536375
effective search space used: 1097536375
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 58 (26.9 bits)
Medicago: description of AC126012.1


