

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC126011.4 + phase: 0 /pseudo
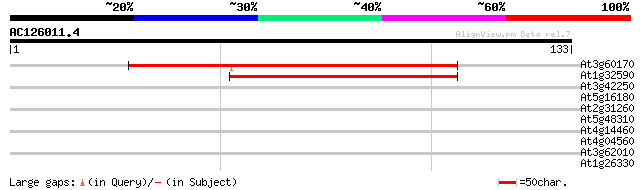
(133 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
At3g60170 putative protein 78 1e-15
At1g32590 hypothetical protein, 5' partial 50 4e-07
At3g42250 hypothetical protein 28 1.1
At5g16180 putative protein 27 3.1
At2g31260 unknown protein 27 3.1
At5g48310 putative protein 26 6.9
At4g14460 retrovirus-related like polyprotein 26 6.9
At4g04560 putative transposon protein 25 9.1
At3g62010 putative protein 25 9.1
At1g26330 hypothetical protein 25 9.1
>At3g60170 putative protein
Length = 1339
Score = 78.2 bits (191), Expect = 1e-15
Identities = 40/79 (50%), Positives = 56/79 (70%), Gaps = 1/79 (1%)
Query: 29 NFLRSKEYWSLVDDGVARLGSGG-PLTAAEQKTPYEMMLKYLKVKNYLLHAIDRKIIETI 87
NFLRS+E W LV++G+ + G P++ A++ E LK LKVKN+L AIDR+I+ETI
Sbjct: 28 NFLRSRELWRLVEEGIPAIVVGTTPVSEAQRSAVEEAKLKDLKVKNFLFQAIDREILETI 87
Query: 88 LEKNTLKKIWASMRGKDKG 106
L+K+T K IW SM+ K +G
Sbjct: 88 LDKSTSKAIWESMKKKYQG 106
>At1g32590 hypothetical protein, 5' partial
Length = 1263
Score = 49.7 bits (117), Expect = 4e-07
Identities = 25/54 (46%), Positives = 36/54 (66%)
Query: 53 LTAAEQKTPYEMMLKYLKVKNYLLHAIDRKIIETILEKNTLKKIWASMRGKDKG 106
LT A++ E +K KVKNYL +ID+ I++TIL+K T K +W SM+ K +G
Sbjct: 10 LTGAQRTELAEKTVKDHKVKNYLFASIDKTILKTILQKETSKDLWESMKRKYQG 63
>At3g42250 hypothetical protein
Length = 100
Score = 28.5 bits (62), Expect = 1.1
Identities = 13/67 (19%), Positives = 30/67 (44%)
Query: 31 LRSKEYWSLVDDGVARLGSGGPLTAAEQKTPYEMMLKYLKVKNYLLHAIDRKIIETILEK 90
L + + W +++ G L + G L+ ++ + + K + +DR E ++E
Sbjct: 26 LGAHDVWEIIEKGFIELENEGSLSQTQKDGLRDSRKRDKKALCLVYQGLDRDTFEKVVEA 85
Query: 91 NTLKKIW 97
+ K+ W
Sbjct: 86 TSAKEAW 92
>At5g16180 putative protein
Length = 718
Score = 26.9 bits (58), Expect = 3.1
Identities = 25/101 (24%), Positives = 41/101 (39%), Gaps = 8/101 (7%)
Query: 4 KKKNVSSIL*LFGNGKQHK*QNLDGNFLRSKEYWSLVDDGVARLGSGGPLTAAEQKTPYE 63
K+++ +L + NGKQ K N G L +E + G P E +
Sbjct: 473 KRESDIELLEVVTNGKQLKETNKSGTLLEFQELQR-------KFGEMDPRNL-ETEAEKA 524
Query: 64 MMLKYLKVKNYLLHAIDRKIIETILEKNTLKKIWASMRGKD 104
+ K LK + + L + KI ++ +E L +W G D
Sbjct: 525 RLEKELKSQEHKLSILKSKIEKSNMELFKLNSLWKPSEGDD 565
>At2g31260 unknown protein
Length = 866
Score = 26.9 bits (58), Expect = 3.1
Identities = 10/48 (20%), Positives = 29/48 (59%)
Query: 37 WSLVDDGVARLGSGGPLTAAEQKTPYEMMLKYLKVKNYLLHAIDRKII 84
W+ V + V +L S L + + ++M+++ ++ +NYL+ +++ ++
Sbjct: 207 WATVLEKVVQLQSSQCLCVVKDLSAHDMVMRLMRKENYLIGMLNKGLL 254
>At5g48310 putative protein
Length = 1156
Score = 25.8 bits (55), Expect = 6.9
Identities = 16/54 (29%), Positives = 29/54 (53%), Gaps = 2/54 (3%)
Query: 53 LTAAEQKTPYEMMLKYLKVKNYLLHAIDRKIIETILEKNT-LKKIWASMRGKDK 105
L EQ ++L+ K +NY+ A+D+ + T KNT LK+I ++ ++
Sbjct: 986 LIFGEQMNVITVLLR-TKYRNYMQAAVDKLVSNTQSNKNTRLKRILEEIKDNER 1038
>At4g14460 retrovirus-related like polyprotein
Length = 1489
Score = 25.8 bits (55), Expect = 6.9
Identities = 10/35 (28%), Positives = 24/35 (68%)
Query: 71 VKNYLLHAIDRKIIETILEKNTLKKIWASMRGKDK 105
V +L++++D+KI +++L T++ IW ++ + K
Sbjct: 101 VSTWLMNSVDKKIGQSLLYIATVQGIWNNLLSRFK 135
>At4g04560 putative transposon protein
Length = 590
Score = 25.4 bits (54), Expect = 9.1
Identities = 22/70 (31%), Positives = 33/70 (46%), Gaps = 4/70 (5%)
Query: 32 RSKEYWSLVDDGVARLGSGGPLT--AAEQKTPYE-MMLKYLKVKNYLLHAIDRKIIETIL 88
++K+ W +VD+GV + + G + A +QKT E LK L L A+ I I
Sbjct: 27 QTKKLWEIVDEGVPKPPAEGDHSPEAVQQKTRCEAASLKDLTALQILQTAVSDSIFPRIA 86
Query: 89 -EKNTLKKIW 97
+ L K W
Sbjct: 87 PASSALGKPW 96
>At3g62010 putative protein
Length = 1254
Score = 25.4 bits (54), Expect = 9.1
Identities = 11/42 (26%), Positives = 22/42 (52%)
Query: 49 SGGPLTAAEQKTPYEMMLKYLKVKNYLLHAIDRKIIETILEK 90
+G P E + PY+ ML+Y + + ++ ++ E IL +
Sbjct: 519 NGKPFVLREVERPYKNMLEYTGIDRDRVEGMEARLKEDILRE 560
>At1g26330 hypothetical protein
Length = 1196
Score = 25.4 bits (54), Expect = 9.1
Identities = 11/22 (50%), Positives = 15/22 (68%), Gaps = 5/22 (22%)
Query: 94 KKIWASMRGKDK-----GYKSI 110
KK W+S RGKD+ GYK++
Sbjct: 31 KKYWSSSRGKDRFPYPVGYKAV 52
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.333 0.147 0.445
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 2,775,847
Number of Sequences: 26719
Number of extensions: 105237
Number of successful extensions: 319
Number of sequences better than 10.0: 10
Number of HSP's better than 10.0 without gapping: 6
Number of HSP's successfully gapped in prelim test: 4
Number of HSP's that attempted gapping in prelim test: 312
Number of HSP's gapped (non-prelim): 10
length of query: 133
length of database: 11,318,596
effective HSP length: 88
effective length of query: 45
effective length of database: 8,967,324
effective search space: 403529580
effective search space used: 403529580
T: 11
A: 40
X1: 15 ( 7.2 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 39 (21.5 bits)
S2: 54 (25.4 bits)
Medicago: description of AC126011.4


