

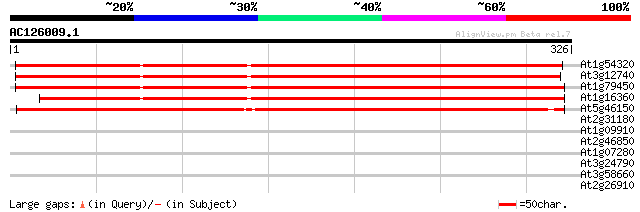
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC126009.1 - phase: 0
(326 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
At1g54320 unknown protein 413 e-116
At3g12740 unknown protein 412 e-115
At1g79450 unknown protein 408 e-114
At1g16360 unknown protein 377 e-105
At5g46150 unknown protein 354 3e-98
At2g31180 putative transcription factor (MYB14) 29 3.0
At1g09910 hypothetical protein 29 3.0
At2g46850 putative Ser/Thr protein kinase 29 4.0
At1g07280 unknown protein 29 4.0
At3g24790 protein kinase, putative 28 6.8
At3g58660 putative protein 28 8.8
At2g26910 putative ABC transporter 28 8.8
>At1g54320 unknown protein
Length = 349
Score = 413 bits (1061), Expect = e-116
Identities = 196/318 (61%), Positives = 240/318 (74%), Gaps = 3/318 (0%)
Query: 4 SRFSQQELHAWQPILTPGWVIAIFTFIGLVFIPIGVASLFASEQVVEVPLRYDDQCLPSL 63
S+F+QQEL A +PILTPGWVI+ F + ++FIP+GV SLFAS+ VVE+ RYD +C+P+
Sbjct: 31 SKFTQQELPACKPILTPGWVISTFLIVSVIFIPLGVISLFASQDVVEIVDRYDTECIPAP 90
Query: 64 YKDDAMTYIKGNRISKTCTKKLTVKSKMKAPIYVYYQLSNFYQNHRHYVKSRDHKQLRSK 123
+ + + YI+G+ K C + L V +MK PIYVYYQL NFYQNHR YVKSR QLRS
Sbjct: 91 ARTNKVAYIQGDG-DKVCNRDLKVTKRMKQPIYVYYQLENFYQNHRRYVKSRSDSQLRST 149
Query: 124 ADENDVGKCFPEDYTANGYLPVVPCGLAAWSLFNDTYRFSNNNKDLVINKKNIAWKSDQK 183
EN + C PED G P+VPCGL AWSLFNDTY S NN L +NKK IAWKSD++
Sbjct: 150 KYENQISACKPEDDV--GGQPIVPCGLIAWSLFNDTYALSRNNVSLAVNKKGIAWKSDKE 207
Query: 184 AKFGSDVYPKNFQTGSLIGGARLNESIPLSEQEDLIVWMRTAALPTFRKLYGKIEVDLEA 243
KFG+ V+PKNFQ G++ GGA L+ IPLSEQEDLIVWMRTAALPTFRKLYGKIE DLE
Sbjct: 208 HKFGNKVFPKNFQKGNITGGATLDPRIPLSEQEDLIVWMRTAALPTFRKLYGKIESDLEM 267
Query: 244 NDEITVVIENNYNTYQFGGTKSVILSTTTWIGGKNDFLGIAYILIGGLSLVYSLVFLLMY 303
D I V + NNYNTY F G K ++LSTT+W+GGKNDFLGIAY+ +GG+ + +L F +MY
Sbjct: 268 GDTIHVKLNNNYNTYSFNGKKKLVLSTTSWLGGKNDFLGIAYLTVGGICFILALAFTIMY 327
Query: 304 LMKPRPLGDPRYLTWNKN 321
L+KPR LGDP YL+WN+N
Sbjct: 328 LVKPRRLGDPSYLSWNRN 345
>At3g12740 unknown protein
Length = 350
Score = 412 bits (1058), Expect = e-115
Identities = 197/317 (62%), Positives = 242/317 (76%), Gaps = 3/317 (0%)
Query: 4 SRFSQQELHAWQPILTPGWVIAIFTFIGLVFIPIGVASLFASEQVVEVPLRYDDQCLPSL 63
S+F+QQEL A +PILTPGWVI+ F I ++FIP+GV SLFAS+ VVE+ RYD C+P
Sbjct: 32 SKFTQQELPACKPILTPGWVISTFLIISVIFIPLGVISLFASQDVVEIVDRYDSACIPLS 91
Query: 64 YKDDAMTYIKGNRISKTCTKKLTVKSKMKAPIYVYYQLSNFYQNHRHYVKSRDHKQLRSK 123
+ + + YI+G +K+CT+ L V +MK PIYVYYQL NFYQNHR YVKSR QLRS
Sbjct: 92 DRANKVAYIQGTG-NKSCTRTLIVPKRMKQPIYVYYQLENFYQNHRRYVKSRSDSQLRSV 150
Query: 124 ADENDVGKCFPEDYTANGYLPVVPCGLAAWSLFNDTYRFSNNNKDLVINKKNIAWKSDQK 183
DEN + C PED G P+VPCGL AWSLFNDTY S NN+ L +NKK IAWKSD++
Sbjct: 151 KDENQIDACKPEDDF--GGQPIVPCGLIAWSLFNDTYVLSRNNQGLTVNKKGIAWKSDKE 208
Query: 184 AKFGSDVYPKNFQTGSLIGGARLNESIPLSEQEDLIVWMRTAALPTFRKLYGKIEVDLEA 243
KFG +V+PKNFQ G+L GGA L+ + PLS+QEDLIVWMRTAALPTFRKLYGKIE DLE
Sbjct: 209 HKFGKNVFPKNFQKGNLTGGASLDPNKPLSDQEDLIVWMRTAALPTFRKLYGKIESDLEK 268
Query: 244 NDEITVVIENNYNTYQFGGTKSVILSTTTWIGGKNDFLGIAYILIGGLSLVYSLVFLLMY 303
+ I V ++NNYNTY F G K ++LSTT+W+GGKNDFLGIAY+ +GG+ V +L F +MY
Sbjct: 269 GENIQVTLQNNYNTYSFSGKKKLVLSTTSWLGGKNDFLGIAYLTVGGICFVLALAFTVMY 328
Query: 304 LMKPRPLGDPRYLTWNK 320
L+KPR LGDP YL+WN+
Sbjct: 329 LVKPRRLGDPTYLSWNR 345
>At1g79450 unknown protein
Length = 350
Score = 408 bits (1048), Expect = e-114
Identities = 194/319 (60%), Positives = 244/319 (75%), Gaps = 3/319 (0%)
Query: 4 SRFSQQELHAWQPILTPGWVIAIFTFIGLVFIPIGVASLFASEQVVEVPLRYDDQCLPSL 63
SRF+QQEL A +PILTP WVI F G+VFIP+GV LFAS+ VVE+ RYD C+P+
Sbjct: 31 SRFTQQELPACKPILTPRWVILTFLVAGVVFIPLGVICLFASQGVVEIVDRYDTDCIPTS 90
Query: 64 YKDDAMTYIKGNRISKTCTKKLTVKSKMKAPIYVYYQLSNFYQNHRHYVKSRDHKQLRSK 123
+++ + YI+G K C + +TV MK P+YVYYQL NFYQNHR YVKSR+ QLRS
Sbjct: 91 SRNNMVAYIQGEG-DKICKRTITVTKAMKHPVYVYYQLENFYQNHRRYVKSRNDAQLRSP 149
Query: 124 ADENDVGKCFPEDYTANGYLPVVPCGLAAWSLFNDTYRFSNNNKDLVINKKNIAWKSDQK 183
+E+DV C PED G P+VPCGL AWSLFNDTY FS N++ L++NKK I+WKSD++
Sbjct: 150 KEEHDVKTCAPEDNV--GGEPIVPCGLVAWSLFNDTYSFSRNSQQLLVNKKGISWKSDRE 207
Query: 184 AKFGSDVYPKNFQTGSLIGGARLNESIPLSEQEDLIVWMRTAALPTFRKLYGKIEVDLEA 243
KFG +V+PKNFQ G+ IGG LN S PLSEQEDLIVWMRTAALPTFRKLYGKIE DL A
Sbjct: 208 NKFGKNVFPKNFQKGAPIGGGTLNISKPLSEQEDLIVWMRTAALPTFRKLYGKIETDLHA 267
Query: 244 NDEITVVIENNYNTYQFGGTKSVILSTTTWIGGKNDFLGIAYILIGGLSLVYSLVFLLMY 303
D ITV+++NNYNTY F G K ++LSTT+W+GG+NDFLGIAY+ +G + L ++ F ++Y
Sbjct: 268 GDTITVLLQNNYNTYSFNGQKKLVLSTTSWLGGRNDFLGIAYLTVGSICLFLAVTFAVLY 327
Query: 304 LMKPRPLGDPRYLTWNKNS 322
L+KPR LGDP YL+WN+++
Sbjct: 328 LVKPRQLGDPSYLSWNRSA 346
>At1g16360 unknown protein
Length = 316
Score = 377 bits (969), Expect = e-105
Identities = 178/305 (58%), Positives = 229/305 (74%), Gaps = 3/305 (0%)
Query: 18 LTPGWVIAIFTFIGLVFIPIGVASLFASEQVVEVPLRYDDQCLPSLYKDDAMTYIKGNRI 77
+T VI F G+VFIP+GV LFAS+ V+E+ RYD C+P +D+ + YI+G
Sbjct: 11 VTVSEVILTFLVSGVVFIPLGVICLFASQGVIEIVDRYDTDCIPLSSRDNKVRYIQGLE- 69
Query: 78 SKTCTKKLTVKSKMKAPIYVYYQLSNFYQNHRHYVKSRDHKQLRSKADENDVGKCFPEDY 137
K C + +TV MK P+YVYYQL N+YQNHR YVKSR QLRS DE++ C PED
Sbjct: 70 DKRCNRTITVTKTMKNPVYVYYQLENYYQNHRRYVKSRQDGQLRSPKDEHETKSCAPEDT 129
Query: 138 TANGYLPVVPCGLAAWSLFNDTYRFSNNNKDLVINKKNIAWKSDQKAKFGSDVYPKNFQT 197
G P+VPCGL AWSLFNDTY F+ NN+ L +NKK+I+WKSD+++KFG +V+PKNFQ
Sbjct: 130 L--GGQPIVPCGLVAWSLFNDTYDFTRNNQKLPVNKKDISWKSDRESKFGKNVFPKNFQK 187
Query: 198 GSLIGGARLNESIPLSEQEDLIVWMRTAALPTFRKLYGKIEVDLEANDEITVVIENNYNT 257
GSLIGG L++ IPLSEQEDLIVWMRTAALPTFRKLYGKI+ DL+A D I V+++NNYNT
Sbjct: 188 GSLIGGKSLDQDIPLSEQEDLIVWMRTAALPTFRKLYGKIDTDLQAGDTIKVLLQNNYNT 247
Query: 258 YQFGGTKSVILSTTTWIGGKNDFLGIAYILIGGLSLVYSLVFLLMYLMKPRPLGDPRYLT 317
Y F G K ++LSTT+W+GG+NDFLGIAY+ +G + L ++ F ++YL KPR LGDP YL+
Sbjct: 248 YSFNGKKKLVLSTTSWLGGRNDFLGIAYLTVGSICLFLAVSFSVLYLAKPRQLGDPSYLS 307
Query: 318 WNKNS 322
WN+++
Sbjct: 308 WNRSA 312
>At5g46150 unknown protein
Length = 343
Score = 354 bits (909), Expect = 3e-98
Identities = 170/318 (53%), Positives = 228/318 (71%), Gaps = 5/318 (1%)
Query: 5 RFSQQELHAWQPILTPGWVIAIFTFIGLVFIPIGVASLFASEQVVEVPLRYDDQCLPSLY 64
+F QQ+L A +P+LTP VI +F +G VFIPIG+ +L AS +E+ RYD +C+P Y
Sbjct: 28 QFKQQKLPACKPVLTPISVITVFMLMGFVFIPIGLITLRASRDAIEIIDRYDVECIPEEY 87
Query: 65 KDDAMTYIKGNRISKTCTKKLTVKSKMKAPIYVYYQLSNFYQNHRHYVKSRDHKQLRSKA 124
+ + + YI + I K CT+ L V+ MKAPI++YYQL N+YQNHR YVKSR +QL
Sbjct: 88 RTNKLLYITDSSIPKNCTRYLKVQKYMKAPIFIYYQLDNYYQNHRRYVKSRSDQQLLHGL 147
Query: 125 DENDVGKCFPEDYTANGYLPVVPCGLAAWSLFNDTYRFSNNNKDLVINKKNIAWKSDQKA 184
+ + C PE+ ++NG LP+VPCGL AWS+FNDT+ FS L +++ NIAWKSD++
Sbjct: 148 EYSHTSSCEPEE-SSNG-LPIVPCGLIAWSMFNDTFTFSRERTKLNVSRNNIAWKSDREH 205
Query: 185 KFGSDVYPKNFQTGSLIGGARLNESIPLSEQEDLIVWMRTAALPTFRKLYGKIEVDLEAN 244
KFG +VYP NFQ G+LIGGA+L+ IPLS+QED IVWMR AAL +FRKLYG+IE DLE
Sbjct: 206 KFGKNVYPINFQNGTLIGGAKLDPKIPLSDQEDFIVWMRAAALLSFRKLYGRIEEDLEPG 265
Query: 245 DEITVVIENNYNTYQFGGTKSVILSTTTWIGGKNDFLGIAYILIGGLSLVYSLVFLLMYL 304
+ V + NNYNTY F G K +ILST+ W+GG+NDFLGI Y+++G S+V S++F+L++L
Sbjct: 266 KVVEVNLMNNYNTYSFSGQKKLILSTSNWLGGRNDFLGITYLVVGSSSIVISIIFMLLHL 325
Query: 305 MKPRPLGDPRYLTWNKNS 322
PRP GD +WNK S
Sbjct: 326 KNPRPYGDN---SWNKKS 340
>At2g31180 putative transcription factor (MYB14)
Length = 249
Score = 29.3 bits (64), Expect = 3.0
Identities = 25/91 (27%), Positives = 40/91 (43%), Gaps = 5/91 (5%)
Query: 100 QLSNFYQNHRHYVKSRDHKQLRSKADENDVGKCFPEDYTANGYLPVVPCGLAAWSLFNDT 159
++ N + H +K R K L + D DV G + V L +S T
Sbjct: 103 EIKNVWHTH---LKKRLSKNLNNGGDTKDVNGINETTNEDKGSVIVDTASLQQFSNSITT 159
Query: 160 YRFSNNNKDLVINKKNIAWKSDQKAKFGSDV 190
+ SN+NKD +++ ++I+ D F SDV
Sbjct: 160 FDISNDNKDDIMSYEDISALIDD--SFWSDV 188
>At1g09910 hypothetical protein
Length = 675
Score = 29.3 bits (64), Expect = 3.0
Identities = 19/64 (29%), Positives = 30/64 (46%), Gaps = 2/64 (3%)
Query: 110 HYVK-SRDHKQLRSKADENDVGKCFPEDYTANGYLPVVPCGLAAWSLFNDTYRFSNNNKD 168
HY+ + D K++ D+ G+C DY L PC +D Y++S NKD
Sbjct: 197 HYMAVADDRKRIMPFPDDLCKGRCQTLDYQEASLL-TAPCDPRLQGEVDDKYQYSCENKD 255
Query: 169 LVIN 172
L ++
Sbjct: 256 LRVH 259
>At2g46850 putative Ser/Thr protein kinase
Length = 633
Score = 28.9 bits (63), Expect = 4.0
Identities = 15/39 (38%), Positives = 22/39 (55%)
Query: 59 CLPSLYKDDAMTYIKGNRISKTCTKKLTVKSKMKAPIYV 97
CL S +KD Y +I K KKLTV + + AP+++
Sbjct: 258 CLKSCFKDGKELYGDKCKIKKHNGKKLTVLAGVLAPLFI 296
>At1g07280 unknown protein
Length = 302
Score = 28.9 bits (63), Expect = 4.0
Identities = 17/70 (24%), Positives = 32/70 (45%), Gaps = 9/70 (12%)
Query: 187 GSDVYPKNFQTGSLIGGARLNESIPLSEQEDLIVW---------MRTAALPTFRKLYGKI 237
GSD + K+ + S +G + + +S QE++ +W M T T + + +
Sbjct: 111 GSDQFRKSQFSSSSLGATSADSDVSVSGQEEIRLWNSILEETAKMETLDHETMKGMVSPV 170
Query: 238 EVDLEANDEI 247
E LEA + +
Sbjct: 171 EARLEAEESM 180
>At3g24790 protein kinase, putative
Length = 379
Score = 28.1 bits (61), Expect = 6.8
Identities = 16/44 (36%), Positives = 24/44 (54%), Gaps = 4/44 (9%)
Query: 188 SDVYPKNFQTGSLIGGARLNESIPLSEQEDLIVWMRTAALPTFR 231
SDVY LI G R+ +++ S +++L+ W ALP FR
Sbjct: 248 SDVYSFGVVLLELISGRRVIDTMRPSHEQNLVTW----ALPIFR 287
>At3g58660 putative protein
Length = 446
Score = 27.7 bits (60), Expect = 8.8
Identities = 20/45 (44%), Positives = 24/45 (52%), Gaps = 1/45 (2%)
Query: 50 EVPLRYDDQCLPSLYKDDAMTYIKGNRISKTCTKKLT-VKSKMKA 93
E+ L DD+ L KDDAM IK I T KL+ +KS KA
Sbjct: 93 EICLIIDDRPKSGLTKDDAMKKIKSENIPITKVIKLSKLKSDYKA 137
>At2g26910 putative ABC transporter
Length = 1420
Score = 27.7 bits (60), Expect = 8.8
Identities = 11/26 (42%), Positives = 19/26 (72%)
Query: 274 IGGKNDFLGIAYILIGGLSLVYSLVF 299
+G K+DFLG++ I++ + +SLVF
Sbjct: 1383 MGYKHDFLGVSAIMVVAFCVFFSLVF 1408
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.320 0.138 0.422
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 7,881,354
Number of Sequences: 26719
Number of extensions: 350236
Number of successful extensions: 717
Number of sequences better than 10.0: 12
Number of HSP's better than 10.0 without gapping: 7
Number of HSP's successfully gapped in prelim test: 5
Number of HSP's that attempted gapping in prelim test: 701
Number of HSP's gapped (non-prelim): 12
length of query: 326
length of database: 11,318,596
effective HSP length: 100
effective length of query: 226
effective length of database: 8,646,696
effective search space: 1954153296
effective search space used: 1954153296
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.8 bits)
S2: 60 (27.7 bits)
Medicago: description of AC126009.1


