

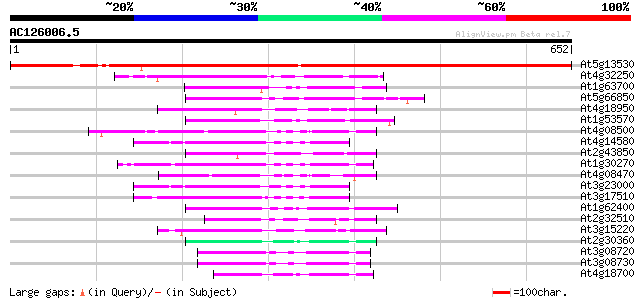
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC126006.5 + phase: 0
(652 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
At5g13530 ankyrin-repeat containing protein 945 0.0
At4g32250 unknown protein 138 1e-32
At1g63700 putative protein kinase 78 1e-14
At5g66850 MAP protein kinase like protein 77 2e-14
At4g18950 protein kinase like protein 77 4e-14
At1g53570 MEK kinase MAP3Ka, putative 76 5e-14
At4g08500 MEKK1/MAP kinase kinase kinase 74 2e-13
At4g14580 CBL-interacting protein kinase 4 (CIPK4) 73 6e-13
At2g43850 ankyrin-kinase like protein (APK1) 72 7e-13
At1g30270 serine/threonine kinase, putative 72 7e-13
At4g08470 putative mitogen-activated protein kinase 72 1e-12
At3g23000 SNF1 related protein kinase (ATSRPK1) 72 1e-12
At3g17510 CBL-interacting protein kinase 1 (CIPK1) 71 2e-12
At1g62400 hypothetical protein 71 2e-12
At2g32510 putative protein kinase 70 4e-12
At3g15220 putative MAP kinase 69 8e-12
At2g30360 putative protein kinase 69 8e-12
At3g08720 putative ribosomal-protein S6 kinase (ATPK19) 68 1e-11
At3g08730 putative ribosomal-protein S6 kinase (ATPK6) 68 2e-11
At4g18700 putative protein kinase 67 2e-11
>At5g13530 ankyrin-repeat containing protein
Length = 834
Score = 945 bits (2443), Expect = 0.0
Identities = 467/661 (70%), Positives = 541/661 (81%), Gaps = 24/661 (3%)
Query: 1 MKIPCCSVCQTRYNEEERVPLLLQCGHGFCKECLSRMFSSSSDANLTCPRCRHVSTVGNS 60
+K+PCCSVC TRYNE+ERVPLLLQCGHGFCK+CLS+MFS+SSD LTCPRCRHVS VGNS
Sbjct: 5 VKVPCCSVCHTRYNEDERVPLLLQCGHGFCKDCLSKMFSTSSDTTLTCPRCRHVSVVGNS 64
Query: 61 VQALRKNYAVLSLILSAADSAAAAGGGGGGDCDFTDDDEDRDDSEVDDGDDQKLDCRKNS 120
VQ LRKNYA+L+LI AA GG DCD+TDD++D D+ +DG D+ D + +
Sbjct: 65 VQGLRKNYAMLALI-------HAASGGANFDCDYTDDEDDDDE---EDGSDE--DGARAA 112
Query: 121 RGSQASSSGG--CAPVIEVGVHQDLKLVRRIGE----GRRAGVEMWSAVI--GGGRCKHQ 172
RG ASSS C PVIEVG H ++KLVR+IGE G GVEMW A + GGGRCKH+
Sbjct: 113 RGFHASSSINSLCGPVIEVGAHPEMKLVRQIGEESSSGGFGGVEMWDATVAGGGGRCKHR 172
Query: 173 VAVKKVVLNEGMDLDWMLGKLEDLRRTSMWCRNVCTFHGAMKVDEGLCLVMDKCFGSVQS 232
VAVKK+ L E MD++WM G+LE LRR SMWCRNVCTFHG +K+D LCL+MD+CFGSVQS
Sbjct: 173 VAVKKMTLTEDMDVEWMQGQLESLRRASMWCRNVCTFHGVVKMDGSLCLLMDRCFGSVQS 232
Query: 233 EMLRNEGRLTLEQVLRYGADIARGVVELHAAGVVCMSLKPSNLLLDANGHAVVSDYGLAT 292
EM RNEGRLTLEQ+LRYGAD+ARGV ELHAAGV+CM++KPSNLLLDA+G+AVVSDYGLA
Sbjct: 233 EMQRNEGRLTLEQILRYGADVARGVAELHAAGVICMNIKPSNLLLDASGNAVVSDYGLAP 292
Query: 293 ILKKPSCWKARPECDSAKIHSCMECIMLSPHYTAPEAWEPVKKSLNLFWDDGIGISPESD 352
ILKKP+C K RPE DS+K+ +C+ LSPHYTAPEAW PVKK LFW+D G+SPESD
Sbjct: 293 ILKKPTCQKTRPEFDSSKVTLYTDCVTLSPHYTAPEAWGPVKK---LFWEDASGVSPESD 349
Query: 353 AWSFGCTLVEMCTGAIPWAGLSAEEIYRQVVKAKKQPPQYASVVGGGIPRELWKMIGECL 412
AWSFGCTLVEMCTG+ PW GLS EEI++ VVKA+K PPQY +VG GIPRELWKMIGECL
Sbjct: 350 AWSFGCTLVEMCTGSTPWDGLSREEIFQAVVKARKVPPQYERIVGVGIPRELWKMIGECL 409
Query: 413 QFKPSKRPTFNAMLAIFLRHLQEIPRSPPASPDNDLVKGSVSNVTEASPVPELEIPQD-P 471
QFKPSKRPTFNAMLA FLRHLQEIPRSP ASPDN + K N+ +A + + QD P
Sbjct: 410 QFKPSKRPTFNAMLATFLRHLQEIPRSPSASPDNGIAKICEVNIVQAPRATNIGVFQDNP 469
Query: 472 NRLHRLVSEGDVTGVRDFLAKAASENESNFISSLLEAQNADGQTALHLACRRGSAELVET 531
N LHR+V EGD GVR+ LAKAA+ + + SLLEAQNADGQ+ALHLACRRGSAELVE
Sbjct: 470 NNLHRVVLEGDFEGVRNILAKAAAGGGGSSVRSLLEAQNADGQSALHLACRRGSAELVEA 529
Query: 532 ILDYPEANVDVLDKDGDPPLVFALAAGSHECVCSLIKRNANVTSRLRDGLGPSVAHVCAY 591
IL+Y EANVD++DKDGDPPLVFALAAGS +CV LIK+ ANV SRLR+G GPSVAHVC+Y
Sbjct: 530 ILEYGEANVDIVDKDGDPPLVFALAAGSPQCVHVLIKKGANVRSRLREGSGPSVAHVCSY 589
Query: 592 HGQPDCMRELLLAGADPNAVDDEGESVLHRAIAKKFTDCALVIVENGGCRSMAISNSKNL 651
HGQPDCMRELL+AGADPNAVDDEGE+VLHRA+AKK+TDCA+VI+ENGG RSM +SN+K L
Sbjct: 590 HGQPDCMRELLVAGADPNAVDDEGETVLHRAVAKKYTDCAIVILENGGSRSMTVSNAKCL 649
Query: 652 T 652
T
Sbjct: 650 T 650
Score = 40.8 bits (94), Expect = 0.002
Identities = 42/150 (28%), Positives = 57/150 (38%), Gaps = 49/150 (32%)
Query: 496 ENESNFISSLLEAQNAD--------GQTALHLACRRGSAELVETILDYPEANVDVLDKDG 547
E E + +L A AD G+TALH A + ELV ILD N ++ +
Sbjct: 701 EKEGRELVQILLAAGADPTAQDAQHGRTALHTAAMANNVELVRVILD-AGVNANIRNVHN 759
Query: 548 DPPLVFALAAGSHECVCSLIKRNANVTSRLRDGLGPSVAHVCAYHGQPDCMRELLLAGAD 607
PL ALA G++ C+ LL +G+D
Sbjct: 760 TIPLHMALARGANS-----------------------------------CVSLLLESGSD 784
Query: 608 PNAVDDEGESVLHRAIAKKFTDCALVIVEN 637
N DDEG++ H A D A +I EN
Sbjct: 785 CNIQDDEGDNAFHIA-----ADAAKMIREN 809
>At4g32250 unknown protein
Length = 611
Score = 138 bits (347), Expect = 1e-32
Identities = 102/317 (32%), Positives = 157/317 (49%), Gaps = 36/317 (11%)
Query: 123 SQASSSGGCAPVIEVGVHQDLKLVRRIGEGRRAGVEMWSAVIGGGRCK----HQVAVKKV 178
S+++ + G +P + LKL RIG G V W A H+VA+K +
Sbjct: 22 SESALAAGTSPWMNSST---LKLRHRIGRGPFGDV--WLATHHQSTEDYDEHHEVAIKML 76
Query: 179 VLNEGMDLDWMLGKLEDLRRTSMWCRNVCTFHGAMKVDEGLCLVMDKCFGSVQSEMLRNE 238
+ ++ K EDL NVC G ++ +C+VM GS+ +M R +
Sbjct: 77 YPIKEDQRRVVVDKFEDLFSKCQGLENVCLLRGVSSINGKICVVMKFYEGSLGDKMARLK 136
Query: 239 G-RLTLEQVLRYGADIARGVVELHAAGVVCMSLKPSNLLLDANGHAVVSDYGLATILKKP 297
G +L+L VLRYG D+A G++ELH+ G + ++LKPSN LL N A++ D G+ +L
Sbjct: 137 GGKLSLPDVLRYGVDLATGILELHSKGFLILNLKPSNFLLSDNDKAILGDVGIPYLLLSI 196
Query: 298 SCWKARPECDSAKIHSCMECIMLSPHYTAPEAWEPVKKSLNLFWDDGIGISPESDAWSFG 357
P D M + +P+Y APE W+P D +S E+D+W FG
Sbjct: 197 PL----PSSD-------MTERLGTPNYMAPEQWQP---------DVRGPMSFETDSWGFG 236
Query: 358 CTLVEMCTGAIPWAGLSAEEIYRQVVKAKKQPPQYASVVGGGIPRELWKMIGECLQFKPS 417
C++VEM TG PW+G SA+EIY VV+ +++ + IP L ++ C +
Sbjct: 237 CSIVEMLTGVQPWSGRSADEIYDLVVRKQEK-----LSIPSSIPPPLENLLRGCFMYDLR 291
Query: 418 KRPTFNAMLAIFLRHLQ 434
RP+ +L + L+ LQ
Sbjct: 292 SRPSMTDILLV-LKSLQ 307
>At1g63700 putative protein kinase
Length = 883
Score = 78.2 bits (191), Expect = 1e-14
Identities = 64/241 (26%), Positives = 107/241 (43%), Gaps = 42/241 (17%)
Query: 204 RNVCTFHGAMKVDEGLCLVMDKCFGSVQSEMLRNEGRLTLEQVLRYGADIARGVVELHAA 263
+N+ ++G+ VD+ L + ++ G ++L+ G+ + Y I G+ LHA
Sbjct: 460 QNIVQYYGSETVDDKLYIYLEYVSGGSIYKLLQEYGQFGENAIRNYTQQILSGLAYLHAK 519
Query: 264 GVVCMSLKPSNLLLDANGHAVVSDYGLA---TILKKPSCWKARPECDSAKIHSCMECIML 320
V +K +N+L+D +G V+D+G+A T P +K
Sbjct: 520 NTVHRDIKGANILVDPHGRVKVADFGMAKHITAQSGPLSFKG------------------ 561
Query: 321 SPHYTAPEAWEPVKKSLNLFWDDGIGISPESDAWSFGCTLVEMCTGAIPWAGLSAEEIYR 380
SP++ APE V K+ N G + D WS GCT++EM T PW+
Sbjct: 562 SPYWMAPE----VIKNSN-------GSNLAVDIWSLGCTVLEMATTKPPWSQYEGVPAMF 610
Query: 381 QVVKAKKQP--PQYASVVGGGIPRELWKMIGECLQFKPSKRPTFNAML-AIFLRHLQEIP 437
++ +K+ P P + S G R +CLQ P+ RPT +L F+R++ +
Sbjct: 611 KIGNSKELPDIPDHLSEEGKDFVR-------KCLQRNPANRPTAAQLLDHAFVRNVMPME 663
Query: 438 R 438
R
Sbjct: 664 R 664
>At5g66850 MAP protein kinase like protein
Length = 533
Score = 77.4 bits (189), Expect = 2e-14
Identities = 73/283 (25%), Positives = 123/283 (42%), Gaps = 35/283 (12%)
Query: 205 NVCTFHGAMKVDEGLCLVMDKCF-GSVQSEMLRNEGRLTLEQVLRYGADIARGVVELHAA 263
N+ + G+ V++ + ++ GS+ + + G +T V + I G+ LH
Sbjct: 224 NIVQYFGSETVEDRFFIYLEYVHPGSINKYIRDHCGTMTESVVRNFTRHILSGLAYLHNK 283
Query: 264 GVVCMSLKPSNLLLDANGHAVVSDYGLATILKKPSCWKARPECDSAKIHSCMECIMLSPH 323
V +K +NLL+DA+G ++D+G+A L D + + SP+
Sbjct: 284 KTVHRDIKGANLLVDASGVVKLADFGMAKHL-------TGQRADLS--------LKGSPY 328
Query: 324 YTAPEAWEPVKKSLNLFWDDGIGISPESDAWSFGCTLVEMCTGAIPWAGLSAEEIYRQVV 383
+ APE + V + D ++ D WS GCT++EM TG PW+ +V+
Sbjct: 329 WMAPELMQAVMQK-----DSNPDLAFAVDIWSLGCTIIEMFTGKPPWSEFEGAAAMFKVM 383
Query: 384 KAKKQPPQYASVVGGGIPRELWKMIGECLQFKPSKRPTFNAMLA-IFLRHLQEIPRSPPA 442
+ P+ S G R C Q P++RPT + +L FL++ + P SP
Sbjct: 384 RDSPPIPESMSPEGKDFLR-------LCFQRNPAERPTASMLLEHRFLKNSLQ-PTSPSN 435
Query: 443 SPDNDLVKGSVSNVTEAS---PVPELEIPQDPNRLHRLVSEGD 482
S + L G N+TE S P ++ Q P + SE +
Sbjct: 436 SDVSQLFNG--MNITEPSSRREKPNFKLDQVPRARNMTSSESE 476
>At4g18950 protein kinase like protein
Length = 459
Score = 76.6 bits (187), Expect = 4e-14
Identities = 71/260 (27%), Positives = 113/260 (43%), Gaps = 34/260 (13%)
Query: 172 QVAVKKVVLNEGMDLDWMLGKLEDLRRTSMWCR--NVCTFHGAMKVDEGLCLVMDKCFGS 229
QVAVKK+ +E + D + K D R N+ F GA+ + +V +
Sbjct: 180 QVAVKKLD-DEVLSDDDQVRKFHDELALLQRLRHPNIVQFLGAVTQSNPMMIVTEYLPRG 238
Query: 230 VQSEMLRNEGRLTLEQVLRYGADIARGVVELH---AAGVVCMSLKPSNLLLDANGHAVVS 286
E+L+ +G+L +RY DIARG+ LH ++ L+PSN+L D +GH V+
Sbjct: 239 DLRELLKRKGQLKPATAVRYALDIARGMSYLHEIKGDPIIHRDLEPSNILRDDSGHLKVA 298
Query: 287 DYGLATILKKPSCWKARPECDSAKIHSCMECIMLSPHYTAPEAWEPVKKSLNLFWDDGIG 346
D+G++ ++ + K C +S Y APE + +
Sbjct: 299 DFGVSKLV-------------TVKEDKPFTCQDISCRYIAPEVFTSEE------------ 333
Query: 347 ISPESDAWSFGCTLVEMCTGAIPWAGLSAEEIYRQVVKAKKQPPQYASVVGGGIPRELWK 406
++D +SF + EM G +P+A E+ A K P + P L
Sbjct: 334 YDTKADVFSFALIVQEMIEGRMPFA--EKEDSEASEAYAGKHRPLF-KAPSKNYPHGLKT 390
Query: 407 MIGECLQFKPSKRPTFNAML 426
+I EC KP+KRPTF ++
Sbjct: 391 LIEECWHEKPAKRPTFREII 410
Score = 41.6 bits (96), Expect = 0.001
Identities = 26/83 (31%), Positives = 43/83 (51%), Gaps = 11/83 (13%)
Query: 473 RLHRLVSEGDVTGVRDFLAKAASENESNFISSLLEAQNADGQTALHLACRRGSAELVETI 532
RL L +EGD+ G+++ + N ++ D +TALH+A +G ++VE +
Sbjct: 46 RLMYLANEGDIEGIKELIDSGIDANY----------RDIDDRTALHVAACQGLKDVVELL 95
Query: 533 LDYPEANVDVLDKDGDPPLVFAL 555
LD +A VD D+ G P A+
Sbjct: 96 LD-RKAEVDPKDRWGSTPFADAI 117
>At1g53570 MEK kinase MAP3Ka, putative
Length = 609
Score = 76.3 bits (186), Expect = 5e-14
Identities = 63/250 (25%), Positives = 104/250 (41%), Gaps = 38/250 (15%)
Query: 205 NVCTFHGAMKVDEGLCLVMDKCFGSVQSEMLRNEGRLTLEQVLRYGADIARGVVELHAAG 264
N+ ++G+ +E L + ++ G ++L++ G T + Y I G+ LH
Sbjct: 275 NIVQYYGSELSEETLSVYLEYVSGGSIHKLLKDYGSFTEPVIQNYTRQILAGLAYLHGRN 334
Query: 265 VVCMSLKPSNLLLDANGHAVVSDYGLATILKKPSCWKARPECDSAKIHSCMECIMLSPHY 324
V +K +N+L+D NG ++D+G+A S M SP++
Sbjct: 335 TVHRDIKGANILVDPNGEIKLADFGMA---------------KHVTAFSTMLSFKGSPYW 379
Query: 325 TAPEAWEPVKKSLNLFWDDGIGISPESDAWSFGCTLVEMCTGAIPWAGLSAEEIYRQVVK 384
APE V S N G + D WS GCT++EM T PW+ ++
Sbjct: 380 MAPE----VVMSQN-------GYTHAVDIWSLGCTILEMATSKPPWSQFEGVAAIFKIGN 428
Query: 385 AKKQPPQYASVVGGGIPRELWKMIGECLQFKPSKRPTFNAMLA-IFLRHLQEIPRS---- 439
+K P + + + I CLQ P+ RPT + +L FLR+ + +
Sbjct: 429 SKDTPE-----IPDHLSNDAKNFIRLCLQRNPTVRPTASQLLEHPFLRNTTRVASTSLPK 483
Query: 440 --PPASPDND 447
PP S D +
Sbjct: 484 DFPPRSYDGN 493
>At4g08500 MEKK1/MAP kinase kinase kinase
Length = 608
Score = 74.3 bits (181), Expect = 2e-13
Identities = 89/350 (25%), Positives = 150/350 (42%), Gaps = 55/350 (15%)
Query: 92 CDFTDDDEDRDDSE------VDDGDDQKLDCR-KNSRGSQASSSGGCAPVIEVG--VHQD 142
CD + E+ ++E + GD C + G +S+ +P+ G +
Sbjct: 273 CDEEEGKEEEAEAEEMGARFIQLGDTADETCSFTTNEGDSSSTVSNTSPIYPDGGAIITS 332
Query: 143 LKLVRRIGEGRRAGVEMWSAVIGGGRCKHQVAVKKV-VLNEGMDLDWMLGKLEDLRR--T 199
+ + +G G V + + G G AVK+V +L++G + +LE + +
Sbjct: 333 WQKGQLLGRGSFGSV--YEGISGDGDF---FAVKEVSLLDQGSQAQECIQQLEGEIKLLS 387
Query: 200 SMWCRNVCTFHGAMKVDEGLCLVMDKCFGSVQSEMLRNEGRLTLEQ--VLRYGADIARGV 257
+ +N+ + G K L + ++ Q +L+ R L V Y I G+
Sbjct: 388 QLQHQNIVRYRGTAKDGSNLYIFLELV---TQGSLLKLYQRYQLRDSVVSLYTRQILDGL 444
Query: 258 VELHAAGVVCMSLKPSNLLLDANGHAVVSDYGLATILKKPSCWKARPECDSAKIHSCMEC 317
LH G + +K +N+L+DANG ++D+GLA + K I SC
Sbjct: 445 KYLHDKGFIHRDIKCANILVDANGAVKLADFGLAKVSKFND------------IKSCKG- 491
Query: 318 IMLSPHYTAPEAWEPVKKSLNLFWDDGIGISPESDAWSFGCTLVEMCTGAIPWAGLS-AE 376
+P + APE +N DG G SP +D WS GCT++EMCTG IP++ L +
Sbjct: 492 ---TPFWMAPEV-------INRKDSDGYG-SP-ADIWSLGCTVLEMCTGQIPYSDLEPVQ 539
Query: 377 EIYRQVVKAKKQPPQYASVVGGGIPRELWKMIGECLQFKPSKRPTFNAML 426
++R + P S+ + I +CL+ P +RPT +L
Sbjct: 540 ALFRIGRGTLPEVPDTLSL-------DARLFILKCLKVNPEERPTAAELL 582
>At4g14580 CBL-interacting protein kinase 4 (CIPK4)
Length = 426
Score = 72.8 bits (177), Expect = 6e-13
Identities = 66/251 (26%), Positives = 111/251 (43%), Gaps = 25/251 (9%)
Query: 144 KLVRRIGEGRRAGVEMWSAVIGGGRCKHQVAVKKVVLNEGMDLDWMLGKLEDLRRTSMWC 203
+L RR+G G A V + ++ G ++ K+ ++ GM+ ++ ++E +RR
Sbjct: 22 ELGRRLGSGSFAKVHVARSISTGELVAIKIIDKQKTIDSGME-PRIIREIEAMRRLHNH- 79
Query: 204 RNVCTFHGAMKVDEGLCLVMDKCFGSVQSEMLRNEGRLTLEQVLRYGADIARGVVELHAA 263
NV H M + LV++ G L GRL RY +A + H
Sbjct: 80 PNVLKIHEVMATKSKIYLVVEYAAGGELFTKLIRFGRLNESAARRYFQQLASALSFCHRD 139
Query: 264 GVVCMSLKPSNLLLDANGHAVVSDYGLATILKKPSCWKARPECDSAKIHSCMECIMLSPH 323
G+ +KP NLLLD G+ VSD+GL+ + + S ++ +H+ +P
Sbjct: 140 GIAHRDVKPQNLLLDKQGNLKVSDFGLSALPEHRS--------NNGLLHTACG----TPA 187
Query: 324 YTAPEAWEPVKKSLNLFWDDGIGISPESDAWSFGCTLVEMCTGAIPWAGLSAEEIYRQVV 383
YTAPE + DG ++DAWS G L + G +P+ + +YR++
Sbjct: 188 YTAPEV-------IAQRGYDG----AKADAWSCGVFLFVLLAGYVPFDDANIVAMYRKIH 236
Query: 384 KAKKQPPQYAS 394
K + P + S
Sbjct: 237 KRDYRFPSWIS 247
>At2g43850 ankyrin-kinase like protein (APK1)
Length = 468
Score = 72.4 bits (176), Expect = 7e-13
Identities = 62/225 (27%), Positives = 101/225 (44%), Gaps = 25/225 (11%)
Query: 205 NVCTFHGAMKVDEGLCLVMDKCFGSVQSEMLRNEGRLTLEQVLRYGADIARGVVELHAA- 263
NV F GA+ + + +V++ S L+ +GRL+ + LR+ DIARG+ LH
Sbjct: 241 NVIQFVGAVTQNIPMMIVVEYNPKGDLSVYLQKKGRLSPSKALRFALDIARGMNYLHECK 300
Query: 264 --GVVCMSLKPSNLLLDANGHAVVSDYGLATILKKPSCWKARPECDSAKIHSCMECIMLS 321
++ LKP N+LLD G +S +G+ + K D AK+ + I LS
Sbjct: 301 PDPIIHCDLKPKNILLDRGGQLKISGFGMIRLSKISQ--------DKAKVANHKAHIDLS 352
Query: 322 PHYTAPEAWEPVKKSLNLFWDDGIGISPESDAWSFGCTLVEMCTGAIPWAGLSAEEIYRQ 381
+Y APE ++ L + DA SFG L E+ G + EE+ R
Sbjct: 353 NYYIAPEVYKDEIFDLRV------------DAHSFGVILYEITEGVPVFHPRPPEEVARM 400
Query: 382 VVKAKKQPPQYASVVGGGIPRELWKMIGECLQFKPSKRPTFNAML 426
+ K+P P ++ ++I +C + RPTF+ ++
Sbjct: 401 MCLEGKRP--VFKTKSRSYPPDIKELIEKCWHPEAGIRPTFSEII 443
Score = 41.2 bits (95), Expect = 0.002
Identities = 38/144 (26%), Positives = 61/144 (41%), Gaps = 28/144 (19%)
Query: 433 LQEIPRSPPASPDNDLVKGSVSNVTEASPVPELEIPQDPNRLHRLV---SEGDVTGVRDF 489
L I RSP +S +D P + +P++ + +L+ S+GDV G+ +
Sbjct: 52 LDPIRRSPDSSKSDD--------------EPHMSVPENLDSTMQLLFMASKGDVRGIEEL 97
Query: 490 LAKAASENESNFISSLLEAQNADGQTALHLACRRGSAELVETILDYPEANVDVLDKDGDP 549
L + N + DG+TALH+A G +V+ +L AN+D D+ G
Sbjct: 98 LDEGIDVNSIDL----------DGRTALHIAACEGHLGVVKALLS-RRANIDARDRWGST 146
Query: 550 PLVFALAAGSHECVCSLIKRNANV 573
A G+ + L R A V
Sbjct: 147 AAADAKYYGNLDVYNLLKARGAKV 170
>At1g30270 serine/threonine kinase, putative
Length = 482
Score = 72.4 bits (176), Expect = 7e-13
Identities = 80/297 (26%), Positives = 123/297 (40%), Gaps = 39/297 (13%)
Query: 126 SSSGGCAPVIEVGVHQDLKLVRRIGEGRRAGVEMWSAVIGGGRCKHQVAVKKVVLNEGMD 185
SSSGG VG ++ L R +GEG A V+ V G +V K+ VL M
Sbjct: 20 SSSGGRT---RVGKYE---LGRTLGEGTFAKVKFARNVENGDNVAIKVIDKEKVLKNKMI 73
Query: 186 LDWMLGKLEDLRRTSMWCRNVCTFHGAMKVDEGLCLVMDKCFGSVQSEMLRNEGRLTLEQ 245
K E + NV M + V++ G + + + GRL ++
Sbjct: 74 AQI---KREISTMKLIKHPNVIRMFEVMASKTKIYFVLEFVTGGELFDKISSNGRLKEDE 130
Query: 246 VLRYGADIARGVVELHAAGVVCMSLKPSNLLLDANGHAVVSDYGLATILKKPSCWKARPE 305
+Y + V H+ GV LKP NLLLDANG VSD+GL+ + ++
Sbjct: 131 ARKYFQQLINAVDYCHSRGVYHRDLKPENLLLDANGALKVSDFGLSALPQQVR------- 183
Query: 306 CDSAKIHSCMECIMLSPHYTAPEAWEPVKKSLNLFWDDGIGISPESDAWSFGCTLVEMCT 365
+ +H+ +P+Y APE +N DG ++D WS G L +
Sbjct: 184 -EDGLLHTTCG----TPNYVAPEV-------INNKGYDG----AKADLWSCGVILFVLMA 227
Query: 366 GAIPWAGLSAEEIYRQVVKAKKQPPQYASVVGGGIPRELWKMIGECLQFKPSKRPTF 422
G +P+ + +Y+++ KA+ P + S K+I L P+ R TF
Sbjct: 228 GYLPFEDSNLTSLYKKIFKAEFTCPPWFSA-------SAKKLIKRILDPNPATRITF 277
>At4g08470 putative mitogen-activated protein kinase
Length = 560
Score = 71.6 bits (174), Expect = 1e-12
Identities = 76/265 (28%), Positives = 120/265 (44%), Gaps = 53/265 (20%)
Query: 174 AVKKV-VLNEGMDLDWMLGKLED--LRRTSMWCRNVCTFHGAMKVDEGLCLVMDKCF-GS 229
AVK+V +L++G+ + +LE + + +N+ + G K L + ++ GS
Sbjct: 329 AVKEVSLLDKGIQAQECIQQLEGEIALLSQLQHQNIVRYRGTAKDVSKLYIFLELVTQGS 388
Query: 230 VQSEMLRNEGRLTLEQVLRYGADIARGVVELHAAGVVCMSLKPSNLLLDANGHAVVSDYG 289
VQ L +L+ V Y I G+ LH G V +K +N+L+DANG ++D+G
Sbjct: 389 VQK--LYERYQLSYTVVSLYTRQILAGLNYLHDKGFVHRDIKCANMLVDANGTVKLADFG 446
Query: 290 LATILKKPSCWKARPECDSAKIHSCMECIMLSPHYTAPEAWEPVKKSLNLFWDDGIGISP 349
LA +++K + M C + + APE +N DG G SP
Sbjct: 447 LA---------------EASKFNDIMSC-KGTLFWMAPEV-------INRKDSDGNG-SP 482
Query: 350 ESDAWSFGCTLVEMCTGAIPWAGLSAEEIYRQVVKAKKQPPQYASVVGGG--------IP 401
+D WS GCT++EMCTG IP++ L +P Q A +G G +
Sbjct: 483 -ADIWSLGCTVLEMCTGQIPYSDL--------------KPIQAAFKIGRGTLPDVPDTLS 527
Query: 402 RELWKMIGECLQFKPSKRPTFNAML 426
+ I CL+ P +RPT +L
Sbjct: 528 LDARHFILTCLKVNPEERPTAAELL 552
>At3g23000 SNF1 related protein kinase (ATSRPK1)
Length = 429
Score = 71.6 bits (174), Expect = 1e-12
Identities = 66/252 (26%), Positives = 109/252 (43%), Gaps = 28/252 (11%)
Query: 144 KLVRRIGEGRRAGVEMWSAVIGGGRCKHQVAVKKVVLNEGMDLDWMLGKLEDLRRTSMWC 203
+L RR+G G A V + ++ ++ KK + GM+ ++ +++ +RR
Sbjct: 26 ELGRRLGSGSFAKVHLARSIESDELVAVKIIEKKKTIESGME-PRIIREIDAMRRLRHH- 83
Query: 204 RNVCTFHGAMKVDEGLCLVMDKCFGSVQSEMLRNEGRLTLEQVLRYGADIARGVVELHAA 263
N+ H M + LVM+ G + GRL RY +A + H
Sbjct: 84 PNILKIHEVMATKSKIYLVMELASGGELFSKVLRRGRLPESTARRYFQQLASALRFSHQD 143
Query: 264 GVVCMSLKPSNLLLDANGHAVVSDYGLATILKKPSCWKARPE-CDSAKIHSCMECIMLSP 322
GV +KP NLLLD G+ VSD+GL+ A PE + +H+ +P
Sbjct: 144 GVAHRDVKPQNLLLDEQGNLKVSDFGLS----------ALPEHLQNGLLHTACG----TP 189
Query: 323 HYTAPEAWEPVKKSLNLFWDDGIGISPESDAWSFGCTLVEMCTGAIPWAGLSAEEIYRQV 382
YTAPE ++ DG ++DAWS G L + G +P+ + +YR++
Sbjct: 190 AYTAPEV-------ISRRGYDG----AKADAWSCGVILFVLLVGDVPFDDSNIAAMYRKI 238
Query: 383 VKAKKQPPQYAS 394
+ + P + S
Sbjct: 239 HRRDYRFPSWIS 250
>At3g17510 CBL-interacting protein kinase 1 (CIPK1)
Length = 444
Score = 70.9 bits (172), Expect = 2e-12
Identities = 63/252 (25%), Positives = 109/252 (43%), Gaps = 28/252 (11%)
Query: 144 KLVRRIGEGRRAGVEMWSAVIGGGRCKHQVAVKKVVLNEGMDLDWMLGKLEDLRRTSMWC 203
+L R +GEG V+ + G H AVK + + DL++ L ++R M
Sbjct: 21 ELGRTLGEGNFGKVKFAKDTVSG----HSFAVKIIDKSRIADLNFSLQIKREIRTLKMLK 76
Query: 204 R-NVCTFHGAMKVDEGLCLVMDKCFGSVQSEMLRNEGRLTLEQVLRYGADIARGVVELHA 262
++ H + + +VM+ G + + + G+LT + + G+ H+
Sbjct: 77 HPHIVRLHEVLASKTKINMVMELVTGGELFDRIVSNGKLTETDGRKMFQQLIDGISYCHS 136
Query: 263 AGVVCMSLKPSNLLLDANGHAVVSDYGLATILKKPSCWKARPECDSAKIHSCMECIMLSP 322
GV LK N+LLDA GH ++D+GL+ + P ++ D +H+ SP
Sbjct: 137 KGVFHRDLKLENVLLDAKGHIKITDFGLSAL---PQHFR-----DDGLLHTTCG----SP 184
Query: 323 HYTAPEAWEPVKKSLNLFWDDGIGISPESDAWSFGCTLVEMCTGAIPWAGLSAEEIYRQV 382
+Y APE N +D SD WS G L + TG +P+ + +Y+++
Sbjct: 185 NYVAPEV------LANRGYD-----GAASDIWSCGVILYVILTGCLPFDDRNLAVLYQKI 233
Query: 383 VKAKKQPPQYAS 394
K P++ S
Sbjct: 234 CKGDPPIPRWLS 245
>At1g62400 hypothetical protein
Length = 345
Score = 70.9 bits (172), Expect = 2e-12
Identities = 64/248 (25%), Positives = 113/248 (44%), Gaps = 34/248 (13%)
Query: 205 NVCTFHGAMKVDEGLCLVMDKCF-GSVQSEMLRNEG-RLTLEQVLRYGADIARGVVELHA 262
N+ F A K C++ + G+++ + + E L++E VLR DI+RG+ LH+
Sbjct: 101 NIVQFIAACKKPPVYCIITEYMSQGNLRMYLNKKEPYSLSIETVLRLALDISRGMEYLHS 160
Query: 263 AGVVCMSLKPSNLLLDANGHAVVSDYGLATILKKPSCWKARPECDSAKIHSCMECIMLSP 322
GV+ LK +NLLL+ V+D+G + + +C AK + M +
Sbjct: 161 QGVIHRDLKSNNLLLNDEMRVKVADFGTSCL---------ETQCREAKGN------MGTY 205
Query: 323 HYTAPEAWEPVKKSLNLFWDDGIGISPESDAWSFGCTLVEMCTGAIPWAGLSAEEIYRQV 382
+ APE +K+ + + D +SFG L E+ T +P+ G++ + V
Sbjct: 206 RWMAPEM---IKEK---------PYTRKVDVYSFGIVLWELTTALLPFQGMTPVQAAFAV 253
Query: 383 VKAKKQPPQYASVVGGGIPRELWKMIGECLQFKPSKRPTFNAMLAIFLRHLQEIPRSPPA 442
+ ++PP AS L +I C PSKRP F+ ++A+ ++ + + P
Sbjct: 254 AEKNERPPLPAS-----CQPALAHLIKRCWSENPSKRPDFSNIVAVLEKYDECVKEGLPL 308
Query: 443 SPDNDLVK 450
+ L K
Sbjct: 309 TSHASLTK 316
>At2g32510 putative protein kinase
Length = 372
Score = 70.1 bits (170), Expect = 4e-12
Identities = 51/205 (24%), Positives = 95/205 (45%), Gaps = 39/205 (19%)
Query: 227 FGSVQSEMLRNEGRLTLEQVLRYGADIARGVVELHAAGVVCMSLKPSNLLLDANGHAVVS 286
+G++ ++ GR+ +V++Y DI +G+ +H+ G+V +K SN+++ G A ++
Sbjct: 84 YGTLTDAAAKDGGRVDETRVVKYTRDILKGLEYIHSKGIVHCDVKGSNVVISEKGEAKIA 143
Query: 287 DYGLATILKKPSCWKARPECDSAKIHSCMECIMLSPHYTAPEAWEPVKKSLNLFWDDGIG 346
D+G A + P +S +M +P + APE K+
Sbjct: 144 DFGCAK--------RVDPVFESP--------VMGTPAFMAPEVARGEKQ----------- 176
Query: 347 ISPESDAWSFGCTLVEMCTGAIPWAGLSAEE-----IYRQVVKAKKQPPQYASVVGGGIP 401
ESD W+ GCT++EM TG+ PW + E +YR V + P+ + +
Sbjct: 177 -GKESDIWAVGCTMIEMVTGSPPWTKADSREDPVSVLYR--VGYSSETPELPCL----LA 229
Query: 402 RELWKMIGECLQFKPSKRPTFNAML 426
E + +CL+ + ++R T +L
Sbjct: 230 EEAKDFLEKCLKREANERWTATQLL 254
>At3g15220 putative MAP kinase
Length = 690
Score = 68.9 bits (167), Expect = 8e-12
Identities = 58/273 (21%), Positives = 122/273 (44%), Gaps = 43/273 (15%)
Query: 172 QVAVKKVVLNEGMDLDWMLGKLEDLRR-----TSMWCRNVCTFHGAMKVDEGLCLVMDKC 226
+VA+K + L E D ++ED+++ + C + ++G+ L ++M+
Sbjct: 40 EVAIKVIDLEESED------EIEDIQKEISVLSQCRCPYITEYYGSYLHQTKLWIIMEYM 93
Query: 227 FGSVQSEMLRNEGRLTLEQVLRYGADIARGVVELHAAGVVCMSLKPSNLLLDANGHAVVS 286
G +++L++ L + D+ V LH G + +K +N+LL NG V+
Sbjct: 94 AGGSVADLLQSNNPLDETSIACITRDLLHAVEYLHNEGKIHRDIKAANILLSENGDVKVA 153
Query: 287 DYGLATILKKPSCWKARPECDSAKIHSCMECIMLSPHYTAPEAWEPVKKSLNLFWDDGIG 346
D+G++ L + S + + +P + APE + + G
Sbjct: 154 DFGVSAQL--------------TRTISRRKTFVGTPFWMAPEVIQ-----------NSEG 188
Query: 347 ISPESDAWSFGCTLVEMCTGAIPWAGLSAEEIYRQVVKAKKQPPQYASVVGGGIPRELWK 406
+ ++D WS G T++EM G P A L + + ++ PPQ + R++ +
Sbjct: 189 YNEKADIWSLGITVIEMAKGEPPLADLHPMRVL--FIIPRETPPQ----LDEHFSRQVKE 242
Query: 407 MIGECLQFKPSKRPTFNAMLA-IFLRHLQEIPR 438
+ CL+ P++RP+ ++ F+++ ++ P+
Sbjct: 243 FVSLCLKKAPAERPSAKELIKHRFIKNARKSPK 275
>At2g30360 putative protein kinase
Length = 435
Score = 68.9 bits (167), Expect = 8e-12
Identities = 54/222 (24%), Positives = 88/222 (39%), Gaps = 30/222 (13%)
Query: 205 NVCTFHGAMKVDEGLCLVMDKCFGSVQSEMLRNEGRLTLEQVLRYGADIARGVVELHAAG 264
N+ H M + M+ G + GRL+ + RY + V HA G
Sbjct: 81 NIVKLHEVMATKSKIFFAMEFVKGGELFNKISKHGRLSEDLSRRYFQQLISAVGYCHARG 140
Query: 265 VVCMSLKPSNLLLDANGHAVVSDYGLATILKKPSCWKARPECDSAKIHSCMECIMLSPHY 324
V LKP NLL+D NG+ VSD+GL+ + D + + + +P Y
Sbjct: 141 VYHRDLKPENLLIDENGNLKVSDFGLSAL------------TDQIRPDGLLHTLCGTPAY 188
Query: 325 TAPEAWEPVKKSLNLFWDDGIGISPESDAWSFGCTLVEMCTGAIPWAGLSAEEIYRQVVK 384
APE KK + D WS G L + G +P+ + +Y+++ K
Sbjct: 189 VAPEILS--KKGYE---------GAKVDVWSCGIVLFVLVAGYLPFNDPNVMNMYKKIYK 237
Query: 385 AKKQPPQYASVVGGGIPRELWKMIGECLQFKPSKRPTFNAML 426
+ + P++ S +L + + L P R T + +L
Sbjct: 238 GEYRFPRWMS-------PDLKRFVSRLLDINPETRITIDEIL 272
>At3g08720 putative ribosomal-protein S6 kinase (ATPK19)
Length = 471
Score = 68.2 bits (165), Expect = 1e-11
Identities = 57/201 (28%), Positives = 90/201 (44%), Gaps = 35/201 (17%)
Query: 219 LCLVMDKCFGSVQSEMLRNEGRLTLEQVLRYGADIARGVVELHAAGVVCMSLKPSNLLLD 278
L LV+D G L ++G + Y A+I V LH G++ LKP N+L+D
Sbjct: 213 LYLVLDFINGGHLFFQLYHQGLFREDLARVYTAEIVSAVSHLHEKGIMHRDLKPENILMD 272
Query: 279 ANGHAVVSDYGLATILKKPSCWKARPECDSAKIHSCMECIMLSPHYTAPEAWEPVKKSLN 338
+GH +++D+GLA ++ + ++ C + + Y APE
Sbjct: 273 VDGHVMLTDFGLAKEFEENT--RSNSMCGTTE-------------YMAPEIVR------- 310
Query: 339 LFWDDGIGISPESDAWSFGCTLVEMCTGAIPWAGLSAEEIYRQVVKAKKQPPQYASVVGG 398
G G +D WS G L EM TG P+ G S +I +++VK K + PQ+ S
Sbjct: 311 -----GKGHDKAADWWSVGILLYEMLTGKPPFLG-SKGKIQQKIVKDKIKLPQFLS---- 360
Query: 399 GIPRELWKMIGECLQFKPSKR 419
E ++ LQ +P +R
Sbjct: 361 ---NEAHALLKGLLQKEPERR 378
>At3g08730 putative ribosomal-protein S6 kinase (ATPK6)
Length = 465
Score = 67.8 bits (164), Expect = 2e-11
Identities = 57/201 (28%), Positives = 90/201 (44%), Gaps = 35/201 (17%)
Query: 219 LCLVMDKCFGSVQSEMLRNEGRLTLEQVLRYGADIARGVVELHAAGVVCMSLKPSNLLLD 278
L LV+D G L ++G + Y A+I V LH G++ LKP N+L+D
Sbjct: 207 LYLVLDFINGGHLFFQLYHQGLFREDLARVYTAEIVSAVSHLHEKGIMHRDLKPENILMD 266
Query: 279 ANGHAVVSDYGLATILKKPSCWKARPECDSAKIHSCMECIMLSPHYTAPEAWEPVKKSLN 338
+GH +++D+GLA ++ + ++ C + + Y APE
Sbjct: 267 TDGHVMLTDFGLAKEFEENT--RSNSMCGTTE-------------YMAPEIVR------- 304
Query: 339 LFWDDGIGISPESDAWSFGCTLVEMCTGAIPWAGLSAEEIYRQVVKAKKQPPQYASVVGG 398
G G +D WS G L EM TG P+ G S +I +++VK K + PQ+ S
Sbjct: 305 -----GKGHDKAADWWSVGILLYEMLTGKPPFLG-SKGKIQQKIVKDKIKLPQFLS---- 354
Query: 399 GIPRELWKMIGECLQFKPSKR 419
E ++ LQ +P +R
Sbjct: 355 ---NEAHAILKGLLQKEPERR 372
>At4g18700 putative protein kinase
Length = 489
Score = 67.4 bits (163), Expect = 2e-11
Identities = 50/185 (27%), Positives = 81/185 (43%), Gaps = 30/185 (16%)
Query: 238 EGRLTLEQVLRYGADIARGVVELHAAGVVCMSLKPSNLLLDANGHAVVSDYGLATILKKP 297
+GRL E +Y + V HA GV LKP NLLLD NG+ VSD+GL+ +
Sbjct: 117 KGRLKEEVARKYFQQLISAVTFCHARGVYHRDLKPENLLLDENGNLKVSDFGLSAV---- 172
Query: 298 SCWKARPECDSAKIHSCMECIMLSPHYTAPEAWEPVKKSLNLFWDDGIGISPESDAWSFG 357
D + +P Y APE +K + + + D WS G
Sbjct: 173 --------SDQIRQDGLFHTFCGTPAYVAPEVL--ARKGYD---------AAKVDIWSCG 213
Query: 358 CTLVEMCTGAIPWAGLSAEEIYRQVVKAKKQPPQYASVVGGGIPRELWKMIGECLQFKPS 417
L + G +P+ + +Y+++ + + + P++ S EL +++ + L+ P
Sbjct: 214 VILFVLMAGYLPFHDRNVMAMYKKIYRGEFRCPRWFST-------ELTRLLSKLLETNPE 266
Query: 418 KRPTF 422
KR TF
Sbjct: 267 KRFTF 271
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.318 0.135 0.415
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 15,917,019
Number of Sequences: 26719
Number of extensions: 731285
Number of successful extensions: 6358
Number of sequences better than 10.0: 1189
Number of HSP's better than 10.0 without gapping: 634
Number of HSP's successfully gapped in prelim test: 558
Number of HSP's that attempted gapping in prelim test: 4050
Number of HSP's gapped (non-prelim): 1975
length of query: 652
length of database: 11,318,596
effective HSP length: 106
effective length of query: 546
effective length of database: 8,486,382
effective search space: 4633564572
effective search space used: 4633564572
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 63 (28.9 bits)
Medicago: description of AC126006.5


