

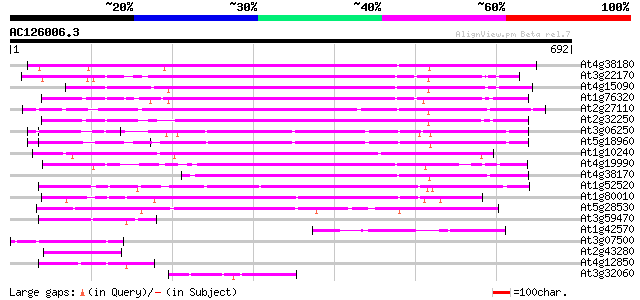
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC126006.3 + phase: 0
(692 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
At4g38180 unknown protein (At4g38180) 372 e-103
At3g22170 far-red impaired response protein, putative 310 2e-84
At4g15090 unknown protein 300 2e-81
At1g76320 putative phytochrome A signaling protein 296 2e-80
At2g27110 Mutator-like transposase 284 1e-76
At2g32250 Mutator-like transposase 257 1e-68
At3g06250 unknown protein 253 2e-67
At5g18960 FAR1 - like protein 246 4e-65
At1g10240 unknown protein 229 4e-60
At4g19990 putative protein 227 1e-59
At4g38170 hypothetical protein 217 2e-56
At1g52520 F6D8.26 198 7e-51
At1g80010 hypothetical protein 197 2e-50
At5g28530 far-red impaired response protein (FAR1) - like 196 4e-50
At3g59470 unknown protein 91 2e-18
At1g42570 hypothetical protein 82 8e-16
At3g07500 hypothetical protein 77 4e-14
At2g43280 unknown protein 72 8e-13
At4g12850 putative protein 70 5e-12
At3g32060 hypothetical protein 59 1e-08
>At4g38180 unknown protein (At4g38180)
Length = 788
Score = 372 bits (955), Expect = e-103
Identities = 221/648 (34%), Positives = 327/648 (50%), Gaps = 23/648 (3%)
Query: 22 NIVQNGEESVVDYI---PHLEMEFESEAAAYEFYNKYSRRIGFGIRREYGNKSKKDGILT 78
N N EE D + P+ +EFESE AA FYN Y+RRIGF R +S++DG +
Sbjct: 56 NYFPNQEEEACDLLDLEPYDGLEFESEEAAKAFYNSYARRIGFSTRVSSSRRSRRDGAII 115
Query: 79 SRRFTCFKEGKRGVDKR---DHLTKEGRVETRTGCDARMVISLDRKIGKYKVVDFVAQHN 135
R+F C KEG R ++++ D K R TR GC A + + + GK+ V FV HN
Sbjct: 116 QRQFVCAKEGFRNMNEKRTKDREIKRPRTITRVGCKASLSVKMQDS-GKWLVSGFVKDHN 174
Query: 136 HLLEPPGYFHTPRSHRQISESRACQVVAADESRLKRKDFQEYVFKQDGGIDDVG------ 189
H L PP H RSHRQIS + + + + + K+ GGI VG
Sbjct: 175 HELVPPDQVHCLRSHRQISGPAKTLIDTLQAAGMGPRRIMSALIKEYGGISKVGFTEVDC 234
Query: 190 --YLQTRRMRSLMYGEVGALLMHFKRQS-ENPSFYYDFQMDVEEKITNVFWADAQMINDY 246
Y++ R +S+ GE+ LL + ++ + +NP+F+Y Q ++ + NVFWAD + I D+
Sbjct: 235 RNYMRNNRQKSIE-GEIQLLLDYLRQMNADNPNFFYSVQGSEDQSVGNVFWADPKAIMDF 293
Query: 247 GCSGDVITFDTTYMTNKDYRPLGVFVGLNNHKQMVVFGATLLYDETIPSFQWLFETFLKA 306
GD +TFDTTY +N+ P F G+N+H Q ++FG + +ET SF WLF T+L A
Sbjct: 294 THFGDTVTFDTTYRSNRYRLPFAPFTGVNHHGQPILFGCAFIINETEASFVWLFNTWLAA 353
Query: 307 MGGEKPKTLLTDQDEAMAKAISVVMPQTFHGLCTWRIRENAQTHVNHLYQKSSKFCSDFE 366
M P ++ TD D + AI V P H C W I + Q ++H++ K F SDF
Sbjct: 354 MSAHPPVSITTDHDAVIRAAIMHVFPGARHRFCKWHILKKCQEKLSHVFLKHPSFESDFH 413
Query: 367 ACIDLHEEEGEFLNSWNVLLVEHNVSEDSWLRMIFQLKEKWAWVYVRKHFTAGMRSTQLS 426
C++L E +F W LL ++ + + WL+ I+ + +W VY+R F A M T S
Sbjct: 414 KCVNLTESVEDFERCWFSLLDKYELRDHEWLQAIYSDRRQWVPVYLRDTFFADMSLTHRS 473
Query: 427 ESFNAELKNYLKSDLNLVQFFSHFGRIVHGIRNNESEADYESRHKLPKLKMKRAPMLVQA 486
+S N+ Y+ + NL QFF + + + E +ADY++ + P LK +PM QA
Sbjct: 474 DSINSYFDGYINASTNLSQFFKLYEKALESRLEKEVKADYDTMNSPPVLKTP-SPMEKQA 532
Query: 487 GNIYTPKTFEEFQEEYEEYLGTCVKNLKEG----LYVVTNY-DNNKERMVIGNLMDQKVA 541
+YT K F FQEE L + Y V Y + +K V N+++ +
Sbjct: 533 SELYTRKLFMRFQEELVGTLTFMASKADDDGDLVTYQVAKYGEAHKAHFVKFNVLEMRAN 592
Query: 542 CDCRKFETHGILCSHALKVLDVMNIKLIPQHYILKRWTRDARLGSNHDLKGQHIELDTKA 601
C C+ FE GI+C H L V V N+ +P +YILKRWTR+A+ D H +
Sbjct: 593 CSCQMFEFSGIICRHILAVFRVTNLLTLPPYYILKRWTRNAKSSVIFDDYNLHAYANYLE 652
Query: 602 HFMRRYNDLCPQMIKLINKASKSHETYTFLSKVFEESDKIIDDMLAKK 649
RYN L + + +A KS T +E+ K + + K+
Sbjct: 653 SHTVRYNTLRHKASNFVQEAGKSLYTCDVAVVALQEAAKTVSLAMNKE 700
>At3g22170 far-red impaired response protein, putative
Length = 814
Score = 310 bits (793), Expect = 2e-84
Identities = 204/650 (31%), Positives = 316/650 (48%), Gaps = 62/650 (9%)
Query: 15 LGNNIDVNIVQNGEESVVDYIPHLE---------------MEFESEAAAYEFYNKYSRRI 59
+G DV++ N ++SV +P E MEFES AY FY +YSR +
Sbjct: 34 IGKIEDVSVEVNTDDSVGMGVPTGELVEYTEGMNLEPLNGMEFESHGEAYSFYQEYSRAM 93
Query: 60 GFGIRREYGNKSKKDGILTSRRFTCFKEG-KRGVDK-----RDHLTKEG-------RVET 106
GF + +SK +F C + G KR DK R +K+ R
Sbjct: 94 GFNTAIQNSRRSKTTREFIDAKFACSRYGTKREYDKSFNRPRARQSKQDPENMAGRRTCA 153
Query: 107 RTGCDARMVISLDRKIGKYKVVDFVAQHNHLLEPPGYFHTPRSHRQISESRACQVVAADE 166
+T C A M + R GK+ + FV +HNH L P Q + ++ AA
Sbjct: 154 KTDCKASMHVKR-RPDGKWVIHSFVREHNHELLPA----------QAVSEQTRKIYAA-- 200
Query: 167 SRLKRKDFQEYVFKQDGGIDDVGYLQTRRMRSLMYGEVGALLMHFKR-QSENPSFYYDFQ 225
K F EY D + R S+ G+ LL R QS N +F+Y
Sbjct: 201 ---MAKQFAEYKTVISLKSDSKSSFEKGRTLSVETGDFKILLDFLSRMQSLNSNFFYAVD 257
Query: 226 MDVEEKITNVFWADAQMINDYGCSGDVITFDTTYMTNKDYRPLGVFVGLNNHKQMVVFGA 285
+ ++++ NVFW DA+ ++YG DV++ DTTY+ NK PL +FVG+N H Q +V G
Sbjct: 258 LGDDQRVKNVFWVDAKSRHNYGSFCDVVSLDTTYVRNKYKMPLAIFVGVNQHYQYMVLGC 317
Query: 286 TLLYDETIPSFQWLFETFLKAMGGEKPKTLLTDQDEAMAKAISVVMPQTFHGLCTWRIRE 345
L+ DE+ ++ WL ET+L+A+GG+ PK L+T+ D M + + P T H L W +
Sbjct: 318 ALISDESAATYSWLMETWLRAIGGQAPKVLITELDVVMNSIVPEIFPNTRHCLFLWHVLM 377
Query: 346 NAQTHVNHLYQKSSKFCSDFEACIDLHEEEGEFLNSWNVLLVEHNVSEDSWLRMIFQLKE 405
++ + ++ F FE CI ++ +F W L + +D W+ +++ ++
Sbjct: 378 KVSENLGQVVKQHDNFMPKFEKCIYKSGKDEDFARKWYKNLARFGLKDDQWMISLYEDRK 437
Query: 406 KWAWVYVRKHFTAGMRSTQLSESFNAELKNYLKSDLNLVQFFSHFGRIVHGIRNNESEAD 465
KWA Y+ AGM ++Q ++S NA Y+ ++ +F + ++ E++AD
Sbjct: 438 KWAPTYMTDVLLAGMSTSQRADSINAFFDKYMHKKTSVQEFVKVYDTVLQDRCEEEAKAD 497
Query: 466 YESRHKLPKLKMKRAPMLVQAGNIYTPKTFEEFQEEYEEYLGTCVKNLKE-------GLY 518
E +K P +K +P +YTP F++FQ E LG + +E +
Sbjct: 498 SEMWNKQPAMK-SPSPFEKSVSEVYTPAVFKKFQ---IEVLGAIACSPREENRDATCSTF 553
Query: 519 VVTNYDNNKERMVIGNLMDQKVACDCRKFETHGILCSHALKVLDVMNIKLIPQHYILKRW 578
V +++NN++ MV N +V+C CR FE G LC H L VL ++ IP YILKRW
Sbjct: 554 RVQDFENNQDFMVTWNQTKAEVSCICRLFEYKGYLCRHTLNVLQCCHLSSIPSQYILKRW 613
Query: 579 TRDARLGSNHDLKGQHIELDTKAHFMRRYNDLCPQMIKLINKASKSHETY 628
T+DA+ S H G+ +L T+ + RYNDLC + +KL +AS S E+Y
Sbjct: 614 TKDAK--SRH-FSGEPQQLQTR---LLRYNDLCERALKLNEEASLSQESY 657
>At4g15090 unknown protein
Length = 768
Score = 300 bits (767), Expect = 2e-81
Identities = 184/592 (31%), Positives = 301/592 (50%), Gaps = 26/592 (4%)
Query: 70 KSKKDGILTSRRFTCFKEGKRGVDKRDHLTKEGRVETRTGCDARMVISLDRKIGKYKVVD 129
+SKK +F C + G + + +T C A M + R GK+ + +
Sbjct: 26 RSKKTKDFIDAKFACSRYGVTPESESSGSSSRRSTVKKTDCKASMHVKR-RPDGKWIIHE 84
Query: 130 FVAQHNHLLEPPGYFHTP-RSHRQISESRACQVVAADESRLKRKDFQEYVFKQDGGIDDV 188
FV HNH L P +H + + +++E ++ A R K+ + + +Q GG ++
Sbjct: 85 FVKDHNHELLPALAYHFRIQRNVKLAEKNNIDILHAVSERTKKMYVE--MSRQSGGYKNI 142
Query: 189 G-YLQTR--------RMRSLMYGEVGALLMHFKR-QSENPSFYYDFQMDVEEKITNVFWA 238
G LQT R +L G+ LL +FKR + ENP F+Y ++ ++++ N+FWA
Sbjct: 143 GSLLQTDVSSQVDKGRYLALEEGDSQVLLEYFKRIKKENPKFFYAIDLNEDQRLRNLFWA 202
Query: 239 DAQMINDYGCSGDVITFDTTYMTNKDYRPLGVFVGLNNHKQMVVFGATLLYDETIPSFQW 298
DA+ +DY DV++FDTTY+ D PL +F+G+N+H Q ++ G L+ DE++ +F W
Sbjct: 203 DAKSRDDYLSFNDVVSFDTTYVKFNDKLPLALFIGVNHHSQPMLLGCALVADESMETFVW 262
Query: 299 LFETFLKAMGGEKPKTLLTDQDEAMAKAISVVMPQTFHGLCTWRIRENAQTHVNHLYQKS 358
L +T+L+AMGG PK +LTDQD+ + A+S ++P T H W + E + +H+ ++
Sbjct: 263 LIKTWLRAMGGRAPKVILTDQDKFLMSAVSELLPNTRHCFALWHVLEKIPEYFSHVMKRH 322
Query: 359 SKFCSDFEACIDLHEEEGEFLNSWNVLLVEHNVSEDSWLRMIFQLKEKWAWVYVRKHFTA 418
F F CI + EF W ++ + + D WL + + ++KW ++ F A
Sbjct: 323 ENFLLKFNKCIFRSWTDDEFDMRWWKMVSQFGLENDEWLLWLHEHRQKWVPTFMSDVFLA 382
Query: 419 GMRSTQLSESFNAELKNYLKSDLNLVQFFSHFGRIVHGIRNNESEADYESRHKLPKLKMK 478
GM ++Q SES N+ Y+ + L +F +G I+ ES AD+++ HK P LK
Sbjct: 383 GMSTSQRSESVNSFFDKYIHKKITLKEFLRQYGVILQNRYEEESVADFDTCHKQPALK-S 441
Query: 479 RAPMLVQAGNIYTPKTFEEFQEEYEEYLGTCVKNLKE----GLYVVTNYDNNKERMVIGN 534
+P Q YT F++FQ E + + KE + V + + + + +V +
Sbjct: 442 PSPWEKQMATTYTHTIFKKFQVEVLGVVACHPRKEKEDENMATFRVQDCEKDDDFLVTWS 501
Query: 535 LMDQKVACDCRKFETHGILCSHALKVLDVMNIKLIPQHYILKRWTRDARLGSNHDLKGQH 594
++ C CR FE G LC HAL +L + IP YILKRWT+DA+ G L G+
Sbjct: 502 KTKSELCCFCRMFEYKGFLCRHALMILQMCGFASIPPQYILKRWTKDAKSGV---LAGEG 558
Query: 595 I-ELDTKAHFMRRYNDLCPQMIKLINKASKSHETYTFLSKVFEESDKIIDDM 645
++ T+ ++RYNDLC + +L + S E Y + E+ K DM
Sbjct: 559 ADQIQTR---VQRYNDLCSRATELSEEGCVSEENYNIALRTLVETLKNCVDM 607
>At1g76320 putative phytochrome A signaling protein
Length = 670
Score = 296 bits (759), Expect = 2e-80
Identities = 194/617 (31%), Positives = 314/617 (50%), Gaps = 37/617 (5%)
Query: 40 MEFESEAAAYEFYNKYSRRIGFGIRREYGNKSKKDGILTSRRFTCFKEGKRGVDKRDHLT 99
MEFE+ AY FY Y++ +GFG + +S+ +F+C + G + ++
Sbjct: 1 MEFETHEDAYLFYKDYAKSVGFGTAKLSSRRSRASKEFIDAKFSCIRYGSK---QQSDDA 57
Query: 100 KEGRVETRTGCDARMVISLDRKIGKYKVVDFVAQHNHLLEPPGYFHTPRSHRQISESRAC 159
R + GC A M + R GK+ V FV +HNH L P H RSHR
Sbjct: 58 INPRASPKIGCKASMHVKR-RPDGKWYVYSFVKEHNHDLLPE-QAHYFRSHRNT------ 109
Query: 160 QVVAADESRLKRK------DFQEYVFKQDGGIDDVGYLQTR----RMRSLMYGEVGALLM 209
++V +++SRL+RK D + D D GY++ + R L G+ LL
Sbjct: 110 ELVKSNDSRLRRKKNTPLTDCKHLSAYHDLDFID-GYMRNQHDKGRRLVLDTGDAEILLE 168
Query: 210 HFKR-QSENPSFYYDFQMDVEEKITNVFWADAQMINDYGCSGDVITFDTTYMTNKDYRPL 268
R Q ENP F++ + + NVFW DA+ I DY DV++F+T+Y +K PL
Sbjct: 169 FLMRMQEENPKFFFAVDFSEDHLLRNVFWVDAKGIEDYKSFSDVVSFETSYFVSKYKVPL 228
Query: 269 GVFVGLNNHKQMVVFGATLLYDETIPSFQWLFETFLKAMGGEKPKTLLTDQDEAMAKAIS 328
+FVG+N+H Q V+ G LL D+T+ ++ WL +++L AMGG+KPK +LTDQ+ A+ AI+
Sbjct: 229 VLFVGVNHHVQPVLLGCGLLADDTVYTYVWLMQSWLVAMGGQKPKVMLTDQNNAIKAAIA 288
Query: 329 VVMPQTFHGLCTWRIRENAQTHVNHLYQKSSKFCSDFEACIDLHEEEGEFLNSWNVLLVE 388
V+P+T H C W + + ++++ F CI E EF W L+ +
Sbjct: 289 AVLPETRHCYCLWHVLDQLPRNLDYWSMWQDTFMKKLFKCIYRSWSEEEFDRRWLKLIDK 348
Query: 389 HNVSEDSWLRMIFQLKEKWAWVYVRKHFTAGMRSTQLSESFNAELKNYLKSDLNLVQFFS 448
++ + W+R +++ ++ WA ++R AG+ SES N+ Y+ + +L +F
Sbjct: 349 FHLRDVPWMRSLYEERKFWAPTFMRGITFAGLSMRCRSESVNSLFDRYVHPETSLKEFLE 408
Query: 449 HFGRIVHGIRNNESEADYESRHKLPKLKMKRAPMLVQAGNIYTPKTFEEFQEEYEEYLGT 508
+G ++ E++AD+++ H+ P+LK +P Q +Y+ + F FQ E LG
Sbjct: 409 GYGLMLEDRYEEEAKADFDAWHEAPELK-SPSPFEKQMLLVYSHEIFRRFQ---LEVLGA 464
Query: 509 C----VKNLKEG-LYVVTNYDNNKERMVIGNLMDQKVACDCRKFETHGILCSHALKVLDV 563
K +EG Y V ++D+ ++ +V + + C CR FE G LC HA+ VL +
Sbjct: 465 AACHLTKESEEGTTYSVKDFDDEQKYLVDWDEFKSDIYCSCRSFEYKGYLCRHAIVVLQM 524
Query: 564 MNIKLIPQHYILKRWTRDARLGSNHDLKGQHIELDTKAHFMRRYNDLCPQMIKLINKASK 623
+ IP +Y+L+RWT AR + H + L+ +RR+NDLC + I L + S
Sbjct: 525 SGVFTIPINYVLQRWTNAAR--NRHQISR---NLELVQSNIRRFNDLCRRAIILGEEGSL 579
Query: 624 SHETYTFLSKVFEESDK 640
S E+Y +E+ K
Sbjct: 580 SQESYDIAMFAMKEAFK 596
>At2g27110 Mutator-like transposase
Length = 851
Score = 284 bits (726), Expect = 1e-76
Identities = 197/651 (30%), Positives = 303/651 (46%), Gaps = 38/651 (5%)
Query: 17 NNIDVNIVQNGEESVVDYIPHLEMEFESEAAAYEFYNKYSRRIGFGIRREYGNKSKKDGI 76
NN+D ++ E + + P + MEF SE A FY++YSR++GF + + DG
Sbjct: 32 NNMDNSLGVQDEIGIAE--PCVGMEFNSEKEAKSFYDEYSRQLGFTSKL----LPRTDGS 85
Query: 77 LTSRRFTCFKEGKRGVDKRDHLTKEGRVETRTGCDARMVISLDRKIGKYKVVDFVAQHNH 136
++ R F C KR + CDA + I L K+ V FV +H H
Sbjct: 86 VSVREFVCSSSSKRSKRRLSE-----------SCDAMVRIELQGH-EKWVVTKFVKEHTH 133
Query: 137 LLEPPGYFHTPRSHRQISESRACQVVAADESRLKRKDFQEYVFKQDGGIDDVGYLQTRRM 196
L H R R + S ++ + + YV
Sbjct: 134 GLASSNMLHCLRPRRHFANSEK----SSYQEGVNVPSGMMYVSMDANSRGARNASMATNT 189
Query: 197 RSLMYGEVGALLMHFKR-QSENPSFYYDFQMDVEEKITNVFWADAQMINDYGCSGDVITF 255
+ + + LL +FKR Q+ENP F+Y Q+D + +++NVFWAD++ Y GD +T
Sbjct: 190 KRTIGRDAHNLLEYFKRMQAENPGFFYAVQLDEDNQMSNVFWADSRSRVAYTHFGDTVTL 249
Query: 256 DTTYMTNKDYRPLGVFVGLNNHKQMVVFGATLLYDETIPSFQWLFETFLKAMGGEKPKTL 315
DT Y N+ P F G+N+H Q ++FG L+ DE+ SF WLF+TFL AM + P +L
Sbjct: 250 DTRYRCNQFRVPFAPFTGVNHHGQAILFGCALILDESDTSFIWLFKTFLTAMRDQPPVSL 309
Query: 316 LTDQDEAMAKAISVVMPQTFHGLCTWRIRENAQTHVNHLYQKSSKFCSDFEACIDLHEEE 375
+TDQD A+ A V P H + W + Q + H+ F + CI+ E
Sbjct: 310 VTDQDRAIQIAAGQVFPGARHCINKWDVLREGQEKLAHVCLAYPSFQVELYNCINFTETI 369
Query: 376 GEFLNSWNVLLVEHNVSEDSWLRMIFQLKEKWAWVYVRKHFTAGMRSTQ-LSESFNAELK 434
EF +SW+ ++ ++++ WL ++ + +W VY R F A + +Q S SF
Sbjct: 370 EEFESSWSSVIDKYDLGRHEWLNSLYNARAQWVPVYFRDSFFAAVFPSQGYSGSF---FD 426
Query: 435 NYLKSDLNLVQFFSHFGRIVHGIRNNESEADYESRHKLPKLKMKRAPMLVQAGNIYTPKT 494
Y+ L FF + R + E EAD ++ + P LK +PM QA N++T K
Sbjct: 427 GYVNQQTTLPMFFRLYERAMESWFEMEIEADLDTVNTPPVLKTP-SPMENQAANLFTRKI 485
Query: 495 FEEFQEEYEEYLGTCVKNLKE----GLYVVTNYDN-NKERMVIGNLMDQKVACDCRKFET 549
F +FQEE E +++ + V N++N NK +V + + C C+ FE
Sbjct: 486 FGKFQEELVETFAHTANRIEDDGTTSTFRVANFENDNKAYIVTFCYPEMRANCSCQMFEH 545
Query: 550 HGILCSHALKVLDVMNIKLIPQHYILKRWTRDARLGSNHDLKGQHIELDTKAHFMRRYND 609
GILC H L V V NI +P HYIL+RWTR+A+ D +H+ + + RYN
Sbjct: 546 SGILCRHVLTVFTVTNILTLPPHYILRRWTRNAKSMVELD---EHVSENGHDSSIHRYNH 602
Query: 610 LCPQMIKLINKASKSHETYTFLSKVFEESDKIIDDMLAKKFVGVESLEMSH 660
LC + IK + + + E Y E K + + +K +G + SH
Sbjct: 603 LCREAIKYAEEGAITAEAYNIALGQLREGGKKVS--VVRKRIGRAAPPSSH 651
>At2g32250 Mutator-like transposase
Length = 684
Score = 257 bits (657), Expect = 1e-68
Identities = 166/607 (27%), Positives = 290/607 (47%), Gaps = 49/607 (8%)
Query: 40 MEFESEAAAYEFYNKYSRRIGFGIRREYGNKSKKDGILTSRRFTCFKEGKRGVDKRDHLT 99
M+FES+ AAY FY +Y+R +GFGI + +SK+ G + C + G KR+ T
Sbjct: 42 MDFESKEAAYYFYREYARSVGFGITIKASRRSKRSGKFIDVKIACSRFGT----KREKAT 97
Query: 100 K-EGRVETRTGCDARMVISLDRKIGKYKVVDFVAQHNHLLEPPGYFHTPRSHRQISESRA 158
R +TGC A + + ++ K+ + +FV +HNH + P ++ + R + + + A
Sbjct: 98 AINPRSCPKTGCKAGLHMKR-KEDEKWVIYNFVKEHNHEICPDDFYVSVRGKNKPAGALA 156
Query: 159 CQVVAADESRLKRKDFQEYVFKQDGGIDDVGYLQTRRMRSLMYGEVGALLMHF-KRQSEN 217
+K Q + ++D + LL HF + Q +
Sbjct: 157 I-----------KKGLQLALEEED---------------------LKLLLEHFMEMQDKQ 184
Query: 218 PSFYYDFQMDVEEKITNVFWADAQMINDYGCSGDVITFDTTYMTNKDYRPLGVFVGLNNH 277
P F+Y D ++++ NVFW DA+ +DY DV+ FDT Y+ N P F+G+++H
Sbjct: 185 PGFFYAVDFDSDKRVRNVFWLDAKAKHDYCSFSDVVLFDTFYVRNGYRIPFAPFIGVSHH 244
Query: 278 KQMVVFGATLLYDETIPSFQWLFETFLKAMGGEKPKTLLTDQDEAMAKAISVVMPQTFHG 337
+Q V+ G L+ + + ++ WLF T+LKA+GG+ P ++TDQD+ ++ + V P H
Sbjct: 245 RQYVLLGCALIGEVSESTYSWLFRTWLKAVGGQAPGVMITDQDKLLSDIVVEVFPDVRHI 304
Query: 338 LCTWRIRENAQTHVNHLYQKSSKFCSDFEACIDLHEEEGEFLNSWNVLLVEHNVSEDSWL 397
C W + +N + F F C+ + F W+ ++ + ++E+ W+
Sbjct: 305 FCLWSVLSKISEMLNPFVSQDDGFMESFGNCVASSWTDEHFERRWSNMIGKFELNENEWV 364
Query: 398 RMIFQLKEKWAWVYVRKHFTAGMRSTQLSESFNAELKNYLKSDLNLVQFFSHFGRIVHGI 457
+++F+ ++KW Y AG+ + S S + Y+ S+ FF + + +
Sbjct: 365 QLLFRDRKKWVPHYFHGICLAGLSGPERSGSIASHFDKYMNSEATFKDFFELYMKFLQYR 424
Query: 458 RNNESEADYESRHKLPKLKMKRAPMLVQAGNIYTPKTFEEFQEEYEEYLGTCVKNLKE-- 515
+ E++ D E + K P L+ A Q IYT F++FQ E + ++ +E
Sbjct: 425 CDVEAKDDLEYQSKQPTLRSSLA-FEKQLSLIYTDAAFKKFQAEVPGVVSCQLQKEREDG 483
Query: 516 --GLYVVTNYDNNKERMVIGNLMDQKVACDCRKFETHGILCSHALKVLDVMNIKLIPQHY 573
++ + +++ + V N C C FE G LC HA+ VL ++ +P Y
Sbjct: 484 TTAIFRIEDFEERQNFFVALNNELLDACCSCHLFEYQGFLCKHAILVLQSADVSRVPSQY 543
Query: 574 ILKRWTRDARLGSNHDLKGQHIELDTKAHFMRRYNDLCPQMIKLINKASKSHETYTFLSK 633
ILKRW++ G+N + K + T + M R++DLC + +KL AS S E K
Sbjct: 544 ILKRWSKK---GNNKEDKND--KCATIDNRMARFDDLCRRFVKLGVVASLSDEACKTALK 598
Query: 634 VFEESDK 640
+ EE+ K
Sbjct: 599 LLEETVK 605
>At3g06250 unknown protein
Length = 764
Score = 253 bits (647), Expect = 2e-67
Identities = 180/624 (28%), Positives = 293/624 (46%), Gaps = 74/624 (11%)
Query: 36 PHLEMEFESEAAAYEFYNKYSRRIGFGIRREYGNKSKKDGILTSRRFTCFKEGKRGVDKR 95
P+ +EF S A +FY Y+ +GF +R +SK DG +TSRRF C KEG +
Sbjct: 190 PYAGLEFNSANEACQFYQAYAEVVGFRVRIGQLFRSKVDGSITSRRFVCSKEGFQH---- 245
Query: 96 DHLTKEGRVETRTGCDARMVISLDRKIGKYKVVDFVAQHNHLLEPPGYFHTPRSHRQISE 155
+R GC A M I G + V HNH LEP
Sbjct: 246 ---------PSRMGCGAYMRIKRQDS-GGWIVDRLNKDHNHDLEPGK------------- 282
Query: 156 SRACQVVAADESRLKRKDFQEYVFKQDGGIDDVGYLQ----TRRMRSLMYGEVG-----A 206
K ++ GG+D V ++ + + S +G
Sbjct: 283 --------------KNAGMKKITDDVTGGLDSVDLIELNDLSNHISSTRENTIGKEWYPV 328
Query: 207 LLMHFK-RQSENPSFYYDFQMDVEEKITNVFWADAQMINDYGCS--GDVITFDTTYMTNK 263
LL +F+ +Q+E+ F+Y ++D ++FWAD++ + + CS GD + FDT+Y
Sbjct: 329 LLDYFQSKQAEDMGFFYAIELDSNGSCMSIFWADSR--SRFACSQFGDAVVFDTSYRKGD 386
Query: 264 DYRPLGVFVGLNNHKQMVVFGATLLYDETIPSFQWLFETFLKAMGGEKPKTLLTDQDEAM 323
P F+G N+H+Q V+ G L+ DE+ +F WLF+T+L+AM G +P++++ DQD +
Sbjct: 387 YSVPFATFIGFNHHRQPVLLGGALVADESKEAFSWLFQTWLRAMSGRRPRSMVADQDLPI 446
Query: 324 AKAISVVMPQTFHGLCTWRIRENAQTHVNHLYQKSSKFCSDFEACIDLHEEEGEFLNSWN 383
+A++ V P T H W+IR + +L ++F ++E C+ + EF W+
Sbjct: 447 QQAVAQVFPGTHHRFSAWQIRSKER---ENLRSFPNEFKYEYEKCLYQSQTTVEFDTMWS 503
Query: 384 VLLVEHNVSEDSWLRMIFQLKEKWAWVYVRKHFTAGMRSTQLSESFNAELKNYLKSDLNL 443
L+ ++ + ++ WLR I++ +EKW Y+R F G+ + F N L S L
Sbjct: 504 SLVNKYGLRDNMWLREIYEKREKWVPAYLRASFFGGIHVDGTFDPFYGTSLNSLTS---L 560
Query: 444 VQFFSHFGRIVHGIRNNESEADYESRHKLPKLKMKRAPMLVQAGNIYTPKTFEEFQEEYE 503
+F S + + + R E + D+ S + P L+ K P+ Q +YT F FQ E
Sbjct: 561 REFISRYEQGLEQRREEERKEDFNSYNLQPFLQTKE-PVEEQCRRLYTLTIFRIFQSELA 619
Query: 504 E---YLGTCVKNLKEGL---YVVTNYDN-NKERMVIGNLMDQKVACDCRKFETHGILCSH 556
+ YLG +K +EG ++V N N++ V + + +C C+ FE G+LC H
Sbjct: 620 QSYNYLG--LKTYEEGAISRFLVRKCGNENEKHAVTFSASNLNASCSCQMFEYEGLLCRH 677
Query: 557 ALKVLDVMNIKLIPQHYILKRWTRDARLGSNHDLKGQHIELDTKAHFMRRYNDLCPQMIK 616
LKV ++++I+ +P YIL RWT++A G D++ D KA + + K
Sbjct: 678 ILKVFNLLDIRELPSRYILHRWTKNAEFGFVRDVESGVTSQDLKALMIWSLREAAS---K 734
Query: 617 LINKASKSHETYTFLSKVFEESDK 640
I + S E Y ++ E K
Sbjct: 735 YIEFGTSSLEKYKLAYEIMREGGK 758
Score = 60.5 bits (145), Expect = 3e-09
Identities = 36/114 (31%), Positives = 61/114 (52%), Gaps = 16/114 (14%)
Query: 23 IVQNGEESVVDYIPHLEMEFESEAAAYEFYNKYSRRIGFGIRREYGNKSKKDGILTSRRF 82
I +N +S ++ P++ +EF++ A ++YN Y+ R GF +R +S+ DG ++SRRF
Sbjct: 17 IAENEGDSGLE--PYVGLEFDTAEEARDYYNSYATRTGFKVRTGQLYRSRTDGTVSSRRF 74
Query: 83 TCFKEGKRGVDKRDHLTKEGRVETRTGCDARMVISLDRKIGKYKVVDFVAQHNH 136
C KEG ++ +RTGC A + + R GK+ + +HNH
Sbjct: 75 VCSKEG-------------FQLNSRTGCPAFIRVQ-RRDTGKWVLDQIQKEHNH 114
>At5g18960 FAR1 - like protein
Length = 788
Score = 246 bits (627), Expect = 4e-65
Identities = 180/616 (29%), Positives = 292/616 (47%), Gaps = 55/616 (8%)
Query: 36 PHLEMEFESEAAAYEFYNKYSRRIGFGIRREYGNKSKKDGILTSRRFTCFKEGKRGVDKR 95
P+ +EF S A +FY Y+ +GF +R +SK DG +TSRRF C +EG +
Sbjct: 211 PYAGLEFGSANEACQFYQAYAEVVGFRVRIGQLFRSKVDGSITSRRFVCSREGFQH---- 266
Query: 96 DHLTKEGRVETRTGCDARMVISLDRKIGKYKVVDFVAQHNHLLEPPGYFHTPRSHRQISE 155
+R GC A M I G + V HNH LEP + ++
Sbjct: 267 ---------PSRMGCGAYMRIKRQDS-GGWIVDRLNKDHNHDLEPG----------KKND 306
Query: 156 SRACQVVAADESRLKRKDFQEYVFKQDGGIDDVGYLQTRRMRSLMYGE--VGALLMHFK- 212
+ ++ L D E ++D G ++ R G+ LL +F+
Sbjct: 307 AGMKKIPDDGTGGLDSVDLIE--------LNDFGNNHIKKTRENRIGKEWYPLLLDYFQS 358
Query: 213 RQSENPSFYYDFQMDVEE-KITNVFWADAQMINDYGCS--GDVITFDTTYMTNKDYRPLG 269
RQ+E+ F+Y ++DV ++FWAD++ + CS GD + FDT+Y P
Sbjct: 359 RQTEDMGFFYAVELDVNNGSCMSIFWADSRA--RFACSQFGDSVVFDTSYRKGSYSVPFA 416
Query: 270 VFVGLNNHKQMVVFGATLLYDETIPSFQWLFETFLKAMGGEKPKTLLTDQDEAMAKAISV 329
+G N+H+Q V+ G ++ DE+ +F WLF+T+L+AM G +P++++ DQD + +A+
Sbjct: 417 TIIGFNHHRQPVLLGCAMVADESKEAFLWLFQTWLRAMSGRRPRSIVADQDLPIQQALVQ 476
Query: 330 VMPQTFHGLCTWRIRENAQTHVNHLYQKSSKFCSDFEACIDLHEEEGEFLNSWNVLLVEH 389
V P H W+IRE + +L S+F ++E CI + EF + W+ L+ ++
Sbjct: 477 VFPGAHHRYSAWQIREKER---ENLIPFPSEFKYEYEKCIYQTQTIVEFDSVWSALINKY 533
Query: 390 NVSEDSWLRMIFQLKEKWAWVYVRKHFTAGMRSTQLSESFNAELKNYLKSDLNLVQFFSH 449
+ +D WLR I++ +E W Y+R F AG+ E F + L L +F S
Sbjct: 534 GLRDDVWLREIYEQRENWVPAYLRASFFAGIPINGTIEPFFGASLDALTP---LREFISR 590
Query: 450 FGRIVHGIRNNESEADYESRHKLPKLKMKRAPMLVQAGNIYTPKTFEEFQEE-YEEYLGT 508
+ + + R E + D+ S + P L+ K P+ Q +YT F FQ E + Y
Sbjct: 591 YEQALEQRREEERKEDFNSYNLQPFLQTKE-PVEEQCRRLYTLTVFRIFQNELVQSYNYL 649
Query: 509 CVKNLKEGL---YVVTNYDNNKER-MVIGNLMDQKVACDCRKFETHGILCSHALKVLDVM 564
C+K +EG ++V N E+ V + + +C C+ FE G+LC H LKV +++
Sbjct: 650 CLKTYEEGAISRFLVRKCGNESEKHAVTFSASNLNSSCSCQMFEHEGLLCRHILKVFNLL 709
Query: 565 NIKLIPQHYILKRWTRDARLGSNHDLKGQHIELDTKAHFMRRYNDLCPQMIKLINKASKS 624
+I+ +P YIL RWT++A G D++ D KA + + K I + S
Sbjct: 710 DIRELPSRYILHRWTKNAEFGFVRDMESGVSAQDLKALMVWSLREAAS---KYIEFGTSS 766
Query: 625 HETYTFLSKVFEESDK 640
E Y ++ E K
Sbjct: 767 LEKYKLAYEIMREGGK 782
Score = 66.6 bits (161), Expect = 4e-11
Identities = 47/154 (30%), Positives = 72/154 (46%), Gaps = 16/154 (10%)
Query: 22 NIVQNGEESVVDYIPHLEMEFESEAAAYEFYNKYSRRIGFGIRREYGNKSKKDGILTSRR 81
N + EE P++ +EF++ A EFYN Y+ R GF +R +S+ DG ++SRR
Sbjct: 29 NGISEDEEGGSGVEPYVGLEFDTAEEAREFYNAYAARTGFKVRTGQLYRSRTDGTVSSRR 88
Query: 82 FTCFKEGKRGVDKRDHLTKEGRVETRTGCDARMVISLDRKIGKYKVVDFVAQHNHLLEPP 141
F C KEG ++ +RTGC A + + R GK+ + +HNH L
Sbjct: 89 FVCSKEG-------------FQLNSRTGCTAFIRVQ-RRDTGKWVLDQIQKEHNHELGGE 134
Query: 142 GYFH--TPRSHRQISESRACQVVAADESRLKRKD 173
G TPR R + ++ V ++K D
Sbjct: 135 GSVEETTPRPSRAPAPTKLGVTVNPHRPKMKVVD 168
>At1g10240 unknown protein
Length = 680
Score = 229 bits (584), Expect = 4e-60
Identities = 160/593 (26%), Positives = 277/593 (45%), Gaps = 31/593 (5%)
Query: 29 ESVVDYIPHLEMEFESEAAAYEFYNKYSRRIGFGIRREYGNKSKKDGI---LTSRRFTCF 85
E+V + IP+L F + AYEFY+ +++R GF IRR KDG+ LT R F C
Sbjct: 41 EAVHNAIPYLGQIFLTHDTAYEFYSTFAKRCGFSIRRH--RTEGKDGVGKGLTRRYFVCH 98
Query: 86 KEGKRGVDK-RDHLTKEGRVETRTGCDARMVISLDRKIG--KYKVVDFVAQHNHLLEPPG 142
+ G + + + R +R GC A + IS ++G +++V F HNH L P
Sbjct: 99 RAGNTPIKTLSEGKPQRNRRSSRCGCQAYLRISKLTELGSTEWRVTGFANHHNHELLEPN 158
Query: 143 YFHTPRSHRQISESRACQVVAADESRLKRKDFQEYVFKQDGGIDDVGYLQ--TRRMRSLM 200
++R IS++ +++ ++ + + + + + G+L + +R+L+
Sbjct: 159 QVRFLPAYRSISDADKSRILMFSKTGISVQQMMRLLELEK--CVEPGFLPFTEKDVRNLL 216
Query: 201 YG---------EVGALLMHFKRQSENPSFYYDFQMDVEEKITNVFWADAQMINDYGCSGD 251
+ L M + ++P+F ++F +D +K+ N+ W+ A I Y GD
Sbjct: 217 QSFKKLDPEDENIDFLRMCQSIKEKDPNFKFEFTLDANDKLENIAWSYASSIQSYELFGD 276
Query: 252 VITFDTTYMTNKDYRPLGVFVGLNNHKQMVVFGATLLYDETIPSFQWLFETFLKAMGGEK 311
+ FDTT+ + PLG++VG+NN+ FG LL DE + S+ W + F M G+
Sbjct: 277 AVVFDTTHRLSAVEMPLGIWVGVNNYGVPCFFGCVLLRDENLRSWSWALQAFTGFMNGKA 336
Query: 312 PKTLLTDQDEAMAKAISVVMPQTFHGLCTWRIRENAQTHVN-HLYQKSSKFCSDFEACID 370
P+T+LTD + + +AI+ MP T H LC W + + N L ++ + + ++F
Sbjct: 337 PQTILTDHNMCLKEAIAGEMPATKHALCIWMVVGKFPSWFNAGLGERYNDWKAEFYRLYH 396
Query: 371 LHEEEGEFLNSWNVLLVEHNVSEDSWLRMIFQLKEKWAWVYVRKHFTAGMRSTQLSESFN 430
L E EF W ++ + + + ++ + W+ Y+R HF AGM T S++ N
Sbjct: 397 LESVE-EFELGWRDMVNSFGLHTNRHINNLYASRSLWSLPYLRSHFLAGMTLTGRSKAIN 455
Query: 431 AELKNYLKSDLNLVQFFSHFGRIVHGIRNNESEADYESRHKLPKLKMKR-APMLVQAGNI 489
A ++ +L + L F +V +++ + L + +K APM A ++
Sbjct: 456 AFIQRFLSAQTRLAHFVEQVAVVVD--FKDQATEQQTMQQNLQNISLKTGAPMESHAASV 513
Query: 490 YTPKTFEEFQEEYEEYLGTCVKNLKEGLYVVTNYDNNKERMVIGNLMDQKVACDCRKFET 549
TP F + QE+ + EG V + + R V + ++C C+ FE
Sbjct: 514 LTPFAFSKLQEQLVLAAHYASFQMDEGYLVRHHTKLDGGRKVYWVPQEGIISCSCQLFEF 573
Query: 550 HGILCSHALKVLDVMNIKLIPQHYILKRWTR-----DARLGSNHDLKGQHIEL 597
G LC HAL+VL N +P Y+ RW R SN + G+ ++L
Sbjct: 574 SGFLCRHALRVLSTGNCFQVPDRYLPLRWRRISTSFSKTFRSNAEDHGERVQL 626
>At4g19990 putative protein
Length = 672
Score = 227 bits (579), Expect = 1e-59
Identities = 164/619 (26%), Positives = 282/619 (45%), Gaps = 85/619 (13%)
Query: 41 EFESEAAAYEFYNKYSRRIGFGIRREYGNKSKKDGILTSRRFTCFKEGKRGVDKRDHLTK 100
EFES+ A+EFY +Y+ +GF + +S+ G +F C + G + D L
Sbjct: 26 EFESKEEAFEFYKEYANSVGFTTIIKASRRSRMTGKFIDAKFVCTRYGSKKEDIDTGLGT 85
Query: 101 EG-------------RVETRTGCDARMVISLDRKIGKYKVVDFVAQHNHLLEPPGYFHTP 147
+G R ++T C A + + R+ G++ V V +HNH
Sbjct: 86 DGFNIPQARKRGRINRSSSKTDCKAFLHVKR-RQDGRWVVRSLVKEHNH----------- 133
Query: 148 RSHRQISESRACQVVAADESRLKRKDFQEYVFKQDGGIDDVGYLQTRRMRSLMYGEVGAL 207
++ L+ + + K +G I ++ + R L G+V L
Sbjct: 134 ------------EIFTGQADSLRELSGRRKLEKLNGAI-----VKEVKSRKLEDGDVERL 176
Query: 208 LMHFKRQSENPSFYYDFQMDVEEKITNVFWADAQMINDYGCSGDVITFDTTYMTNKDYRP 267
L +F+ D Q + N+FW DA+ DY C DV++ DTT++ N+ P
Sbjct: 177 L----------NFFTDMQ-----SLRNIFWVDAKGRFDYTCFSDVVSIDTTFIKNEYKLP 221
Query: 268 LGVFVGLNNHKQMVVFG-ATLLYDETIPSFQWLFETFLKAMGGEKPKTLLTDQDEAMAKA 326
L F G+N+H Q ++ G LL DE+ F WLF +LKAM G +P+ +LT D+ + +A
Sbjct: 222 LVAFTGVNHHGQFLLLGFGLLLTDESKSGFVWLFRAWLKAMHGCRPRVILTKHDQMLKEA 281
Query: 327 ISVVMPQTFHGLCTWRIRENAQTHVNHLYQKSSKFCSDFEACIDLHEEEGEFLNSWNVLL 386
+ V P + H W + H+ + K + I + +F +W ++
Sbjct: 282 VLEVFPSSRHCFYMWDTLGQMPEKLGHVIRLEKKLVDEINDAIYGSCQSEDFEKNWWEVV 341
Query: 387 VEHNVSEDSWLRMIFQLKEKWAWVYVRKHFTAGMRSTQLSESFNAELKNYLKSDLNLVQF 446
++ ++ WL+ +++ +E W VY++ AGM + Q S+S N+ L Y++ F
Sbjct: 342 DRFHMRDNVWLQSLYEDREYWVPVYMKDVSLAGMCTAQRSDSVNSGLDKYIQRKTTFKAF 401
Query: 447 FSHFGRIVHGIRNNESEADYESRHKLPKLKMKRAPMLVQAGNIYTPKTFEEFQEEYEEYL 506
+ +++ E +++ E+ +K P LK +P Q +YT + F++FQ E +
Sbjct: 402 LEQYKKMIQERYEEEEKSEIETLYKQPGLK-SPSPFGKQMAEVYTREMFKKFQVEVLGGV 460
Query: 507 GTCVK------NLKEGLYVVTNYDNNKERMVIGNLMDQKVACDCRKFETHGILCSHALKV 560
K + + + V +Y+ N+ +V+ N +V C CR FE G L
Sbjct: 461 ACHPKKESEEDGVNKRTFRVQDYEQNRSFVVVWNSESSEVVCSCRLFELKGELS------ 514
Query: 561 LDVMNIKLIPQHYILKRWTRDARLGSNHDLKGQHIELD-TKAHFMRRYNDLCPQMIKLIN 619
IP Y+LKRWT+DA+ S ++ +++ TKA +RY DLC + +KL
Sbjct: 515 --------IPSQYVLKRWTKDAK--SREVMESDQTDVESTKA---QRYKDLCLRSLKLSE 561
Query: 620 KASKSHETYTFLSKVFEES 638
+AS S E+Y + V E+
Sbjct: 562 EASLSEESYNAVVNVLNEA 580
>At4g38170 hypothetical protein
Length = 531
Score = 217 bits (552), Expect = 2e-56
Identities = 125/435 (28%), Positives = 213/435 (48%), Gaps = 13/435 (2%)
Query: 212 KRQSENPSFYYDFQMDVEEKITNVFWADAQMINDYGCSGDVITFDTTYMTNKDYR-PLGV 270
+RQ ENP F Y +E+ NVFWAD +Y GD + FDTTY K Y+ P
Sbjct: 13 RRQLENPGFLYA----IEDDCGNVFWADPTCRLNYTYFGDTLVFDTTYRRGKRYQVPFAA 68
Query: 271 FVGLNNHKQMVVFGATLLYDETIPSFQWLFETFLKAMGGEKPKTLLTDQDEAMAKAISVV 330
F G N+H Q V+FG L+ +E+ SF WLF+T+L+AM P ++ + D + A+S V
Sbjct: 69 FTGFNHHGQPVLFGCALILNESESSFAWLFQTWLQAMSAPPPPSITVEPDRLIQVAVSRV 128
Query: 331 MPQTFHGLCTWRIRENAQTHVNHLYQKSSKFCSDFEACIDLHEEEGEFLNSWNVLLVEHN 390
QT I E + + H++Q F S+F C+ E EF SW+ ++ +
Sbjct: 129 FSQTRLRFSQPLIFEETEEKLAHVFQAHPTFESEFINCVTETETAAEFEASWDSIVRRYY 188
Query: 391 VSEDSWLRMIFQLKEKWAWVYVRKHFTAGMRSTQLSESFNAELKNYLKSDLNLVQFFSHF 450
+ ++ WL+ I+ +++W V++R F + + + S N+ + ++ + + +
Sbjct: 189 MEDNDWLQSIYNARQQWVRVFIRDTFYGELSTNEGSSILNSFFQGFVDASTTMQMLIKQY 248
Query: 451 GRIVHGIRNNESEADYESRHKLPKLKMKRAPMLVQAGNIYTPKTFEEFQEEYEEYLGTCV 510
+ + R E +ADYE+ + P +K +PM QA ++YT F +FQEE+ E L
Sbjct: 249 EKAIDSWREKELKADYEATNSTPVMKTP-SPMEKQAASLYTRAAFIKFQEEFVETLAIPA 307
Query: 511 KNLKEG----LYVVTNY-DNNKERMVIGNLMDQKVACDCRKFETHGILCSHALKVLDVMN 565
+ + Y V + + +K V + ++ K C C+ FE GI+C H L V N
Sbjct: 308 NIISDSGTHTTYRVAKFGEVHKGHTVSFDSLEVKANCSCQMFEYSGIICRHILAVFSAKN 367
Query: 566 IKLIPQHYILKRWTRDARLGSNHDLKGQHIELDTKAHFMRRYNDLCPQMIKLINKASKSH 625
+ +P Y+L+RWT++A++ + + +N L + K + + +KS
Sbjct: 368 VLALPSRYLLRRWTKEAKIRGTEE--QPEFSNGCQESLNLCFNSLRQEATKYVEEGAKSI 425
Query: 626 ETYTFLSKVFEESDK 640
+ Y +E+ K
Sbjct: 426 QIYKVAMDALDEAAK 440
>At1g52520 F6D8.26
Length = 703
Score = 198 bits (504), Expect = 7e-51
Identities = 168/636 (26%), Positives = 290/636 (45%), Gaps = 55/636 (8%)
Query: 36 PHLEMEFESEAAAYEFYNKYSRRIGFGIRREYGNKSKKDGILTSRRFTCFKEGKRGVDKR 95
P + MEFES AY +YN Y+ +GF +R + ++ C +G + ++
Sbjct: 85 PAVGMEFESYDDAYNYYNCYASEVGFRVRVKNSWFKRRSKEKYGAVLCCSSQGFKRINDV 144
Query: 96 DHLTKEGRVETRTGCDARMVI-SLDRKIGKYKVVDFVAQHNHLLEPPGYFHTPRSHRQIS 154
+ + KE TRTGC A + + +D K +++VV+ HNHLL Y R + +S
Sbjct: 145 NRVRKE----TRTGCPAMIRMRQVDSK--RWRVVEVTLDHNHLLGCKLYKSVKRKRKCVS 198
Query: 155 ES----------RACQVVAADESRLKRKDFQEYVFKQDGGIDDVGYLQTRRMRSLMYGEV 204
RAC V + S + F+ G D + +L G+
Sbjct: 199 SPVSDAKTIKLYRAC--VVDNGSNVNPNSTLNKKFQNSTGSPD--------LLNLKRGDS 248
Query: 205 GALLMHFKR-QSENPSFYYDFQMDVEEKITNVFWADAQMINDYGCS--GDVITFDTTYMT 261
A+ +F R Q NP+F+Y ++ E ++ NVFWADA + CS GDVI D++Y++
Sbjct: 249 AAIYNYFCRMQLTNPNFFYLMDVNDEGQLRNVFWADA--FSKVSCSYFGDVIFIDSSYIS 306
Query: 262 NKDYRPLGVFVGLNNHKQMVVFGATLLYDETIPSFQWLFETFLKAMGGEKPKTLLTDQDE 321
K PL F G+N+H + + L ET+ S+ WL + +L M P+T++TD+ +
Sbjct: 307 GKFEIPLVTFTGVNHHGKTTLLSCGFLAGETMESYHWLLKVWLSVMK-RSPQTIVTDRCK 365
Query: 322 AMAKAISVVMPQTFHGLCTWRIRENAQTHVNHLYQKSSKFCSDFEACIDLHEEEGEFLNS 381
+ AIS V P++ I + L+ + + +A + + EF +
Sbjct: 366 PLEAAISQVFPRSHQRFSLTHIMRKIPEKLGGLHNYDAVRKAFTKAVYETLKVV-EFEAA 424
Query: 382 WNVLLVEHNVSEDSWLRMIFQLKEKWAWVYVRKHFTAGMRSTQLSESFNAELKNYLKSDL 441
W ++ V E+ WLR +++ + KWA VY++ F AG+ + E+ + Y+
Sbjct: 425 WGFMVHNFGVIENEWLRSLYEERAKWAPVYLKDTFFAGIAAAHPGETLKPFFERYVHKQT 484
Query: 442 NLVQFFSHFGRIVHGIRNNESEADYESRHKLPKLKMKRAPMLVQAGNIYTPKTFEEFQEE 501
L +F + + E+ +D ES+ + Q IYT F++FQ E
Sbjct: 485 PLKEFLDKYELALQKKHREETLSDIESQTLNTAELKTKCSFETQLSRIYTRDMFKKFQIE 544
Query: 502 YEEYLGTCVKNLK---EGLYVV----------TNYDNNKERMVIGNLMDQKVACDCRKFE 548
EE + +C + +G +V+ ++ ++ V+ N +V C C F
Sbjct: 545 VEE-MYSCFSTTQVHVDGPFVIFLVKERVRGESSRREIRDFEVLYNRSVGEVRCICSCFN 603
Query: 549 THGILCSHALKVLDVMNIKLIPQHYILKRWTRDARL--GSNHDLKGQHIELDTKAHFMRR 606
+G LC HAL VL+ ++ IP YIL RW +D + +++ L G D F +
Sbjct: 604 FYGYLCRHALCVLNFNGVEEIPLRYILPRWRKDYKRLHFADNGLTGFVDGTDRVQWFDQL 663
Query: 607 YNDLCPQMIKLINKASKSHETYTFLSKVFEES-DKI 641
Y + ++++ + + S + Y +V +ES DK+
Sbjct: 664 YKN----SLQVVEEGAVSLDHYKVAMQVLQESLDKV 695
>At1g80010 hypothetical protein
Length = 696
Score = 197 bits (500), Expect = 2e-50
Identities = 159/576 (27%), Positives = 248/576 (42%), Gaps = 45/576 (7%)
Query: 40 MEFESEAAAYEFYNKYSRRIGFGIRREYG----NKSKKDGILTSRRFTCFKEGKRGVDKR 95
MEFES AY FYN Y+R +GF IR + N +K G + FK K +R
Sbjct: 70 MEFESYDDAYSFYNSYARELGFAIRVKSSWTKRNSKEKRGAVLCCNCQGFKLLKDAHSRR 129
Query: 96 DHLTKEGRVETRTGCDARMVISLDRKIGKYKVVDFVAQHNHLLEPPGYFHTPRSHRQISE 155
ETRTGC A + + L ++KV HNH +P H +SH++ S
Sbjct: 130 K--------ETRTGCQAMIRLRLIH-FDRWKVDQVKLDHNHSFDPQRA-HNSKSHKKSSS 179
Query: 156 SRACQVVAADES--RLKRKDFQEY------------VFKQDGGIDDVG--YLQTRRMRSL 199
S + E ++ + + Y G D+ + Q+ R L
Sbjct: 180 SASPATKTNPEPPPHVQVRTIKLYRTLALDTPPALGTSLSSGETSDLSLDHFQSSRRLEL 239
Query: 200 MYGEVGALLMHFKRQSENPSFYYDFQMDVEEKITNVFWADAQMINDYGCSGDVITFDTTY 259
G F+ Q +P+F Y + + + NVFW DA+ Y GDV+ FDTT
Sbjct: 240 RGGFRALQDFFFQIQLSSPNFLYLMDLADDGSLRNVFWIDARARAAYSHFGDVLLFDTTC 299
Query: 260 MTNKDYRPLGVFVGLNNHKQMVVFGATLLYDETIPSFQWLFETFLKAMGGEKPKTLLTDQ 319
++N PL FVG+N+H ++ G LL D++ ++ WLF +L M G P+ +T+Q
Sbjct: 300 LSNAYELPLVAFVGINHHGDTILLGCGLLADQSFETYVWLFRAWLTCMLGRPPQIFITEQ 359
Query: 320 DEAMAKAISVVMPQTFHGLCTWRIRENAQTHVNHLYQKSSKFCSDFEACIDLHEEEGEFL 379
+AM A+S V P+ H L + N V L Q S F + + EF
Sbjct: 360 CKAMRTAVSEVFPRAHHRLSLTHVLHNICQSVVQL-QDSDLFPMALNRVVYGCLKVEEFE 418
Query: 380 NSWNVLLVEHNVSEDSWLRMIFQLKEKWAWVYVRKHFTAGMRSTQLSE-SFNAELKNYLK 438
+W +++ ++ + +R +FQ +E WA VY++ F AG + L + Y+
Sbjct: 419 TAWEEMIIRFGMTNNETIRDMFQDRELWAPVYLKDTFLAGALTFPLGNVAAPFIFSGYVH 478
Query: 439 SDLNLVQFFSHFGRIVHGIRNNESEADYESRHKLPKLKMKRAPMLVQAGNIYTPKTFEEF 498
+ +L +F + + E+ D ES +PKLK P Q ++T + F F
Sbjct: 479 ENTSLREFLEGYESFLDKKYTREALCDSESLKLIPKLKTTH-PYESQMAKVFTMEIFRRF 537
Query: 499 QEEYEEYLGTCV------KNLKEGLYVVTNYDNNKER-----MVIGNLMDQKVACDCRKF 547
Q+E + +C N YVV + +K R + C C F
Sbjct: 538 QDEVSA-MSSCFGVTQVHSNGSASSYVVKEREGDKVRDFEVIYETSAAAQVRCFCVCGGF 596
Query: 548 ETHGILCSHALKVLDVMNIKLIPQHYILKRWTRDAR 583
+G C H L +L ++ +P YIL+RW +D +
Sbjct: 597 SFNGYQCRHVLLLLSHNGLQEVPPQYILQRWRKDVK 632
>At5g28530 far-red impaired response protein (FAR1) - like
Length = 700
Score = 196 bits (498), Expect = 4e-50
Identities = 158/609 (25%), Positives = 263/609 (42%), Gaps = 59/609 (9%)
Query: 34 YIPHLEMEFESEAAAYEFYNKYSRRIGFGIRREYGNKSKKDGILTSRRFTCFKEGKRGVD 93
+ P++ F ++ A+E+Y+ ++R+ GF IR+ +S+ G+ R F C++ G
Sbjct: 68 FTPYVGQIFTTDDEAFEYYSTFARKSGFSIRKARSTESQNLGVYR-RDFVCYRSGFNQPR 126
Query: 94 KRDHLTK-EGRVETRTGCDARMVISLD--RKIGKYKVVDFVAQHNHLLEPPGYFHTPRSH 150
K+ ++ R R GCD ++ ++ + + + V F HNH L ++
Sbjct: 127 KKANVEHPRERKSVRCGCDGKLYLTKEVVDGVSHWYVSQFSNVHNHELLEDDQVRLLPAY 186
Query: 151 RQISESRACQV----------------------VAADESRLKRKDFQEYVFKQDGGIDDV 188
R+I +S ++ V + + KD + +V + +
Sbjct: 187 RKIQQSDQERILLLSKAGFPVNRIVKLLELEKGVVSGQLPFIEKDVRNFVRACKKSVQEN 246
Query: 189 GYLQTRRMRSLMYGEVGALLMHFKRQSENP-SFYYDFQMDVEEKITNVFWADAQMINDYG 247
T + S + LL K +E F YD D +K+ N+ WA + Y
Sbjct: 247 DAFMTEKRES----DTLELLECCKGLAERDMDFVYDCTSDENQKVENIAWAYGDSVRGYS 302
Query: 248 CSGDVITFDTTYMTNKDYRPLGVFVGLNNHKQMVVFGATLLYDETIPSFQWLFETFLKAM 307
GDV+ FDT+Y + LGVF G++N+ + ++ G LL DE+ SF W +TF++ M
Sbjct: 303 LFGDVVVFDTSYRSVPYGLLLGVFFGIDNNGKAMLLGCVLLQDESCRSFTWALQTFVRFM 362
Query: 308 GGEKPKTLLTDQDEAMAKAISVVMPQTFHGLCTWRIRENAQTHVNHLYQKSSKFCSDFEA 367
G P+T+LTD D + AI MP T H + I + + Q +F A
Sbjct: 363 RGRHPQTILTDIDTGLKDAIGREMPNTNHVVFMSHIVSKLASWFS---QTLGSHYEEFRA 419
Query: 368 CIDLHEEEG---EFLNSWNVLLVEHNVSEDSWLRMIFQLKEKWAWVYVRKHFTAGMRSTQ 424
D+ G EF W++L+ + D +++ + W +R+HF A T
Sbjct: 420 GFDMLCRAGNVDEFEQQWDLLVTRFGLVPDRHAALLYSCRASWLPCCIREHFVA---QTM 476
Query: 425 LSESFNAELKNYLKSDLNLVQFFSHFGRIVHGIRNNESEADYESRHKLPKLKMKR----- 479
SE FN + ++LK ++ G + ES + L K + R
Sbjct: 477 TSE-FNLSIDSFLKRVVD--------GATCMQLLLEESALQVSAAASLAKQILPRFTYPS 527
Query: 480 ----APMLVQAGNIYTPKTFEEFQEEYEEYLGTCVKNLKEGLYVVTNYDNNK-ERMVIGN 534
PM A I TP F Q E + V + G ++V +Y + E VI N
Sbjct: 528 LKTCMPMEDHARGILTPYAFSVLQNEMVLSVQYAVAEMANGPFIVHHYKKMEGECCVIWN 587
Query: 535 LMDQKVACDCRKFETHGILCSHALKVLDVMNIKLIPQHYILKRWTRDARLGSNHDLKGQH 594
++++ C C++FE GILC H L+VL V N IP+ Y L RW +++ + + GQ
Sbjct: 588 PENEEIQCSCKEFEHSGILCRHTLRVLTVKNCFHIPEQYFLLRWRQESPHVATENQNGQG 647
Query: 595 IELDTKAHF 603
I D+ F
Sbjct: 648 IGDDSAQTF 656
>At3g59470 unknown protein
Length = 251
Score = 90.9 bits (224), Expect = 2e-18
Identities = 59/153 (38%), Positives = 84/153 (54%), Gaps = 11/153 (7%)
Query: 36 PHLEMEFESEAAAYEFYNKYSRRIGFGIRREYGNKSKKDGILTSRRFTCFKEGKRGVDKR 95
P++ EFESEAAA+ FYN Y+ ++GF IR ++S+ DG R+ C KEG R KR
Sbjct: 70 PYVGQEFESEAAAHGFYNAYATKVGFVIRVSKLSRSRHDGSPIGRQLVCNKEGYRLPSKR 129
Query: 96 DHLTKEGRVETRTGCDARMVISLDRKIGKYKVVDFVAQHNHLLEPPG------YFHTPRS 149
D + ++ R ETR GC A ++I + GK+ + FV +HNH L P Y P
Sbjct: 130 DKVIRQ-RAETRVGCKAMILIRKENS-GKWVITKFVKEHNHSLMPGRVRRGCIYDQYPNE 187
Query: 150 HRQISESRACQVVAADESRLKR-KDFQEYVFKQ 181
H +I E Q +AA++ R K E +F+Q
Sbjct: 188 HDKIQE--LMQQLAAEKKRAATYKRHLEMLFEQ 218
>At1g42570 hypothetical protein
Length = 224
Score = 82.4 bits (202), Expect = 8e-16
Identities = 62/240 (25%), Positives = 100/240 (40%), Gaps = 59/240 (24%)
Query: 374 EEGEFLNSWNVLLVEHNVSEDSWLRMIFQLKEKWAWVYVRKHFTAGMRSTQLSESFNAEL 433
EE + L +W+ +L +HN++ D WL+ +F+L+EK A +
Sbjct: 21 EEEDLLLAWSNMLQKHNLTNDKWLKNLFELREKCA------------------------M 56
Query: 434 KNYLKSDLNLVQFFSHFGRIVHGIRNNESEADYESRHKLPKL--KMKRAPMLVQAGNIYT 491
+Y NL+ F H R++ R E D+ H P L ++ ML A +Y
Sbjct: 57 HSY-----NLLCFSEHSERVLEDQRYKELVGDFNMMHTSPVLCATVEMLEMLQHAEEVYI 111
Query: 492 PKTFEEFQEEYEEYLGTCVKNLKEGLYVVTNYDNNKERMVIGNLMDQKVACDCRKFETHG 551
P+ FQ++Y +VIG+ + KV+ F
Sbjct: 112 PEICSLFQKQY---------------------------IVIGDYVANKVS-KSEMFSFAA 143
Query: 552 ILCSHALKVLDVMNIKLIPQHYILKRWTRDARLGSNHDLKGQHIELDTKAHFMRRYNDLC 611
ILC HA KVLD N++ IP YIL RW+++A+ + + K +Y+ +C
Sbjct: 144 ILCRHASKVLDKKNVRRIPSTYILNRWSKEAKAQKISSYHSESPDETVKQSIGNQYSHIC 203
>At3g07500 hypothetical protein
Length = 214
Score = 76.6 bits (187), Expect = 4e-14
Identities = 50/140 (35%), Positives = 76/140 (53%), Gaps = 4/140 (2%)
Query: 1 MNGATIPIVEGVEKLGNNIDVNIVQNGEESVVDYIPHLEMEFESEAAAYEFYNKYSRRIG 60
+ G ++P E VE + N+ + V + +V+ P + MEFESE AA FY+ Y+ +G
Sbjct: 9 VEGNSLP-EEDVEMMENSDVMKTVTDEASPMVE--PFIGMEFESEEAAKSFYDNYATCMG 65
Query: 61 FGIRREYGNKSKKDGILTSRRFTCFKEGKRGVDKRDHLTKEGRVETRTGCDARMVISLDR 120
F +R + +S +DG + RR C KEG R R +++ R TR GC A +V+ +
Sbjct: 66 FVMRVDAFRRSMRDGTVVWRRLVCNKEGFRRSRPRRSESRKPRAITREGCKALIVVKRE- 124
Query: 121 KIGKYKVVDFVAQHNHLLEP 140
K G + V F +HNH L P
Sbjct: 125 KSGTWLVTKFEKEHNHPLLP 144
>At2g43280 unknown protein
Length = 206
Score = 72.4 bits (176), Expect = 8e-13
Identities = 42/98 (42%), Positives = 58/98 (58%), Gaps = 2/98 (2%)
Query: 42 FESEAAAYEFYNKYSRRIGFGIRREYGNKSKKDGILTSRRFTCFKEGK-RGVDKRDHLTK 100
FESE AA FY+ YSRR+GF +R +S+KDG + +RRF C KEG + + +
Sbjct: 28 FESEDAAKMFYDDYSRRLGFVMRVMSCRRSEKDGRILARRFGCNKEGHCVSIRGKFGSVR 87
Query: 101 EGRVETRTGCDARMVISLDRKIGKYKVVDFVAQHNHLL 138
+ R TR GC A + + DR GK+ + FV +HNH L
Sbjct: 88 KPRPSTREGCKAMIHVKYDRS-GKWVITKFVKEHNHPL 124
>At4g12850 putative protein
Length = 183
Score = 69.7 bits (169), Expect = 5e-12
Identities = 44/149 (29%), Positives = 75/149 (49%), Gaps = 10/149 (6%)
Query: 36 PHLEMEFESEAAAYEFYNKYSRRIGFGIRREYGNKSKKDGILTSRRFTCFKEGKRGVDKR 95
P++ ++FESE A +FY +YS+R+GF +R +S DG +RR C K+G
Sbjct: 10 PYVGLKFESEEEAKDFYVEYSKRLGFVVRMMQRRRSGIDGRTLARRLGC---NKQGFGPN 66
Query: 96 DHLTKEGRVETRTGCDARMVISLDRKIGKYKVVDFVAQHNHLLEPPG------YFHTPRS 149
+ + +R GC A +++ ++ K GK+ V F+ +HNH L+ G + R
Sbjct: 67 NQRSSSSSSSSREGCKATILVKME-KSGKWVVTRFIKEHNHSLQFIGSSSYDSFADKERK 125
Query: 150 HRQISESRACQVVAADESRLKRKDFQEYV 178
++++E CQ D R + F + V
Sbjct: 126 IKELTEEIECQDRLCDVYRDRLVSFIDNV 154
>At3g32060 hypothetical protein
Length = 487
Score = 58.5 bits (140), Expect = 1e-08
Identities = 40/160 (25%), Positives = 67/160 (41%), Gaps = 8/160 (5%)
Query: 197 RSLMYGEVGALLMHFKRQSENPSFYYDFQMDVEEKITNVFWADAQMINDYGCSGDVITFD 256
R Y ++ L K+ ++ Y ++D +K VF A I + V+ D
Sbjct: 213 REESYKDINCYLYMLKKVNDGTVTY--LKLDENDKFQYVFVALGASIEGFRVMRKVLIVD 270
Query: 257 TTYMTNKDYRPLGVFVGL---NNHKQMVVFGATLLYDETIPSFQWLFETFLKAMGGEKPK 313
T++ N Y + VF N H ++ F +L E S++W FE +
Sbjct: 271 ATHLKN-GYGGVLVFASAQDPNRHHYIIAFA--VLDGENDASWEWFFEKLKTVVPDTSEL 327
Query: 314 TLLTDQDEAMAKAISVVMPQTFHGLCTWRIRENAQTHVNH 353
+TD++ ++ KAI V HG C W + +N + H H
Sbjct: 328 VFMTDRNASLIKAIRNVYTAAHHGYCIWHLSQNVKGHATH 367
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.320 0.137 0.406
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 15,825,594
Number of Sequences: 26719
Number of extensions: 688062
Number of successful extensions: 1838
Number of sequences better than 10.0: 82
Number of HSP's better than 10.0 without gapping: 57
Number of HSP's successfully gapped in prelim test: 25
Number of HSP's that attempted gapping in prelim test: 1668
Number of HSP's gapped (non-prelim): 123
length of query: 692
length of database: 11,318,596
effective HSP length: 106
effective length of query: 586
effective length of database: 8,486,382
effective search space: 4973019852
effective search space used: 4973019852
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.8 bits)
S2: 64 (29.3 bits)
Medicago: description of AC126006.3


