

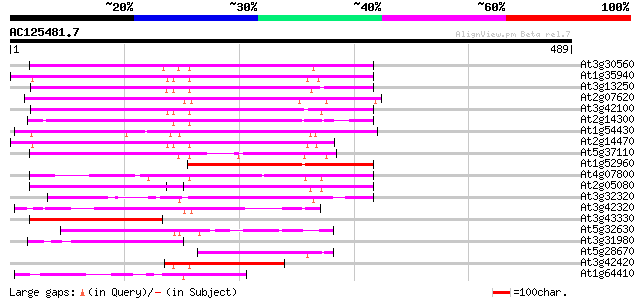
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC125481.7 + phase: 0 /pseudo
(489 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
At3g30560 hypothetical protein 269 3e-72
At1g35940 hypothetical protein 252 3e-67
At3g13250 hypothetical protein 252 4e-67
At2g07620 putative helicase 247 9e-66
At3g42100 putative protein 238 7e-63
At2g14300 pseudogene; similar to MURA transposase of maize Muta... 228 8e-60
At1g54430 hypothetical protein 217 1e-56
At2g14470 pseudogene 214 7e-56
At5g37110 putative helicase 189 2e-48
At1g52960 hypothetical protein 182 5e-46
At4g07800 hypothetical protein 171 1e-42
At2g05080 putative helicase 162 5e-40
At3g32320 hypothetical protein 159 3e-39
At3g42320 putative protein 130 1e-30
At3g43330 putative protein 129 5e-30
At5g32630 putative protein 123 3e-28
At3g31980 hypothetical protein 107 2e-23
At5g28670 putative protein 100 2e-21
At3g42420 putative protein 88 1e-17
At1g64410 unknown protein 75 8e-14
>At3g30560 hypothetical protein
Length = 1473
Score = 269 bits (687), Expect = 3e-72
Identities = 147/370 (39%), Positives = 198/370 (52%), Gaps = 70/370 (18%)
Query: 18 VDCYTMIESQRLRYIRKNQKTIRCGILNGLHEAMDMGETDASNVGQKIVLQPSFAGGRHY 77
VD +TMIES RLR+I+KNQ +R + +A D G+ D SN G+ +++ PSF GG Y
Sbjct: 415 VDAFTMIESNRLRFIKKNQTKLRSTNKQAVQDASDAGDNDLSNKGKSVIIPPSFTGGPAY 474
Query: 78 MFNNCQDAMSICKKYGYPDLFVTIACNTNWREIQDFLKERNLKASDRPDIVCRVF----- 132
M N DAM CK +G+PDLF+T CN+ W +I F+KER L A D+PDI+CR++
Sbjct: 475 MQQNYLDAMGTCKHFGFPDLFITFTCNSKWPKITRFVKERKLNAEDKPDIICRIYKMKLE 534
Query: 133 -------------KMKLDKLMDDFEK---------------------------------- 145
K KL +F+K
Sbjct: 535 NLMDDLTKNHIFGKTKLATYTIEFQKRGLPHAHIIVWMDPRYKFPIADHVDKIIFAEIPD 594
Query: 146 EECLAKLMQR------HVPCGGARPFSPCMDRGRCTKYYRKNYKGTTTID-EGYPRYKRR 198
+E KL Q H PC P SPCM+ G+C+KYY KN+ T +D EGYP Y+RR
Sbjct: 595 KENDPKLYQAVSECMIHGPCRLVNPNSPCMENGKCSKYYPKNHVENTFLDNEGYPIYRRR 654
Query: 199 DFGIHVDKQRVLLDNRYVVPYNPHLLIRYGGHVNVECCNKSNSIKYLFKYVNKVPDKALM 258
D G + K + DNRYVVPYN LL +Y H+NVE CN+S S+KYLFKYVNK PD+ +
Sbjct: 655 DTGRFIKKNKYPCDNRYVVPYNDFLLRKYRAHINVEWCNQSVSVKYLFKYVNKGPDRVTV 714
Query: 259 QLSVD-----------GDNRDKSKPVDEIKQYYDCRYVSPCEAVWRIFAFDIHHK*PHVL 307
L G+ + + ++++ Y+DCRYVS CEA+WRI + IH++ V
Sbjct: 715 SLEPHRKEVVSEENNVGETNNDPQEQNQVEDYFDCRYVSACEAMWRIKGYPIHYRQTLVT 774
Query: 308 KLLFHLHNEQ 317
KL FH +Q
Sbjct: 775 KLTFHEKGKQ 784
>At1g35940 hypothetical protein
Length = 1678
Score = 252 bits (644), Expect = 3e-67
Identities = 150/393 (38%), Positives = 200/393 (50%), Gaps = 76/393 (19%)
Query: 1 MQDRVVDYGNVVNSRRSV-----DCYTMIESQRLRYIRKNQKTIRCGILNGLHEAMDMGE 55
+Q+R V+ ++ S+R D YT IES RL YI+ NQ +RC + EA G
Sbjct: 628 IQEREVECQTLLRSKRLFQQCLCDAYTTIESNRLNYIKFNQSKLRCENYTSVKEAAAAGA 687
Query: 56 TDASNVGQKIVLQPSFAGGRHYMFNNCQDAMSICKKYGYPDLFVTIACNTNWREIQDFLK 115
T G ++++ SF GG YM + DAM+ICK YG+PDLF+T CN W EI +
Sbjct: 688 TTMEEEGNQLLIPASFTGGPRYMVQSYYDAMAICKHYGFPDLFITFTCNPKWPEITRYYD 747
Query: 116 ERNLKASDRPDIVCRVFKMKL--------------------------------------- 136
+R L DRP+I+ R+FK+KL
Sbjct: 748 KRGLNPEDRPNIIARIFKIKLDSLMNDLTVKKMLGKTVASMYTVEFQKRGLPHAHILLFM 807
Query: 137 ------------DKLMD----DFEKEECLAKLMQR---HVPCGGARPFSPCMDRGRCTKY 177
DKL+ D EKE L ++++ H PCG A SPCM G C+K
Sbjct: 808 HAKSKLPTADDIDKLISAEIPDKEKEPDLYEVIKNSMIHGPCGSANVNSPCMVDGECSKL 867
Query: 178 YRKNYKGTTTI-DEGYPRYKRRDFGIHVDKQRVLLDNRYVVPYNPHLLIRYGGHVNVECC 236
Y K ++ T I +GYP Y+RR +V+K + DNRYVVPYN L +RY H+NVE C
Sbjct: 868 YPKKHQDITKIGSDGYPIYRRRKTDDYVEKGGIKCDNRYVVPYNKKLSLRYNAHINVEWC 927
Query: 237 NKSNSIKYLFKYVNKVPDKALM-----QLSVDGDNR-------DKSKPVDEIKQYYDCRY 284
N++ SIKYLFKY+NK PDK + Q + GD+ K +EIK ++DCRY
Sbjct: 928 NQNGSIKYLFKYINKGPDKVVFIVEPTQQTTAGDSETPQQEQGSAEKKKNEIKDWFDCRY 987
Query: 285 VSPCEAVWRIFAFDIHHK*PHVLKLLFHLHNEQ 317
VS EAVWRIF + I H V KL FH+ +Q
Sbjct: 988 VSASEAVWRIFKYPIQHISTPVQKLSFHVEGKQ 1020
>At3g13250 hypothetical protein
Length = 1419
Score = 252 bits (643), Expect = 4e-67
Identities = 143/372 (38%), Positives = 194/372 (51%), Gaps = 75/372 (20%)
Query: 19 DCYTMIESQRLRYIRKNQKTIRCGILNGLHEAMDMGETDASNVGQKIVLQPSFAGGRHYM 78
D YT IES RL YI+ NQ +RC N + E+ G T S G ++++ SF GG YM
Sbjct: 309 DAYTTIESNRLAYIKFNQSKLRCENFNSIKESASSGSTTMSEEGNQVLIPSSFTGGPRYM 368
Query: 79 FNNCQDAMSICKKYGYPDLFVTIACNTNWREIQDFLKERNLKASDRPDIVCRVFKMK--- 135
DAM+ICK +G+PDLF+T CN W EI + ++R L A DRPDIV R+FK+K
Sbjct: 369 LQTYYDAMAICKHFGFPDLFITFTCNPKWPEITRYCEKRGLTADDRPDIVARIFKIKLDS 428
Query: 136 ------------------------------------------------LDKLMD----DF 143
+DK++ +
Sbjct: 429 LMKDLTERHLLGKTVASMYTVEFQKRGLPHAHILLFMAANSKLPTADDIDKIISAEIPNK 488
Query: 144 EKEECLAKLMQR---HVPCGGARPFSPCMDRGRCTKYYRKNYKGTTTID-EGYPRYKRRD 199
+KE L ++++ H PCG A SPCM G+C+K Y K ++ T + +GYP Y+RR
Sbjct: 489 DKEPELYEVIKNSMMHGPCGSANTSSPCMVDGQCSKLYPKKHQEITKVGADGYPIYRRRL 548
Query: 200 FGIHVDKQRVLLDNRYVVPYNPHLLIRYGGHVNVECCNKSNSIKYLFKYVNKVPDKALMQ 259
+++K V DNRYVVPYN L +RY H+NVE CN++ SIKYLFKY+NK PD+ +
Sbjct: 549 TDDYIEKGGVKCDNRYVVPYNKKLSLRYQAHINVEWCNQNGSIKYLFKYINKGPDRVVFI 608
Query: 260 LS--------------VDGDNRDKSKPVDEIKQYYDCRYVSPCEAVWRIFAFDIHHK*PH 305
+ V+ D +K K DEIK ++DCRYVS EA+WRIF F I H+
Sbjct: 609 VEPIKEATSSDTTAPVVESDTTEKKK--DEIKDWFDCRYVSASEAIWRIFKFPIQHRSTP 666
Query: 306 VLKLLFHLHNEQ 317
V KL FH +Q
Sbjct: 667 VQKLSFHDKGKQ 678
>At2g07620 putative helicase
Length = 1241
Score = 247 bits (631), Expect = 9e-66
Identities = 146/384 (38%), Positives = 195/384 (50%), Gaps = 73/384 (19%)
Query: 14 SRRSVDCYTMIESQRLRYIRKNQKTIRCGILNGLHEAMDMGETDASNVGQKIVLQPSFAG 73
+ + ++ YT IES RL YI+ NQ +RC + + A + G T+ S G+ I + SF G
Sbjct: 268 ANKCINGYTTIESNRLAYIKFNQSNLRCKNYDTVKAAREAGNTNMSEQGKSIRIPQSFTG 327
Query: 74 GRHYMFNNCQDAMSICKKYGYPDLFVTIACNTNWREIQDFLKERNLKASDRPDIVCRVFK 133
G +M N+ DAM+ICK YG+P+LF+T CN EI + KER L A DRPD+VC +FK
Sbjct: 328 GPRHMLNSYYDAMAICKMYGFPNLFITFTCNPKLPEITSYCKERKLNAEDRPDVVCWIFK 387
Query: 134 MKLDKLMDDFEKEECLAK--------LMQRHV---------------------------- 157
MKLD LM D ++ L K L H+
Sbjct: 388 MKLDSLMKDLTEDHLLGKTVMFQKRGLPHAHILLFMDKSCKFPTSDDIDNILSAEIPDKA 447
Query: 158 ----------------PCGGARPFSPCMDRGRCTKYYRKNYKGTTTI-DEGYPRYKRRDF 200
PCG A SPC+ G+C+K++ K TT+ +GYP Y+RR+
Sbjct: 448 KDPKLYEVVNDCMIHGPCGAANKESPCIVDGKCSKFFPKKLVEQTTVGKDGYPIYRRRES 507
Query: 201 GIHVDKQRVLLDNRYVVPYNPHLLIRYGGHVNVECCNKSNSIKYLFKYVNK--------- 251
V+K + DN YVVPYN L +RY H+NVE C +S SIKYLFKY+NK
Sbjct: 508 EHFVEKGGIKCDNTYVVPYNRMLSLRYRAHINVEWCKQSGSIKYLFKYINKGQDRVAIVV 567
Query: 252 -VPDKALMQLSVDGDNRDKSKPVD---EIKQYYDCRYVSPCEAVWRIFAFDIHHK*PHVL 307
DK + G + +D EIK Y+DCRYVS EAVWRIF F I ++ V+
Sbjct: 568 EPKDKTSNMVLFSGSQKLLVAVIDDDKEIKDYFDCRYVSASEAVWRIFKFPIQYRTTPVM 627
Query: 308 KLLFHLHNEQ-------GNIDHVS 324
KL +HL +Q NID +S
Sbjct: 628 KLSYHLPGKQPLCFEDTQNIDELS 651
>At3g42100 putative protein
Length = 1752
Score = 238 bits (606), Expect = 7e-63
Identities = 140/372 (37%), Positives = 188/372 (49%), Gaps = 75/372 (20%)
Query: 19 DCYTMIESQRLRYIRKNQKTIRCGILNGLHEAMDMGETDASNVGQKIVLQPSFAGGRHYM 78
D YT IES RL YI+ Q +RC N L +A + G T + G ++++ S GG YM
Sbjct: 641 DAYTTIESNRLSYIKFKQSKLRCENYNSLKKASEAGTTSMNEEGNQVLIPSSLTGGPRYM 700
Query: 79 FNNCQDAMSICKKYGYPDLFVTIACNTNWREIQDFLKERNLKASDRPDIVCRVFKMKL-- 136
N DAM+ICK YG+PDLF+T CN W EI + R L DRPDIV R+FK+KL
Sbjct: 701 VQNYYDAMAICKHYGFPDLFITFTCNPKWPEITRHCQARGLSVDDRPDIVARIFKIKLDS 760
Query: 137 -------------------------------------------------DKLMD----DF 143
DK++ D
Sbjct: 761 LMKDLTDGKMLGKTVASMHTVEFQKRGLPHAHILLFMDAKSKLPTADDIDKIISAEIPDK 820
Query: 144 EKEECLAKLMQR---HVPCGGARPFSPCMDRGRCTKYYRKNYKGTTTID-EGYPRYKRRD 199
+KE L ++++ H PCG A SPCM G+C+K Y K ++ T + +GYP Y+RR
Sbjct: 821 DKEPELYEVIKNSMIHGPCGAANMNSPCMVEGKCSKQYPKKHQDITKVGKDGYPIYRRRM 880
Query: 200 FGIHVDKQRVLLDNRYVVPYNPHLLIRYGGHVNVECCNKSNSIKYLFKYVNKVPDKALMQ 259
+++K DN YVVPYN L +RY H+NVE CN+S SIKYLFKY+NK D+ +
Sbjct: 881 TEDYIEKGGFKCDNGYVVPYNKKLSLRYQAHINVEWCNQSGSIKYLFKYINKGADRVV-- 938
Query: 260 LSVDGDNRDKS--------------KPVDEIKQYYDCRYVSPCEAVWRIFAFDIHHK*PH 305
V+ N+DK+ K DEIK ++DCRYVS EAVWRI+ F + +
Sbjct: 939 FIVEPVNQDKTTENATSGEPPNSTEKKKDEIKDWFDCRYVSASEAVWRIYKFPLQDRSTA 998
Query: 306 VLKLLFHLHNEQ 317
V +L FH +Q
Sbjct: 999 VQRLSFHDEGKQ 1010
>At2g14300 pseudogene; similar to MURA transposase of maize Mutator
transposon
Length = 1230
Score = 228 bits (580), Expect = 8e-60
Identities = 133/356 (37%), Positives = 185/356 (51%), Gaps = 70/356 (19%)
Query: 16 RSVDCYTMIESQRLRYIRKNQKTIRCGILNGLHEAMDMGETDASNVGQKIVLQPSFAGGR 75
R + + ++ S+RL +++ +R + +A D G+ D SN G+ ++ PSF GG
Sbjct: 272 RKDEKHILLRSKRL--LQQFMTKLRSTNKQAVQDASDAGDNDLSNKGKSYIIPPSFTGGP 329
Query: 76 HYMFNNCQDAMSICKKYGYPDLFVTIACNTNWREIQDFLKERNLKASDRPDIVCRVFKMK 135
YM N DA+ CK +G+PDLF+T CN W EI F+KER L A DRPDI+CR++KMK
Sbjct: 330 AYMQQNYLDAIGTCKHFGFPDLFITFTCNPKWPEITRFVKERKLNAEDRPDIICRIYKMK 389
Query: 136 LDKLMD--------------------------------------------------DFEK 145
LD LMD D EK
Sbjct: 390 LDNLMDDLTKNHIFAMYTVEFQKRGLPHAHIIVWMDPRYKFHTADHVDKIIFAEIPDKEK 449
Query: 146 EECLAKLMQR---HVPCGGARPFSPCMDRGRCTKYYRKNYKGTTTID-EGYPRYKRRDFG 201
L + + H PC P SPCM+ G+C+KYY KN+ T++D +GYP Y+RRD G
Sbjct: 450 HPELYQAVSECMIHGPCRLVNPNSPCMENGKCSKYYPKNHVENTSLDYKGYPIYRRRDSG 509
Query: 202 IHVDKQRVLLDNRYVVPYNPHLLIRYGGHVNVECCNKSNSIKYLFKYVNKVPDKALMQLS 261
++K + DN YVVPYN LL +Y H+NVE CN+S SIKYLFKYVNK PD+ + Q +
Sbjct: 510 RFIEKNKYQCDNWYVVPYNDVLLRKYRAHINVEWCNQSVSIKYLFKYVNKGPDR-VTQNN 568
Query: 262 VDGDNRDKSKPVDEIKQYYDCRYVSPCEAVWRIFAFDIHHK*PHVLKLLFHLHNEQ 317
V N D + ++++ Y+DCR + IH++ V KL FH +Q
Sbjct: 569 VGEINNDPQER-NQVQDYFDCR------------GYPIHYRQTSVTKLTFHEKGKQ 611
>At1g54430 hypothetical protein
Length = 1639
Score = 217 bits (553), Expect = 1e-56
Identities = 139/390 (35%), Positives = 199/390 (50%), Gaps = 75/390 (19%)
Query: 5 VVDYG-NVVNSRRS-----VDCYTMIESQRLRYIRKNQKTIRCGILNGLHEAMDMGETDA 58
VV+ G +V S+R VD YT IE +RLR+ R NQK +R N + +A+ G+TDA
Sbjct: 541 VVEKGMTIVRSKRLLHQYIVDAYTSIEQERLRWYRLNQKKLRADQYNNVKDAVARGDTDA 600
Query: 59 SNVGQKIVLQPSFAGGRHYMFNNCQDAMSICKKYGYPDLFVT------------------ 100
++G++++L S+ G YM DAM+IC+ YG PDLF+T
Sbjct: 601 KSIGKRVILPASYTGSPRYMVEKYHDAMAICRWYGNPDLFITMTTNPKWEEISEHLKTYG 660
Query: 101 ---------IACNTNWREIQDFLKERNLKASDRPDIVCRVFKMKLDK-----------LM 140
I C ++ + L + N K P + V+ ++ K L
Sbjct: 661 NDAANVRPDIECRVFKIKLDELLADFN-KGLFFPKPIAIVYTIEFQKRGLPHAHILLWLQ 719
Query: 141 DDFEKE----------------------ECLAKLMQRHVPCGGARPFSPCMDRGRCTKYY 178
D +K L + H PCG RP SPCM++G C+K +
Sbjct: 720 GDLKKPTPNDIDKYISAEIPVKDKDPEGHTLVEQHMMHGPCGKDRPSSPCMEKGICSKKF 779
Query: 179 RKNYKGTTTIDE-GYPRYKRRDFGIHVDKQRVLLDNRYVVPYNPHLLIRYGGHVNVECCN 237
+ + T ++E G+ Y+RR+ +V K + LDNR+VVP+N +L +Y H+NVE CN
Sbjct: 780 PREFVNHTKMNESGFILYRRRNDQRYVLKGQTRLDNRFVVPHNLEILKKYKAHINVEWCN 839
Query: 238 KSNSIKYLFKYVNKVPDKALMQL----SVDGD---NRDKSKPVDEIKQYYDCRYVSPCEA 290
KS++IKYLFKY+ K DKA + SV+G N + KP +EI +Y DCRY+S CEA
Sbjct: 840 KSSAIKYLFKYITKGVDKATFIIQKGNSVNGQGSGNGFEEKPRNEINEYLDCRYLSACEA 899
Query: 291 VWRIFAFDIHHK*PHVLKLLFHLHNEQGNI 320
+WRIF F+IHH P V +L HL EQ I
Sbjct: 900 MWRIFMFNIHHHNPPVQRLPLHLPGEQSTI 929
>At2g14470 pseudogene
Length = 1265
Score = 214 bits (546), Expect = 7e-56
Identities = 129/359 (35%), Positives = 178/359 (48%), Gaps = 76/359 (21%)
Query: 1 MQDRVVDYGNVVNSRRSV-----DCYTMIESQRLRYIRKNQKTIRCGILNGLHEAMDMGE 55
+Q+R V+ ++ S+R D YT IES RL YI+ NQ +RC + EA G
Sbjct: 287 IQEREVECQTLLRSKRLFQQFLCDAYTTIESNRLNYIKFNQSKLRCENYTSVKEAAAAGA 346
Query: 56 TDASNVGQKIVLQPSFAGGRHYMFNNCQDAMSICKKYGYPDLFVTIACNTNWREIQDFLK 115
T G ++++ SF GG YM + DAM+ICK YG+PDLF+T CN W I +
Sbjct: 347 TTMEEEGNQLLIPASFNGGPRYMVQSYYDAMAICKLYGFPDLFITFTCNPKWPHITRYCD 406
Query: 116 ERNLKASDRPDIVCRVFKMKL--------------------------------------- 136
+R L DR DI+ R+FK+KL
Sbjct: 407 KRGLNPKDRLDIIARIFKIKLDSLMNDLTVKKMLGKTVASMYTVEFQKRGLPHAHILLFM 466
Query: 137 ------------DKLMD----DFEKEECLAKLMQR---HVPCGGARPFSPCMDRGRCTKY 177
DKL+ D EKE L ++++ H PCG A SPCM G C+K
Sbjct: 467 HAKSKLPTSDDIDKLISAEIPDKEKEPELYEVIKNSMIHGPCGSANVKSPCMVDGECSKL 526
Query: 178 YRKNYKGTTTI-DEGYPRYKRRDFGIHVDKQRVLLDNRYVVPYNPHLLIRYGGHVNVECC 236
Y K ++ T + +GYP Y+RR +V+K + DNRYV+PYN +RY H+NVE C
Sbjct: 527 YPKKHQDITKVGSDGYPIYRRRKIDDYVEKGGIKCDNRYVMPYNKKFSLRYNAHINVEWC 586
Query: 237 NKSNSIKYLFKYVNKVPDKALM-----QLSVDGDN-------RDKSKPVDEIKQYYDCR 283
N+++SIKYLFKY+NK PDK + Q + GD+ R K +EIK ++DCR
Sbjct: 587 NQNDSIKYLFKYINKGPDKVIFIVEPTQQATAGDSETPQQEQRSAEKKKNEIKDWFDCR 645
>At5g37110 putative helicase
Length = 1307
Score = 189 bits (481), Expect = 2e-48
Identities = 113/319 (35%), Positives = 160/319 (49%), Gaps = 77/319 (24%)
Query: 18 VDCYTMIESQRLRYIRKNQKTIRCGILNGLHEAMDMGETDASNVGQKIVLQPSFAGGRHY 77
VD YT IES L YI+ NQ ++R N + +A + G++D + G L +F GG Y
Sbjct: 382 VDSYTAIESNWLGYIKLNQTSLRAYNYNSVQKASEEGKSDLKDQGLACYLPATFTGGPRY 441
Query: 78 MFNNCQDAMSICKKYGYPDLFVTIACNTNWREIQDFLKERNLKASDRPDIVCRVFKMKLD 137
M N DAM++CK +G+PD F+T CN W E+ F ERNL+A DRP+I+C++ KMKLD
Sbjct: 442 MRNMYLDAMALCKHFGFPDYFITFTCNPKWPELIRFCGERNLRADDRPEIICKILKMKLD 501
Query: 138 KLMDDFEK-----EECLAKLMQR-----------------------------HVPCGGAR 163
LM D K E+ KL + H PCG
Sbjct: 502 SLMLDLTKKKPIREDKYIKLTKAEHIDKVISAEIPDKLKDPELFEVIKESMVHGPCGVVN 561
Query: 164 PFSPCMDRGRCTKYYRKNYKGTTTIDEGYPRYKRR--DFGIHVDKQRVLLDNRYVVPYNP 221
P PCM+ G KRR DF V+K+ DNRYV+PYN
Sbjct: 562 PKCPCMENG-----------------------KRRTDDF---VEKKDFKCDNRYVIPYNR 595
Query: 222 HLLIRYGGHVNVECCNKSNSIKYLFKYVNKVPDK--ALMQLSVDGDNRDKSKPVD----- 274
L +RY H+NVE CN+S S+KY+FKY++K PD+ +++ S++ N++ K D
Sbjct: 596 SLSLRYRAHINVEWCNQSGSVKYIFKYIHKGPDRVTVVVESSLNSKNKENGKQKDNADTD 655
Query: 275 --------EIKQYYDCRYV 285
E++ Y++CR +
Sbjct: 656 GSETKKKNEVEDYFNCRRI 674
>At1g52960 hypothetical protein
Length = 924
Score = 182 bits (461), Expect = 5e-46
Identities = 85/164 (51%), Positives = 117/164 (70%), Gaps = 4/164 (2%)
Query: 156 HVPCGGARPFSPCMDRGRCTKYYRKNYKGTTTID-EGYPRYKRRDFGIHVDKQRVLLDNR 214
H PCG P SPCM+ G+C KY+ K+Y TT +D +G+P Y+RRD GI+V+K DNR
Sbjct: 40 HGPCGVGHPNSPCMENGKCKKYFPKSYSDTTKVDNDGFPVYRRRDTGIYVEKNGFQCDNR 99
Query: 215 YVVPYNPHLLIRYGGHVNVECCNKSNSIKYLFKYVNKVPDKALMQLSVDGDNRDKS-KPV 273
YV+PYN + +RY H+NVE CN+S SIKYLFKYV+K D+ + ++V+ +++D + K
Sbjct: 100 YVIPYNEKVSLRYQAHINVELCNQSGSIKYLFKYVHKGHDR--VTVTVEPNDQDTAKKEK 157
Query: 274 DEIKQYYDCRYVSPCEAVWRIFAFDIHHK*PHVLKLLFHLHNEQ 317
DE+K Y+DCRYVS CEA+WRIF F IH++ V+KL FH +Q
Sbjct: 158 DEVKDYFDCRYVSACEAMWRIFKFPIHYRTTPVVKLFFHEEGKQ 201
>At4g07800 hypothetical protein
Length = 448
Score = 171 bits (432), Expect = 1e-42
Identities = 114/327 (34%), Positives = 161/327 (48%), Gaps = 60/327 (18%)
Query: 18 VDCYTMIESQRLRYIRKNQKTIRCGILNGLHEAMDMGETDASNVGQKIVLQPSFAGGRHY 77
+D YT+IES RL YI+ NQ +R SF Y
Sbjct: 56 IDAYTIIESNRLAYIKFNQSKLR----------------------------SSFTSASRY 87
Query: 78 MFNNCQDAMSICKKYGYPDLFVTIACNTNWREIQDFLKERNL-----------KASDRPD 126
M DAM+ICK YG+PD + ++ R+ L ERNL + R
Sbjct: 88 MLQTYYDAMAICKHYGFPDFLSRLRLDSLMRD----LTERNLLRKTVAFKFKKRGLPRAR 143
Query: 127 IVCRVFKMKLDKLMDDFEKEECLAKLMQR---HVPCGGARPFSPCMDRGRCTKYYRKNYK 183
I+ + + + DD E E+ L +L++ H CG A S CM G+C+K Y K ++
Sbjct: 144 ILLFMEANRKLPIADDIETEQELYELIKNSMIHGLCGSANTNSLCMVDGQCSKLYPKKHQ 203
Query: 184 GTTTID-EGYPRYKRRDFGIHVDKQRVLLDNRYVVPYNPHLLIRYGGHVNVECCNKSNSI 242
T + +GYP Y+RR ++K V DN YVVPYN ++Y H+NVE CN++ SI
Sbjct: 204 ELTKVGADGYPVYRRRPIDGCIEKGGVKCDNMYVVPYN-RFSLKYQAHINVEWCNQNGSI 262
Query: 243 KYLFKYVNKVPDKA--LMQLSVDGDNRDKS----------KPVDEIKQYYDCRYVSPCEA 290
KYLFKY+NK PD+ +++L + N + + K D IK ++D YVS EA
Sbjct: 263 KYLFKYINKGPDRVVFIVELVKEETNSNTTTLGDETVTTKKKKDGIKDWFDYIYVSASEA 322
Query: 291 VWRIFAFDIHHK*PHVLKLLFHLHNEQ 317
+WRIF F I H+ V KL FH+ +Q
Sbjct: 323 IWRIFKFPIQHRSTPVQKLSFHVEGKQ 349
>At2g05080 putative helicase
Length = 1219
Score = 162 bits (409), Expect = 5e-40
Identities = 84/197 (42%), Positives = 119/197 (59%), Gaps = 18/197 (9%)
Query: 137 DKLMDDFEKEECLAKLMQRHVPCGGARPFSPCMDRGRCTKYYRKNYKGTTTID-EGYPRY 195
DKL D E + K M H PCG P PCM+ G+C+K+Y K++ T ID EG+P Y
Sbjct: 305 DKLKDPELFE--VIKEMMVHGPCGVVNPKCPCMENGKCSKFYPKDHVPKTIIDKEGFPIY 362
Query: 196 KRRDFGIHVDKQRVLLDNRYVVPYNPHLLIRYGGHVNVECCNKSNSIKYLFKYVNKVPDK 255
+RR V K+ DNRYV+PYN L +RY H+NVE CN+S S+KY+FKY++K PD+
Sbjct: 363 RRRRIDDFVQKKDFKCDNRYVIPYNRSLSLRYRAHINVEWCNQSGSVKYIFKYIHKGPDR 422
Query: 256 ALMQL--SVDGDNRDKS-------------KPVDEIKQYYDCRYVSPCEAVWRIFAFDIH 300
+ + S++ N++K K +E++ +++CRYVS CEA WRI + IH
Sbjct: 423 VTVVVGSSLNSKNKEKGKQKVNADTDGSEPKKKNEVEDFFNCRYVSACEAAWRILKYPIH 482
Query: 301 HK*PHVLKLLFHLHNEQ 317
++ V+KL FHL EQ
Sbjct: 483 YRSTSVMKLSFHLPGEQ 499
Score = 128 bits (321), Expect = 8e-30
Identities = 61/134 (45%), Positives = 80/134 (59%)
Query: 18 VDCYTMIESQRLRYIRKNQKTIRCGILNGLHEAMDMGETDASNVGQKIVLQPSFAGGRHY 77
VD Y IES RL YI+ NQ ++R N + +A + G+ D G L +F GG Y
Sbjct: 126 VDSYIAIESNRLGYIKLNQSSLRADNYNSVQKASEEGKCDLKYQGLACYLPATFTGGPRY 185
Query: 78 MFNNCQDAMSICKKYGYPDLFVTIACNTNWREIQDFLKERNLKASDRPDIVCRVFKMKLD 137
M N DAM++CK +G+PD F+T CN W E+ F ERNL+ DRP+I+C++FKMKLD
Sbjct: 186 MRNMYLDAMAVCKHFGFPDYFITFTCNPKWPELIRFCGERNLRVDDRPEIICKIFKMKLD 245
Query: 138 KLMDDFEKEECLAK 151
LM D K L K
Sbjct: 246 SLMLDLTKRNILGK 259
>At3g32320 hypothetical protein
Length = 494
Score = 159 bits (403), Expect = 3e-39
Identities = 101/299 (33%), Positives = 145/299 (47%), Gaps = 62/299 (20%)
Query: 34 KNQKTIRCGILNGLHEAMDMGETDASNVGQKIVLQPSFAGGRHYMFNNCQDAMSICKKYG 93
KNQ +R + + D G+ D SN G+ ++ PSF GG YM N DA+ Y
Sbjct: 115 KNQTKLRNTNKQAVQDTSDAGDNDLSNKGKSCIIPPSFTGGPAYMQQNYLDAI-----YK 169
Query: 94 YPDLFVTIACNTNWREIQDFLKERNLKASDRPDIVCRVFKMKLDKLMDDFEKE---ECLA 150
+P +D D++ +F DK D + + EC+
Sbjct: 170 FP-------------------------TADHVDMI--IFAEIPDKEKDPEQYQAVSECII 202
Query: 151 KLMQRHVPCGGARPFSPCMDRGRCTKYYRKNYKGTTTID-EGYPRYKRRDFGIHVDKQRV 209
H PCG P SPCM G+C+KYY KN+ T++D EGYP Y+RRD G + K +
Sbjct: 203 -----HGPCGLVNPNSPCMKNGKCSKYYPKNHVENTSLDNEGYPIYRRRDTGRFIKKNKY 257
Query: 210 LLDNRYVVPYNPHLLIRYGGHVNVECCNKSNSIKYLFKYVNKVPDKALMQLSVD------ 263
DN YVVPYN LL +Y H+NVE CN+S S+KYLFKYVNK PD+ + +
Sbjct: 258 QCDNWYVVPYNDVLLRKYKAHINVEWCNQSVSVKYLFKYVNKGPDRVTVSVEPHRKEVVS 317
Query: 264 -----GDNRDKSKPVDEIKQYYDCRYVSPCEAVWRIFAFDIHHK*PHVLKLLFHLHNEQ 317
G+ + + ++++ Y+DC RI + IH++ + KL FH +Q
Sbjct: 318 EQNNVGETNNDQQERNQVQDYFDC----------RIRGYPIHYRQTSITKLTFHEKGKQ 366
>At3g42320 putative protein
Length = 541
Score = 130 bits (328), Expect = 1e-30
Identities = 95/326 (29%), Positives = 134/326 (40%), Gaps = 118/326 (36%)
Query: 5 VVDYGNVVNSRRSVDCYTMIESQRLRYIRKNQKTIRCGILNGLHEAMDMGETDASNVGQK 64
+ D N VN+R Y ++ R+ Y + I G HE MD
Sbjct: 94 IADTENEVNNR-----YNIMSDPRI-YNLPTAYEVAALIPGGFHEDMDKR---------- 137
Query: 65 IVLQPSFAGGRHYMFNNCQDAMSICKKYGYPDLFVTIACNTNWREIQDFLKERNLKASDR 124
G YM NN DAM+IC+ +G P +F+T CN W EI F K RNLK+ DR
Sbjct: 138 ---------GPRYMANNYLDAMAICQHFGLPSVFITFTCNPKWHEIVRFYKARNLKSEDR 188
Query: 125 PDIVCRVFKMKLDKLMDDFEKEECLAK--------------LMQRHV------------- 157
PDI+C++FKMKLD LM D + L K L+ H+
Sbjct: 189 PDIICQIFKMKLDNLMYDLTTKYLLGKTVSCMYTIEFQKRGLLHAHILLFLHATNKLYTA 248
Query: 158 -------------------------------PCGGARPFSPCMDRGRCTKYYRKNYKGTT 186
PCG P SPCM G+C KY+ K+Y TT
Sbjct: 249 EDTDKVITAEIPDKKKKLELYVLVKDCMIHGPCGVGHPNSPCMVNGQCKKYFSKSYYDTT 308
Query: 187 TID-EGYPRYKRRDFGIHVDKQRVLLDNRYVVPYNPHLLIRYGGHVNVECCNKSNSIKYL 245
+D +G+P Y+R + GI+ + K+ IKYL
Sbjct: 309 KVDNDGFPVYRRHNTGIYAE--------------------------------KNGLIKYL 336
Query: 246 FKYVNKVPDKALMQLSVDGDNRDKSK 271
FKYV+K D+ + ++++ +N+D +K
Sbjct: 337 FKYVHKEHDR--VTVTIEPNNQDTTK 360
>At3g43330 putative protein
Length = 489
Score = 129 bits (323), Expect = 5e-30
Identities = 59/116 (50%), Positives = 74/116 (62%)
Query: 18 VDCYTMIESQRLRYIRKNQKTIRCGILNGLHEAMDMGETDASNVGQKIVLQPSFAGGRHY 77
VD YTMIES RLRYI+KNQ R + A + G D N G+++ + SF GG Y
Sbjct: 374 VDAYTMIESNRLRYIKKNQPKFRSSTREPIQNASNSGNNDLENQGKEVKIPASFTGGPRY 433
Query: 78 MFNNCQDAMSICKKYGYPDLFVTIACNTNWREIQDFLKERNLKASDRPDIVCRVFK 133
M NN DAM+ICK +G+P LF+T CN W EI F KERNLK+ DR DI+CR+ +
Sbjct: 434 MTNNYLDAMAICKHFGFPSLFITFTCNPKWPEIVRFCKERNLKSEDRQDIICRILR 489
>At5g32630 putative protein
Length = 856
Score = 123 bits (308), Expect = 3e-28
Identities = 84/278 (30%), Positives = 118/278 (42%), Gaps = 69/278 (24%)
Query: 45 NGLHEAMDMGETDASNVGQKIVLQPSFAGGRHYMFNNCQDAMSICKKYGYPDLFVTIACN 104
+ L +A + G T + G K+++ SF GG YM + DAM+IC+ YG+PDLF+T CN
Sbjct: 167 SSLKKASEAGTTSINEEGNKVLIPTSFTGGPRYMVQSYYDAMAICEHYGFPDLFITFTCN 226
Query: 105 TNWREIQDFLKERNLKASDRPDIVCRVFKMKLDKLMDD---------------FEKE--- 146
W I + + L DRPDIV R+FK+KLD LM D F+K
Sbjct: 227 PKWPGINRYCQAIGLSVDDRPDIVARIFKIKLDSLMKDLTDGNAGKDSMHTVEFQKRGLL 286
Query: 147 -----------------ECLAKLMQRHVPCGGARP-----FSPCMDRGRCTKYYRKNYKG 184
+ + KL+ +P P M G C N
Sbjct: 287 HAPTLLFMDAKSKLPTTDDIDKLISAEIPDKDKEPEFYEVIKNSMIHGPCG---AANMNS 343
Query: 185 TTTIDEGYPRYKRRDFGIHVDKQRVLLDNRYVVPYNPHLLIRYGGHVNVECCNKSNSIKY 244
+++ Y ++K DN YVVPYN L +RY H+NVEC +SN IK
Sbjct: 344 PCMVEDDY-----------IEKGGFKCDNSYVVPYNQKLSLRYQAHINVEC--QSNRIKQ 390
Query: 245 LFKYVNKVPDKALMQLSVDGDNRDKSKPVDEIKQYYDC 282
+ + P+ K DEIK ++DC
Sbjct: 391 KAATLGEPPNST-------------EKKKDEIKDWFDC 415
>At3g31980 hypothetical protein
Length = 1099
Score = 107 bits (266), Expect = 2e-23
Identities = 59/136 (43%), Positives = 76/136 (55%), Gaps = 12/136 (8%)
Query: 16 RSVDCYTMIESQRLRYIRKNQKTIRCGILNGLHEAMDMGETDASNVGQKIVLQPSFAGGR 75
RS + T++ S+RL Q+ + G G G T+ S G+ I + SF GG
Sbjct: 137 RSDEAQTLLRSKRLF-----QQFLVDGYTTG-------GNTNMSEQGKSIRIPQSFTGGP 184
Query: 76 HYMFNNCQDAMSICKKYGYPDLFVTIACNTNWREIQDFLKERNLKASDRPDIVCRVFKMK 135
YM N+ D M+I K YG+P+LF+T CN W EI + KER L A DRPD VC +FKMK
Sbjct: 185 RYMLNSYYDVMAITKMYGFPNLFITFTCNPKWPEITRYCKERKLNAEDRPDGVCWIFKMK 244
Query: 136 LDKLMDDFEKEECLAK 151
LD LM D +E L K
Sbjct: 245 LDSLMKDLTEEHLLGK 260
Score = 31.2 bits (69), Expect = 1.4
Identities = 12/31 (38%), Positives = 15/31 (47%)
Query: 253 PDKALMQLSVDGDNRDKSKPVDEIKQYYDCR 283
P + + R K K DEIK Y+DCR
Sbjct: 349 PKDKTSNMQTGSETRSKEKSTDEIKDYFDCR 379
>At5g28670 putative protein
Length = 647
Score = 100 bits (249), Expect = 2e-21
Identities = 53/132 (40%), Positives = 77/132 (58%), Gaps = 14/132 (10%)
Query: 164 PFSPCMDRGRCTKYYRKNYKGTTTID-EGYPRYKRRDFGIHVDKQRVLLDNRYVVPYNPH 222
P S CM+ +C+K+Y KN+ T++D EGY Y+R D G ++K + DN YV+PYN
Sbjct: 297 PNSLCMENDKCSKFYPKNHVENTSLDNEGYQIYRRIDTGRFIEKNKFQYDNWYVIPYNDV 356
Query: 223 LLIRYGGHVNVECCNKSNSIKYLFKYVNKVPDKALM------------QLSVDGDNRDKS 270
L +Y H+NVE CN+S +KYLFKYVNK D+ + Q +V N+D
Sbjct: 357 FLRKYRAHINVEWCNQSVFVKYLFKYVNKDLDRVTVSIKPHRKEDVTEQNNVGETNKDPQ 416
Query: 271 KPVDEIKQYYDC 282
+ +++ YYDC
Sbjct: 417 ER-NQVHDYYDC 427
>At3g42420 putative protein
Length = 1018
Score = 87.8 bits (216), Expect = 1e-17
Identities = 46/112 (41%), Positives = 69/112 (61%), Gaps = 8/112 (7%)
Query: 136 LDKLMD----DFEKEECLAKLMQR---HVPCGGARPFSPCMDRGRCTKYYRKNYKGTTTI 188
+DK++ D EKE L +++++ H PCG A SPCM G+C+K Y K ++ T +
Sbjct: 422 IDKMISAEIPDKEKEPELYEVIKKCMIHGPCGAANKNSPCMVDGKCSKNYPKKFEEITKV 481
Query: 189 D-EGYPRYKRRDFGIHVDKQRVLLDNRYVVPYNPHLLIRYGGHVNVECCNKS 239
+G+P Y+R+ +V+K DNRYVV YN L +RYG H+NVE CN++
Sbjct: 482 GKDGFPVYRRKQSHNYVEKSGFRCDNRYVVSYNKILSVRYGAHINVEWCNQN 533
>At1g64410 unknown protein
Length = 753
Score = 75.1 bits (183), Expect = 8e-14
Identities = 58/205 (28%), Positives = 87/205 (42%), Gaps = 48/205 (23%)
Query: 5 VVDYGNVVNSRRSVDCYTMIESQRLRYIRKNQKTIRCGILNGLHEAMDMGETDASNVGQK 64
+VD N VN+R ++ RLRYI+ NQ R + A + G D N G
Sbjct: 129 IVDTENEVNNRYNI--------MRLRYIKNNQPQFRSSNKESIQNASNSGNNDLENQGMY 180
Query: 65 IVLQPSFAGGRHYMFNNCQDAMSICKKYGYPDLFVTIACNTNWREIQDFLKERNLKASDR 124
+ +K G P + + + + L ++
Sbjct: 181 TIE---------------------FQKRGLPHAHILLFMHPTSK----------LSTAED 209
Query: 125 PDIVCRVFKMKLDKLMDDFEKEECLA--KLMQRHVPCGGARPFSPCMDRGRCTKYYRKNY 182
D K+ ++ D +K E A K H PCG P SPCM+ G+C KY+ K+Y
Sbjct: 210 TD------KVITAEIPDKKKKPELYAVVKDCMIHGPCGVGHPNSPCMENGKCKKYFPKSY 263
Query: 183 KGTTTID-EGYPRYKRRDFGIHVDK 206
TT +D +G+P Y+RRD GI+V+K
Sbjct: 264 SDTTKVDNDGFPVYRRRDTGIYVEK 288
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.342 0.150 0.514
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 10,614,852
Number of Sequences: 26719
Number of extensions: 449312
Number of successful extensions: 1286
Number of sequences better than 10.0: 31
Number of HSP's better than 10.0 without gapping: 25
Number of HSP's successfully gapped in prelim test: 6
Number of HSP's that attempted gapping in prelim test: 1198
Number of HSP's gapped (non-prelim): 56
length of query: 489
length of database: 11,318,596
effective HSP length: 103
effective length of query: 386
effective length of database: 8,566,539
effective search space: 3306684054
effective search space used: 3306684054
T: 11
A: 40
X1: 15 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 39 (22.0 bits)
S2: 62 (28.5 bits)
Medicago: description of AC125481.7


