

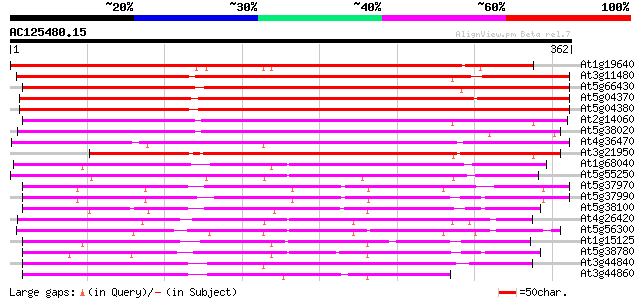
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC125480.15 - phase: 0
(362 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
At1g19640 S-adenosyl-L-methionine:jasmonic acid carboxyl methylt... 305 3e-83
At3g11480 unknown protein 304 6e-83
At5g66430 S-adenosyl-L-methionine:salicylic acid carboxyl methyl... 291 4e-79
At5g04370 S-adenosyl-L-methionine:salicylic acid carboxyl methyl... 291 4e-79
At5g04380 S-adenosyl-L-methionine:salicylic acid carboxyl methyl... 290 6e-79
At2g14060 hypothetical protein 280 1e-75
At5g38020 SAMT-like protein 275 3e-74
At4g36470 hypothetical protein 266 1e-71
At3g21950 salicylic acid carboxyl methyltransferase, putative 250 7e-67
At1g68040 putative S-adenosyl-L-methionine:salicylic acid carbox... 203 1e-52
At5g55250 S-adenosyl-L-methionine:salicylic acid carboxyl methyl... 190 1e-48
At5g37970 putative protein 174 7e-44
At5g37990 putative protein 173 1e-43
At5g38100 unknown protein 168 4e-42
At4g26420 putative protein 164 9e-41
At5g56300 S-adenosyl-L-methionine:salicylic acid carboxyl methyl... 160 1e-39
At1g15125 putative protein 158 5e-39
At5g38780 AtPP - like protein 157 7e-39
At3g44840 proteinkinase AtPP -like protein 149 3e-36
At3g44860 AtPP -like protein 148 5e-36
>At1g19640 S-adenosyl-L-methionine:jasmonic acid carboxyl
methyltransferase (JMT)
Length = 389
Score = 305 bits (781), Expect = 3e-83
Identities = 174/366 (47%), Positives = 228/366 (61%), Gaps = 29/366 (7%)
Query: 1 MEVAKVLHMNGGVGETSYANNSLVQRKVIYLTKPLRDEAITS-MYNNTLSKSLTIADLGC 59
MEV +VLHMN G GETSYA NS Q +I L + + DEA+ M +N+ S+ IADLGC
Sbjct: 1 MEVMRVLHMNKGNGETSYAKNSTAQSNIISLGRRVMDEALKKLMMSNSEISSIGIADLGC 60
Query: 60 SSGSNTLLVILDIIKVVEKLCRKLNHKSPEYMIYLNDLPGNDFNTIFTSLDIFKEKLLDE 119
SSG N+LL I +I+ + LC L+ PE + LNDLP NDFN I SL F +++ +
Sbjct: 61 SSGPNSLLSISNIVDTIHNLCPDLDRPVPELRVSLNDLPSNDFNYICASLPEFYDRVNNN 120
Query: 120 ---MGTEMG---PCFFSGVPGSFHGRIFPLQSLHFVHSSYSLHWLSKVP----EGADN-- 167
+G G CF S VPGSF+GR+FP +SLHFVHSS SLHWLS+VP E D
Sbjct: 121 KEGLGFGRGGGESCFVSAVPGSFYGRLFPRRSLHFVHSSSSLHWLSQVPCREAEKEDRTI 180
Query: 168 -----NKGNIYISSTSPLNVVKAYYKQFQIDFSLFLKCRAEEIVEGGCMIITFVGRKSDN 222
N G IYIS TSP + KAY QFQ DF +FL+ R+EE+V GG M+++F+GR+S +
Sbjct: 181 TADLENMGKIYISKTSPKSAHKAYALQFQTDFWVFLRSRSEELVPGGRMVLSFLGRRSLD 240
Query: 223 PTSKECCYIWELLAMALNDMVLEGIIKEEKLNTFNIPIYYPSPSEVKLEVITEGSFVMNQ 282
PT++E CY WELLA AL M EGII+EEK++ FN P Y S E+K+ + EGSF +++
Sbjct: 241 PTTEESCYQWELLAQALMSMAKEGIIEEEKIDAFNAPYYAASSEELKMVIEKEGSFSIDR 300
Query: 283 LEISEVNWNARDDFESESFG----------DDGYNVAQCMRALAEPLLVSHFGEGVVKEI 332
LEIS ++W ES+ G V+ +RA+ EP+L FGE V+ E+
Sbjct: 301 LEISPIDWEG-GSISEESYDLVIRSKPEALASGRRVSNTIRAVVEPMLEPTFGENVMDEL 359
Query: 333 FNRYKK 338
F RY K
Sbjct: 360 FERYAK 365
>At3g11480 unknown protein
Length = 379
Score = 304 bits (778), Expect = 6e-83
Identities = 156/362 (43%), Positives = 229/362 (63%), Gaps = 14/362 (3%)
Query: 5 KVLHMNGGVGETSYANNSLVQRKVIYLTKP-LRDEAITSMYNNTLSKSLTIADLGCSSGS 63
K L M+GG G SY+ NS +Q+KV+ + KP L M N + +A+LGCSSG
Sbjct: 27 KALCMSGGDGANSYSANSRLQKKVLSMAKPVLVRNTEEMMMNLDFPTYIKVAELGCSSGQ 86
Query: 64 NTLLVILDIIKVVEKLCRKLNHKSPEYMIYLNDLPGNDFNTIFTSLDIFKEKLLDEMGTE 123
N+ L I +II + LC+ +N SPE LNDLP NDFNT F + F ++L M T
Sbjct: 87 NSFLAIFEIINTINVLCQHVNKNSPEIDCCLNDLPENDFNTTFKFVPFFNKEL---MITN 143
Query: 124 MGPCFFSGVPGSFHGRIFPLQSLHFVHSSYSLHWLSKVPEGADNNKGNIYISSTSPLNVV 183
CF G PGSF+ R+F SLH +HSSY+LHWLSKVPE +NNKGN+YI+S+SP +
Sbjct: 144 KSSCFVYGAPGSFYSRLFSRNSLHLIHSSYALHWLSKVPEKLENNKGNLYITSSSPQSAY 203
Query: 184 KAYYKQFQIDFSLFLKCRAEEIVEGGCMIITFVGRKS-DNPTSKECCYIWELLAMALNDM 242
KAY QFQ DF++FL+ R+EEIV G M++TF+GR + ++P ++CC+ W LL+ +L D+
Sbjct: 204 KAYLNQFQKDFTMFLRLRSEEIVSNGRMVLTFIGRNTLNDPLYRDCCHFWTLLSNSLRDL 263
Query: 243 VLEGIIKEEKLNTFNIPIYYPSPSEVKLEVITEGSFVMNQLE---ISEVNWNARDDFESE 299
V EG++ E KL+ FN+P Y P+ E+K + EGSF +N+LE ++ DDFE+
Sbjct: 264 VFEGLVSESKLDAFNMPFYDPNVQELKEVIQKEGSFEINELESHGFDLGHYYEEDDFEA- 322
Query: 300 SFGDDGYNVAQCMRALAEPLLVSHFGEGVVKEIFNRYKKYLTDRNSKGRTKFINITILLA 359
G N A +RA++EP+L++HFGE ++ +F++Y ++T + +++ + L
Sbjct: 323 -----GRNEANGIRAVSEPMLIAHFGEEIIDTLFDKYAYHVTQHANCRNKTTVSLVVSLT 377
Query: 360 KR 361
K+
Sbjct: 378 KK 379
>At5g66430 S-adenosyl-L-methionine:salicylic acid carboxyl
methyltransferase-like protein
Length = 354
Score = 291 bits (745), Expect = 4e-79
Identities = 149/359 (41%), Positives = 221/359 (61%), Gaps = 11/359 (3%)
Query: 9 MNGGVGETSYANNSLVQRKVIYLTKP-LRDEAITSMYNNTLSKSLTIADLGCSSGSNTLL 67
M+GG G+ SY+ NSL+Q+KV+ KP L M N + +ADLGC++G NT L
Sbjct: 1 MSGGDGDNSYSTNSLLQKKVLSKAKPVLVKNTKGMMINLNFPNYIKVADLGCATGENTFL 60
Query: 68 VILDIIKVVEKLCRKLNHKSPEYMIYLNDLPGNDFNTIFTSLDIFKEKLLDEMGTEMGPC 127
+ +I+ + LC++ N K PE LNDLP NDFNT F + F +++ + C
Sbjct: 61 TMAEIVNTINVLCQQCNQKPPEIDCCLNDLPDNDFNTTFKFVPFFNKRVKSKR-----LC 115
Query: 128 FFSGVPGSFHGRIFPLQSLHFVHSSYSLHWLSKVPEGADNNKGNIYISSTSPLNVVKAYY 187
F SGVPGSF+ R+FP +SLHFVHSSYSLHWLSKVP+G + N ++YI+++SP N KAY
Sbjct: 116 FVSGVPGSFYSRLFPRKSLHFVHSSYSLHWLSKVPKGLEKNSSSVYITTSSPPNAYKAYL 175
Query: 188 KQFQIDFSLFLKCRAEEIVEGGCMIITFVGRKS-DNPTSKECCYIWELLAMALNDMVLEG 246
QFQ DF FL+ R+EE+V G M++TF+GRK+ D+P ++CC+ W LL+ +L D+V EG
Sbjct: 176 NQFQSDFKSFLEMRSEEMVSNGRMVLTFIGRKTLDDPLHRDCCHFWTLLSTSLRDLVYEG 235
Query: 247 IIKEEKLNTFNIPIYYPSPSEVKLEVITEGSFVMNQLEISEVNW---NARDDFESES-FG 302
++ K+++FNIP Y PS EV + EGSF +N LEI N +D+ S
Sbjct: 236 LVSASKVDSFNIPFYDPSKEEVMEMIRNEGSFEINDLEIHGFELGLSNHDEDYMLHSQIS 295
Query: 303 DDGYNVAQCMRALAEPLLVSHFGEGVVKEIFNRYKKYLTDRNSKGRTKFINITILLAKR 361
G A C+RA++E +LV+ FG ++ +F ++ +++ S + + + L ++
Sbjct: 296 KAGQREANCIRAVSESMLVADFGVDIMDTLFKKFAYHVSQHASCTNKTTVTLVVSLIRK 354
>At5g04370 S-adenosyl-L-methionine:salicylic acid carboxyl
methyltransferase-like protein
Length = 396
Score = 291 bits (745), Expect = 4e-79
Identities = 152/357 (42%), Positives = 218/357 (60%), Gaps = 7/357 (1%)
Query: 7 LHMNGGVGETSYANNSLVQRKVIYLTKP-LRDEAITSMYNNTLSKSLTIADLGCSSGSNT 65
L M GG G SY++NSL+QR+V+ KP L M N + +ADLGCSSG NT
Sbjct: 39 LCMRGGDGYNSYSSNSLLQRRVLSKAKPVLVKNTKDLMINLNFPTYIKVADLGCSSGQNT 98
Query: 66 LLVILDIIKVVEKLCRKLNHKSPEYMIYLNDLPGNDFNTIFTSLDIFKEKLLDEMGTEMG 125
L + +II + C++ N PE LNDLP NDFNT F + F + T
Sbjct: 99 FLAMSEIINTINVFCQQRNQNPPEIDCCLNDLPSNDFNTTFKFIQFFNGMNI----TSKE 154
Query: 126 PCFFSGVPGSFHGRIFPLQSLHFVHSSYSLHWLSKVPEGADNNKGNIYISSTSPLNVVKA 185
F GVPGSF+ R+FP +SLHFVHSSY LHWLSKVPEG + NK ++YI+++SPL+ KA
Sbjct: 155 SYFVYGVPGSFYSRLFPRRSLHFVHSSYGLHWLSKVPEGLEKNKMSVYITNSSPLSTYKA 214
Query: 186 YYKQFQIDFSLFLKCRAEEIVEGGCMIITFVGRKS-DNPTSKECCYIWELLAMALNDMVL 244
Y QFQ DF+ FLK R+EE+V G M++TF+GR + DNP ++CC+ W LL+ +L D+V
Sbjct: 215 YLNQFQRDFATFLKLRSEEMVSNGRMVLTFIGRNTIDNPLHRDCCHFWTLLSKSLRDLVA 274
Query: 245 EGIIKEEKLNTFNIPIYYPSPSEVKLEVITEGSFVMNQLEISEVNWNARDDFESESFGDD 304
EG++ K+++F +P Y P+ E+K V EGSF + LE + + ES+
Sbjct: 275 EGLVSASKVDSFYLPFYDPNEKEIKEMVQKEGSFEIRDLETHGYDLGHCNQDESKR-SKS 333
Query: 305 GYNVAQCMRALAEPLLVSHFGEGVVKEIFNRYKKYLTDRNSKGRTKFINITILLAKR 361
G N A +RA++EPLL +HFG+ ++ +FN++ +++ S ++I + L K+
Sbjct: 334 GQNEANYIRAVSEPLLAAHFGDAIINILFNKFACHVSQHVSCRNKTTVSIVVSLTKK 390
>At5g04380 S-adenosyl-L-methionine:salicylic acid carboxyl
methyltransferase-like protein
Length = 385
Score = 290 bits (743), Expect = 6e-79
Identities = 147/359 (40%), Positives = 226/359 (62%), Gaps = 9/359 (2%)
Query: 7 LHMNGGVGETSYANNSLVQRKVIYLTKPLRDEAITSMYNNT-LSKSLTIADLGCSSGSNT 65
L MNGG + SY SL+Q++V+ +T P+ + M N K + +ADLGCSSG NT
Sbjct: 32 LCMNGGDVDNSYTTKSLLQKRVLSITNPILVKNTEEMLTNLDFPKCIKVADLGCSSGQNT 91
Query: 66 LLVILDIIKVVEKLCRKLNHKSPEYMIYLNDLPGNDFNTIFTSLDIFKEKLLDEMGTEMG 125
L + +I+ + LC+K N PE LNDLP NDFNT F + F +KL T G
Sbjct: 92 FLAMSEIVNTINVLCQKWNQSRPEIDCCLNDLPTNDFNTTFKFITFFNKKL-----TSNG 146
Query: 126 PCFFSGVPGSFHGRIFPLQSLHFVHSSYSLHWLSKVPEGADNNKGNIYISSTSPLNVVKA 185
CF SGVPGSF+ R+FP +SLHF++S YS+H+LSKVP+G + NK ++YI+S+SPL+ KA
Sbjct: 147 SCFVSGVPGSFYSRLFPRKSLHFIYSIYSIHFLSKVPDGLEKNKMSVYITSSSPLSEYKA 206
Query: 186 YYKQFQIDFSLFLKCRAEEIVEGGCMIITFVGRKS-DNPTSKECCYIWELLAMALNDMVL 244
Y QF+ DF+ FL+ R+EE+V G M++T +GR + DNP ++CC+ W LL+ +L D+V
Sbjct: 207 YLNQFKRDFTTFLRMRSEEMVHNGRMVLTLIGRNTLDNPLYRDCCHCWTLLSNSLRDLVF 266
Query: 245 EGIIKEEKLNTFNIPIYYPSPSEVKLEVITEGSFVMNQLEISEVN-WNARDDFESESF-G 302
EG++ K+ +F +P Y P+ EVK + EGSF +N LE+ E + ++++ +S
Sbjct: 267 EGLLSASKVYSFKMPFYDPNEEEVKEIIRNEGSFQINDLEMHEFDLGHSKEKCSLQSHKA 326
Query: 303 DDGYNVAQCMRALAEPLLVSHFGEGVVKEIFNRYKKYLTDRNSKGRTKFINITILLAKR 361
G A C+RA+ E +LV+HFG+ ++ +F++Y +++ S + + + L ++
Sbjct: 327 KAGQKEASCIRAVTETMLVAHFGDDIIDALFHKYAHHVSQHASCRVKTSVTLIVSLVRK 385
>At2g14060 hypothetical protein
Length = 359
Score = 280 bits (715), Expect = 1e-75
Identities = 148/362 (40%), Positives = 218/362 (59%), Gaps = 13/362 (3%)
Query: 9 MNGGVGETSYANNSLVQRKVIYLTKPLRDEAITSMY-NNTLSKSLTIADLGCSSGSNTLL 67
M GG G+ SYA NS QR V Y +PL E + M N + +ADLGCS+G NT+L
Sbjct: 1 MKGGTGDHSYATNSHYQRSVFYEIQPLVIENVREMLLKNGFPGCIKVADLGCSTGQNTVL 60
Query: 68 VILDIIKVVEKLCRKLNHKSPEYMIYLNDLPGNDFNTIFTSLDIFKEKLLDEMGTEMGPC 127
+ I + + ++++ PE YLNDLP NDFNT F F+EKL E+ G
Sbjct: 61 AMSAIAYTIMESYQQMSKNPPEIDCYLNDLPENDFNTTFKLFHSFQEKLKPEV---KGKW 117
Query: 128 FFSGVPGSFHGRIFPLQSLHFVHSSYSLHWLSKVPEGADNNKGNIYISSTSPLNVVKAYY 187
F SGVPGSF+ R+FP +SLHFVHS++S+HWLS++P+G ++N +I+I P NV K+Y
Sbjct: 118 FVSGVPGSFYSRLFPRKSLHFVHSAFSIHWLSRIPDGLESNTKSIHIKYPYPSNVYKSYL 177
Query: 188 KQFQIDFSLFLKCRAEEIVEGGCMIITFVGRKSDNPTSKECCYIWELLAMALNDMVLEGI 247
QF+IDFSLFLK R+EE+V G M++TFVGRK + SK+C +W LL+ L D+ EG
Sbjct: 178 NQFKIDFSLFLKMRSEEVVHNGHMVLTFVGRKVSDTLSKDCFQVWSLLSDCLLDLASEGF 237
Query: 248 IKEEKLNTFNIPIYYPSPSEVKLEVITEGSFVMNQLE----ISEVNWNARDDFESESFG- 302
+ + + +FN+P Y P+ EV+ ++ EGSF + ++E + + ++ E +S
Sbjct: 238 VNDSMVKSFNMPFYNPNEEEVREFILKEGSFEITKIEKFDHVVPYKIDREEEDEEQSLQL 297
Query: 303 DDGYNVAQCMRALAEPLLVSHFGEGVVKEIFNRY----KKYLTDRNSKGRTKFINITILL 358
+ G A R + EPLLV+HFG+ +++ +FN+Y KYL+ N + + + L
Sbjct: 298 EAGIKHASWARCITEPLLVAHFGDAIIEPVFNKYAHYMAKYLSVSNHRRNMTLVIVVSLT 357
Query: 359 AK 360
K
Sbjct: 358 RK 359
>At5g38020 SAMT-like protein
Length = 368
Score = 275 bits (703), Expect = 3e-74
Identities = 155/367 (42%), Positives = 220/367 (59%), Gaps = 20/367 (5%)
Query: 6 VLHMNGGVGETSYANNSLVQRKVIYLTKPLRDEAITSMYNNT-LSKSLTIADLGCSSGSN 64
VL M GG GE SYANNS Q+++ KP+ E + M T + +ADLGCSSG N
Sbjct: 3 VLSMKGGDGEHSYANNSEGQKRLASDAKPVVVETVKEMIVKTDFPGCIKVADLGCSSGEN 62
Query: 65 TLLVILDIIKVVEKLCRKLNHKSPEYMIYLNDLPGNDFNTIFTSLDIFKEKLLDEMGTEM 124
TLLV+ +I+ + ++ PE LNDLP NDFNT F + F + L ++
Sbjct: 63 TLLVMSEIVNTIITSYQQKGKNLPEINCCLNDLPDNDFNTTFKLVPAFHKLLKMDV---K 119
Query: 125 GPCFFSGVPGSFHGRIFPLQSLHFVHSSYSLHWLSKVPEGADNNKGNIYISSTSPLNVVK 184
G CF SGVPGSF+ R+FP +SLHFVHSS LHWLSKVP+G ++NK N+Y+ S P NV K
Sbjct: 120 GKCFISGVPGSFYSRLFPSKSLHFVHSSLCLHWLSKVPDGLEDNKKNVYLRSPCPPNVYK 179
Query: 185 AYYKQFQIDFSLFLKCRAEEIVEGGCMIITFVGRKSDNPTSKECCYIWELLAMALNDMVL 244
+Y QF+ DFSLFL+ RA+E V G M +TFVGRKS +P SK+C W ++ +L D+V
Sbjct: 180 SYLTQFKNDFSLFLRLRADETVPNGRMALTFVGRKSLDPLSKDCFQNWSSISDSLLDLVS 239
Query: 245 EGIIKEEKLNTFNIPIYYPSPSEVKLEVITEGSFVMNQLE-ISEVNWNARDDFESESFGD 303
EGI+KE +++FN+P Y P SEV+ + +EGSF ++ E I + ++ + D
Sbjct: 240 EGIVKESDVDSFNLPFYNPDESEVREVIESEGSFKISNFETIFGLLFSYKTGRTEVKDDD 299
Query: 304 DGYNV----------AQCMRALAEPLLVSHFGEGVVKEIFNRYKKYLTDRNSKGRTK--- 350
D + A +R++ EP+L +HFG+ ++ +F RY +L +R R K
Sbjct: 300 DNLDQSCRFEVIRKRASIIRSITEPMLGAHFGDAIMDRLFERYTYHLAERYDTLRNKPTV 359
Query: 351 --FINIT 355
F+++T
Sbjct: 360 QFFVSLT 366
>At4g36470 hypothetical protein
Length = 371
Score = 266 bits (681), Expect = 1e-71
Identities = 151/373 (40%), Positives = 225/373 (59%), Gaps = 20/373 (5%)
Query: 2 EVAKVLHMNGGVGETSYANNSLVQRKVIYLTKPLRDEAITSMYNNTLSKSLTIADLGCSS 61
++ + +M GG G+TSYA NS +Q+K K + E + +Y T KSL IADLGCSS
Sbjct: 5 DMEREFYMTGGDGKTSYARNSSLQKKASDTAKHITLETLQQLYKETRPKSLGIADLGCSS 64
Query: 62 GSNTLLVILDIIKVVEKLCRKLNHKS------PEYMIYLNDLPGNDFNTIFTSLDIFKEK 115
G NTL I D IK V+ +H+ PE+ I+LNDLPGNDFN IF SL F +
Sbjct: 65 GPNTLSTITDFIKTVQVA----HHREIPIQPLPEFSIFLNDLPGNDFNFIFKSLPDFHIE 120
Query: 116 LL-DEMGTEMGPCFFSGVPGSFHGRIFPLQSLHFVHSSYSLHWLSKVP------EGADNN 168
L D + F + PGSF+GR+FP ++HFV++S+SLHWLSKVP +G N
Sbjct: 121 LKRDNNNGDCPSVFIAAYPGSFYGRLFPENTIHFVYASHSLHWLSKVPTALYDEQGKSIN 180
Query: 169 KGNIYISSTSPLNVVKAYYKQFQIDFSLFLKCRAEEIVEGGCMIITFVGRKSDNPTSKEC 228
KG + I S S V KAY QF+ DFS+FL+CR++E+V G M++ +GR+ + +
Sbjct: 181 KGCVSICSLSSEAVSKAYCSQFKEDFSIFLRCRSKEMVSAGRMVLIILGREGPDHVDRGN 240
Query: 229 CYIWELLAMALNDMVLEGIIKEEKLNTFNIPIYYPSPSEVKLEVITEGSFVMNQLEISEV 288
+ WELL+ ++ D+V +G +EEKL+++++ Y PS E++ EV EGSF + +LE+ EV
Sbjct: 241 SFFWELLSRSIADLVAQGETEEEKLDSYDMHFYAPSADEIEGEVDKEGSFELERLEMLEV 300
Query: 289 NWNARDDFESESFGDDGYNVAQCMRALAEPLLVSHFGEGVVKEIFNRYKKYLTDRNSKGR 348
+D +E G VA+ +RA+ E +LV HFGE ++ ++F+ Y + + D +K
Sbjct: 301 K---KDKGNTEGDISYGKAVAKTVRAVQESMLVQHFGEKILDKLFDTYCRMVDDELAKED 357
Query: 349 TKFINITILLAKR 361
+ I ++L K+
Sbjct: 358 IRPITFVVVLRKK 370
>At3g21950 salicylic acid carboxyl methyltransferase, putative
Length = 335
Score = 250 bits (639), Expect = 7e-67
Identities = 137/321 (42%), Positives = 195/321 (60%), Gaps = 21/321 (6%)
Query: 52 LTIADLGCSSGSNTLLVILDIIKVVEKLCRKLNHKSPEYMIYLNDLPGNDFNTIFTSLDI 111
+ +ADLGCSSG NT LV+ +I+ + ++ PE LNDLP NDFNT F +
Sbjct: 17 IKVADLGCSSGENTFLVMSEIVNTIITTYQQNGQNLPEIDCCLNDLPENDFNTTFKLIPS 76
Query: 112 FKEKLLDEMGTEMGPCFFSGVPGSFHGRIFPLQSLHFVHSSYSLHWLSKVPEGADNNKGN 171
F EKL +M + G C+ SG PGSF+ R+FP +SLHFVHSS+ LHWLSKVP+G + NK N
Sbjct: 77 FHEKL--KMNVK-GNCYVSGCPGSFYTRLFPSKSLHFVHSSFCLHWLSKVPDGLEENKKN 133
Query: 172 IYISSTSPLNVVKAYYKQFQIDFSLFLKCRAEEIVEGGCMIITFVGRKSDNPTSKECCYI 231
+Y+ S P N+ ++Y+ QF+ DFS+FL+ RAEE + G M +T VGRK+ +P SKEC
Sbjct: 134 VYLRSPCPPNLYESYWNQFKKDFSMFLRMRAEETMPSGRMALTLVGRKTLDPLSKECFKD 193
Query: 232 WELLAMALNDMVLEGIIKEEKLNTFNIPIYYPSPSEVKLEVITEGSFVMNQLEI------ 285
W L++ +L D+V EG++KE L +FN+P Y P SEVK + EGSF + E
Sbjct: 194 WSLVSDSLLDLVSEGVVKESDLESFNLPYYSPDESEVKEVIENEGSFEIKNFETIFGLLF 253
Query: 286 ------SEVNWNARDDFESESFGDDGYNVAQCMRALAEPLLVSHFGEGVVKEIFNRY--- 336
SEV + DD + + A R++ EP+LV+HFGE ++ +F++Y
Sbjct: 254 SYKTGHSEVK-DDDDDVDHSRRFEVVKTRANMTRSIIEPMLVAHFGEAIIDRLFDKYIYH 312
Query: 337 --KKYLTDRNSKGRTKFINIT 355
++Y T RN F+++T
Sbjct: 313 ACQRYDTLRNKPTVNFFVSLT 333
>At1g68040 putative S-adenosyl-L-methionine:salicylic acid carboxyl
methyltransferase
Length = 363
Score = 203 bits (516), Expect = 1e-52
Identities = 128/357 (35%), Positives = 189/357 (52%), Gaps = 31/357 (8%)
Query: 3 VAKVLHMNGGVGETSYANNSLVQRKVIYLTKPLRDEAITSMYN-----NTLSKSLTIADL 57
V L M+GG G SY+ NS +QRK L K D+ + N ++ S + IADL
Sbjct: 7 VRNSLPMSGGDGPNSYSKNSHLQRKTTSLLKEKIDKLVLEKLNAKTLISSDSNTFRIADL 66
Query: 58 GCSSGSNTLLVILDIIKVVEKLCRKLNHKSPEYMIYLNDLPGNDFNTIFTSLDIFKEKLL 117
GC++G NT ++ +IIK +E RK N PE++++ NDLP NDFNT+FTSL + L
Sbjct: 67 GCATGPNTFFLVDNIIKSIETSLRKSNSSKPEFLVFFNDLPQNDFNTLFTSLPQDRSYLA 126
Query: 118 DEMGTEMGPCFFSGVPGSFHGRIFPLQSLHFVHSSYSLHWLSKVPEGADN------NKGN 171
GVPGSF+GR+ P S+H V + + HWLS VP+ + NKG
Sbjct: 127 ------------VGVPGSFYGRVLPQSSVHIVVTMGATHWLSSVPKEVLDKSSKAWNKGK 174
Query: 172 IYISSTSPLNVVKAYYKQFQIDFSLFLKCRAEEIVEGGCMIITFVGRKSDNPTSKEC-CY 230
++ S+ + VVKAY QF D FL+ RA EIV GG +++ G P S
Sbjct: 175 VHYSNAAD-EVVKAYRDQFGRDMEKFLEARATEIVSGGLLVVGMCGIPKGMPFSNLADSI 233
Query: 231 IWELLAMALNDMVLEGIIKEEKLNTFNIPIYYPSPSEVKLEVITEGSFVMNQLEISE-VN 289
++ +A L M EG+I EE+++TFNIPIY +P EV + V+ G F + +E+ +
Sbjct: 234 MYTSMADVLTQMHSEGLISEEQVDTFNIPIYSATPEEVTVLVVKNGCFTVESMELMDPTA 293
Query: 290 WNARDDFESESFGDDGYNVAQCMRALAEPLLVSHFGEGVVKEIFNRYKKYLTDRNSK 346
W R + +D + C++A L ++HFGE ++ ++F+R L K
Sbjct: 294 WLKR-----PTNVEDVRHWMVCIKATMGSLFINHFGEHLLDDVFDRLTAKLVGLTEK 345
>At5g55250 S-adenosyl-L-methionine:salicylic acid carboxyl
methyltransferase-like protein
Length = 386
Score = 190 bits (482), Expect = 1e-48
Identities = 124/358 (34%), Positives = 190/358 (52%), Gaps = 23/358 (6%)
Query: 1 MEVAKVLHMNGGVGETSYANNSLVQRKVIYLTKPLRDEAITSMYNNTLSKS--LTIADLG 58
M++ ++L M GG G+ SYANNS Q L +E + +++ N+ + T DLG
Sbjct: 13 MKLERLLSMKGGKGQDSYANNSQAQAMHARSMLHLLEETLENVHLNSSASPPPFTAVDLG 72
Query: 59 CSSGSNTLLVILDIIKVVEKLCRKLNHKSPEYMIYLNDLPGNDFNTIFTSLD-IFKEKLL 117
CSSG+NT+ +I I+K + K PE+ + +DLP NDFNT+F L + +
Sbjct: 73 CSSGANTVHIIDFIVKHISKRFDAAGIDPPEFTAFFSDLPSNDFNTLFQLLPPLVSNTCM 132
Query: 118 DEMGTEMG--PCFFSGVPGSFHGRIFPLQSLHFVHSSYSLHWLSKVPE------GADNNK 169
+E G F +GVPGSF+ R+FP +++ F HS++SLHWLS+VPE A N+
Sbjct: 133 EECLAADGNRSYFVAGVPGSFYRRLFPARTIDFFHSAFSLHWLSQVPESVTDRRSAAYNR 192
Query: 170 GNIYISSTSPLNVVKAYYKQFQIDFSLFLKCRAEEIVEGGCMIITFVGRKSDNPTSK--E 227
G ++I AY +QFQ D + FL+ RA E+ GG M + +GR S +PT +
Sbjct: 193 GRVFIHGAGE-KTTTAYKRQFQADLAEFLRARAAEVKRGGAMFLVCLGRTSVDPTDQGGA 251
Query: 228 CCYIWELLAMALNDMVLEGIIKEEKLNTFNIPIYYPSPSEVKLEVITEGSFVMNQLEI-- 285
A +D+V EG++ EK + FNIP+Y PS + K V GSF +++L +
Sbjct: 252 GLLFGTHFQDAWDDLVREGLVAAEKRDGFNIPVYAPSLQDFKEVVDANGSFAIDKLVVYK 311
Query: 286 --SEVNWNARDDFESESFGDDGYNVAQCMRALAEPLLVSHFGEGVVKEIFNRYKKYLT 341
S + N DD + G A R++A L+ +H GE + ++F+R + T
Sbjct: 312 GGSPLVVNEPDD-----ASEVGRAFASSCRSVAGVLVEAHIGEELSNKLFSRVESRAT 364
>At5g37970 putative protein
Length = 412
Score = 174 bits (441), Expect = 7e-44
Identities = 129/376 (34%), Positives = 181/376 (47%), Gaps = 46/376 (12%)
Query: 9 MNGGVGETSYANNSLVQRKVIYLTKPLRDEAITS------MYNNTLSKSLTIADLGCSSG 62
MNGG G SY +NS Q+ I K EAI + N+ L I D GCS G
Sbjct: 60 MNGGDGPHSYIHNSSYQKVAIDGVKERTSEAILEKLDLEFLNRNSEENILRIVDFGCSIG 119
Query: 63 SNTLLVILDIIKVVEKLCRKLNHK---SP-EYMIYLNDLPGNDFNTIFTSLDIFKEKLLD 118
NT V+ +II V++ K N +P E+ + ND P NDFNT+F + F K
Sbjct: 120 PNTFDVVQNIIDTVKQKRLKENKTYIGAPLEFQVCFNDQPNNDFNTLFRTQPFFSRK--- 176
Query: 119 EMGTEMGPCFFSGVPGSFHGRIFPLQSLHFVHSSYSLHWLSKVPEGADNNKGNIYISSTS 178
F GVPGSFHGR+ P SLH H+SY+LHWLS VP+ + K S
Sbjct: 177 -------EYFSVGVPGSFHGRVLPKNSLHIGHTSYTLHWLSNVPQHVCDKKSPALNKSYI 229
Query: 179 PLN-----VVKAYYKQFQIDFSLFLKCRAEEIVEGGCMIITFVGRKSDNPTSKECCY--- 230
N V KAY QF+ DF FL+ RAEE+V GG MI++ G+ + K +
Sbjct: 230 QCNNLVDEVTKAYKIQFRKDFGGFLEARAEELVSGGLMILS--GQCLPDGIPKALTWQGV 287
Query: 231 IWELLAMALNDMVLEGIIKEEKLNTFNIPIYYPSPSEVKLEVITEGSF---VMNQLEISE 287
+ +++ L D+ GI +EK+ F++P Y P SE K + +F M ++
Sbjct: 288 VIDMIGDCLMDLAKLGITSKEKIELFSLPTYIPHISEFKANIEQNENFNVETMEEISHPM 347
Query: 288 VNWNARDDFESESFGDDGYNVAQCMRALAEPLLVSHFGEGVVKEIFNRYKKYLTDR--NS 345
+DF + F RA+ ++ HFGEGVV E+F+R K L +
Sbjct: 348 DYMPLTNDFITSMF-----------RAILNTIIEEHFGEGVVNELFSRLAKRLDKYPIDF 396
Query: 346 KGRTKFINITILLAKR 361
K K++N I+L ++
Sbjct: 397 KRCKKYVNYFIVLKRK 412
>At5g37990 putative protein
Length = 362
Score = 173 bits (439), Expect = 1e-43
Identities = 125/373 (33%), Positives = 185/373 (49%), Gaps = 40/373 (10%)
Query: 9 MNGGVGETSYANNSLVQRKVIYLTKPLRDEAITS------MYNNTLSKSLTIADLGCSSG 62
MNGG G SY +NS Q+ I K EAI + N+ L IAD GCS G
Sbjct: 10 MNGGDGPHSYIHNSSYQKVAIDGAKEKTSEAILKNLDLELLNRNSDENILRIADFGCSIG 69
Query: 63 SNTLLVILDIIKVVEKLCRKLNHK---SP-EYMIYLNDLPGNDFNTIFTSLDIFKEKLLD 118
NT V+ +II V++ K N+ +P E+ + ND P NDFNT+F + I ++
Sbjct: 70 PNTFEVVQNIIDTVKQKNLKENNAYIGAPLEFQVCFNDQPNNDFNTLFRTQPISSKQAYL 129
Query: 119 EMGTEMGPCFFSGVPGSFHGRIFPLQSLHFVHSSYSLHWLSKVPEGADNNKGNIYISSTS 178
+G VPGSFHGR+ P SLH H +Y+LHWLS VP+ + K S
Sbjct: 130 SVG----------VPGSFHGRVLPKNSLHIGHITYALHWLSTVPQHVCDKKSPALNKSYI 179
Query: 179 PLN-----VVKAYYKQFQIDFSLFLKCRAEEIVEGGCMIITFVGRKSDNPTSKECCY--- 230
N V +AY QF+ D FL RAEE+V GG MI++ G+ + K +
Sbjct: 180 QCNNLVEEVTEAYRVQFKKDMGDFLGARAEELVSGGLMILS--GQCLPDGVPKALTWQGV 237
Query: 231 IWELLAMALNDMVLEGIIKEEKLNTFNIPIYYPSPSEVKLEVITEGSFVMNQLEISEVNW 290
+ +++ L DM +GI +EK+ F++PIY P SE K E+ +F + +E
Sbjct: 238 VIDMIGDCLMDMAKQGITTKEKIELFSLPIYIPHISEFKAEIERNENFSIETME------ 291
Query: 291 NARDDFESESFGDDGYNVAQCMRALAEPLLVSHFGEGVVKEIFNRYKKYLTDR--NSKGR 348
+ + +D + RA+ ++ HFG+GVV E+F+R+ K L + K
Sbjct: 292 KISHPMDYKPLTND--FITSMFRAILNTIIEEHFGDGVVNELFDRFAKKLNKYPIDFKRC 349
Query: 349 TKFINITILLAKR 361
K++N I+L ++
Sbjct: 350 KKYVNYFIVLKRK 362
>At5g38100 unknown protein
Length = 359
Score = 168 bits (426), Expect = 4e-42
Identities = 119/351 (33%), Positives = 172/351 (48%), Gaps = 39/351 (11%)
Query: 9 MNGGVGETSYANNSLVQRKVIYLTKPLRDEAITSMYNNTLSK---SLTIADLGCSSGSNT 65
M+ G + SY +NS Q+ I I + LS + IAD GCS G NT
Sbjct: 10 MSSGHDQHSYIHNSSYQKAAISSAVEKTRRCIFEKLDLQLSSDFGTFRIADFGCSIGPNT 69
Query: 66 LLVILDIIKVVEKLCRKLNHKSP------EYMIYLNDLPGNDFNTIFTSLDIFKEKLLDE 119
V II V+ ++L + E+ ++ ND P NDFNT+F + + E+
Sbjct: 70 FHVAQSIIDTVKS--KRLEESTENSLVPLEFQVFFNDQPTNDFNTLFRTQPLSPER---- 123
Query: 120 MGTEMGPCFFSGVPGSFHGRIFPLQSLHFVHSSYSLHWLSKVPEGADNNK----GNIYIS 175
F GVPGSF+GR+ P S+H H+SY+ HWLSKVP+ + K YI
Sbjct: 124 ------EYFSVGVPGSFYGRVLPRNSIHIGHTSYTTHWLSKVPDNVCDKKSMAWNKNYIQ 177
Query: 176 STSPL-NVVKAYYKQFQIDFSLFLKCRAEEIVEGGCMIITFVGR-KSDNPTSKECC--YI 231
+ L V KAY QF D +FL RAEE+V GG MI+ +G D + E Y+
Sbjct: 178 CNNLLEEVTKAYKVQFIKDMEIFLDARAEELVPGGLMIV--IGECLPDGVSLYETWQGYV 235
Query: 232 WELLAMALNDMVLEGIIKEEKLNTFNIPIYYPSPSEVKLEVITEGSFVMNQLEISEVNWN 291
+ + L DM GI EEK++ F++P+Y+P SE+K E+ GSF + +E +
Sbjct: 236 MDTIGDCLMDMAKSGITSEEKIDLFSLPVYFPQFSELKGEIEKNGSFTIELMETTS---- 291
Query: 292 ARDDFESESFGDDGYNVAQCMRALAEPLLVSHFGEGVVKEIFNRYKKYLTD 342
E + +D + RA ++ HFG+GVV E+F R K L++
Sbjct: 292 --HPLEGKPLTND--FITSTFRAFLTTIIEKHFGDGVVDELFYRLAKKLSN 338
>At4g26420 putative protein
Length = 619
Score = 164 bits (414), Expect = 9e-41
Identities = 117/353 (33%), Positives = 180/353 (50%), Gaps = 32/353 (9%)
Query: 6 VLHMNGGVGETSYANNSLVQRKVIYLTKPLRDEAITSM-YNNTLSKSLTIADLGCSSGSN 64
VL M GG + SY N + L+KP+ AI S+ S L IADLGC+ G N
Sbjct: 10 VLSMQGGEDDASYVKNCYGPAARLALSKPMLTTAINSIKLTEGCSSHLKIADLGCAIGDN 69
Query: 65 TLLVILDIIKVVEKLCRKLN---HKSPEYMIYLNDLPGNDFNTIFTSLDIFKEKLLDEMG 121
T + +++V+ K ++ E+ ++ +DL NDFN +F SLD +++
Sbjct: 70 TFSTVETVVEVLGKKLAVIDGGTEPEMEFEVFFSDLSSNDFNALFRSLD-------EKVN 122
Query: 122 TEMGPCFFSGVPGSFHGRIFPLQSLHFVHSSYSLHWLSKVPE-----GADN-NKGNIYIS 175
F +GVPGSF+ R+FP LH V + +L WLS+VPE G+ + NKG ++I
Sbjct: 123 GSSRKYFAAGVPGSFYKRLFPKGELHVVVTMSALQWLSQVPEKVMEKGSKSWNKGGVWIE 182
Query: 176 STSPLNVVKAYYKQFQIDFSLFLKCRAEEIVEGGCMIITFVGRKS------DNPTSKECC 229
VV+AY +Q D FLKCR EEIV GG + + GR S +P S
Sbjct: 183 GAEK-EVVEAYAEQADKDLVEFLKCRKEEIVVGGVLFMLMGGRPSGSVNQIGDPDSSLKH 241
Query: 230 YIWELLAMALNDMVLEGIIKEEKLNTFNIPIYYPSPSEVKLEVITEGSFVMNQLE--ISE 287
L+ A D+V EG+I+EEK + FNIP+Y+ + E+ + G F + + E I
Sbjct: 242 PFTTLMDQAWQDLVDEGLIEEEKRDGFNIPVYFRTTEEIAAAIDRCGGFKIEKTENLIIA 301
Query: 288 VNWNARDD---FESESFGDDGYNVAQCMRALAEPLLVSHFGEGVVKEIFNRYK 337
+ N + + + +S+G D N AQ A +P++ ++ G + ++F R +
Sbjct: 302 DHMNGKQEELMKDPDSYGRDRANYAQ---AGLKPIVQAYLGPDLTHKLFKRMR 351
>At5g56300 S-adenosyl-L-methionine:salicylic acid carboxyl
methyltransferase-like protein
Length = 387
Score = 160 bits (404), Expect = 1e-39
Identities = 118/373 (31%), Positives = 185/373 (48%), Gaps = 40/373 (10%)
Query: 5 KVLHMNGGVGETSYANNSLVQRKVIYLTKPLRDEAITSMYNNTLSKSLTIADLGCSSGSN 64
+V M GG + SY NNS Q I L+KP+ ++ S+ + + I DLGC++GSN
Sbjct: 20 RVFAMQGGEDDLSYVNNSDSQALAITLSKPILISSLQSIKLFSDQTPIKITDLGCATGSN 79
Query: 65 TLLVILDIIKVVEK--LCRKLNHKSPEYMIYLNDLPGNDFNTIFTSLDIFKEKLLDEMGT 122
T + +++ +++ R SPE+ + DLP NDFN +F KLL E
Sbjct: 80 TFTTVDTVVETLQRRYTARCGGGGSPEFEAFFCDLPSNDFNMLF--------KLLAEKQK 131
Query: 123 EMGPC--FFSGVPGSFHGRIFPLQSLHFVHSSYSLHWLSKVP------EGADNNKGNIYI 174
P F GV GSF+ R+FP ++H S +LHWLS++P E NKG +I
Sbjct: 132 VDSPAKYFAGGVAGSFYDRLFPRGTIHVAVSLSALHWLSQIPEKVLEKESRTWNKGKTWI 191
Query: 175 SSTSPLNVVKAYYKQFQIDFSLFLKCRAEEIVEGGCMIITFVGRKS-------DNPTSKE 227
VV+AY +Q D F+ CR EE+V+GG + + GR S D T +
Sbjct: 192 EGAKK-EVVEAYAEQSDKDLDDFMSCRKEEMVKGGVLFVLMAGRPSGSSSQFGDQDTRAK 250
Query: 228 CCYIWELLAMALNDMVLEGIIKEEKLNTFNIPIYYPSPSEVKLEVITEGSF---VMNQLE 284
+ + A D++ EG+I EE + FNIP Y SP EV + G F M+ L+
Sbjct: 251 HPFT-TTMEQAWQDLIEEGLIDEETRDGFNIPAYMRSPEEVTAGIDRCGGFKIGKMDFLK 309
Query: 285 ISEVNWNARDDFESE--SFGDDGYNVAQCMRALAEPLLVSHFGEGVVKEIFNRYKKYLTD 342
I E + +++++ + S+G N+ Q A P++ ++ G + E+F RY+ ++
Sbjct: 310 IVEYSDEKQEEWKKDPVSYGRARTNLVQ---AAIRPMVDAYLGPDLSHELFKRYENRVST 366
Query: 343 RNSKGRTKFINIT 355
+F++IT
Sbjct: 367 NQ-----EFLHIT 374
>At1g15125 putative protein
Length = 347
Score = 158 bits (399), Expect = 5e-39
Identities = 109/340 (32%), Positives = 172/340 (50%), Gaps = 36/340 (10%)
Query: 9 MNGGVGETSYANNSLVQRKVIYLTKPLRDEAITSMYN--NTLSKSLTIADLGCSSGSNTL 66
MNGG G +SYA NS QR I + L I + + N S TIAD GCSSG NT+
Sbjct: 1 MNGGDGASSYARNSSYQRGAIEAAEALLRNEINARLDITNHSFSSFTIADFGCSSGPNTV 60
Query: 67 LVILDIIKVV-EKLCRKL-NHKSPEYMIYLNDLPGNDFNTIFTSLDIFKEKLLDEMGTEM 124
+ + II+ + K L N +P++ ++ ND+ DFN +F L
Sbjct: 61 IAVDIIIQALYHKFTSSLPNTTTPQFQVFFNDVSHTDFNALFALLP------------PQ 108
Query: 125 GPCFFSGVPGSFHGRIFPLQSLHFVHSSYSLHWLSKVPEGADN------NKGNIYISSTS 178
P F +GVPGSF+G +FP L+ +SS +L WLS +P + N+G I+ + S
Sbjct: 109 RPYFVAGVPGSFYGNLFPKAHLNLAYSSCALCWLSDLPSELTDTSSPAYNRGRIHYTGAS 168
Query: 179 PLNVVKAYYKQFQIDFSLFLKCRAEEIVEGGCMIITFVGRKSDNPTSKECC--YIWELLA 236
V +AY Q++ D LFL R++E+ E G M + G +E ++LL
Sbjct: 169 -AEVAQAYSSQYKKDIKLFLHARSQELAENGLMALIVPGVPDGFLDCQEASTGSEFDLLG 227
Query: 237 MALNDMVLEGIIKEEKLNTFNIPIYYPSPSEVKLEVITEGSFVMNQLEISEVNWNARDDF 296
L DM E EE++N+FN+PIYY +P E++ + + G ++++E
Sbjct: 228 SCLMDMAKE----EEEVNSFNLPIYYTTPKELEDIIRSNGELKIDKME-------TLGSM 276
Query: 297 ESESFGDDGYNVAQCMRALAEPLLVSHFGEGVVKEIFNRY 336
+++ D + +RA+ E L+ +HFG ++ ++F+RY
Sbjct: 277 DAQDTMPDLESRVLYLRAVLEGLVRTHFGHQILDDLFDRY 316
>At5g38780 AtPP - like protein
Length = 361
Score = 157 bits (398), Expect = 7e-39
Identities = 114/351 (32%), Positives = 176/351 (49%), Gaps = 37/351 (10%)
Query: 9 MNGGVGETSYANNSLVQRKVIY-LTKPLRD---EAITSMYNNTLSKSLTIADLGCSSGSN 64
M+GG + SY +NS Q+ I + + R E + + N + TIAD GCS G N
Sbjct: 10 MSGGDDQHSYIHNSSYQKAGIDGVQEKARQYILENLDLLNMNPNLSTFTIADFGCSIGPN 69
Query: 65 TLLVILDIIKVVE----KLCRKLNHKSP-EYMIYLNDLPGNDFNTIFTSLDIFKEKLLDE 119
T + +II +V+ K ++ + +P E+ +Y NDLP NDFNT+F + +
Sbjct: 70 TFHAVQNIIDIVKLKHLKESQEDSRVAPLEFQVYFNDLPNNDFNTLFRT----------Q 119
Query: 120 MGTEMGPCFFSGVPGSFHGRIFPLQSLHFVHSSYSLHWLSKVPEGADN------NKGNIY 173
+ F GVPGSF+GR+ P S+H ++S++ HWLSKVPE + NK I+
Sbjct: 120 PPSSKQEYFSVGVPGSFYGRVLPRNSIHIGNTSFTTHWLSKVPEEVCDKNSLAWNKNYIH 179
Query: 174 ISSTSPLNVVKAYYKQFQIDFSLFLKCRAEEIVEGGCMIITFVGRKSDNPTSKECC--YI 231
++ V +AY QF+ D +FLK RAEE+V GG M IT D E +
Sbjct: 180 CNNLIE-EVTEAYKVQFEKDMGVFLKARAEELVPGGLM-ITLGQCLPDGVAMYETWSGIV 237
Query: 232 WELLAMALNDMVLEGIIKEEKLNTFNIPIYYPSPSEVKLEVITEGSFVMNQLEISEVNWN 291
+ + L DM G+ EEK+ FN+P+Y+P SE+K + F + +EI
Sbjct: 238 KDTIGDCLQDMATLGVTTEEKIEMFNLPVYFPQVSELKGAIEQNIRFTIEMMEI------ 291
Query: 292 ARDDFESESFGDDGYNVAQCMRALAEPLLVSHFGEGVVKEIFNRYKKYLTD 342
E+ ++ + RA+ ++ HFG VV E+F ++ K L++
Sbjct: 292 VSHPLEAVQLSNN--FITSMYRAILSTVIERHFGGSVVDELFRQFAKKLSE 340
>At3g44840 proteinkinase AtPP -like protein
Length = 348
Score = 149 bits (375), Expect = 3e-36
Identities = 107/336 (31%), Positives = 168/336 (49%), Gaps = 27/336 (8%)
Query: 9 MNGGVGETSYANNSLVQRKVIYLTKPLRDEAITSMYNNTLSKSLT-IADLGCSSGSNTLL 67
M GG G SY +S Q ++ +EAI++ N L+ +L IAD GCS+G NT
Sbjct: 7 MIGGEGPESYRQHSKYQGGLLEAATEKINEAISTKLNIDLASNLVNIADFGCSTGPNTFR 66
Query: 68 VILDIIKVVE-KLCRKLNHKSPEYMIYLNDLPGNDFNTIFTSLDIFKEKLLDEMGTEMGP 126
+ II VE K ++ N + E+ ++ ND NDFNT+F +L ++
Sbjct: 67 AVQTIIDAVEHKYQQENNLEEIEFQVFFNDSSNNDFNTLFKTLPPARKY----------- 115
Query: 127 CFFSGVPGSFHGRIFPLQSLHFVHSSYSLHWLSKVP---EGADNNKGNIYISSTS-PLNV 182
F +GVP SF GR+ P SLH SSYSLH+LSK+P + D++ N I T V
Sbjct: 116 -FATGVPASFFGRVLPRSSLHVGVSSYSLHFLSKIPKKIKDCDSHAWNKDIHCTGFSKEV 174
Query: 183 VKAYYKQFQIDFSLFLKCRAEEIVEGGCMIITFVGRKSDNPTSKEC-CYIWELLAMALND 241
V+AY Q++ID FL RA+E+V GG + + + S+ + + + +LND
Sbjct: 175 VRAYLDQYKIDMESFLTARAQELVSGGLLFLLGSCLPNGVQMSETLNGMMIDCIGSSLND 234
Query: 242 MVLEGIIKEEKLNTFNIPIYYPSPSEVKLEVITEGSFVMNQLEISEVNWNARDDFESESF 301
+ +G+I +EKL+TF +PIY E+K + + + + +I E+E
Sbjct: 235 IAKQGLIDQEKLDTFKLPIYVAYAGEIKQIIEDNVYYTIERFDIISQ--------ENEEI 286
Query: 302 GDDGYNVAQCMRALAEPLLVSHFGEGVVKEIFNRYK 337
D + + ++ SHFG+ V+++ F K
Sbjct: 287 PLDPEFLTVSFKVTVGGIVASHFGQHVMEKTFEVVK 322
>At3g44860 AtPP -like protein
Length = 348
Score = 148 bits (373), Expect = 5e-36
Identities = 98/287 (34%), Positives = 149/287 (51%), Gaps = 27/287 (9%)
Query: 9 MNGGVGETSYANNSLVQRKVIYLTKPLRDEAITSMYNNTLSKSLT-IADLGCSSGSNTLL 67
M GG G SY +S Q ++ K +EAI++ + + +L IAD GCSSG NT
Sbjct: 7 MIGGEGPNSYREHSKYQGALVIAAKEKINEAISTKLDIDFTSNLVNIADFGCSSGPNTFT 66
Query: 68 VILDIIKVVE-KLCRKLNHKSPEYMIYLNDLPGNDFNTIFTSLDIFKEKLLDEMGTEMGP 126
+ +I VE K ++ N + E+ ++ ND NDFNT+F +L +
Sbjct: 67 AVQTLIDAVENKYKKESNIEGIEFQVFFNDSSNNDFNTLFKTLPPARLY----------- 115
Query: 127 CFFSGVPGSFHGRIFPLQSLHFVHSSYSLHWLSKVPEGADNNKGNIYIS----STSPLNV 182
F SGVPGSF GR+ P SLH SSYSLH++SKVP+ + ++ S S V
Sbjct: 116 -FASGVPGSFFGRVLPKNSLHVGVSSYSLHFVSKVPKEIKDRDSLVWNKDIHCSGSSKEV 174
Query: 183 VKAYYKQFQIDFSLFLKCRAEEIVEGGCMIITFVGRKSDNPTSKECC-----YIWELLAM 237
VK Y Q++ID FL RA+E+V GG +++ S PT + + + +
Sbjct: 175 VKLYLGQYKIDVGSFLTARAQELVSGGLLLLL----GSCRPTGVQMFETVEGMMIDFIGS 230
Query: 238 ALNDMVLEGIIKEEKLNTFNIPIYYPSPSEVKLEVITEGSFVMNQLE 284
+LN++ +G+I ++KL+TF +PIY P+ E+K + F + E
Sbjct: 231 SLNEIANQGLIDQQKLDTFKLPIYAPNVDELKQIIEDNKCFTIEAFE 277
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.319 0.137 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 8,330,949
Number of Sequences: 26719
Number of extensions: 367353
Number of successful extensions: 976
Number of sequences better than 10.0: 34
Number of HSP's better than 10.0 without gapping: 27
Number of HSP's successfully gapped in prelim test: 7
Number of HSP's that attempted gapping in prelim test: 841
Number of HSP's gapped (non-prelim): 34
length of query: 362
length of database: 11,318,596
effective HSP length: 101
effective length of query: 261
effective length of database: 8,619,977
effective search space: 2249813997
effective search space used: 2249813997
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.8 bits)
S2: 61 (28.1 bits)
Medicago: description of AC125480.15


