

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC125479.4 - phase: 0
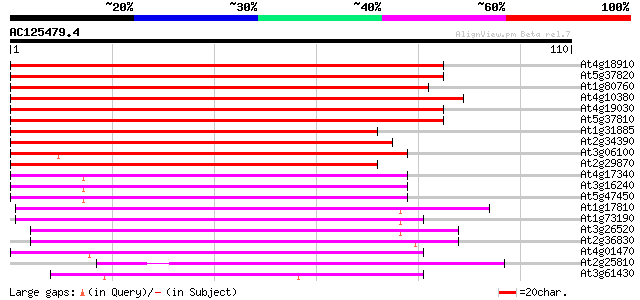
(110 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
At4g18910 major intrinsic protein (MIP)- like 108 3e-25
At5g37820 Membrane integral protein (MIP) -like 104 6e-24
At1g80760 nodulin-like protein 104 6e-24
At4g10380 major intrinsic protein (MIP) - like 103 1e-23
At4g19030 nodulin-26 like protein 102 2e-23
At5g37810 Membrane integral protein (MIP) -like 101 7e-23
At1g31885 major intrinsic protein - like 90 2e-19
At2g34390 putative aquaporin (plasma membrane intrinsic protein) 81 1e-16
At3g06100 putative major intrinsic protein 77 2e-15
At2g29870 putative aquaporin (plasma membrane intrinsic protein) 76 2e-15
At4g17340 membrane channel like protein 71 1e-13
At3g16240 delta tonoplast integral protein (delta-TIP) 70 2e-13
At5g47450 membrane channel protein-like; aquaporin (tonoplast in... 69 3e-13
At1g17810 putative protein 63 3e-11
At1g73190 tonoplast intrinsic protein, alpha (alpha-TIP) 61 1e-10
At3g26520 salt-stress induced tonoplast intrinsic protein 59 5e-10
At2g36830 putative aquaporin (tonoplast intrinsic protein gamma) 55 6e-09
At4g01470 putative water channel protein 55 7e-09
At2g25810 putative aquaporin (tonoplast intrinsic protein) 54 2e-08
At3g61430 plasma membrane intrinsic protein 1a 50 2e-07
>At4g18910 major intrinsic protein (MIP)- like
Length = 294
Score = 108 bits (271), Expect = 3e-25
Identities = 48/85 (56%), Positives = 68/85 (79%)
Query: 1 MVFISTAVATDSKATGELAGVAVGSSVTIASIVAGPISGGSMNPARTLGPAIATSSYKGI 60
++F+ + VATD++A GELAG+AVGS+V + I+AGP+SG SMNP R+LGPA+ S Y+G+
Sbjct: 188 LMFVISGVATDNRAIGELAGLAVGSTVLLNVIIAGPVSGASMNPGRSLGPAMVYSCYRGL 247
Query: 61 WIYMVGPITGALLGAWSYVVIQETD 85
WIY+V PI GA+ GAW Y +++ TD
Sbjct: 248 WIYIVSPIVGAVSGAWVYNMVRYTD 272
Score = 28.9 bits (63), Expect = 0.44
Identities = 21/74 (28%), Positives = 34/74 (45%), Gaps = 3/74 (4%)
Query: 5 STAVATDSKATGELAGVAVGSSVTIASIV--AGPISGGSMNPARTLG-PAIATSSYKGIW 61
+ AV T L G+A+ +T+ +V G ISG NPA T+ + K +
Sbjct: 71 AVAVNTQHDKAVTLPGIAIVWGLTVMVLVYSLGHISGAHFNPAVTIAFASCGRFPLKQVP 130
Query: 62 IYMVGPITGALLGA 75
Y++ + G+ L A
Sbjct: 131 AYVISQVIGSTLAA 144
>At5g37820 Membrane integral protein (MIP) -like
Length = 283
Score = 104 bits (260), Expect = 6e-24
Identities = 48/85 (56%), Positives = 63/85 (73%)
Query: 1 MVFISTAVATDSKATGELAGVAVGSSVTIASIVAGPISGGSMNPARTLGPAIATSSYKGI 60
++F+ + VATDS+ATGELAG+AVG ++ + VAGPISG SMNPAR+LGPAI YKGI
Sbjct: 172 LMFVISGVATDSRATGELAGIAVGMTIILNVFVAGPISGASMNPARSLGPAIVMGRYKGI 231
Query: 61 WIYMVGPITGALLGAWSYVVIQETD 85
W+Y+VGP G G + Y ++ TD
Sbjct: 232 WVYIVGPFVGIFAGGFVYNFMRFTD 256
>At1g80760 nodulin-like protein
Length = 305
Score = 104 bits (260), Expect = 6e-24
Identities = 44/82 (53%), Positives = 66/82 (79%)
Query: 1 MVFISTAVATDSKATGELAGVAVGSSVTIASIVAGPISGGSMNPARTLGPAIATSSYKGI 60
++F+ TAVATD++A GELAG+AVG++V + ++AGP + SMNP RTLGPAIA ++Y+ I
Sbjct: 208 LMFVVTAVATDTRAVGELAGIAVGATVMLNILIAGPATSASMNPVRTLGPAIAANNYRAI 267
Query: 61 WIYMVGPITGALLGAWSYVVIQ 82
W+Y+ PI GAL+GA +Y +++
Sbjct: 268 WVYLTAPILGALIGAGTYTIVK 289
Score = 31.2 bits (69), Expect = 0.089
Identities = 23/89 (25%), Positives = 41/89 (45%), Gaps = 6/89 (6%)
Query: 1 MVFISTAVA-----TDSKATGELAGVAVGSSVTIASIVAGPISGGSMNPARTLG-PAIAT 54
++F TA A TD T + G +V I + G ISG +NPA T+ A+
Sbjct: 92 LIFAGTATAIVNQKTDGAETLIGCAASAGLAVMIVILSTGHISGAHLNPAVTIAFAALKH 151
Query: 55 SSYKGIWIYMVGPITGALLGAWSYVVIQE 83
+K + +Y+ + ++ A++ + E
Sbjct: 152 FPWKHVPVYIGAQVMASVSAAFALKAVFE 180
>At4g10380 major intrinsic protein (MIP) - like
Length = 304
Score = 103 bits (257), Expect = 1e-23
Identities = 46/89 (51%), Positives = 67/89 (74%)
Query: 1 MVFISTAVATDSKATGELAGVAVGSSVTIASIVAGPISGGSMNPARTLGPAIATSSYKGI 60
++F+ TAVATD++A GELAG+AVG++V + +VAGP +GGSMNP RTLGPA+A+ +Y+ +
Sbjct: 206 LLFVVTAVATDTRAVGELAGIAVGATVMLNILVAGPSTGGSMNPVRTLGPAVASGNYRSL 265
Query: 61 WIYMVGPITGALLGAWSYVVIQETDHKQD 89
W+Y+V P GA+ GA Y ++ D D
Sbjct: 266 WVYLVAPTLGAISGAAVYTGVKLNDSVTD 294
Score = 26.9 bits (58), Expect = 1.7
Identities = 19/65 (29%), Positives = 31/65 (47%), Gaps = 3/65 (4%)
Query: 11 DSKATGELAGVAVGSSVTIASIVAGPISGGSMNPARTLGPAIATSSYKGIWIYMVGPITG 70
D T +L VG+ + I + AGPI + A TL I ++ G+ + ++ TG
Sbjct: 73 DVSLTRKLGAEFVGTFILIFTATAGPIVNQKYDGAETL---IGNAACAGLAVMIIILSTG 129
Query: 71 ALLGA 75
+ GA
Sbjct: 130 HISGA 134
>At4g19030 nodulin-26 like protein
Length = 296
Score = 102 bits (255), Expect = 2e-23
Identities = 45/85 (52%), Positives = 63/85 (73%)
Query: 1 MVFISTAVATDSKATGELAGVAVGSSVTIASIVAGPISGGSMNPARTLGPAIATSSYKGI 60
++FI + VATD++A GELAG+A+GS+V + ++A P+S SMNP R+LGPA+ YKGI
Sbjct: 191 LMFIISGVATDNRAIGELAGLAIGSTVLLNVLIAAPVSSASMNPGRSLGPALVYGCYKGI 250
Query: 61 WIYMVGPITGALLGAWSYVVIQETD 85
WIY+V P GA+ GAW Y ++ TD
Sbjct: 251 WIYLVAPTLGAIAGAWVYNTVRYTD 275
Score = 31.2 bits (69), Expect = 0.089
Identities = 24/88 (27%), Positives = 42/88 (47%), Gaps = 8/88 (9%)
Query: 18 LAGVAVGSSVTIASIV--AGPISGGSMNPARTLG-PAIATSSYKGIWIYMVGPITGALLG 74
L G+A+ +TI ++ G ISG +NPA T+ + K + Y++ + G+ L
Sbjct: 87 LPGIAIVWGLTIMVLIYSLGHISGAHINPAVTIAFASCGRFPLKQVPAYVISQVIGSTLA 146
Query: 75 AWSYVVIQETDH-----KQDLATSQSPL 97
A + ++ DH K D+ SP+
Sbjct: 147 AATLRLLFGLDHDVCSGKHDVFIGSSPV 174
>At5g37810 Membrane integral protein (MIP) -like
Length = 283
Score = 101 bits (251), Expect = 7e-23
Identities = 45/85 (52%), Positives = 64/85 (74%)
Query: 1 MVFISTAVATDSKATGELAGVAVGSSVTIASIVAGPISGGSMNPARTLGPAIATSSYKGI 60
++F+ + VATD++A GELAG+AVG ++ + VAGPISG SMNPAR+LGPA+ YK I
Sbjct: 172 LMFVISGVATDNRAVGELAGIAVGMTIMVNVFVAGPISGASMNPARSLGPALVMGVYKHI 231
Query: 61 WIYMVGPITGALLGAWSYVVIQETD 85
W+Y+VGP+ G + G + Y +I+ TD
Sbjct: 232 WVYIVGPVLGVISGGFVYNLIRFTD 256
>At1g31885 major intrinsic protein - like
Length = 294
Score = 89.7 bits (221), Expect = 2e-19
Identities = 37/72 (51%), Positives = 56/72 (77%)
Query: 1 MVFISTAVATDSKATGELAGVAVGSSVTIASIVAGPISGGSMNPARTLGPAIATSSYKGI 60
++F+ +AVATD +ATG AG+A+G+++ + + +GPISG SMNPAR+LGPA+ YK +
Sbjct: 179 LMFVISAVATDKRATGSFAGIAIGATIVLDILFSGPISGASMNPARSLGPALIWGCYKDL 238
Query: 61 WIYMVGPITGAL 72
W+Y+V P+ GAL
Sbjct: 239 WLYIVSPVIGAL 250
>At2g34390 putative aquaporin (plasma membrane intrinsic protein)
Length = 288
Score = 80.9 bits (198), Expect = 1e-16
Identities = 36/75 (48%), Positives = 52/75 (69%)
Query: 1 MVFISTAVATDSKATGELAGVAVGSSVTIASIVAGPISGGSMNPARTLGPAIATSSYKGI 60
++ + AV T + T EL G+ +G++VT+ I AG +SG SMNPAR++GPA+ YKGI
Sbjct: 183 LMLVVCAVTTTKRTTEELEGLIIGATVTLNVIFAGEVSGASMNPARSIGPALVWGCYKGI 242
Query: 61 WIYMVGPITGALLGA 75
WIY++ P GA+ GA
Sbjct: 243 WIYLLAPTLGAVSGA 257
>At3g06100 putative major intrinsic protein
Length = 275
Score = 76.6 bits (187), Expect = 2e-15
Identities = 32/79 (40%), Positives = 54/79 (67%), Gaps = 1/79 (1%)
Query: 1 MVFISTAV-ATDSKATGELAGVAVGSSVTIASIVAGPISGGSMNPARTLGPAIATSSYKG 59
+VF+++A+ + G L G +G+ +++ ++ GPISGGSMNPAR+LGPA+ ++
Sbjct: 174 VVFLASALHCGPHQNLGNLTGFVIGTVISLGVLITGPISGGSMNPARSLGPAVVAWDFED 233
Query: 60 IWIYMVGPITGALLGAWSY 78
+WIYM P+ GA++G +Y
Sbjct: 234 LWIYMTAPVIGAIIGVLTY 252
Score = 26.2 bits (56), Expect = 2.8
Identities = 24/87 (27%), Positives = 39/87 (44%), Gaps = 10/87 (11%)
Query: 1 MVFISTAVATDSKATGELAG-----VAVGSSVTIASIVAGPISGGSMNPARTLGPAIATS 55
++F V + ++ +G G V G SV + G ISG +NP+ T IA +
Sbjct: 58 LMFSVCGVISSTQLSGGHVGLLEYAVTAGLSVVVVVYSIGHISGAHLNPSIT----IAFA 113
Query: 56 SYKGI-WIYMVGPITGALLGAWSYVVI 81
+ G W + IT LGA + ++
Sbjct: 114 VFGGFPWSQVPLYITAQTLGATAATLV 140
>At2g29870 putative aquaporin (plasma membrane intrinsic protein)
Length = 139
Score = 76.3 bits (186), Expect = 2e-15
Identities = 33/72 (45%), Positives = 49/72 (67%)
Query: 1 MVFISTAVATDSKATGELAGVAVGSSVTIASIVAGPISGGSMNPARTLGPAIATSSYKGI 60
++ + AV T + T EL G+ +G++VT+ I G +SG SMNPAR++GPA+ YKGI
Sbjct: 34 LMLVVCAVTTTKRTTEELEGLIIGATVTLNVIFVGEVSGASMNPARSIGPALVWGCYKGI 93
Query: 61 WIYMVGPITGAL 72
WIY++ P GA+
Sbjct: 94 WIYLLAPTLGAV 105
>At4g17340 membrane channel like protein
Length = 250
Score = 70.9 bits (172), Expect = 1e-13
Identities = 35/80 (43%), Positives = 47/80 (58%), Gaps = 2/80 (2%)
Query: 1 MVFISTAVATDSK--ATGELAGVAVGSSVTIASIVAGPISGGSMNPARTLGPAIATSSYK 58
+V+ A A D K + G +A +A+G V + AGP SGGSMNPAR+ GPA+ + +
Sbjct: 153 LVYTVYATAADPKKGSLGTIAPIAIGFIVGANILAAGPFSGGSMNPARSFGPAVVSGDFS 212
Query: 59 GIWIYMVGPITGALLGAWSY 78
IWIY VGP+ G L Y
Sbjct: 213 QIWIYWVGPLVGGALAGLIY 232
Score = 28.1 bits (61), Expect = 0.75
Identities = 14/32 (43%), Positives = 20/32 (61%)
Query: 21 VAVGSSVTIASIVAGPISGGSMNPARTLGPAI 52
VA ++ + +A ISGG +NPA TLG A+
Sbjct: 61 VAHAFALFVGVSIAANISGGHLNPAVTLGLAV 92
>At3g16240 delta tonoplast integral protein (delta-TIP)
Length = 250
Score = 69.7 bits (169), Expect = 2e-13
Identities = 35/80 (43%), Positives = 47/80 (58%), Gaps = 2/80 (2%)
Query: 1 MVFISTAVATDSK--ATGELAGVAVGSSVTIASIVAGPISGGSMNPARTLGPAIATSSYK 58
+V+ A A D K + G +A +A+G V + AGP SGGSMNPAR+ GPA+A +
Sbjct: 153 LVYTVYATAADPKKGSLGTIAPLAIGLIVGANILAAGPFSGGSMNPARSFGPAVAAGDFS 212
Query: 59 GIWIYMVGPITGALLGAWSY 78
G W+Y VGP+ G L Y
Sbjct: 213 GHWVYWVGPLIGGGLAGLIY 232
Score = 28.1 bits (61), Expect = 0.75
Identities = 16/40 (40%), Positives = 23/40 (57%), Gaps = 2/40 (5%)
Query: 15 TGELAGVAV--GSSVTIASIVAGPISGGSMNPARTLGPAI 52
T L +AV G ++ +A + ISGG +NPA T G A+
Sbjct: 53 TPGLVAIAVCHGFALFVAVAIGANISGGHVNPAVTFGLAV 92
>At5g47450 membrane channel protein-like; aquaporin (tonoplast
intrinsic protein)-like
Length = 250
Score = 69.3 bits (168), Expect = 3e-13
Identities = 35/80 (43%), Positives = 46/80 (56%), Gaps = 2/80 (2%)
Query: 1 MVFISTAVATDSK--ATGELAGVAVGSSVTIASIVAGPISGGSMNPARTLGPAIATSSYK 58
+V+ A A D K + G +A +A+G V + AGP SGGSMNPAR+ GPA+ +
Sbjct: 153 LVYTVYATAADPKKGSLGTIAPIAIGFIVGANILAAGPFSGGSMNPARSFGPAVVSGDLS 212
Query: 59 GIWIYMVGPITGALLGAWSY 78
IWIY VGP+ G L Y
Sbjct: 213 QIWIYWVGPLVGGALAGLIY 232
Score = 28.1 bits (61), Expect = 0.75
Identities = 14/32 (43%), Positives = 20/32 (61%)
Query: 21 VAVGSSVTIASIVAGPISGGSMNPARTLGPAI 52
+A ++ + +A ISGG +NPA TLG AI
Sbjct: 61 IAHAFALFVGVSIAANISGGHLNPAVTLGLAI 92
>At1g17810 putative protein
Length = 267
Score = 62.8 bits (151), Expect = 3e-11
Identities = 35/95 (36%), Positives = 48/95 (49%), Gaps = 2/95 (2%)
Query: 2 VFISTAVATDSKATGELAGVAVGSSVTIASIVAGPISGGSMNPARTLGPAIATSSYKGIW 61
V STA+ + G +A +A+G V +V GP G SMNPAR GPA+ + W
Sbjct: 166 VVYSTAIDPKRGSIGIIAPLAIGLIVGANILVGGPFDGASMNPARAFGPALVGWRWSNHW 225
Query: 62 IYMVGPITGALLGA--WSYVVIQETDHKQDLATSQ 94
IY VGP G L A + Y++I + +T Q
Sbjct: 226 IYWVGPFIGGALAALIYEYMIIPSVNEPPHHSTHQ 260
>At1g73190 tonoplast intrinsic protein, alpha (alpha-TIP)
Length = 268
Score = 60.8 bits (146), Expect = 1e-10
Identities = 34/82 (41%), Positives = 45/82 (54%), Gaps = 2/82 (2%)
Query: 2 VFISTAVATDSKATGELAGVAVGSSVTIASIVAGPISGGSMNPARTLGPAIATSSYKGIW 61
V ST + + G +A +A+G V +V GP SG SMNPAR GPA+ + W
Sbjct: 166 VVYSTLIDPKRGSLGIIAPLAIGLIVGANILVGGPFSGASMNPARAFGPALVGWRWHDHW 225
Query: 62 IYMVGPITGALLGA--WSYVVI 81
IY VGP G+ L A + Y+VI
Sbjct: 226 IYWVGPFIGSALAALIYEYMVI 247
Score = 26.9 bits (58), Expect = 1.7
Identities = 20/71 (28%), Positives = 35/71 (49%), Gaps = 5/71 (7%)
Query: 7 AVATDSKATGELAGVAVGSSVTIASIVAGPI--SGGSMNPARTLGPAIA--TSSYKGIWI 62
A + G L VA+ + + + V+ I SGG +NPA T G + ++ + I+
Sbjct: 55 AAHAGTNTPGGLILVALAHAFALFAAVSAAINVSGGHVNPAVTFGALVGGRVTAIRAIY- 113
Query: 63 YMVGPITGALL 73
Y + + GA+L
Sbjct: 114 YWIAQLLGAIL 124
>At3g26520 salt-stress induced tonoplast intrinsic protein
Length = 253
Score = 58.5 bits (140), Expect = 5e-10
Identities = 30/86 (34%), Positives = 46/86 (52%), Gaps = 2/86 (2%)
Query: 5 STAVATDSKATGELAGVAVGSSVTIASIVAGPISGGSMNPARTLGPAIATSSYKGIWIYM 64
+TAV + + G +A +A+G V + G SG SMNPA GPA+ + ++ W+Y
Sbjct: 162 ATAVDPKNGSLGTIAPIAIGFIVGANILAGGAFSGASMNPAVAFGPAVVSWTWTNHWVYW 221
Query: 65 VGPITGALLGA--WSYVVIQETDHKQ 88
GP+ G L + +V I E H+Q
Sbjct: 222 AGPLIGGGLAGIIYDFVFIDENAHEQ 247
Score = 25.8 bits (55), Expect = 3.7
Identities = 17/56 (30%), Positives = 29/56 (51%), Gaps = 3/56 (5%)
Query: 19 AGVAVGSSVTIASIVAGPISGGSMNPARTLGPAIA--TSSYKGIWIYMVGPITGAL 72
A +A + +A V ISGG +NPA T G + + +GI +Y + + G++
Sbjct: 62 AALAHAFGLFVAVSVGANISGGHVNPAVTFGVLLGGNITLLRGI-LYWIAQLLGSV 116
>At2g36830 putative aquaporin (tonoplast intrinsic protein gamma)
Length = 251
Score = 55.1 bits (131), Expect = 6e-09
Identities = 28/85 (32%), Positives = 44/85 (50%), Gaps = 1/85 (1%)
Query: 5 STAVATDSKATGELAGVAVGSSVTIASIVAGPISGGSMNPARTLGPAIATSSYKGIWIYM 64
+TA+ + + G +A +A+G V + G SG SMNPA GPA+ + ++ W+Y
Sbjct: 161 ATAIDPKNGSLGTIAPIAIGFIVGANILAGGAFSGASMNPAVAFGPAVVSWTWTNHWVYW 220
Query: 65 VGPITGALLGAWSY-VVIQETDHKQ 88
GP+ G + Y V T H+Q
Sbjct: 221 AGPLVGGGIAGLIYEVFFINTTHEQ 245
Score = 28.1 bits (61), Expect = 0.75
Identities = 19/57 (33%), Positives = 30/57 (52%), Gaps = 3/57 (5%)
Query: 19 AGVAVGSSVTIASIVAGPISGGSMNPARTLGPAIA--TSSYKGIWIYMVGPITGALL 73
A VA + +A V ISGG +NPA T G I + +GI +Y + + G+++
Sbjct: 61 AAVAHAFGLFVAVSVGANISGGHVNPAVTFGAFIGGNITLLRGI-LYWIAQLLGSVV 116
>At4g01470 putative water channel protein
Length = 252
Score = 54.7 bits (130), Expect = 7e-09
Identities = 31/83 (37%), Positives = 43/83 (51%), Gaps = 2/83 (2%)
Query: 1 MVFISTAVATDSKA--TGELAGVAVGSSVTIASIVAGPISGGSMNPARTLGPAIATSSYK 58
+V+ A A D K G +A +A+G V +V G G SMNPA + GPA+ + +
Sbjct: 155 LVYTVYATAVDPKKGDIGIIAPLAIGLIVGANILVGGAFDGASMNPAVSFGPAVVSWIWT 214
Query: 59 GIWIYMVGPITGALLGAWSYVVI 81
W+Y VGP GA + A Y I
Sbjct: 215 NHWVYWVGPFIGAAIAAIVYDTI 237
Score = 25.0 bits (53), Expect = 6.3
Identities = 15/56 (26%), Positives = 29/56 (51%), Gaps = 1/56 (1%)
Query: 19 AGVAVGSSVTIASIVAGPISGGSMNPARTLGPAIATS-SYKGIWIYMVGPITGALL 73
A ++ ++ +A V +SGG +NPA T G I + + +Y + + GA++
Sbjct: 61 ASLSHAFALFVAVSVGANVSGGHVNPAVTFGAFIGGNITLLRAILYWIAQLLGAVV 116
>At2g25810 putative aquaporin (tonoplast intrinsic protein)
Length = 249
Score = 53.5 bits (127), Expect = 2e-08
Identities = 27/80 (33%), Positives = 42/80 (51%), Gaps = 4/80 (5%)
Query: 18 LAGVAVGSSVTIASIVAGPISGGSMNPARTLGPAIATSSYKGIWIYMVGPITGALLGAWS 77
L G VG+++ + G SG SMNPAR+ GPA+ + ++ W+Y VGP+ G L +
Sbjct: 172 LTGFVVGANI----LAGGAFSGASMNPARSFGPALVSGNWTDHWVYWVGPLIGGGLAGFI 227
Query: 78 YVVIQETDHKQDLATSQSPL 97
Y + +A + PL
Sbjct: 228 YENVLIDRPHVPVADDEQPL 247
Score = 37.4 bits (85), Expect = 0.001
Identities = 23/50 (46%), Positives = 31/50 (62%), Gaps = 4/50 (8%)
Query: 4 ISTAVATDSKATGELAG---VAVGSSVTIASIV-AGPISGGSMNPARTLG 49
+ +A+ATDS L G VAV + +A ++ AG ISGG +NPA TLG
Sbjct: 36 VGSAMATDSLVGNTLVGLFAVAVAHAFVVAVMISAGHISGGHLNPAVTLG 85
>At3g61430 plasma membrane intrinsic protein 1a
Length = 286
Score = 50.1 bits (118), Expect = 2e-07
Identities = 30/82 (36%), Positives = 45/82 (54%), Gaps = 9/82 (10%)
Query: 9 ATDSKATGE------LAGVAVGSSVTIASIVAGPISGGSMNPARTLGPAIATS---SYKG 59
ATD+K LA + +G +V + + PI+G +NPAR+LG AI + S+
Sbjct: 195 ATDAKRNARDSHVPILAPLPIGFAVFLVHLATIPITGTGINPARSLGAAIIYNKDHSWDD 254
Query: 60 IWIYMVGPITGALLGAWSYVVI 81
W++ VGP GA L A +VV+
Sbjct: 255 HWVFWVGPFIGAALAALYHVVV 276
Score = 26.6 bits (57), Expect = 2.2
Identities = 19/55 (34%), Positives = 24/55 (43%), Gaps = 1/55 (1%)
Query: 22 AVGSSVTIASIVAGPISGGSMNPARTLGPAIATS-SYKGIWIYMVGPITGALLGA 75
A G + ISGG +NPA T G +A S Y+V GA+ GA
Sbjct: 93 AFGGMIFALVYCTAGISGGHINPAVTFGLFLARKLSLTRALYYIVMQCLGAICGA 147
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.313 0.129 0.365
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 2,113,370
Number of Sequences: 26719
Number of extensions: 69914
Number of successful extensions: 234
Number of sequences better than 10.0: 43
Number of HSP's better than 10.0 without gapping: 36
Number of HSP's successfully gapped in prelim test: 7
Number of HSP's that attempted gapping in prelim test: 149
Number of HSP's gapped (non-prelim): 78
length of query: 110
length of database: 11,318,596
effective HSP length: 86
effective length of query: 24
effective length of database: 9,020,762
effective search space: 216498288
effective search space used: 216498288
T: 11
A: 40
X1: 16 ( 7.2 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 42 (21.9 bits)
S2: 52 (24.6 bits)
Medicago: description of AC125479.4


