

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC125479.3 - phase: 0
(68 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
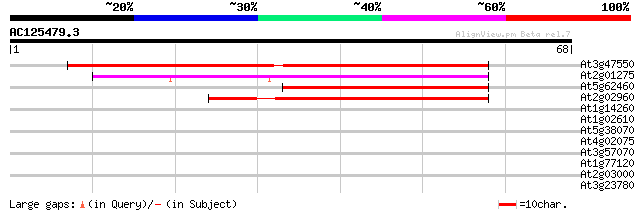
Score E
Sequences producing significant alignments: (bits) Value
At3g47550 unknown protein 46 4e-06
At2g01275 unknown protein 42 4e-05
At5g62460 unknown protein 39 4e-04
At2g02960 unknown protein 39 4e-04
At1g14260 hypothetical protein, 5' partial 37 0.002
At1g02610 hypothetical protein 37 0.002
At5g38070 putative protein 35 0.005
At4g02075 PIT1 (Pit1) 34 0.012
At3g57070 unknown protein 31 0.100
At1g77120 alcohol dehydrogenase 28 1.1
At2g03000 hypothetical protein 26 4.2
At3g23780 DNA-directed RNA polymerase subunit, putative 25 9.3
>At3g47550 unknown protein
Length = 288
Score = 45.8 bits (107), Expect = 4e-06
Identities = 22/51 (43%), Positives = 33/51 (64%), Gaps = 1/51 (1%)
Query: 8 TVHQVEIPIEELSAALREQLVIGKMGECRYCHEEEWIYRLEAPCRCDGSLK 58
+V++ E+P E + A E+ ++ + ECR C EE+ LEAPC C+GSLK
Sbjct: 43 SVNETEVPREYYAVADEEEPLLQSV-ECRICQEEDSTKNLEAPCACNGSLK 92
>At2g01275 unknown protein
Length = 259
Score = 42.4 bits (98), Expect = 4e-05
Identities = 23/53 (43%), Positives = 28/53 (52%), Gaps = 5/53 (9%)
Query: 11 QVEIPIEE---LSAALREQLVIG--KMGECRYCHEEEWIYRLEAPCRCDGSLK 58
Q IP+EE L E L G M ECR CH+E+ +E PC C GS+K
Sbjct: 30 QANIPVEECTILDDKTLEMLRNGDLSMAECRICHDEDLDSNMETPCSCSGSVK 82
>At5g62460 unknown protein
Length = 307
Score = 39.3 bits (90), Expect = 4e-04
Identities = 15/25 (60%), Positives = 18/25 (72%)
Query: 34 ECRYCHEEEWIYRLEAPCRCDGSLK 58
ECR C EE+ + LE+PC C GSLK
Sbjct: 77 ECRICQEEDSVKNLESPCSCSGSLK 101
>At2g02960 unknown protein
Length = 271
Score = 39.3 bits (90), Expect = 4e-04
Identities = 17/34 (50%), Positives = 22/34 (64%), Gaps = 2/34 (5%)
Query: 25 EQLVIGKMGECRYCHEEEWIYRLEAPCRCDGSLK 58
E L++ GECR C +E + LE+PC C GSLK
Sbjct: 35 EPLIVS--GECRICSDESPVENLESPCACSGSLK 66
>At1g14260 hypothetical protein, 5' partial
Length = 669
Score = 37.0 bits (84), Expect = 0.002
Identities = 15/27 (55%), Positives = 19/27 (69%)
Query: 34 ECRYCHEEEWIYRLEAPCRCDGSLKVF 60
ECR C +E I LE+PC C+GSLK +
Sbjct: 481 ECRICQDECDIKNLESPCACNGSLKFY 507
>At1g02610 hypothetical protein
Length = 146
Score = 36.6 bits (83), Expect = 0.002
Identities = 15/34 (44%), Positives = 18/34 (52%)
Query: 25 EQLVIGKMGECRYCHEEEWIYRLEAPCRCDGSLK 58
E + CR CHEEE EAPC C G++K
Sbjct: 10 ETYLKSSFNRCRICHEEEAESYFEAPCSCSGTIK 43
>At5g38070 putative protein
Length = 259
Score = 35.4 bits (80), Expect = 0.005
Identities = 16/42 (38%), Positives = 23/42 (54%), Gaps = 2/42 (4%)
Query: 17 EELSAALREQLVIGKMGECRYCHEEEWIYRLEAPCRCDGSLK 58
E +SA L K +CR CH+E+ ++ PC C G+LK
Sbjct: 38 ESISAGA--DLCESKFVQCRICHDEDEDTNMDTPCSCSGTLK 77
>At4g02075 PIT1 (Pit1)
Length = 218
Score = 34.3 bits (77), Expect = 0.012
Identities = 13/28 (46%), Positives = 17/28 (60%)
Query: 31 KMGECRYCHEEEWIYRLEAPCRCDGSLK 58
++ CR CHEEE E PC C G++K
Sbjct: 16 RITRCRICHEEEEESFFEVPCACSGTVK 43
>At3g57070 unknown protein
Length = 417
Score = 31.2 bits (69), Expect = 0.100
Identities = 16/45 (35%), Positives = 25/45 (55%), Gaps = 6/45 (13%)
Query: 18 ELSAALREQLVIGKMGECRYCHEEEWIYRLEAPC-RCDGSLKVFQ 61
EL+ L++ ++G CR C + ++ PC CDGS KVF+
Sbjct: 353 ELAEMLKDFPACERLGTCRSCGDARFV-----PCTNCDGSTKVFE 392
>At1g77120 alcohol dehydrogenase
Length = 379
Score = 27.7 bits (60), Expect = 1.1
Identities = 9/13 (69%), Positives = 11/13 (84%)
Query: 30 GKMGECRYCHEEE 42
G+ GECR+CH EE
Sbjct: 97 GECGECRHCHSEE 109
>At2g03000 hypothetical protein
Length = 535
Score = 25.8 bits (55), Expect = 4.2
Identities = 17/59 (28%), Positives = 28/59 (46%), Gaps = 4/59 (6%)
Query: 13 EIPIEELSAALREQLVIGKMGECRYCHEEEWI---YRLEAPCRCDGSLKVFQFYLKHHS 68
E P + A+ ++ + + GEC C EEW E PC+ L+ + +LK H+
Sbjct: 459 ESPATIRAVAMLPRVAMVEKGECVICF-EEWSKSDMETELPCKHKYHLECVEKWLKIHT 516
>At3g23780 DNA-directed RNA polymerase subunit, putative
Length = 946
Score = 24.6 bits (52), Expect = 9.3
Identities = 9/26 (34%), Positives = 14/26 (53%)
Query: 18 ELSAALREQLVIGKMGECRYCHEEEW 43
+L ++ L GK+G+ RY H W
Sbjct: 267 DLRRTRQQVLYTGKVGDARYPHPSHW 292
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.324 0.140 0.446
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 1,590,534
Number of Sequences: 26719
Number of extensions: 51822
Number of successful extensions: 146
Number of sequences better than 10.0: 12
Number of HSP's better than 10.0 without gapping: 10
Number of HSP's successfully gapped in prelim test: 2
Number of HSP's that attempted gapping in prelim test: 135
Number of HSP's gapped (non-prelim): 12
length of query: 68
length of database: 11,318,596
effective HSP length: 44
effective length of query: 24
effective length of database: 10,142,960
effective search space: 243431040
effective search space used: 243431040
T: 11
A: 40
X1: 15 ( 7.0 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 40 (21.5 bits)
S2: 52 (24.6 bits)
Medicago: description of AC125479.3


