

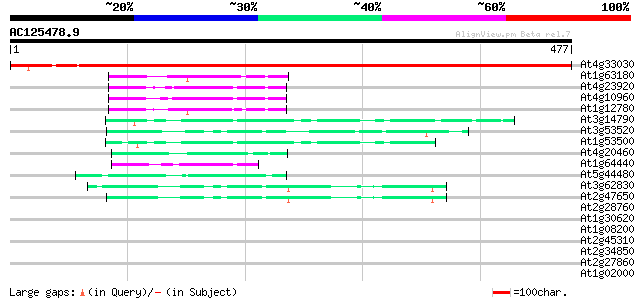
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC125478.9 + phase: 0
(477 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
At4g33030 sulfolipid biosynthesis protein SQD1 756 0.0
At1g63180 uridine diphosphate glucose epimerase like protein 53 3e-07
At4g23920 UDPglucose 4-epimerase - like protein 50 2e-06
At4g10960 UDP-galactose 4-epimerase - like protein 50 2e-06
At1g12780 uridine diphosphate glucose epimerase 50 3e-06
At3g14790 dTDP-glucose 4,6-dehydratase, putative 49 5e-06
At3g53520 UDP-glucuronic acid decarboxylase (UXS1) 47 2e-05
At1g53500 dTDP-D-glucose 4,6-dehydratase, putative 46 5e-05
At4g20460 UDP-glucose 4-epimerase - like protein 43 3e-04
At1g64440 UDP-galactose 4-epimerase, putative 43 3e-04
At5g44480 putative protein 42 6e-04
At3g62830 dTDP-glucose 4-6-dehydratase homolog D18 42 6e-04
At2g47650 putative nucleotide-sugar dehydratase 42 7e-04
At2g28760 putative nucleotide-sugar dehydratase 39 0.005
At1g30620 UDP-galactose 4-epimerase-like protein 37 0.024
At1g08200 unknown protein 37 0.031
At2g45310 putative nucleotide sugar epimerase 36 0.041
At2g34850 putative UDP-galactose-4-epimerase 36 0.041
At2g27860 putative dTDP-glucose 4-6-dehydratase 35 0.091
At1g02000 nucleotide sugar epimerase, putative 35 0.12
>At4g33030 sulfolipid biosynthesis protein SQD1
Length = 477
Score = 756 bits (1951), Expect = 0.0
Identities = 373/481 (77%), Positives = 421/481 (86%), Gaps = 8/481 (1%)
Query: 1 MAQLLSSSCSLTFS---ASNKPCLKPFHQCSTSFSNTVVCDNSKTPFRQLFLREQKPRKS 57
MA LLS+SC S +S+K +KPF T F+ ++ S++ + L +E+KPRKS
Sbjct: 1 MAHLLSASCPSVISLSSSSSKNSVKPFVSGQTFFNAQLL---SRSSLKGLLFQEKKPRKS 57
Query: 58 LAVVNASTISTGQEAPVQTSSGDPF-KPKRVMVIGGDGYCGWATALHLSNKGYEVAIVDN 116
V A+ + Q+AP +TS+ + KPKRVMVIGGDGYCGWATALHLS K YEV IVDN
Sbjct: 58 -CVFRATAVPITQQAPPETSTNNSSSKPKRVMVIGGDGYCGWATALHLSKKNYEVCIVDN 116
Query: 117 LVRRLFDHQLGLDSLTPISSIQDRIQCWKSLTGKSIELYIGDICEFEFLSETFKSYEPDA 176
LVRRLFDHQLGL+SLTPI+SI DRI WK+LTGKSIELY+GDIC+FEFL+E+FKS+EPD+
Sbjct: 117 LVRRLFDHQLGLESLTPIASIHDRISRWKALTGKSIELYVGDICDFEFLAESFKSFEPDS 176
Query: 177 VVHFGEQRSAPYSMIDRSRAVYTQQNNVVGTLNVLFAIKEYREDCHLVKLGTMGEYGTPN 236
VVHFGEQRSAPYSMIDRSRAVYTQ NNV+GTLNVLFAIKE+ E+CHLVKLGTMGEYGTPN
Sbjct: 177 VVHFGEQRSAPYSMIDRSRAVYTQHNNVIGTLNVLFAIKEFGEECHLVKLGTMGEYGTPN 236
Query: 237 IDIEEGYITITHNGRTDTLPYPKQASSFYHLSKVHDSHNIAFTCKAWGIRATDLNQGVVY 296
IDIEEGYITITHNGRTDTLPYPKQASSFYHLSKVHDSHNIAFTCKAWGIRATDLNQGVVY
Sbjct: 237 IDIEEGYITITHNGRTDTLPYPKQASSFYHLSKVHDSHNIAFTCKAWGIRATDLNQGVVY 296
Query: 297 GVRTDETAMHEELCNRFDYDAIFGTALNRFCVQAAVGHPLTVYGKGGQTRAFLDIRDTVQ 356
GV+TDET MHEEL NR DYDA+FGTALNRFCVQAAVGHPLTVYGKGGQTR +LDIRDTVQ
Sbjct: 297 GVKTDETEMHEELRNRLDYDAVFGTALNRFCVQAAVGHPLTVYGKGGQTRGYLDIRDTVQ 356
Query: 357 CVELAIANPANPGEFRVFNQFTEQFKVTELAELVTKAGEKLGLDVKTISVPNPRVELEEH 416
CVE+AIANPA GEFRVFNQFTEQF V ELA LVTKAG KLGLDVK ++VPNPRVE EEH
Sbjct: 357 CVEIAIANPAKAGEFRVFNQFTEQFSVNELASLVTKAGSKLGLDVKKMTVPNPRVEAEEH 416
Query: 417 YYNCKNTKLVDLGLKPHFLSDSLIDSLLNFAVQYKDRVDTKQIMPGVSWRKVGVKTKTLT 476
YYN K+TKL++LGL+PH+LSDSL+DSLLNFAVQ+KDRVDTKQIMP VSW+K+GVKTK++T
Sbjct: 417 YYNAKHTKLMELGLEPHYLSDSLLDSLLNFAVQFKDRVDTKQIMPSVSWKKIGVKTKSMT 476
Query: 477 S 477
+
Sbjct: 477 T 477
>At1g63180 uridine diphosphate glucose epimerase like protein
Length = 351
Score = 53.1 bits (126), Expect = 3e-07
Identities = 40/158 (25%), Positives = 74/158 (46%), Gaps = 26/158 (16%)
Query: 85 KRVMVIGGDGYCGWATALHLSNKGYEVAIVDNLVRRLFDHQLGLDSLTPISSIQDRIQCW 144
+ ++V GG G+ G T + L N+G++V I+DNL +S+ + +
Sbjct: 7 QNILVTGGAGFIGTHTVVQLLNQGFKVTIIDNL----------------DNSVVEAVHRV 50
Query: 145 KSLTG----KSIELYIGDICEFEFLSETFKSYEPDAVVHFGEQRSAPYSMIDRSRAVYTQ 200
+ L G +E +GD+ + + F + DAV+HF ++ S+ + R
Sbjct: 51 RELVGPDLSTKLEFNLGDLRNKGDIEKLFSNQRFDAVIHFAGLKAVGESVGNPRRYF--- 107
Query: 201 QNNVVGTLNVLFAIKEYREDCHLVKLGTMGE-YGTPNI 237
NN+VGT+N+ + +Y +C ++ + YG P I
Sbjct: 108 DNNLVGTINLYETMAKY--NCKMMVFSSSATVYGQPEI 143
>At4g23920 UDPglucose 4-epimerase - like protein
Length = 350
Score = 50.4 bits (119), Expect = 2e-06
Identities = 43/152 (28%), Positives = 71/152 (46%), Gaps = 19/152 (12%)
Query: 85 KRVMVIGGDGYCGWATALHLSNKGYEVAIVDNLVRRLFDHQLGLDSLTPISSIQDRIQCW 144
K V+V GG GY G T L L GY +VDN +D+ + +S+Q R++
Sbjct: 3 KSVLVTGGAGYIGSHTVLQLLEGGYSAVVVDN-----YDN-------SSAASLQ-RVKKL 49
Query: 145 KSLTGKSIELYIGDICEFEFLSETFKSYEPDAVVHFGEQRSAPYSMIDRSRAVYTQQNNV 204
G + + D+ + L + F + DAV+HF ++ S+ + + NN+
Sbjct: 50 AGENGNRLSFHQVDLRDRPALEKIFSETKFDAVIHFAGLKAVGESV---EKPLLYYNNNI 106
Query: 205 VGTLNVLFAIKEYREDC-HLVKLGTMGEYGTP 235
VGT+ +L + +Y C +LV + YG P
Sbjct: 107 VGTVTLLEVMAQY--GCKNLVFSSSATVYGWP 136
>At4g10960 UDP-galactose 4-epimerase - like protein
Length = 351
Score = 50.4 bits (119), Expect = 2e-06
Identities = 42/152 (27%), Positives = 73/152 (47%), Gaps = 19/152 (12%)
Query: 85 KRVMVIGGDGYCGWATALHLSNKGYEVAIVDNLVRRLFDHQLGLDSLTPISSIQDRIQCW 144
+ V+V GG GY G T L L GY V +VDN LD+ + +S R++
Sbjct: 4 RNVLVSGGAGYIGSHTVLQLLLGGYSVVVVDN-----------LDNSSAVS--LQRVKKL 50
Query: 145 KSLTGKSIELYIGDICEFEFLSETFKSYEPDAVVHFGEQRSAPYSMIDRSRAVYTQQNNV 204
+ G+ + + D+ + L + F + DAV+HF ++ S+ + + NN+
Sbjct: 51 AAEHGERLSFHQVDLRDRSALEKIFSETKFDAVIHFAGLKAVGESV---EKPLLYYNNNL 107
Query: 205 VGTLNVLFAIKEYREDC-HLVKLGTMGEYGTP 235
VGT+ +L + ++ C +LV + YG+P
Sbjct: 108 VGTITLLEVMAQH--GCKNLVFSSSATVYGSP 137
>At1g12780 uridine diphosphate glucose epimerase
Length = 351
Score = 50.1 bits (118), Expect = 3e-06
Identities = 39/156 (25%), Positives = 74/156 (47%), Gaps = 26/156 (16%)
Query: 85 KRVMVIGGDGYCGWATALHLSNKGYEVAIVDNLVRRLFDHQLGLDSLTPISSIQDRIQCW 144
+ ++V GG G+ G T + L G++V+I+DN FD +S+ + +
Sbjct: 7 QNILVTGGAGFIGTHTVVQLLKDGFKVSIIDN-----FD-----------NSVIEAVDRV 50
Query: 145 KSLTG----KSIELYIGDICEFEFLSETFKSYEPDAVVHFGEQRSAPYSMIDRSRAVYTQ 200
+ L G K ++ +GD+ + + F DAV+HF ++ S ++ R +
Sbjct: 51 RELVGPDLSKKLDFNLGDLRNKGDIEKLFSKQRFDAVIHFAGLKAVGES-VENPRRYF-- 107
Query: 201 QNNVVGTLNVLFAIKEYREDCHLVKLGTMGE-YGTP 235
NN+VGT+N+ + +Y +C ++ + YG P
Sbjct: 108 DNNLVGTINLYETMAKY--NCKMMVFSSSATVYGQP 141
>At3g14790 dTDP-glucose 4,6-dehydratase, putative
Length = 664
Score = 49.3 bits (116), Expect = 5e-06
Identities = 77/353 (21%), Positives = 139/353 (38%), Gaps = 60/353 (16%)
Query: 82 FKPKRVMVIGGDGYCGWATALHL--SNKGYEVAIVDNLVRRLFDHQLGLDSLTPISSIQD 139
+KPK +++ G G+ A L S Y++ ++D L D+ L +L P
Sbjct: 4 YKPKNILITGAAGFIASHVANRLVRSYPDYKIVVLDKL-----DYCSNLKNLNP------ 52
Query: 140 RIQCWKSLTGKSIELYIGDICEFEFLSETFKSYEPDAVVHFGEQRSAPYSMIDRSRAVYT 199
S + + + GDI + ++ + E D ++HF Q S +
Sbjct: 53 ------SKSSPNFKFVKGDIASADLVNYLLITEEIDTIMHFAAQTHVDNSF---GNSFEF 103
Query: 200 QQNNVVGTLNVLFAIKEYREDCHLVKLGTMGEYGTPNIDIEEGYITITHNGRTDTLPYPK 259
+NN+ GT +L A K + + + T YG + D G ++ + LP
Sbjct: 104 TKNNIYGTHVLLEACKVTGQIRRFIHVSTDEVYGETDEDASVG-----NHEASQLLP--- 155
Query: 260 QASSFYHLSKVHDSHNIAFTCKAWGIRATDLNQGVVYGVRTDETAMHEELCNRFDYDAIF 319
++ Y +K + +++G+ VYG N+F I
Sbjct: 156 --TNPYSATKAGAEMLVMAYGRSYGLPVITTRGNNVYGP------------NQFPEKLI- 200
Query: 320 GTALNRFCVQAAVGHPLTVYGKGGQTRAFLDIRDTVQCVELAIANPANPGEF-RVFN-QF 377
+F + A G PL ++G G R++L D + E+ + + GE V+N
Sbjct: 201 ----PKFILLAMNGKPLPIHGDGSNVRSYLYCEDVAEAFEVVL----HKGEVNHVYNIGT 252
Query: 378 TEQFKVTELAELVTKAGEKLGLDV-KTISVPNPRVELEEHYYNCKNTKLVDLG 429
T + +V ++A ++K G+D TI R ++ Y+ + KL LG
Sbjct: 253 TRERRVIDVANDISKL---FGIDPDSTIQYVENRPFNDQRYF-LDDQKLKKLG 301
>At3g53520 UDP-glucuronic acid decarboxylase (UXS1)
Length = 433
Score = 47.4 bits (111), Expect = 2e-05
Identities = 75/314 (23%), Positives = 117/314 (36%), Gaps = 70/314 (22%)
Query: 83 KPKRVMVIGGDGYCGWATALHLSNKGYEVAIVDNLVRRLFDHQLGLDSLTPISSIQDRIQ 142
K R++V GG G+ G L +G EV ++DN ++ + L S
Sbjct: 118 KRLRIVVTGGAGFVGSHLVDKLIGRGDEVIVIDNFFTGRKENLVHLFS------------ 165
Query: 143 CWKSLTGKSIELYIGDICEFEFLSETFKSYEPDAVVHFGEQRSAPYSMID-RSRAVYTQQ 201
EL D+ E L E D + H + P S + + V T +
Sbjct: 166 ------NPRFELIRHDVVEPILL-------EVDQIYHL----ACPASPVHYKYNPVKTIK 208
Query: 202 NNVVGTLNVLFAIKEYREDCHLVKLGTMGEYGTPNIDIEEGYITITHNGRTDTLPYPKQA 261
NV+GTLN+L K R + T YG P L +P++
Sbjct: 209 TNVMGTLNMLGLAK--RVGARFLLTSTSEVYGDP-------------------LEHPQKE 247
Query: 262 SSFYHLSKVHDSHNIAFTCKAWGIRATDLNQGVVYGVRTDETAMHEELCNRFDYDAIFGT 321
+ + +++ + + + A D ++G GV + R D G
Sbjct: 248 TYWGNVNPIGERSCYDEGKRTAETLAMDYHRGA--GVEVRIARIFNTYGPRMCLDD--GR 303
Query: 322 ALNRFCVQAAVGHPLTVYGKGGQTRAFLDIRD-----TVQCVELAIANPANPGEFRVFNQ 376
++ F Q HP+TVYG G QTR+F + D ++ + N NPGEF +
Sbjct: 304 VVSNFVAQTIRKHPMTVYGDGKQTRSFQYVSDLGLVALMENDHVGPFNLGNPGEFTML-- 361
Query: 377 FTEQFKVTELAELV 390
ELAE+V
Sbjct: 362 --------ELAEVV 367
>At1g53500 dTDP-D-glucose 4,6-dehydratase, putative
Length = 667
Score = 45.8 bits (107), Expect = 5e-05
Identities = 58/287 (20%), Positives = 110/287 (38%), Gaps = 57/287 (19%)
Query: 82 FKPKRVMVIGGDGYCGWATALHLSNK------GYEVAIVDNLVRRLFDHQLGLDSLTPIS 135
+KPK +++ G G+ A H++N+ Y++ ++D L D+ L +L P
Sbjct: 6 YKPKNILITGAAGFI----ASHVANRLIRNYPDYKIVVLDKL-----DYCSDLKNLDP-- 54
Query: 136 SIQDRIQCWKSLTGKSIELYIGDICEFEFLSETFKSYEPDAVVHFGEQRSAPYSMIDRSR 195
S + + + GDI + ++ + D ++HF Q S
Sbjct: 55 ----------SFSSPNFKFVKGDIASDDLVNYLLITENIDTIMHFAAQTHVDNSF---GN 101
Query: 196 AVYTQQNNVVGTLNVLFAIKEYREDCHLVKLGTMGEYGTPNIDIEEGYITITHNGRTDTL 255
+ +NN+ GT +L A K + + + T YG + D G ++ + L
Sbjct: 102 SFEFTKNNIYGTHVLLEACKVTGQIRRFIHVSTDEVYGETDEDAAVG-----NHEASQLL 156
Query: 256 PYPKQASSFYHLSKVHDSHNIAFTCKAWGIRATDLNQGVVYGVRTDETAMHEELCNRFDY 315
P ++ Y +K + +++G+ VYG N+F
Sbjct: 157 P-----TNPYSATKAGAEMLVMAYGRSYGLPVITTRGNNVYGP------------NQFPE 199
Query: 316 DAIFGTALNRFCVQAAVGHPLTVYGKGGQTRAFLDIRDTVQCVELAI 362
I +F + A G PL ++G G R++L D + E+ +
Sbjct: 200 KMI-----PKFILLAMSGKPLPIHGDGSNVRSYLYCEDVAEAFEVVL 241
>At4g20460 UDP-glucose 4-epimerase - like protein
Length = 379
Score = 43.1 bits (100), Expect = 3e-04
Identities = 41/150 (27%), Positives = 59/150 (39%), Gaps = 18/150 (12%)
Query: 87 VMVIGGDGYCGWATALHLSNKGYEVAIVDNLVRRLFDHQLGLDSLTPISSIQDRIQCWKS 146
V+V GG GY G AL L Y V IVDNL R L L P
Sbjct: 40 VLVTGGAGYIGSHAALRLLKDSYRVTIVDNLSRGNLGAVKVLQGLFPEPG---------- 89
Query: 147 LTGKSIELYIGDICEFEFLSETFKSYEPDAVVHFGEQRSAPYSMIDRSRAVYTQQNNVVG 206
++ D+ + + + + F DAV+HF S +D + + +N
Sbjct: 90 ----RLQFIYADLGDAKAVDKIFSENAFDAVMHFAAVAYVGESTLDPLKYYHNITSN--- 142
Query: 207 TLNVLFAIKEYREDCHLVKLGTMGEYGTPN 236
TL VL A+ ++ L+ T YG P+
Sbjct: 143 TLVVLEAVARHKVK-KLIYSSTCATYGEPD 171
>At1g64440 UDP-galactose 4-epimerase, putative
Length = 348
Score = 43.1 bits (100), Expect = 3e-04
Identities = 32/125 (25%), Positives = 58/125 (45%), Gaps = 16/125 (12%)
Query: 87 VMVIGGDGYCGWATALHLSNKGYEVAIVDNLVRRLFDHQLGLDSLTPISSIQDRIQCWKS 146
++V GG GY G T L L GY ++DNL SL I ++D
Sbjct: 5 ILVTGGAGYIGSHTVLQLLLGGYNTVVIDNLDN---------SSLVSIQRVKD----LAG 51
Query: 147 LTGKSIELYIGDICEFEFLSETFKSYEPDAVVHFGEQRSAPYSMIDRSRAVYTQQNNVVG 206
G+++ ++ D+ + L + F + DAV+HF ++ S+ ++ + NN++
Sbjct: 52 DHGQNLTVHQVDLRDKPALEKVFSETKFDAVMHFAGLKAVGESV---AKPLLYYNNNLIA 108
Query: 207 TLNVL 211
T+ +L
Sbjct: 109 TITLL 113
>At5g44480 putative protein
Length = 436
Score = 42.4 bits (98), Expect = 6e-04
Identities = 46/179 (25%), Positives = 68/179 (37%), Gaps = 22/179 (12%)
Query: 57 SLAVVNASTISTGQEAPVQTSSGDPFKPKRVMVIGGDGYCGWATALHLSNKGYEVAIVDN 116
++ V++ S+ T A Q G V+V GG GY G AL L Y V IVDN
Sbjct: 71 AIIVISQSSSFTSPSAFSQREEG----VTHVLVTGGAGYIGSHAALRLLRDSYRVTIVDN 126
Query: 117 LVRRLFDHQLGLDSLTPISSIQDRIQCWKSLTGKSIELYIGDICEFEFLSETFKSYEPDA 176
L R L L P TG+ ++ D+ + + + F DA
Sbjct: 127 LSRGNLGAVKTLQQLFP-------------QTGR-LQFIYADLGDPLAVEKIFSENAFDA 172
Query: 177 VVHFGEQRSAPYSMIDRSRAVYTQQNNVVGTLNVLFAIKEYREDCHLVKLGTMGEYGTP 235
V+HF S + + + +N +G L + K + L+ T YG P
Sbjct: 173 VMHFAAVAYVGESTLYPLKYYHNITSNTLGVLEAMARHKVKK----LIYSSTCATYGEP 227
>At3g62830 dTDP-glucose 4-6-dehydratase homolog D18
Length = 445
Score = 42.4 bits (98), Expect = 6e-04
Identities = 76/327 (23%), Positives = 115/327 (34%), Gaps = 94/327 (28%)
Query: 67 STGQEAPVQTSSGDPFKPKRVMVIGGDGYCGWATALHLSNKGYEVAIVDNLVRRLFDHQL 126
+TG + P+ G K RV+V GG G+ G L +G V +VDN ++ +
Sbjct: 105 ATGGKIPL----GLKRKGLRVVVTGGAGFVGSHLVDRLMARGDTVIVVDNFFTGRKENVM 160
Query: 127 GLDSLTPISSIQDRIQCWKSLTGKSIELYIGDICEFEFLSETFKSYEPDAVVHFGEQRSA 186
+ + E+ D+ E L E D + H +
Sbjct: 161 ------------------HHFSNPNFEMIRHDVVEPILL-------EVDQIYHL----AC 191
Query: 187 PYSMID-RSRAVYTQQNNVVGTLNVLFAIKEYREDCHLVKLGTMGEYGTP---------- 235
P S + + V T + NVVGTLN+L K R + T YG P
Sbjct: 192 PASPVHYKFNPVKTIKTNVVGTLNMLGLAK--RVGARFLLTSTSEVYGDPLQHPQVETYW 249
Query: 236 ----NIDIEEGYITITHNGRTDTLPYPKQASSFYHLSKVHDSHNIAFTCKAWGIRATDLN 291
I + Y T T+ Y + A+ ++++ ++
Sbjct: 250 GNVNPIGVRSCYDEGKRTAETLTMDYHRGANVEVRIARIFNT------------------ 291
Query: 292 QGVVYGVRTDETAMHEELCNRFDYDAIFGTALNRFCVQAAVGHPLTVYGKGGQTRAFLDI 351
YG R +C G ++ F QA PLTVYG G QTR+F +
Sbjct: 292 ----YGPR---------MC------IDDGRVVSNFVAQALRKEPLTVYGDGKQTRSFQFV 332
Query: 352 RDTVQCV-------ELAIANPANPGEF 371
D V+ + + N NPGEF
Sbjct: 333 SDLVEGLMRLMEGEHVGPFNLGNPGEF 359
>At2g47650 putative nucleotide-sugar dehydratase
Length = 443
Score = 42.0 bits (97), Expect = 7e-04
Identities = 72/311 (23%), Positives = 107/311 (34%), Gaps = 90/311 (28%)
Query: 83 KPKRVMVIGGDGYCGWATALHLSNKGYEVAIVDNLVRRLFDHQLGLDSLTPISSIQDRIQ 142
K RV+V GG G+ G L +G V +VDN ++ +
Sbjct: 119 KVLRVVVTGGAGFVGSHLVDRLMARGDNVIVVDNFFTGRKENVM---------------- 162
Query: 143 CWKSLTGKSIELYIGDICEFEFLSETFKSYEPDAVVHFGEQRSAPYSMID-RSRAVYTQQ 201
+ E+ D+ E L E D + H + P S + + V T +
Sbjct: 163 --HHFNNPNFEMIRHDVVEPILL-------EVDQIYHL----ACPASPVHYKFNPVKTIK 209
Query: 202 NNVVGTLNVLFAIKEYREDCHLVKLGTMGEYGTP--------------NIDIEEGYITIT 247
NVVGTLN+L K R + T YG P I + Y
Sbjct: 210 TNVVGTLNMLGLAK--RVGARFLLTSTSEVYGDPLQHPQVETYWGNVNPIGVRSCYDEGK 267
Query: 248 HNGRTDTLPYPKQASSFYHLSKVHDSHNIAFTCKAWGIRATDLNQGVVYGVRTDETAMHE 307
T T+ Y + A+ ++++ ++ YG R
Sbjct: 268 RTAETLTMDYHRGANVEVRIARIFNT----------------------YGPR-------- 297
Query: 308 ELCNRFDYDAIFGTALNRFCVQAAVGHPLTVYGKGGQTRAFLDIRDTVQCV-------EL 360
+C G ++ F QA PLTVYG G QTR+F + D V+ + +
Sbjct: 298 -MC------IDDGRVVSNFVAQALRKEPLTVYGDGKQTRSFQFVSDLVEGLMRLMEGEHV 350
Query: 361 AIANPANPGEF 371
N NPGEF
Sbjct: 351 GPFNLGNPGEF 361
>At2g28760 putative nucleotide-sugar dehydratase
Length = 343
Score = 39.3 bits (90), Expect = 0.005
Identities = 70/298 (23%), Positives = 111/298 (36%), Gaps = 69/298 (23%)
Query: 86 RVMVIGGDGYCGWATALHL-SNKGYEVAIVDNLVRRLFDHQLGLDSLTPISSIQDRIQCW 144
R++V GG G+ G L N+ EV + DN + +D ++ W
Sbjct: 32 RILVTGGAGFIGSHLVDKLMQNEKNEVIVADNY----------------FTGSKDNLKKW 75
Query: 145 KSLTGKSIELYIGDICEFEFLSETFKSYEPDAVVHFGEQRSAPYSMID-RSRAVYTQQNN 203
+ EL D+ E F+ E D + H + P S I + V T + N
Sbjct: 76 --IGHPRFELIRHDVTEPLFV-------EVDQIYHL----ACPASPIFYKYNPVKTIKTN 122
Query: 204 VVGTLNVLFAIKEYREDCHLVKLGTMGEYGTPNIDIEEGYITITHNGRTDTLPYPKQASS 263
V+GTLN+L K R ++ T YG P L +P+ S
Sbjct: 123 VIGTLNMLGLAK--RVGARILLTSTSEVYGDP-------------------LVHPQTESY 161
Query: 264 FYHLSKVHDSHNIAFTCKAWGIRATD---LNQGVVYGVRTDETAMHEELCNRFDYDAIFG 320
+ +++ + +C G R + + +G+ + R + D G
Sbjct: 162 WGNVNPIG-----VRSCYDEGKRVAETLMFDYHRQHGIEIRIARIFNTYGPRMNIDD--G 214
Query: 321 TALNRFCVQAAVGHPLTVYGKGGQTRAFLDIRDTVQCV-------ELAIANPANPGEF 371
++ F QA G LTV G QTR+F + D V+ + + N NPGEF
Sbjct: 215 RVVSNFIAQALRGEALTVQKPGTQTRSFCYVSDMVEGLMRLMEGDQTGPINIGNPGEF 272
>At1g30620 UDP-galactose 4-epimerase-like protein
Length = 419
Score = 37.0 bits (84), Expect = 0.024
Identities = 36/151 (23%), Positives = 61/151 (39%), Gaps = 18/151 (11%)
Query: 87 VMVIGGDGYCGWATALHLSNKGYEVAIVDNLVRRLFDHQLGLDSLTPISSIQDRIQCWKS 146
V+V GG GY G AL L + Y V IVDNL R +L + +Q+
Sbjct: 73 VLVTGGAGYIGSHAALRLLKESYRVTIVDNLSR---------GNLAAVRILQELFP---- 119
Query: 147 LTGKSIELYIGDICEFEFLSETFKSYEPDAVVHFGEQRSAPYSMIDRSRAVYTQQNNVVG 206
++ D+ + + +++ F DAV+HF +A + + ++ +N+
Sbjct: 120 -EPGRLQFIYADLGDAKAVNKIFTENAFDAVMHF----AAVAYVGESTQFPLKYYHNITS 174
Query: 207 TLNVLFAIKEYREDCHLVKLGTMGEYGTPNI 237
V+ L+ T YG P+I
Sbjct: 175 NTLVVLETMAAHGVKTLIYSSTCATYGEPDI 205
>At1g08200 unknown protein
Length = 389
Score = 36.6 bits (83), Expect = 0.031
Identities = 36/124 (29%), Positives = 56/124 (45%), Gaps = 14/124 (11%)
Query: 335 PLTVYGKGGQTRAFLDIRDTVQCVELAIANP--ANPGEFRVFNQFTEQFKVTELAELVTK 392
PL + G R F+ I+D ++ V L I NP AN F V N + V +LAE++T+
Sbjct: 249 PLKLVDGGESQRTFIYIKDAIEAVLLMIENPERANGHIFNVGNP-NNEVTVRQLAEMMTE 307
Query: 393 AGEKLGLDVKTISVPNPRVELEEHY---YNCKNTKLVD-------LGLKPHFLSDSLIDS 442
K+ + I P V +E Y Y+ + ++ D LG P L++S
Sbjct: 308 VYAKVSGET-AIESPTIDVSSKEFYGEGYDDSDKRIPDMTIINRQLGWNPKTSLWDLLES 366
Query: 443 LLNF 446
L +
Sbjct: 367 TLTY 370
>At2g45310 putative nucleotide sugar epimerase
Length = 437
Score = 36.2 bits (82), Expect = 0.041
Identities = 50/212 (23%), Positives = 77/212 (35%), Gaps = 31/212 (14%)
Query: 87 VMVIGGDGYCGWATALHLSNKGYEVAIVDNLVRRLFDHQLGLDSLTPISSIQDRIQCWKS 146
V+V G G+ G + L +G D +GLD+ +
Sbjct: 99 VLVTGAAGFVGTHVSAALKRRG--------------DGVIGLDNFNDYYDPSLKRARRAL 144
Query: 147 LTGKSIELYIGDICEFEFLSETFKSYEPDAVVHFGEQRSAPYSMIDRSRAVYTQQNNVVG 206
L I + GDI + E L + FK V+H Q Y+M + S V+ +N+ G
Sbjct: 145 LERSGIFIVEGDINDVELLRKLFKIVSFTHVMHLAAQAGVRYAMENPSSYVH---SNIAG 201
Query: 207 TLNVLFAIKEYREDCHLVKLGTMGEYGTPNIDIEEGYITITHNGRTDTLPYPKQASSFYH 266
+N+L K +V + YG + + +TD Q +S Y
Sbjct: 202 FVNLLEICKSVNPQPAIVWASSSSVYGL------NTKVPFSEKDKTD------QPASLYA 249
Query: 267 LSKVHDSHNIAFTCK-AWGIRATDLNQGVVYG 297
+K IA T +G+ T L VYG
Sbjct: 250 ATK-KAGEEIAHTYNHIYGLSLTGLRFFTVYG 280
>At2g34850 putative UDP-galactose-4-epimerase
Length = 385
Score = 36.2 bits (82), Expect = 0.041
Identities = 28/94 (29%), Positives = 37/94 (38%), Gaps = 14/94 (14%)
Query: 87 VMVIGGDGYCGWATALHLSNKGYEVAIVDNLVRRLFDHQLGLDSLTPISSIQDRIQCWKS 146
V+V GG GY G AL L Y V IVDNL R L L P
Sbjct: 40 VLVTGGAGYIGSHAALRLLKDSYRVTIVDNLSRGNLGAVKILQQLFPEPG---------- 89
Query: 147 LTGKSIELYIGDICEFEFLSETFKSYEPDAVVHF 180
++ D+ + +++ F DAV+HF
Sbjct: 90 ----KLQFIYADLGDANAVNKIFSENAFDAVMHF 119
>At2g27860 putative dTDP-glucose 4-6-dehydratase
Length = 389
Score = 35.0 bits (79), Expect = 0.091
Identities = 36/124 (29%), Positives = 55/124 (44%), Gaps = 14/124 (11%)
Query: 335 PLTVYGKGGQTRAFLDIRDTVQCVELAIANP--ANPGEFRVFNQFTEQFKVTELAELVTK 392
PL + G R F+ I D ++ V L I NP AN F V N + V +LAE++T+
Sbjct: 249 PLKLVDGGESQRTFVYINDAIEAVLLMIENPERANGHIFNVGNP-NNEVTVRQLAEMMTE 307
Query: 393 AGEKLGLDVKTISVPNPRVELEEHY---YNCKNTKLVD-------LGLKPHFLSDSLIDS 442
K+ + I P V +E Y Y+ + ++ D LG P L++S
Sbjct: 308 VYAKVSGE-GAIESPTVDVSSKEFYGEGYDDSDKRIPDMTIINRQLGWNPKTSLWDLLES 366
Query: 443 LLNF 446
L +
Sbjct: 367 TLTY 370
>At1g02000 nucleotide sugar epimerase, putative
Length = 434
Score = 34.7 bits (78), Expect = 0.12
Identities = 48/212 (22%), Positives = 75/212 (34%), Gaps = 31/212 (14%)
Query: 87 VMVIGGDGYCGWATALHLSNKGYEVAIVDNLVRRLFDHQLGLDSLTPISSIQDRIQCWKS 146
V+V G G+ G + L +G D LGLD+ +
Sbjct: 94 VLVTGAAGFVGTHVSAALKRRG--------------DGVLGLDNFNDYYDTSLKRSRQAL 139
Query: 147 LTGKSIELYIGDICEFEFLSETFKSYEPDAVVHFGEQRSAPYSMIDRSRAVYTQQNNVVG 206
L + + GDI + L + F+ V+H Q Y+M + V+ +N+ G
Sbjct: 140 LERSGVFIVEGDINDLSLLKKLFEVVPFTHVMHLAAQAGVRYAMENPGSYVH---SNIAG 196
Query: 207 TLNVLFAIKEYREDCHLVKLGTMGEYGTPNIDIEEGYITITHNGRTDTLPYPKQASSFYH 266
+N+L K +V + YG + + RTD Q +S Y
Sbjct: 197 FVNLLEVCKSANPQPAIVWASSSSVYGL------NTKVPFSEKDRTD------QPASLYA 244
Query: 267 LSKVHDSHNIAFTCK-AWGIRATDLNQGVVYG 297
+K IA T +G+ T L VYG
Sbjct: 245 ATK-KAGEEIAHTYNHIYGLSLTGLRFFTVYG 275
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.319 0.135 0.404
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 10,681,504
Number of Sequences: 26719
Number of extensions: 454003
Number of successful extensions: 1044
Number of sequences better than 10.0: 31
Number of HSP's better than 10.0 without gapping: 20
Number of HSP's successfully gapped in prelim test: 11
Number of HSP's that attempted gapping in prelim test: 1000
Number of HSP's gapped (non-prelim): 45
length of query: 477
length of database: 11,318,596
effective HSP length: 103
effective length of query: 374
effective length of database: 8,566,539
effective search space: 3203885586
effective search space used: 3203885586
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.8 bits)
S2: 62 (28.5 bits)
Medicago: description of AC125478.9


