

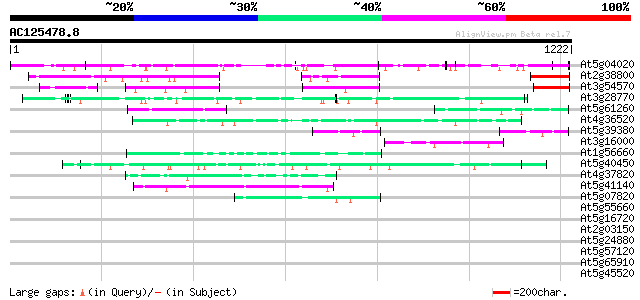
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC125478.8 + phase: 0
(1222 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
At5g04020 pathogen-induced calmodulin-binding protein (PICBP) 226 6e-59
At2g38800 unknown protein 153 7e-37
At3g54570 putative protein 92 2e-18
At3g28770 hypothetical protein 83 1e-15
At5g61260 putative protein 54 5e-07
At4g36520 trichohyalin like protein 52 2e-06
At5g39380 putative protein 49 2e-05
At3g16000 myosin heavy chain like protein 48 3e-05
At1g56660 hypothetical protein 47 5e-05
At5g40450 unknown protein 47 7e-05
At4g37820 unknown protein 46 1e-04
At5g41140 putative protein 44 4e-04
At5g07820 putative protein 44 6e-04
At5g55660 putative protein 43 0.001
At5g16720 putative protein 42 0.002
At2g03150 Unknown protein (T18E12.18; At2g03150) 42 0.002
At5g24880 glutamic acid-rich protein 42 0.003
At5g57120 unknown protein 41 0.004
At5g65910 unknown protein 39 0.014
At5g45520 putative protein 39 0.018
>At5g04020 pathogen-induced calmodulin-binding protein (PICBP)
Length = 1495
Score = 226 bits (576), Expect = 6e-59
Identities = 335/1319 (25%), Positives = 543/1319 (40%), Gaps = 231/1319 (17%)
Query: 1 MKSSRSIKLLTVKGPKSTTKLYSESTDG--IDGNNRNSTSDAGNKSQRVMTRRLSLKPVR 58
+K RS+K LT + K + D + + TS + +K+ + L ++
Sbjct: 139 LKKMRSMKRLTSNSRQILRKKNLDRGDFGLLQPHYLRPTSSSASKNVENNQKNLGAARLK 198
Query: 59 --ISAKKPSLHKATCSSTIKDSHFPNHIDLPQEGSSSQGVSAVKVCTYAYCSLHGHHHGD 116
S + L KATCSS +K GSSS+ + VCTY YCSLHG H
Sbjct: 199 RIASLRYNGLLKATCSSAMK-------------GSSSKRSN--DVCTYRYCSLHGRRHSH 243
Query: 117 -------LPPLKRFVSMRRRQLKSQKSTKK---------DGRSKQVGNARKGTQKTKTVH 160
+P LKRFVSMRR+ + QKS + + +G Q++K V
Sbjct: 244 AADNNAGVPSLKRFVSMRRKFMNRQKSVNRRLVLLKRTLSRKRGPLGGRVVTDQESKEVD 303
Query: 161 SE-DGNSQQNVKNVSMESSPFKPHDAPPSTVNECDTSTKDKHMVTDYEVLQKSSTQEEPK 219
DG+S + V + SS +D NE + + MV + + + E
Sbjct: 304 DNVDGDSDEEVFEEEVSSSVDGGND------NESIGRSSETVMVDVDDNVDRGMDAME-- 355
Query: 220 PGSTTSVAYGVQERDQKYIKK--WHLMYKHAVLSNT---GKCDNKVPLVEKEKEGGEEDN 274
+VA VQE + + W + + V + GK D +G DN
Sbjct: 356 -----TVASKVQESKTETVGATLWRAICEQTVTGHDHDDGKVDGTT------SDGTVGDN 404
Query: 275 EGNNSYRNYSETDSDMDDEKKN------VIELVQKAFDEILLPEVEDLSSEGHSKSRGNE 328
E R S + +D KK + LV++AFDEIL +D SS+ S ++
Sbjct: 405 E--EVCREGSSGELREEDGKKTEYVWNETVTLVKQAFDEILAEITDDDSSDDISMTKDEA 462
Query: 329 TDEVLLEKSGGK------------IEERNT----TTFTESPKEVPKMESKQKSWSHLKKV 372
+ L E+ G + ER+T T ++ + K WS+LK+V
Sbjct: 463 LEVGLGEEDVGADSSDSSCSDMQPVIERDTHLSVIASTFHMRDEFGHQRGPKKWSYLKRV 522
Query: 373 ILLKRFVKALEKVRNINSRRPRQLPSDANFEAEKVLLNRQTSEERKKSEEWMLDYALQKV 432
ILLKRF+K+L+ R+ R+ SD + L R+ ERK +EEWMLD+AL++V
Sbjct: 523 ILLKRFLKSLD-------RKERRKLSDGKESETIMRLRRELVGERKNAEEWMLDHALRQV 575
Query: 433 ISKLAPAQRQRVTLLVEAFETIRPVQDAENG--------PQTSATVESHANLI--QSLDA 482
IS LAP+Q+++V LV+AFE++ P+ G + + TV S I + DA
Sbjct: 576 ISTLAPSQKKKVKHLVKAFESLIPMDGGSRGHDDLVSPAREENETVNSQTQTILRDNKDA 635
Query: 483 SSNHSKEEINDRRDFEVTERARNDKNMDACKKNDESATVKSTATKAVKFPVCDTGIMEEE 542
+ D + +T A + ++D K ++E+ TV S T + E
Sbjct: 636 TDILEVSPAKDLEETNLTCEASSFLSID-MKSDEENETVNS-----------PTIWRDNE 683
Query: 543 VTAEGEYKVQEKSIVKEDLKHGTSTTDVPYGVQERDQKYIKKWHLMYKQAVLSNTGKYDN 602
T + V K + + +L +S++ + + L AV
Sbjct: 684 DTTDLLEVVPAKDLEETNLTSESSSSLCIGMKSDEALESTADASLCNHLAVEEEVDGLAL 743
Query: 603 KLPVVGKDKEGREQGDAVFNGGN---------NSSCHNYNETDSD------------MDE 641
+ ++K+G + + N ++S N NET+ + +D+
Sbjct: 744 GSFIEEEEKKGESEKQNLSTWRNLIQKHMVMRDNSEGNRNETEQEHKWSYGTDQMTGIDD 803
Query: 642 EKKNVIELVQKAFDEILLPETEDLSSDDRSKSRSYGSDELLEKSEGEREEMNATSFTETP 701
++ +Q AF E +L E D SSD+ S S S S+ L E+ E + E
Sbjct: 804 ANAAAVKSIQLAF-ETILSEIPDSSSDEESVSES--SNSLKEEKEHQGE----------- 849
Query: 702 KEAKKTENKPKSWSHLKKLIMLKRFVKALDKVRNINPRRPRELPSDANFEGEKVFLNRQT 761
+SW+ L+K+I+LKRFVK+L+KV+ NPR+ R LP ++ FE E VFL ++
Sbjct: 850 --------TKRSWNSLRKVILLKRFVKSLEKVQVPNPRKMRNLPVESAFEAENVFLRHRS 901
Query: 762 SEE--RKKSEEWMLDYALQKVISKLAPAQRQRVTLLIEAFETLRPIQDAENGLRSSAT-- 817
E R + EE MLDYAL++ IS+LAP QR++V LL++AF+ + D ++S T
Sbjct: 902 IMEGTRTEGEEMMLDYALRQAISRLAPIQRKKVDLLVQAFDIVLDGHDTPKQTKNSDTPR 961
Query: 818 -----------------VESLENPLQSLDASSVLSAKTLLGKVSFSNDSTMEFSDKASDN 860
V E ++++ A + K L G+ N + K S N
Sbjct: 962 NNDETKEGKPRVEEGCEVNKDEQKIKNVFARFQVHQKDLKGEEEVHN------TPKESRN 1015
Query: 861 --PMPELCKPIKPVETISSCHEEAPTKRMV------DEVPEDLVSDLNTKTKDVIGGHGE 912
P+ + I + S + K MV D + V+ + ++ D G G
Sbjct: 1016 LPPIRNFKQRIVVEKGKDSRMWKLIYKHMVTEKEGIDSANAESVASVESEYDDEAG--GL 1073
Query: 913 QFSVTKSLILNGIVRSLRSNL--VVPEAPSNRLDEPTTDIKDVVEKDQL---------EK 961
Q +S G V +R L ++ E P N D+ + D D+ + +L EK
Sbjct: 1074 QIDARRS----GTVTLVREALEKILSEIPDNSSDDQSMD-SDITTEQELFERNSQVSEEK 1128
Query: 962 SEAPTSAVVESKNQLEKQGSTGLWFTV-FKHMVSD---MTENNSKTSTDVADEKDSKYED 1017
SE ++ + +G + + K VSD +T + KT + E D + E
Sbjct: 1129 SEVSSATFKPKFTEKRVKGWNNVKKVILLKRFVSDLGSLTRLSPKTPRVLPWEPDPETEK 1188
Query: 1018 ITTREISV-SYENTPVVIQDMPFKDRAVVDAEVELRQIEAIKMVEDAID----------S 1066
I R + N+ + D + A + R++ + D I
Sbjct: 1189 IRLRHQEIGGKRNSEEWMLDYALRQAISTLAPSQKRKVSLLAQAFDTISLQDMGSGSTPG 1248
Query: 1067 ILPDTQPLPDNSTIDRTGGIYSEGLNQKEQKMESGN---GIVEERKEESVSKEVNKPNQK 1123
++ + S+I Y N + + + N + E K + VSK++ + Q+
Sbjct: 1249 SAASSRNISRQSSISSMAAHYENEANAEIIRGKLRNLQEDLKESAKLDGVSKDLEE-KQQ 1307
Query: 1124 LSRNWSNLKKVVLLRRFIKAL-EKVRKFNPREPRYLPLEPDSEDEKVQLRHQDMAERKG 1181
S W L K + + L E+ RK E L + + EK++L + E G
Sbjct: 1308 CSSLWRILCKQMEDNEKNQTLPEETRK--EEEEEELKEDTSVDGEKMELYQTEAVELLG 1364
Score = 209 bits (531), Expect = 1e-53
Identities = 186/660 (28%), Positives = 300/660 (45%), Gaps = 140/660 (21%)
Query: 165 NSQQNVKNVSMESSPFKPHDAPPSTVNECDTST---KDKHMVTDYEVLQKSSTQEE---- 217
++ + KN + + + P C+ + K K++ ++V QK EE
Sbjct: 949 DTPKQTKNSDTPRNNDETKEGKPRVEEGCEVNKDEQKIKNVFARFQVHQKDLKGEEEVHN 1008
Query: 218 -PKPGSTTSVAYGVQER----DQKYIKKWHLMYKHAVLSNTGKCDNKVPLVEKEKEGGEE 272
PK ++R K + W L+YKH V EKEG +
Sbjct: 1009 TPKESRNLPPIRNFKQRIVVEKGKDSRMWKLIYKHMVT---------------EKEGIDS 1053
Query: 273 DNEGNNSYRNYSETDSDMDDE---------KKNVIELVQKAFDEILLPEVEDLSSEGHSK 323
N + + +S+ DDE + + LV++A ++IL E+ D SS+ S
Sbjct: 1054 ANA-----ESVASVESEYDDEAGGLQIDARRSGTVTLVREALEKIL-SEIPDNSSDDQSM 1107
Query: 324 SRGNETDEVLLEKSGGKIEERN-TTTFTESPKEVPKMESKQKSWSHLKKVILLKRFVKAL 382
T++ L E++ EE++ ++ T PK E + K W+++KKVILLKRFV L
Sbjct: 1108 DSDITTEQELFERNSQVSEEKSEVSSATFKPKFT---EKRVKGWNNVKKVILLKRFVSDL 1164
Query: 383 EKVRNINSRRPRQLPSDANFEAEKVLLNRQTSEERKKSEEWMLDYALQKVISKLAPAQRQ 442
+ ++ + PR LP + + E EK+ L Q ++ SEEWMLDYAL++ IS LAP+Q++
Sbjct: 1165 GSLTRLSPKTPRVLPWEPDPETEKIRLRHQEIGGKRNSEEWMLDYALRQAISTLAPSQKR 1224
Query: 443 RVTLLVEAFETIRPVQDAENGPQTSATVESHANLIQSLDASSNHSKEEINDRRDFEVTER 502
+V+LL +AF+TI +QD +G T + S N+ + SS + E N+ + +
Sbjct: 1225 KVSLLAQAFDTIS-LQDMGSG-STPGSAASSRNISRQSSISSMAAHYE-NEANAEIIRGK 1281
Query: 503 ARNDKNMDACKKNDESATVKSTATKAVKFPVCDTGIMEEEVTAEGEYKVQEKSIVKEDLK 562
RN ++EDLK
Sbjct: 1282 LRN---------------------------------------------------LQEDLK 1290
Query: 563 HGTSTTDVPYGVQERDQKYIKKWHLMYKQAVLSNTGKYDNKLPVVGKDKEGREQGDAVFN 622
V ++E+ Q+ W ++ KQ + + LP + +E E+
Sbjct: 1291 ESAKLDGVSKDLEEK-QQCSSLWRILCKQM---EDNEKNQTLPEETRKEEEEEE------ 1340
Query: 623 GGNNSSCHNYNETDSDMDEEKKNVIELVQKAFDEILLPETEDLSSDDRSKSRSYGSDELL 682
+ D+ +D EK +EL Q E LL E D S + S+ ++ ++E
Sbjct: 1341 ----------LKEDTSVDGEK---MELYQTEAVE-LLGEVIDGISLEESQDQNLNNEETR 1386
Query: 683 EKSEGEREEMNATSFTETPKEAKKTENKPKSWSHLKKLIMLKRFVKALDKVRNINPRRPR 742
+KSE + ++ + WS+LK+ I+L+RFVKAL+ VR NPR PR
Sbjct: 1387 QKSE----------------TLQVSKVRIDRWSNLKRAILLRRFVKALENVRKFNPREPR 1430
Query: 743 ELPSDANFEGEKVFLNRQTSEERKKSEEWMLDYALQKVISKLAPAQRQRVTLLIEAFETL 802
LP + E EKV L Q ++ +K +EWM+D ALQ V+SKL PA++ +V LL++AFE+L
Sbjct: 1431 FLPPNPEVEAEKVNLRHQETQNKKNGDEWMVDNALQGVVSKLTPARKLKVQLLVQAFESL 1490
Score = 191 bits (485), Expect = 2e-48
Identities = 177/632 (28%), Positives = 283/632 (44%), Gaps = 125/632 (19%)
Query: 623 GGNNSSCHNYNETDSDMDEEKKN------VIELVQKAFDEILLPETEDLSSDDRSKSRSY 676
G N C + + ++ KK + LV++AFDEIL T+D SSDD S ++
Sbjct: 402 GDNEEVCREGSSGELREEDGKKTEYVWNETVTLVKQAFDEILAEITDDDSSDDISMTK-- 459
Query: 677 GSDELLEKSEGEREEMNATSFTETPKEAKKT----------------------ENKPKSW 714
DE LE GE E++ A S + + + + PK W
Sbjct: 460 --DEALEVGLGE-EDVGADSSDSSCSDMQPVIERDTHLSVIASTFHMRDEFGHQRGPKKW 516
Query: 715 SHLKKLIMLKRFVKALDKVRNINPRRPRELPSDANFEGEKVFLNRQTSEERKKSEEWMLD 774
S+LK++I+LKRF+K+LD R+ R SD + L R+ ERK +EEWMLD
Sbjct: 517 SYLKRVILLKRFLKSLD-------RKERRKLSDGKESETIMRLRRELVGERKNAEEWMLD 569
Query: 775 YALQKVISKLAPAQRQRVTLLIEAFETLRPIQDAENGLRSSATVESLENPLQSLDASSVL 834
+AL++VIS LAP+Q+++V L++AFE+L P+ G + EN + ++L
Sbjct: 570 HALRQVISTLAPSQKKKVKHLVKAFESLIPMDGGSRGHDDLVSPAREENETVNSQTQTIL 629
Query: 835 SAKTLLGKVSFSNDSTMEFSDKASDNPMPELCKPIKPVETISSCHEEAPTKRMVDEVPED 894
+ D+T D +P +L + E S + + + V
Sbjct: 630 RD---------NKDAT----DILEVSPAKDLEETNLTCEASSFLSIDMKSDEENETVNSP 676
Query: 895 LVSDLNTKTKDVIGGHGEQFSVTKSLILNGIVRSLRSNLVVPEAPSNRLDEPTTDIKDVV 954
+ N T D++ + K L + S+L + + DE D
Sbjct: 677 TIWRDNEDTTDLL-----EVVPAKDLEETNLTSESSSSLCI----GMKSDEALESTADAS 727
Query: 955 EKDQLEKSEAPTSAVV-------ESKNQLEKQGSTGLWFTVFKHMVSDMTENNSKTSTDV 1007
+ L E + E K + EKQ + + KHMV ++ T
Sbjct: 728 LCNHLAVEEEVDGLALGSFIEEEEKKGESEKQNLSTWRNLIQKHMVMRDNSEGNRNET-- 785
Query: 1008 ADEKDSKYEDITTREISVSYENTPVVIQDMPFKDRAVVDAEVELRQIEAIKMVEDAIDSI 1067
E++ K+ T + + N A+K ++ A ++I
Sbjct: 786 --EQEHKWSYGTDQMTGIDDANAA------------------------AVKSIQLAFETI 819
Query: 1068 LPDTQPLPDNSTIDRTGGIYSEGLNQKEQKMESGNGIVEERKEESVSKEVNKPNQKLSRN 1127
L + +PD+S+ +E ES N + EE++ + +K R+
Sbjct: 820 LSE---IPDSSS-------------DEESVSESSNSLKEEKEHQGETK----------RS 853
Query: 1128 WSNLKKVVLLRRFIKALEKVRKFNPREPRYLPLEPDSEDEKVQLRHQDMAE--RKGTEEW 1185
W++L+KV+LL+RF+K+LEKV+ NPR+ R LP+E E E V LRH+ + E R EE
Sbjct: 854 WNSLRKVILLKRFVKSLEKVQVPNPRKMRNLPVESAFEAENVFLRHRSIMEGTRTEGEEM 913
Query: 1186 MLDYALRQVVSKLTPARKRKVELLVEAFETVV 1217
MLDYALRQ +S+L P +++KV+LLV+AF+ V+
Sbjct: 914 MLDYALRQAISRLAPIQRKKVDLLVQAFDIVL 945
Score = 143 bits (361), Expect = 5e-34
Identities = 81/266 (30%), Positives = 143/266 (53%), Gaps = 32/266 (12%)
Query: 951 KDVVEKDQLEKSEAPTSAVVESKNQLEKQGSTGLWFTVFKHMVSDMTENNSKTSTDVADE 1010
++ V E P + + +EK + +W ++KHMV++ +S + VA
Sbjct: 1003 EEEVHNTPKESRNLPPIRNFKQRIVVEKGKDSRMWKLIYKHMVTEKEGIDSANAESVASV 1062
Query: 1011 KDSKYEDITTREISVSYENTPVVIQDMPFKDRAVVDAEVELRQIEAIKMVEDAIDSILPD 1070
+S+Y+D +++ R+ + +V +A++ IL +
Sbjct: 1063 -ESEYDD-------------------------EAGGLQIDARRSGTVTLVREALEKILSE 1096
Query: 1071 TQPLPDNSTIDRTGGIYSEGLNQKEQKMESGNGIVEERKEESVSKEVNKPNQKLSRNWSN 1130
+PDNS+ D++ + +++ E + + EE+ E S + K +K + W+N
Sbjct: 1097 ---IPDNSSDDQS---MDSDITTEQELFERNSQVSEEKSEVSSATFKPKFTEKRVKGWNN 1150
Query: 1131 LKKVVLLRRFIKALEKVRKFNPREPRYLPLEPDSEDEKVQLRHQDMAERKGTEEWMLDYA 1190
+KKV+LL+RF+ L + + +P+ PR LP EPD E EK++LRHQ++ ++ +EEWMLDYA
Sbjct: 1151 VKKVILLKRFVSDLGSLTRLSPKTPRVLPWEPDPETEKIRLRHQEIGGKRNSEEWMLDYA 1210
Query: 1191 LRQVVSKLTPARKRKVELLVEAFETV 1216
LRQ +S L P++KRKV LL +AF+T+
Sbjct: 1211 LRQAISTLAPSQKRKVSLLAQAFDTI 1236
Score = 142 bits (358), Expect = 1e-33
Identities = 94/248 (37%), Positives = 138/248 (54%), Gaps = 53/248 (21%)
Query: 972 SKNQLEKQGSTGLWFTVFKHMVSDMTENNSKTSTDVADEKDSKYEDITTREISVSYENTP 1031
SK+ EKQ + LW + K M E+N K T + + + E+ + SV E
Sbjct: 1299 SKDLEEKQQCSSLWRILCKQM-----EDNEKNQTLPEETRKEEEEEELKEDTSVDGE--- 1350
Query: 1032 VVIQDMPFKDRAVVDAEVELRQIEAIKMVEDAIDSILPDTQPLPDNSTIDRTGGIYSEGL 1091
++EL Q EA++++ + ID G+
Sbjct: 1351 ----------------KMELYQTEAVELLGEVID------------------------GI 1370
Query: 1092 NQKEQKMESGNGIVEERKEESVSKEVNKPNQKLSRNWSNLKKVVLLRRFIKALEKVRKFN 1151
+ +E + ++ N EE +++S + +V+K ++ R WSNLK+ +LLRRF+KALE VRKFN
Sbjct: 1371 SLEESQDQNLNN--EETRQKSETLQVSKV--RIDR-WSNLKRAILLRRFVKALENVRKFN 1425
Query: 1152 PREPRYLPLEPDSEDEKVQLRHQDMAERKGTEEWMLDYALRQVVSKLTPARKRKVELLVE 1211
PREPR+LP P+ E EKV LRHQ+ +K +EWM+D AL+ VVSKLTPARK KV+LLV+
Sbjct: 1426 PREPRFLPPNPEVEAEKVNLRHQETQNKKNGDEWMVDNALQGVVSKLTPARKLKVQLLVQ 1485
Query: 1212 AFETVVPT 1219
AFE++ T
Sbjct: 1486 AFESLSAT 1493
Score = 90.9 bits (224), Expect = 4e-18
Identities = 84/283 (29%), Positives = 139/283 (48%), Gaps = 38/283 (13%)
Query: 949 DIKDVVEKDQLEKSEAPTSAVVESKNQLEKQGSTGLWFTVFKHMVSDMTENNSKTSTDVA 1008
D+ D V++ ++ E S V ESK E G+T LW + + V+ ++ K
Sbjct: 342 DVDDNVDRG-MDAMETVASKVQESKT--ETVGAT-LWRAICEQTVTGHDHDDGKV----- 392
Query: 1009 DEKDSKYEDITTREISVSYENTPVVIQDMPFKDRAVVDAEVELRQIEAIKMVEDAIDSIL 1068
D TT + +V +N V + + R + E E + +V+ A D IL
Sbjct: 393 --------DGTTSDGTVG-DNEEVCREGSSGELREEDGKKTEYVWNETVTLVKQAFDEIL 443
Query: 1069 PDTQPLPDNSTIDRTGGIYSE-GLNQKEQKMESGNG-------IVEERKEESVSKEV--- 1117
+ + I T E GL +++ +S + ++E SV
Sbjct: 444 AEITDDDSSDDISMTKDEALEVGLGEEDVGADSSDSSCSDMQPVIERDTHLSVIASTFHM 503
Query: 1118 --NKPNQKLSRNWSNLKKVVLLRRFIKALEKVRKFNPREPRYLPLEPDSEDEKVQLRHQD 1175
+Q+ + WS LK+V+LL+RF+K+L++ +E R L +SE ++LR +
Sbjct: 504 RDEFGHQRGPKKWSYLKRVILLKRFLKSLDR------KERRKLSDGKESETI-MRLRREL 556
Query: 1176 MAERKGTEEWMLDYALRQVVSKLTPARKRKVELLVEAFETVVP 1218
+ ERK EEWMLD+ALRQV+S L P++K+KV+ LV+AFE+++P
Sbjct: 557 VGERKNAEEWMLDHALRQVISTLAPSQKKKVKHLVKAFESLIP 599
Score = 30.8 bits (68), Expect = 5.0
Identities = 25/69 (36%), Positives = 36/69 (51%), Gaps = 11/69 (15%)
Query: 661 ETEDLSSDDRSK-SRSYGSDELLEKSEGEREEMNATSFT---ETPKEAKKTENKPKSWSH 716
E+ED SS + S SR+Y KS E EE++ S T E P+ K+ K KS S
Sbjct: 65 ESEDSSSSEMSDDSRTYS------KSSDENEELDQASLTICSERPRSVKR-RAKSKSISR 117
Query: 717 LKKLIMLKR 725
+ + +L+R
Sbjct: 118 ISSMKVLRR 126
>At2g38800 unknown protein
Length = 612
Score = 153 bits (386), Expect = 7e-37
Identities = 136/480 (28%), Positives = 218/480 (45%), Gaps = 73/480 (15%)
Query: 42 NKSQRVMTRRLSLKPVRISAKKPSLHKATCSSTIKDSHFPNHIDLPQEGSSSQ--GVSAV 99
NKS R L+ P+ K +ATCSST+KDS FP ++ L + Q G S +
Sbjct: 122 NKSSSRNGRGLTKAPIF----KRCSQRATCSSTLKDSKFPEYLMLNHGETFDQVNGTSVL 177
Query: 100 KVCTYAYCSLHGH-HHGDLPPLKRFVSMRRRQLKSQKSTKKDGRSKQVGNARKGTQKTKT 158
KVC Y YCSL+GH H PPLK F+S+RR+ LKSQKS K + ++ +K +
Sbjct: 178 KVCPYTYCSLNGHLHAAQYPPLKSFISLRRQSLKSQKSVKMEASEEEFVKMDDLEEKKEF 237
Query: 159 VHSEDGNSQQNVKNVSMESSPFKPHDAPPSTVNECDTSTKDKHMVTDY------EVLQKS 212
+ G+ + ++ + E+ + P + + D + MV + E L
Sbjct: 238 ENGNGGSCEVDIDSQISET----VSEGAPRSETDSDDYSDSAEMVIELKESCLEETLVDD 293
Query: 213 STQEEPKPGSTTSVAYGVQERDQKYIKKWHLM-----YKHAVLSNTGKCDNKVPLVEKEK 267
S E + + Y ++E D + M + ++G D++V + K
Sbjct: 294 SENEVQEKANRDGDTYLLKESDLEETLVEDSMNQDEGNRDGDADHSGCFDSEVIGIIKNS 353
Query: 268 EGGEEDNEG--NNSYRNYSET-------------DSDMDDEKKNV-------IELVQKAF 305
E +E ++S +++ +T +S++ D KN LV ++
Sbjct: 354 EADIVIDETLIDDSVKDFEDTTNIYGDANEFGCFNSEVIDMMKNTEADTAIGETLVDESM 413
Query: 306 DEILLPEVEDLSSEGHSKSRGNETDEVLLEKSGGKIEERNTT------------------ 347
EI E +D ++ S D + + IE+++ T
Sbjct: 414 KEIQEKENKDEDADESSCFVSELIDMIKNSAASDAIEDKDATGEETLKDKAEDCKEVSKG 473
Query: 348 -----TFTESPKEVP-----KMESKQKSWSHLK-KVILLKRFVKALEKVRNINSRRPRQL 396
TE +VP K +++S S + +I K+ V E +R N R P L
Sbjct: 474 QTEVILMTEENAKVPFNRTRKPCKQEESGSTISWTIIKCKKPVAETEDLREFNPREPNYL 533
Query: 397 PSDANFEAEKVLLNRQTSEERKKSEEWMLDYALQKVISKLAPAQRQRVTLLVEAFETIRP 456
P+ + +AEKV L Q +ER+ SE+WM DYALQ+ +SKLAPA++++V LLVEAFET++P
Sbjct: 534 PAVMDEDAEKVDLKHQDIDERRNSEDWMFDYALQRAVSKLAPARKRKVALLVEAFETVQP 593
Score = 105 bits (263), Expect = 1e-22
Identities = 49/85 (57%), Positives = 64/85 (74%)
Query: 1134 VVLLRRFIKALEKVRKFNPREPRYLPLEPDSEDEKVQLRHQDMAERKGTEEWMLDYALRQ 1193
++ ++ + E +R+FNPREP YLP D + EKV L+HQD+ ER+ +E+WM DYAL++
Sbjct: 509 IIKCKKPVAETEDLREFNPREPNYLPAVMDEDAEKVDLKHQDIDERRNSEDWMFDYALQR 568
Query: 1194 VVSKLTPARKRKVELLVEAFETVVP 1218
VSKL PARKRKV LLVEAFETV P
Sbjct: 569 AVSKLAPARKRKVALLVEAFETVQP 593
Score = 100 bits (248), Expect = 7e-21
Identities = 68/181 (37%), Positives = 101/181 (55%), Gaps = 22/181 (12%)
Query: 636 DSDMDEEKKNVIELVQK-----AFDEILLPETEDLSSDDRSKSRSYGSDELLEKSEGERE 690
D D DE V EL+ A D I E +D + ++ K ++ ++ E S+G+ E
Sbjct: 423 DEDADESSCFVSELIDMIKNSAASDAI---EDKDATGEETLKDKA---EDCKEVSKGQTE 476
Query: 691 -----EMNA-TSFTETPKEAKKTENKPK-SWSHLKKLIMLKRFVKALDKVRNINPRRPRE 743
E NA F T K K+ E+ SW+ +I K+ V + +R NPR P
Sbjct: 477 VILMTEENAKVPFNRTRKPCKQEESGSTISWT----IIKCKKPVAETEDLREFNPREPNY 532
Query: 744 LPSDANFEGEKVFLNRQTSEERKKSEEWMLDYALQKVISKLAPAQRQRVTLLIEAFETLR 803
LP+ + + EKV L Q +ER+ SE+WM DYALQ+ +SKLAPA++++V LL+EAFET++
Sbjct: 533 LPAVMDEDAEKVDLKHQDIDERRNSEDWMFDYALQRAVSKLAPARKRKVALLVEAFETVQ 592
Query: 804 P 804
P
Sbjct: 593 P 593
>At3g54570 putative protein
Length = 417
Score = 92.0 bits (227), Expect = 2e-18
Identities = 44/77 (57%), Positives = 53/77 (68%)
Query: 1142 KALEKVRKFNPREPRYLPLEPDSEDEKVQLRHQDMAERKGTEEWMLDYALRQVVSKLTPA 1201
+ +E R+ NPREP Y+ + +E V LRHQDM ERK EEWM+DYAL+ VSKL
Sbjct: 340 ECIEDCRRLNPREPNYIQTTVEPSNETVDLRHQDMDERKKAEEWMIDYALQHTVSKLVVE 399
Query: 1202 RKRKVELLVEAFETVVP 1218
RK+ V LLVEAFET VP
Sbjct: 400 RKKDVALLVEAFETTVP 416
Score = 82.8 bits (203), Expect = 1e-15
Identities = 69/236 (29%), Positives = 106/236 (44%), Gaps = 33/236 (13%)
Query: 253 TGKCDNKVPLV----EKEKEGGEE------DNEGNNSYRNYSETDSDMDDEKKNVIELVQ 302
+G+CD+ + +KE E D+E N E D+D E ++ L++
Sbjct: 182 SGECDDGCVEIYVDEKKENRSTRETDIKVIDSEAENIEMELGEV-KDLDSESAEIVSLLE 240
Query: 303 KAFDE---ILLPEVEDLSSEGHSKSRG----NETDEVLLEKS--------GGKIEERNTT 347
E + E + SSE + G N T+ +L E+S G ++E++ +
Sbjct: 241 GEGIESCNFAVLEQSENSSEDQEREEGGFSNNTTNSLLFEQSIIQDDIILGNAVDEKHES 300
Query: 348 TFTESPKEVPKMESKQKSWSHLKKVILLKRFVK-------ALEKVRNINSRRPRQLPSDA 400
E KE + K++ LK L + +E R +N R P + +
Sbjct: 301 KEAEDWKEADGEKVKERIKLVLKTEEALLSLAQKPCNREECIEDCRRLNPREPNYIQTTV 360
Query: 401 NFEAEKVLLNRQTSEERKKSEEWMLDYALQKVISKLAPAQRQRVTLLVEAFETIRP 456
E V L Q +ERKK+EEWM+DYALQ +SKL +++ V LLVEAFET P
Sbjct: 361 EPSNETVDLRHQDMDERKKAEEWMIDYALQHTVSKLVVERKKDVALLVEAFETTVP 416
Score = 74.7 bits (182), Expect = 3e-13
Identities = 54/190 (28%), Positives = 94/190 (49%), Gaps = 23/190 (12%)
Query: 638 DMDEEKKNVIELVQ----KAFDEILLPETEDLSSDDRSKSRSYGSDE----LLEKSEGER 689
D+D E ++ L++ ++ + +L ++E+ S D + + ++ L E+S +
Sbjct: 227 DLDSESAEIVSLLEGEGIESCNFAVLEQSENSSEDQEREEGGFSNNTTNSLLFEQSIIQD 286
Query: 690 EEM--NATSFTETPKEAK--KTENKPKSWSHLKKLIMLKRFVKAL-----------DKVR 734
+ + NA KEA+ K + K +K ++ + + +L + R
Sbjct: 287 DIILGNAVDEKHESKEAEDWKEADGEKVKERIKLVLKTEEALLSLAQKPCNREECIEDCR 346
Query: 735 NINPRRPRELPSDANFEGEKVFLNRQTSEERKKSEEWMLDYALQKVISKLAPAQRQRVTL 794
+NPR P + + E V L Q +ERKK+EEWM+DYALQ +SKL +++ V L
Sbjct: 347 RLNPREPNYIQTTVEPSNETVDLRHQDMDERKKAEEWMIDYALQHTVSKLVVERKKDVAL 406
Query: 795 LIEAFETLRP 804
L+EAFET P
Sbjct: 407 LVEAFETTVP 416
Score = 65.5 bits (158), Expect = 2e-10
Identities = 41/127 (32%), Positives = 64/127 (50%), Gaps = 10/127 (7%)
Query: 65 SLHKATCSSTIKDSHFPNHIDLPQEGSSSQGVSAVKVCTYAYCSLHGHHHGDLPPLKRFV 124
++H+ATCSS +K+S F + +KVC Y YCSL+ H H PPL F+
Sbjct: 118 NVHRATCSSLLKNSKFTEDLMFTSP-------HILKVCPYTYCSLNAHLHSQFPPLLSFI 170
Query: 125 SMRRRQLKSQKSTK-KDGRSKQVGNARKGTQKTKTVHSEDGNSQQNVKNVSMESSPFKPH 183
S RRR LKS S + DG + + +K + T+ + +S+ +N+ ME K
Sbjct: 171 SERRRSLKSHASGECDDGCVEIYVDEKKENRSTRETDIKVIDSE--AENIEMELGEVKDL 228
Query: 184 DAPPSTV 190
D+ + +
Sbjct: 229 DSESAEI 235
>At3g28770 hypothetical protein
Length = 2081
Score = 82.8 bits (203), Expect = 1e-15
Identities = 132/688 (19%), Positives = 273/688 (39%), Gaps = 80/688 (11%)
Query: 28 GIDGNNRNSTSDAGNKSQRVMTRRLSLKPVRISAKKPSLHKATCSSTIKDSHFPNHIDLP 87
G+D N N K R + + + K + K+ + + SST + F N++D+
Sbjct: 847 GVDTNVGNKEDSKDLKDDRSVEVKAN-KEESMKKKREEVQRNDKSSTKEVRDFANNMDID 905
Query: 88 QEGSSSQGVSAVKVCTYAYCSLHGHHHGDLPPLKRFVSMRRRQLKSQKSTKKDGRSKQVG 147
+ S + V K G+ K ++ +Q K + KK SK
Sbjct: 906 VQKGSGESVKYKK---------DEKKEGNKEENKDTINTSSKQ-KGKDKKKKKKESKNSN 955
Query: 148 NARKGTQKTKTVHSEDGNSQQNVKNVSM-ESSPFKPHDAPPSTVNECDTSTKDKHMVTDY 206
+K K + V++E + N K + E+S K + E + S +Y
Sbjct: 956 MKKKEEDKKEYVNNELKKQEDNKKETTKSENSKLKEENKDNKEKKESEDSASKNREKKEY 1015
Query: 207 EVLQKSSTQEEPKPGSTTSVAYGVQERDQKYIKKWHLMYKHAVLSNTGKCDNKVPLVEKE 266
E +KS T+EE K S +E+D + K S K +++ +K+
Sbjct: 1016 EE-KKSKTKEEAKKEKKKSQDKKREEKDSEERK-----------SKKEKEESRDLKAKKK 1063
Query: 267 KEGGEEDNEGNNSYRNYSETDSDMDDEKKNVIELVQKAFDEILLPEVEDLSSEGHSKSRG 326
+E +E E N E + +D K ++K D+ ++ SKSR
Sbjct: 1064 EEETKEKKESENHKSKKKEDKKEHEDNKS-----MKKEEDK------KEKKKHEESKSRK 1112
Query: 327 NETDEVLLEKSGGKIEERNTTTFTESPKEVPKMESKQKSWSHLKKVILLKRFVKALEKVR 386
E D+ +EK +E++N+ E E K+KS H+K L+K+ EK
Sbjct: 1113 KEEDKKDMEK----LEDQNSNKKKEDKNE------KKKS-QHVK---LVKKESDKKEKKE 1158
Query: 387 NINSRRPRQLPSDANFEAEKVLLNRQTSEERKKSEEWMLDYALQKVISKLAPAQRQRVTL 446
N +++ S + + E +++S++++K +E + + +K + K ++++ +
Sbjct: 1159 NEEKSETKEIESSKSQKNEVDKKEKKSSKDQQKKKEKEMKESEEKKLKKNEEDRKKQTS- 1217
Query: 447 LVEAFETIRPVQDAENGPQTSA--TVESHANLIQSLDASSNHSKEEINDRRDFEVTERAR 504
VE + + + +N P+ T + +S+++ S +E +++ + T +A
Sbjct: 1218 -VEENKKQKETKKEKNKPKDDKKNTTKQSGGKKESMESES----KEAENQQKSQATTQAD 1272
Query: 505 NDKNMDACKKNDESATVKSTATKAVKFPVCDTGIMEEEVTAEGEYKVQEKSIVKEDLKHG 564
+D++ + +S + ++A D+ + E+ + + + + +ED K
Sbjct: 1273 SDESKNEILMQADSQADSHSDSQA------DSDESKNEILMQADSQATTQRNNEEDRKKQ 1326
Query: 565 TSTTDVPYGVQERDQKYIKKWHLMYKQAVLSNTGKYDNKLPVVGKDKEGREQGDAVFNGG 624
TS + + +++K K K+ +G + K+ E +++ A
Sbjct: 1327 TSVAENKKQKETKEEKNKPKDD---KKNTTKQSGGKKESMESESKEAENQQKSQAT---- 1379
Query: 625 NNSSCHNYNETDSDMDEEKKNVIELVQKAFDEILLPETEDLSSDDRSKSRSYGSDELLEK 684
T +D DE K ++ D + + S + ++
Sbjct: 1380 ----------TQADSDESKNEILMQADSQADSHSDSQADSDESKNEILMQADSQATTQRN 1429
Query: 685 SEGEREEMNATSFTETPKEAKKTENKPK 712
+E +R++ + + + KE K+ +NKPK
Sbjct: 1430 NEEDRKKQTSVAENKKQKETKEEKNKPK 1457
Score = 67.0 bits (162), Expect = 6e-11
Identities = 197/1063 (18%), Positives = 387/1063 (35%), Gaps = 141/1063 (13%)
Query: 128 RRQLKSQKSTKKDGRSKQVGNARK-----GTQKTKTVHSEDGNSQQNVKNVSMESSPFKP 182
+ +KS+ K+ S G + G +T++SE+ S ++ + + ++
Sbjct: 354 KEDVKSEVEAAKNAGSSMTGKLEEAQRNNGVSTNETMNSENKGSGESTNDKMVNATT--- 410
Query: 183 HDAPPSTVNECDTSTKDKHMVTDYEVLQKSSTQEEPKPGSTTSVAYGVQERDQKYIKKWH 242
+D N+ +T + V E L+ + EE G G +E
Sbjct: 411 NDEDHKKENKEETHENNGESVKG-ENLENKAGNEESMKGENLENKVGNEE---------- 459
Query: 243 LMYKHAVLSNTGKCDNKVPLVEKEKEGGEEDNEGNNSYRNYSET---------DSDMDDE 293
L + K ++E EE N Y N T +S D
Sbjct: 460 -------LKGNASVEAKTNNESSKEEKREESQRSNEVYMNKETTKGENVNIQGESIGDST 512
Query: 294 KKNVIELVQKAFDEILLPEVEDLSSEGHSKSRGNETDEVLLEKSGGKIEERNTTTF--TE 351
K N +E E + P+V+ S+G+S ++ +V +G E++N E
Sbjct: 513 KDNSLEN-----KEDVKPKVDANESDGNSTKERHQEAQV---NNGVSTEDKNLDNIGADE 564
Query: 352 SPKEVPKMESKQKSWSHLKKVILLKRFVKALEKVRNINSRRPRQLPSDANFEAEKVLLNR 411
K +E H K+ + E V+N N + E+ N
Sbjct: 565 QKKNDKSVEVTTNDGDHTKEKREETQGNNG-ESVKNENLENKEDKKELKDDESVGAKTNN 623
Query: 412 QTS--EERKKSEEWMLDYALQKVISKL---APAQRQRVTLLVEAF---------ETIRPV 457
+TS E+R+++++ + K++ A + +++ + ++ +T V
Sbjct: 624 ETSLEEKREQTQKGHDNSINSKIVDNKGGNADSNKEKEVHVGDSTNDNNMESKEDTKSEV 683
Query: 458 QDAENGPQTSATVESHANLIQSLDASSNHSKEEINDRRDFEVTE-----------RARND 506
+ +N + E N S++ +KE D +D + + +++D
Sbjct: 684 EVKKNDGSSEKGEEGKENNKDSMEDKKLENKESQTDSKDDKSVDDKQEEAQIYGGESKDD 743
Query: 507 KNMDACKKNDESATVKSTATKAVKFPVCDTGIMEEEVTAEGEYKVQEKSIVKEDLKHGTS 566
K+++A K ES K T T + + +E +G K EK + K + K
Sbjct: 744 KSVEAKGKKKESKENKKTKTN-------ENRVRNKEENVQGNKKESEK-VEKGEKKESKD 795
Query: 567 TTDVPYGVQERDQKYIKKWHLMYKQAVLSNTGKYDNKLPVVGKDKEGREQGDAVFNGGNN 626
V + +D K + + +G+ + + KD + E + NGG +
Sbjct: 796 AKSV----ETKDNKKLSSTE--NRDEAKERSGEDNKEDKEESKDYQSVEAKEKNENGGVD 849
Query: 627 SSCHNYNETDSDMDEEKKNVIELVQKAFDEILLPETEDLSSDDRSKSRSY---------- 676
++ N E D+ +++ ++ + +E + + E++ +D+S ++
Sbjct: 850 TNVGN-KEDSKDLKDDRSVEVKANK---EESMKKKREEVQRNDKSSTKEVRDFANNMDID 905
Query: 677 ---GSDELL-----EKSEGEREEMNATSFTETPKEAKKTENKPKSWSHLKKLIMLKRFVK 728
GS E + EK EG +EE T T + ++ K + K K K M K+
Sbjct: 906 VQKGSGESVKYKKDEKKEGNKEENKDTINTSSKQKGKDKKKKKKE---SKNSNMKKKEED 962
Query: 729 ALDKVRNINPRRPRELPSDANFEGEKVFLNRQTSEERKKSEEWMLDYALQKVISKLAPAQ 788
+ V N ++ E K+ + ++E+K+SE+ +K +
Sbjct: 963 KKEYVNNELKKQEDNKKETTKSENSKLKEENKDNKEKKESEDSASKNREKKEYEEKKSKT 1022
Query: 789 RQRVTLLIEAFETLRPIQDAENGLRSSATVESLENPLQSLDASSVLSAKTLLGKVSFSND 848
++ EA + + QD + + S +S + +S D L AK + +
Sbjct: 1023 KE------EAKKEKKKSQDKKREEKDSEERKSKKEKEESRD----LKAKKKEEETKEKKE 1072
Query: 849 STMEFSDKASDNPMPELCKPIKPVE--TISSCHEEAPT-KRMVDEVPEDLVSDLNTKTKD 905
S S K D E K +K E HEE+ + K+ D+ + + D N+ K
Sbjct: 1073 SENHKSKKKEDKKEHEDNKSMKKEEDKKEKKKHEESKSRKKEEDKKDMEKLEDQNSNKKK 1132
Query: 906 VIGGHGEQFSVTKSLILNGIVRSLRSN---LVVPEAPSNRLDEPTTDIKD-VVEKDQLEK 961
++ K + + + N E S++ + D K+ KDQ +K
Sbjct: 1133 EDKNEKKKSQHVKLVKKESDKKEKKENEEKSETKEIESSKSQKNEVDKKEKKSSKDQQKK 1192
Query: 962 SEAPTSAVVESKNQLEKQGSTGLWFTVFKHMVSDMTENNSKTSTDVADEKDSKYEDITTR 1021
E E K + ++ + + + EN + T K + TT+
Sbjct: 1193 KEKEMKESEEKKLKKNEED---------RKKQTSVEENKKQKETKKEKNKPKDDKKNTTK 1243
Query: 1022 EISVSYENTPVVIQDM--PFKDRAVVDAEVELRQIEAIKMVEDAIDSILPDTQPLPDNST 1079
+ E+ ++ K +A A+ + + E + + DS D+Q D S
Sbjct: 1244 QSGGKKESMESESKEAENQQKSQATTQADSDESKNEILMQADSQADS-HSDSQADSDESK 1302
Query: 1080 IDRTGGIYSEGLNQK--EQKMESGNGIVEERKEESVSKEVNKP 1120
+ S+ Q+ E+ + + E +K++ +E NKP
Sbjct: 1303 NEILMQADSQATTQRNNEEDRKKQTSVAENKKQKETKEEKNKP 1345
Score = 66.2 bits (160), Expect = 1e-10
Identities = 192/1033 (18%), Positives = 363/1033 (34%), Gaps = 167/1033 (16%)
Query: 132 KSQKSTKKDGRSKQVGNARKGTQKTKTVHSEDGNSQQNVKNVSMESSPFKPHDAPPSTVN 191
K KS + G+ K+ K +KTKT + N ++NV+ ES + + S +
Sbjct: 741 KDDKSVEAKGKKKE----SKENKKTKTNENRVRNKEENVQGNKKESEKVEKGEKKESK-D 795
Query: 192 ECDTSTKDKHMVTDYEVLQKSSTQEEPKPGSTTSVAYGVQERDQKYIKKWHLMYKHAVLS 251
TKD + L + ++E K S G ++ K K +
Sbjct: 796 AKSVETKDN------KKLSSTENRDEAKERS------GEDNKEDKEESKDY--------- 834
Query: 252 NTGKCDNKVPLVEKEKEGGEEDNEGNNSYRNYSETDSDMDDE-----KKNVIELVQKAFD 306
V EK + GG + N GN E D+ D+ K N E ++K +
Sbjct: 835 ------QSVEAKEKNENGGVDTNVGNK------EDSKDLKDDRSVEVKANKEESMKKKRE 882
Query: 307 EILLPEVEDLSSEGHSKSRGNETDEVLLEKSGGKIEERNTTTFTESPKEVPKMESKQKSW 366
E+ + D SS + N D + + SG ++ + + KE K E+K
Sbjct: 883 EV---QRNDKSSTKEVRDFANNMDIDVQKGSGESVKYKK-----DEKKEGNKEENKDT-- 932
Query: 367 SHLKKVILLKRFVKALEKVRNINSRRPRQLPSDANFEAEKVLLNRQTSEERKKSEEWMLD 426
+ K + + ++ ++ ++N + ++ + E KK E+
Sbjct: 933 ------------INTSSKQKGKDKKKKKKESKNSNMKKKEEDKKEYVNNELKKQED---- 976
Query: 427 YALQKVISKLAPAQRQRVTLLVEAFETIRPVQDAENGPQTSATVESHANLIQSLDASSNH 486
+ +EN ++ AS N
Sbjct: 977 --------------------------NKKETTKSENSKLKEENKDNKEKKESEDSASKNR 1010
Query: 487 SKEEINDRRDFEVTERARNDKNMDACKKNDESATVKSTATKAVKFPVCDTGIMEEEVTAE 546
K+E +++ + E A+ +K KK +E + + + K + +EE T E
Sbjct: 1011 EKKEYEEKKS-KTKEEAKKEKKKSQDKKREEKDSEERKSKKEKEESRDLKAKKKEEETKE 1069
Query: 547 GEYKVQEKSIVKEDLKHGTSTTDVPYGVQERDQKYIKKWHLMYKQAVLSNTGKYDNKLPV 606
+ KS KED K + +E D+K KK H K K KL
Sbjct: 1070 KKESENHKSKKKEDKKEHEDNKSMK---KEEDKKEKKK-HEESKSRKKEEDKKDMEKLED 1125
Query: 607 VGKDKEGREQGDAVFNGGNNSSCHNYNETDSDMDEEKKNVIELVQKAFDEILLPETEDLS 666
+K+ ++ + + ++ + +EEK E+ + + + E S
Sbjct: 1126 QNSNKKKEDKNEKKKSQHVKLVKKESDKKEKKENEEKSETKEIESSKSQKNEVDKKEKKS 1185
Query: 667 SDDRSKSRSY----GSDELLEKSEGEREEMNATSFTETPKEAKKTENKPK-------SWS 715
S D+ K + ++ L+K+E +R++ + + KE KK +NKPK S
Sbjct: 1186 SKDQQKKKEKEMKESEEKKLKKNEEDRKKQTSVEENKKQKETKKEKNKPKDDKKNTTKQS 1245
Query: 716 HLKKLIMLKRFVKALDKVRN------INPRRPRELPSDANFEGEKVFLNRQTSEERKKSE 769
KK M +A ++ ++ + E+ A+ + + ++ S+E K
Sbjct: 1246 GGKKESMESESKEAENQQKSQATTQADSDESKNEILMQADSQADSHSDSQADSDESK--N 1303
Query: 770 EWMLDYALQKVISKLAPAQRQRVTLLIE-------AFETLRPIQDAENGLR-SSATVESL 821
E ++ Q + R++ T + E E +P D +N + S ES+
Sbjct: 1304 EILMQADSQATTQRNNEEDRKKQTSVAENKKQKETKEEKNKPKDDKKNTTKQSGGKKESM 1363
Query: 822 ENPLQSLDASSVLSAKTLLGKVSFSNDSTMEFSDKASDNPMPELCKPIKPVETISSCHEE 881
E+ + + A T N+ M+ +A + + E + +
Sbjct: 1364 ESESKEAENQQKSQATTQADSDESKNEILMQADSQADSHSDSQADSDESKNEILMQADSQ 1423
Query: 882 APTKRMVDEVPEDLVSDLNTKTKDVIGGHGEQFSVTKSLILNGIVRSLRSNLVVPEAPSN 941
A T+R +E D +T S+ N E
Sbjct: 1424 ATTQRNNEE-------DRKKQT------------------------SVAENKKQKETKEE 1452
Query: 942 RLDEPTTDIKDVVEKDQLEKSEAPTSAVVESKNQLEKQGSTGLWFTVFKHMVSDMTENNS 1001
+ ++P D K+ E+ K E+ S E++NQ + Q +T K+ + ++ +
Sbjct: 1453 K-NKPKDDKKNTTEQSG-GKKESMESESKEAENQQKSQATTQGESDESKNEILMQADSQA 1510
Query: 1002 KTSTDVADEKDSKYEDITTREISV------SYENTPVVIQDMPFKDRAVVDAEVELRQIE 1055
T + + D +I + S S E+ ++ + + D++ +I
Sbjct: 1511 DTHANSQGDSDESKNEILMQADSQADSQTDSDESKNEILMQADSQADSQTDSDESKNEIL 1570
Query: 1056 AIKMVEDAIDSILPDTQPLPDNSTIDRTGGIYSEGLNQKEQKMESGNGIVEERKEESVS- 1114
+ I L D + D G S+ + K + ES +G E + VS
Sbjct: 1571 MQADSQAKIGESLEDNKVKGKEDNGDEVGKENSKTIEVKGRHEESKDGKTNENGGKEVST 1630
Query: 1115 KEVNKPNQKLSRN 1127
+E +K + + RN
Sbjct: 1631 EEGSKDSNIVERN 1643
Score = 60.1 bits (144), Expect = 8e-09
Identities = 102/594 (17%), Positives = 224/594 (37%), Gaps = 72/594 (12%)
Query: 127 RRRQLKSQKSTKKDGRSKQVGNARKGTQKTKTVHSEDGNSQQNVKNVSMESSPFKPHDAP 186
+ R + K K++ + Q A++ +E+G NV N +S K +
Sbjct: 817 KERSGEDNKEDKEESKDYQSVEAKE--------KNENGGVDTNVGNKE-DSKDLKDDRSV 867
Query: 187 PSTVNECDTSTKDKHMVTDYEVLQKSSTQEEPKPGSTTSVAYGVQERDQKYIKKWHLMYK 246
N+ ++ K + V + KSST+E + + VQ+ + +K YK
Sbjct: 868 EVKANKEESMKKKREEVQRND---KSSTKEVRDFANNMDI--DVQKGSGESVK-----YK 917
Query: 247 HAVLSNTGKCDNKVPLVEKEKEGGEEDNEGNNSYRNYSETDSDMDDEKKNVIELVQKAFD 306
K +NK + K+ G++ + +N S +D+K+ V ++K D
Sbjct: 918 KDEKKEGNKEENKDTINTSSKQKGKDKKKKKKESKN-SNMKKKEEDKKEYVNNELKKQED 976
Query: 307 ---EILLPEVEDLSSEGH-SKSRGNETDEVLLEKSGGKIEERNTTTFTESPKEVPKMESK 362
E E L E +K + D + + EE+ + T E+ KE K + K
Sbjct: 977 NKKETTKSENSKLKEENKDNKEKKESEDSASKNREKKEYEEKKSKTKEEAKKEKKKSQDK 1036
Query: 363 QKSWSHLKKVILLKRFVKALEKVRNINSRRPRQLPSDANFEAEKVLLNRQTSEERKKSEE 422
++ ++ ++ K E+ R++ +++ + E+ +++ + K +E
Sbjct: 1037 KREEKDSEE----RKSKKEKEESRDLKAKK----------KEEETKEKKESENHKSKKKE 1082
Query: 423 WMLDYALQKVISKLAPAQRQRVTLLVEAFETIRPVQDAENGPQTSATVESHANLIQSLDA 482
++ K + K E + + ++ E ++ + D
Sbjct: 1083 DKKEHEDNKSMKK----------------EEDKKEKKKHEESKSRKKEEDKKDMEKLEDQ 1126
Query: 483 SSNHSKEEINDRRDFEVTERARNDKNMDACKKNDESATVKSTATKAVKFPVCDT----GI 538
+SN KE+ N+++ + + + + + K+N+E + K + + D
Sbjct: 1127 NSNKKKEDKNEKKKSQHVKLVKKESDKKEKKENEEKSETKEIESSKSQKNEVDKKEKKSS 1186
Query: 539 MEEEVTAEGEYKVQEKSIVKEDLKHGTSTTDVPYGVQERDQKYIKKWHLMYKQAVLSNTG 598
+++ E E K E+ +K++ + T V ++++ K K K+ +G
Sbjct: 1187 KDQQKKKEKEMKESEEKKLKKNEEDRKKQTSVEENKKQKETKKEKNKPKDDKKNTTKQSG 1246
Query: 599 KYDNKLPVVGKDKEGREQGDAVFNGGNNSSCHNYNETDSDMDEEKKNVIELVQKAFDEIL 658
+ K+ E +++ A T +D DE K ++ D
Sbjct: 1247 GKKESMESESKEAENQQKSQAT--------------TQADSDESKNEILMQADSQADSHS 1292
Query: 659 LPETEDLSSDDRSKSRSYGSDELLEKSEGEREEMNATSFTETPKEAKKTENKPK 712
+ + S + ++ +E +R++ + + + KE K+ +NKPK
Sbjct: 1293 DSQADSDESKNEILMQADSQATTQRNNEEDRKKQTSVAENKKQKETKEEKNKPK 1346
Score = 55.8 bits (133), Expect = 1e-07
Identities = 110/603 (18%), Positives = 230/603 (37%), Gaps = 57/603 (9%)
Query: 121 KRFVSMRRRQLKSQKSTKKDGRSKQVGNARKGTQ--KTKTVHSEDGNSQQNVKNVSMESS 178
K +++ + +K +KD ++ ++ ++ K K E +++ + S +
Sbjct: 1023 KEEAKKEKKKSQDKKREEKDSEERKSKKEKEESRDLKAKKKEEETKEKKESENHKSKKKE 1082
Query: 179 PFKPHDAPPSTVNECDTSTKDKHMVTDYEVLQKSSTQEEPKPGSTTSVAYGVQERDQKYI 238
K H+ S E D K KH + KS +EE K ++ +
Sbjct: 1083 DKKEHEDNKSMKKEEDKKEKKKHEES------KSRKKEEDKKDMEKLEDQNSNKKKEDKN 1136
Query: 239 KKWHLMYKHAVLSNTGKCDNKVPLVEKEKEGGEEDNEGNNSYRNY---SETDSDMDDEKK 295
+K + V + K + K E E++ ++ E + S +N E S D +KK
Sbjct: 1137 EKKKSQHVKLVKKESDKKEKK----ENEEKSETKEIESSKSQKNEVDKKEKKSSKDQQKK 1192
Query: 296 NVIELVQKAFDEILLPEVEDLSSEGHSKSRGNETDEVLLEKSGGKIEERNTTTFTESPKE 355
E+ K +E L + E+ + S + E EK+ K +++NTT + KE
Sbjct: 1193 KEKEM--KESEEKKLKKNEEDRKKQTSVEENKKQKETKKEKNKPKDDKKNTTKQSGGKKE 1250
Query: 356 VPKMESK-----QKSWSHLKKVILLKRFVKALEKVRNINSRRPRQLPSDANFEAEKVLLN 410
+ ESK QKS + + + ++ +S Q SD + + +
Sbjct: 1251 SMESESKEAENQQKSQATTQADSDESKNEILMQADSQADSHSDSQADSDESKNEILMQAD 1310
Query: 411 RQTSEERKKSEEWMLDYALQKVISKLAPAQRQRVTLLVEAFETIRPVQDAENGPQTSATV 470
Q + +R E+ +K + +A ++Q+ T E +P D +N + S
Sbjct: 1311 SQATTQRNNEED-------RKKQTSVAENKKQKET----KEEKNKPKDDKKNTTKQSGGK 1359
Query: 471 ESHANLIQSLDASSNHSKEEINDRRDFEVTERARNDKNMDACKKNDESATVKSTATKAVK 530
+ ++ + SKE N ++ T+ ++ + + D A S +
Sbjct: 1360 K---------ESMESESKEAENQQKSQATTQADSDESKNEILMQADSQADSHSDSQ---- 1406
Query: 531 FPVCDTGIMEEEVTAEGEYKVQEKSIVKEDLKHGTSTTDVPYGVQERDQKYIKKWHLMYK 590
D+ + E+ + + + + +ED K TS + ++++ K K K
Sbjct: 1407 ---ADSDESKNEILMQADSQATTQRNNEEDRKKQTSVAE---NKKQKETKEEKNKPKDDK 1460
Query: 591 QAVLSNTGKYDNKLPVVGKDKEGREQGDAVFNGGNNSSCHN-YNETDSDMDEEKKNVIEL 649
+ +G + K+ E +++ A G ++ S + + DS D + +
Sbjct: 1461 KNTTEQSGGKKESMESESKEAENQQKSQATTQGESDESKNEILMQADSQADTHANSQGD- 1519
Query: 650 VQKAFDEILL---PETEDLSSDDRSKSRSYGSDELLEKSEGEREEMNATSFTETPKEAKK 706
++ +EIL+ + + + D SK+ + S+ + +E + +AK
Sbjct: 1520 SDESKNEILMQADSQADSQTDSDESKNEILMQADSQADSQTDSDESKNEILMQADSQAKI 1579
Query: 707 TEN 709
E+
Sbjct: 1580 GES 1582
>At5g61260 putative protein
Length = 496
Score = 53.9 bits (128), Expect = 5e-07
Identities = 48/222 (21%), Positives = 95/222 (42%), Gaps = 12/222 (5%)
Query: 257 DNKVPLVEKEKEGGEEDNEGNNSYRNYSETDSDMDDEKKNVIELVQKAFDEILLPEVEDL 316
++ V V+ EK+ E + ++ S T K + + L + ++
Sbjct: 271 ESSVKGVKSEKQPSSEKKTMKSGNKSLSTTPKRGSSPTKQIPGKISTG-----LTKKKET 325
Query: 317 SSEGHSKSRGNETDEVLLEKSGGKIEERNTTTFTESPKEVPKMESKQKSWSHLKKVILLK 376
S ++ +V +K+G K+ TF + PK E W KK ++ +
Sbjct: 326 GSADVVEANPKPEKKVRPKKTGVKVSLAQQMTFKKGKVLDPKPEDSSPRWIKFKKRVVQE 385
Query: 377 RFVKALEKVRNINSRR---PRQLPSDANFEAEKVLLNRQTSEERKKSEEWMLDYALQKVI 433
++ K +N+ RR + S + EKV+L + E +KK + + +++ +
Sbjct: 386 LKTQSEGKKKNLKDRRLGVETKTDSCEGSKREKVVLRHRKVEGKKKMIT-LFNNVIEETV 444
Query: 434 SKLAPAQRQRVTLLVEAFETIRPVQD---AENGPQTSATVES 472
+KL ++ +V L+ AFET+ +QD + PQ+ AT +
Sbjct: 445 NKLTKVRKHKVKALIGAFETVISLQDTNKTSHKPQSKATTSA 486
Score = 48.5 bits (114), Expect = 2e-05
Identities = 74/324 (22%), Positives = 132/324 (39%), Gaps = 46/324 (14%)
Query: 926 VRSLRSNLVVPEAPSNRLDEPTTDIKDVVEKDQLEKSEAPTSAVVESKNQLEKQGSTGLW 985
V + R L V + P ++ ++ E ++ KS S + + K+ +GL
Sbjct: 157 VSAKRDALAVKKKPCASVNSESSS----KEGSEIAKSVDGLSVKSNDRARKNKETESGLS 212
Query: 986 FTVFKHMVSDMTENNSKTSTDVADEKDSKYEDITTREISVSYENTPVVIQDMPFKDRAVV 1045
+ V + + S TST V K +++ +V T I K++ V
Sbjct: 213 GSAVVKKVPALRTDKSSTSTGVGSSKVCAPKNLK----NVEKAKTTQTISGEDVKEKTVC 268
Query: 1046 DAEVELRQIEAIKMVEDAIDSILP-----DTQPLPDNSTIDRTGGIYSEGLNQKEQKMES 1100
E ++ +++ K ++ T P +S + G S GL +K+ E+
Sbjct: 269 VVESSVKGVKSEKQPSSEKKTMKSGNKSLSTTPKRGSSPTKQIPGKISTGLTKKK---ET 325
Query: 1101 GNGIVEE---RKEESV-------------------SKEVNKPNQKLSRNWSNLKKVVLLR 1138
G+ V E + E+ V K ++ + S W KK V+
Sbjct: 326 GSADVVEANPKPEKKVRPKKTGVKVSLAQQMTFKKGKVLDPKPEDSSPRWIKFKKRVVQE 385
Query: 1139 RFIKALEKVRKFNPREPRY-LPLEPDS----EDEKVQLRHQDMAERKGTEEWMLDYALRQ 1193
+K + +K N ++ R + + DS + EKV LRH+ + E K + + + +
Sbjct: 386 --LKTQSEGKKKNLKDRRLGVETKTDSCEGSKREKVVLRHRKV-EGKKKMITLFNNVIEE 442
Query: 1194 VVSKLTPARKRKVELLVEAFETVV 1217
V+KLT RK KV+ L+ AFETV+
Sbjct: 443 TVNKLTKVRKHKVKALIGAFETVI 466
Score = 42.0 bits (97), Expect = 0.002
Identities = 29/105 (27%), Positives = 52/105 (48%), Gaps = 4/105 (3%)
Query: 706 KTENKPKSWSHLKKLIMLKRFVKALDKVRNINPRRPRELPSDANFEG---EKVFLNRQTS 762
K E+ W KK ++ + ++ K +N+ RR + EG EKV L +
Sbjct: 367 KPEDSSPRWIKFKKRVVQELKTQSEGKKKNLKDRRLGVETKTDSCEGSKREKVVLRHRKV 426
Query: 763 EERKKSEEWMLDYALQKVISKLAPAQRQRVTLLIEAFETLRPIQD 807
E +KK + + +++ ++KL ++ +V LI AFET+ +QD
Sbjct: 427 EGKKKMIT-LFNNVIEETVNKLTKVRKHKVKALIGAFETVISLQD 470
>At4g36520 trichohyalin like protein
Length = 1432
Score = 52.4 bits (124), Expect = 2e-06
Identities = 182/914 (19%), Positives = 339/914 (36%), Gaps = 147/914 (16%)
Query: 268 EGGEEDNEGNNSYRNYSETD----SDMDDEKKNV-IELVQKAFDEILLPEVEDLSSEGHS 322
+G +E E NN Y T+ +D + K+V I L + FD+ L +
Sbjct: 405 DGNDEWKEANNQYVELVRTELPRNADENSGGKDVSIPLNAEFFDQELTWAGNVDWEKQRR 464
Query: 323 KSRGNETDEVL--LEKSGGKIE-----ERNTTTFTESPKEVPKMESKQKSWSHLKKVILL 375
+++G+ D L K G + +R+ E E PK+E + H++ L
Sbjct: 465 RAKGDREDHEARKLPKHSGTRKLSSRHKRHENKLAEKAPEEPKIEKSR----HVEMGNDL 520
Query: 376 KRFVKALEKVRNINSRRPRQLPSDANFEAEKVLLNRQTSEERKKSEEWMLDYALQKVISK 435
+ K RN+ +L ++ + +K LL E+ K+ + LD +K K
Sbjct: 521 PDH-GGIVKHRNLLKPEENKLFTEKPAKQKKELL---CEEKTKRIQNQQLD---KKTHQK 573
Query: 436 LAPAQRQRVTLLVEAFETIRPVQDAE-----------NGPQTSATVESHANLIQSLDASS 484
A ++ V + +R E NG + +S L + L
Sbjct: 574 AAETNQECVYDWEQNARKLREALGNESTLEVSVELNGNGKKMEMRSQSETKLNEPLKRME 633
Query: 485 NHSK------EEINDRRDFEVTERARNDKNMDACKKNDESAT-VKSTATKAVKFPVCDTG 537
++ E NDRR+ E+A N+K + A + +E +K KA +
Sbjct: 634 EETRIKEARLREENDRRERVAVEKAENEKRLKAALEQEEKERKIKEAREKAEN----ERR 689
Query: 538 IMEEEVTAEGEYKVQEKSIVKEDLKHGTSTTDVPYGVQERDQKYIKKWHLMYKQAVLSNT 597
+E AE E K++E+ ++ LK + ++E +K ++ +
Sbjct: 690 AVEAREKAEQERKMKEQQELELQLKEAFEKEEENRRMREAFALEQEK-----ERRIKEAR 744
Query: 598 GKYDNKLPVVGKDKEGREQGDAVFNGGNNSSCHNYNETDSDMDEEKKNVIELVQKAFDEI 657
K +N+ + KE RE+ +++++ K +E +K E
Sbjct: 745 EKEENERRI----KEAREK--------------------AELEQRLKATLEQEEK---ER 777
Query: 658 LLPETEDLSSDDRSKSRSYGSDELLEKSEGEREEMNATSFTETPKEAKKTENKPKSWSHL 717
+ E ++ ++R + E+LE++E ER+ A E + K+T K ++ L
Sbjct: 778 QIKERQEREENERR------AKEVLEQAENERKLKEALEQKENERRLKETREKEENKKKL 831
Query: 718 KKLIML----KRFVKALDKVRNINPRRPRELPSDANFEGEKVFLNRQTSEER----KKSE 769
++ I L KR ++A ++ RR +E + E E++ + Q ++ER ++++
Sbjct: 832 REAIELEEKEKRLIEAFERAE--IERRLKE-----DLEQEEMRMRLQEAKERERLHRENQ 884
Query: 770 EWMLDYALQKVISKLAPAQRQRVTLLIE--------------AFETLRPIQDAENGLRSS 815
E + Q S +++R +E + E+L + + +
Sbjct: 885 EHQENERKQHEYSGEESDEKERDACEMEKTCETTKEAHGEQSSNESLSDTLEENESIDND 944
Query: 816 ATVESLENPLQSLDASSVLSAKTLLGKVSFSNDSTMEFSDKASDNPMPELCKPIKPVETI 875
+V + + +SA+T KV + + S K N M + + E
Sbjct: 945 VSVNKQKKEEEGTRQRESMSAETCPWKV--FEKTLKDASQKEGTNEMDADTRLFERNEET 1002
Query: 876 SSCHEEAPTKRMVDEVPEDLVSDLNTKTKDVIGGHGEQFSVTKSLILNGIVRSLRSNLVV 935
E + E E+ S T+++IGG EQ S + V S L
Sbjct: 1003 PRLGENGGCNQQNGESGEESTS----VTENIIGGKLEQKSKNSETSKDASVLKRVSGL-- 1056
Query: 936 PEAPSNRLDEPTTDIKDV--VEKDQLEKSEAPTSAV------------VESKNQLEKQGS 981
++E D+ V +++ E AP ++ E N E Q
Sbjct: 1057 ----KTEVEERLEDVVGVGRDQRNPEESKSAPKTSYGFRNHEYKFTHQQERGNIYETQAG 1112
Query: 982 TGLWFTVFKHMVSDMTENNSKTSTDVADEKDSKYEDITTREISVSYENTPVVIQD-MPFK 1040
V + + S ++ K + + E+D + E + R++ E +D M F
Sbjct: 1113 LNQDAKVERPLPSRVSVQREKEAERLKRERDLEMEQL--RKVEEEREREREREKDRMAFD 1170
Query: 1041 DRAVVDAEVELRQIEAIKMVEDAIDSILPDTQPLPDNSTIDRTGGIYSEGLNQKEQKMES 1100
RA+ DA L K +A + LPD + +R + +++ E
Sbjct: 1171 QRALADARERLE-----KACAEAREKSLPDKLSMEARLRAER-AAVERATSEARDRAAEK 1224
Query: 1101 GNGIVEERKEESVS 1114
ER E SVS
Sbjct: 1225 AAFEARERMERSVS 1238
Score = 39.3 bits (90), Expect = 0.014
Identities = 108/540 (20%), Positives = 194/540 (35%), Gaps = 83/540 (15%)
Query: 664 DLSSDDRSKSRSYGSDELLEKS-----EGEREEMNATSFTETP------------KEAK- 705
D + R + G++ LE S G++ EM + S T+ KEA+
Sbjct: 584 DWEQNARKLREALGNESTLEVSVELNGNGKKMEMRSQSETKLNEPLKRMEEETRIKEARL 643
Query: 706 KTENKPKSWSHLKKLIMLKRFVKALDKVRNINPRRPRELPSDANFEGEKVFLNRQTSEER 765
+ EN + ++K KR AL++ R+ +E A E V + +ER
Sbjct: 644 REENDRRERVAVEKAENEKRLKAALEQEEK--ERKIKEAREKAENERRAVEAREKAEQER 701
Query: 766 KKSEEWMLDYALQKVISKLAPAQRQRVTLLIEAFETLRPIQDAENGLRSSATVESLENPL 825
K E+ L+ L++ K +R R +E Q+ E ++ + E E +
Sbjct: 702 KMKEQQELELQLKEAFEKEEENRRMREAFALE--------QEKERRIKEAREKEENERRI 753
Query: 826 QSLDASSVLSAKTLLGKVSFSNDSTMEFSDKASDNPM--PELCKPIKPVETISSCHEEAP 883
+ + L + + ++ + +N E+ + + + E+
Sbjct: 754 KEAREKAELEQRLKATLEQEEKERQIKERQEREENERRAKEVLEQAENERKLKEALEQKE 813
Query: 884 TKRMVDEVPEDLVSDLNTKTKDVIGGHGEQFSVTKSLILNGIVRSLRSNLVVPEA----- 938
+R + E E + K ++ I ++ + ++ I R L+ +L E
Sbjct: 814 NERRLKETREK--EENKKKLREAIELEEKEKRLIEAFERAEIERRLKEDLEQEEMRMRLQ 871
Query: 939 ---PSNRLDEPTTDIKDVVEK---------DQLEKSEAPTSAVVESKNQLEKQGSTGLWF 986
RL + ++ K D+ E+ E+ + + S+
Sbjct: 872 EAKERERLHRENQEHQENERKQHEYSGEESDEKERDACEMEKTCETTKEAHGEQSSN--- 928
Query: 987 TVFKHMVSDMTENNSKTSTDVADEKDSKYEDITTREISVSYENTPVVIQDMPFKDRAVVD 1046
+SD E N DV+ K K E+ T + S+S E P + + KD + +
Sbjct: 929 ----ESLSDTLEENESIDNDVSVNKQKKEEEGTRQRESMSAETCPWKVFEKTLKDASQKE 984
Query: 1047 AEVELRQIEAIKMVEDAIDSILPDTQPLPDNSTIDRTGGIYSEGLNQKEQKMESGNGIVE 1106
E+ DA DT+ N R G + G NQ Q ESG
Sbjct: 985 GTNEM----------DA------DTRLFERNEETPRLG--ENGGCNQ--QNGESGEESTS 1024
Query: 1107 ERKEESVSK-EVNKPNQKLSRNWSNLKKVVLLRRFI-KALEKV-----RKFNPREPRYLP 1159
+ K E N + S++ S LK+V L+ + + LE V + NP E + P
Sbjct: 1025 VTENIIGGKLEQKSKNSETSKDASVLKRVSGLKTEVEERLEDVVGVGRDQRNPEESKSAP 1084
>At5g39380 putative protein
Length = 507
Score = 48.5 bits (114), Expect = 2e-05
Identities = 52/178 (29%), Positives = 74/178 (41%), Gaps = 37/178 (20%)
Query: 1068 LPDTQPLP--DNSTIDRTGGI-YSEGLNQKEQKMESGNGIVEERKEESVSKEVNKPNQKL 1124
LP TQ P D T+ T Y+ G N+ E + E E+K + KE + ++
Sbjct: 334 LPPTQSTPKDDECTVSETEEYEYTSGSNEAESEEEEIGLSNGEKKPRAARKEGDSADEAA 393
Query: 1125 SRNWSNLKKVVLLRRFI----KALEKVRKFNPREPRYL---------------------P 1159
+K+ R I EK RK R R L
Sbjct: 394 -------RKLRFRRGTIVDPDTVGEKARKLKFRRGRGLGEDKAQDAQVRRSFKKREDIRE 446
Query: 1160 LEPDSEDEKVQLRHQDMAERKGTEEWMLDYALRQVVSKLTPARKRKVELLVEAFETVV 1217
E + + EKV LRHQD+ E+ + + + + + SKL ARK KV+ LV AFETV+
Sbjct: 447 EEVNEDGEKVVLRHQDVQEKDA--QGLFNNVIEETASKLVEARKSKVKALVGAFETVI 502
Score = 46.6 bits (109), Expect = 9e-05
Identities = 44/178 (24%), Positives = 77/178 (42%), Gaps = 32/178 (17%)
Query: 659 LPETEDLSSDDR---SKSRSYGSDELLEKSEGEREEMNATSFTETPKEAKKTENKPKSWS 715
LP T+ DD S++ Y ++E E EE+ ++ + P+ A+K + +
Sbjct: 334 LPPTQSTPKDDECTVSETEEYEYTSGSNEAESEEEEIGLSNGEKKPRAARKEGDSADEAA 393
Query: 716 HLKKLIMLKRFVKALD----KVRNINPRRPRELPSD---------------------ANF 750
+KL + + D K R + RR R L D N
Sbjct: 394 --RKLRFRRGTIVDPDTVGEKARKLKFRRGRGLGEDKAQDAQVRRSFKKREDIREEEVNE 451
Query: 751 EGEKVFLNRQTSEERKKSEEWMLDYALQKVISKLAPAQRQRVTLLIEAFETLRPIQDA 808
+GEKV L Q +E K + + + +++ SKL A++ +V L+ AFET+ +Q++
Sbjct: 452 DGEKVVLRHQDVQE--KDAQGLFNNVIEETASKLVEARKSKVKALVGAFETVISLQES 507
Score = 39.7 bits (91), Expect = 0.011
Identities = 23/77 (29%), Positives = 42/77 (53%), Gaps = 2/77 (2%)
Query: 384 KVRNINSRRPRQLPSDANFEAEKVLLNRQTSEERKKSEEWMLDYALQKVISKLAPAQRQR 443
+VR +R + N + EKV+L Q +E K + + + +++ SKL A++ +
Sbjct: 433 QVRRSFKKREDIREEEVNEDGEKVVLRHQDVQE--KDAQGLFNNVIEETASKLVEARKSK 490
Query: 444 VTLLVEAFETIRPVQDA 460
V LV AFET+ +Q++
Sbjct: 491 VKALVGAFETVISLQES 507
>At3g16000 myosin heavy chain like protein
Length = 727
Score = 48.1 bits (113), Expect = 3e-05
Identities = 68/279 (24%), Positives = 117/279 (41%), Gaps = 41/279 (14%)
Query: 817 TVESLENPLQSLDASSVLSAKTLLGKVSFSNDSTMEFSDKASDNPMPELCKPIKPVETIS 876
T+ESL+N L+ + + VL K K+ + + +KA + E +S
Sbjct: 158 TIESLKNQLKDRERALVLKEKDFEAKLQHEQEERKKEVEKAKE-------------EQLS 204
Query: 877 SCHEEAPTKRMVDEVPEDLVSD--LNTKTKDVIGGHGEQFSVTKSLILNG-----IVRSL 929
++ K +V E+ +L S+ L K KD I S+ SL G + L
Sbjct: 205 LINQLNSAKDLVTELGRELSSEKKLCEKLKDQIE------SLENSLSKAGEDKEALETKL 258
Query: 930 RSNLVVPEAPSNRLDEPTTDIKDVVEKDQ-LEKSEAPTSAVVESKNQLEKQGSTGLWFTV 988
R L + E +R++ + ++KD EK Q S A A ++ N + Q S L
Sbjct: 259 REKLDLVEGLQDRINLLSLELKDSEEKAQRFNASLAKKEAELKELNSIYTQTSRDL--AE 316
Query: 989 FKHMVSDMTENNSKTSTDVADEKDSKYEDITTREISVSYENTPVVIQ-DMPFKD------ 1041
K + E +T +++ D K+S E++ TR ++ E + + D KD
Sbjct: 317 AKLEIKQQKEELIRTQSEL-DSKNSAIEELNTRITTLVAEKESYIQKLDSISKDYSALKL 375
Query: 1042 ----RAVVDAEVELRQIEAIKMVEDAIDSILPDTQPLPD 1076
+A DAE+ R+ + I+ + + +D L D D
Sbjct: 376 TSETQAAADAELISRKEQEIQQLNENLDRALDDVNKSKD 414
>At1g56660 hypothetical protein
Length = 522
Score = 47.4 bits (111), Expect = 5e-05
Identities = 115/567 (20%), Positives = 202/567 (35%), Gaps = 99/567 (17%)
Query: 255 KCDNKVPLVEKEKEGGE--EDNEGNNSYRNYSETDSDMDDEKKNVIELVQKAFDE---IL 309
K KV + E+ G+ +D E SE D DD+KK ++V K +E L
Sbjct: 40 KSIEKVKAKKDEESSGKSKKDKEKKKGKNVDSEVKEDKDDDKKKDGKMVSKKHEEGHGDL 99
Query: 310 LPEVEDLSSEGHSK--SRGNE-TDEVLLEKSGGKIEERNTTTFTESPKEVPKMESKQKSW 366
+ D+ E H K +G E E L E+ GK ++ P+E K K+K
Sbjct: 100 EVKESDVKVEEHEKEHKKGKEKKHEELEEEKEGKKKKNKKEKDESGPEEKNKKADKEKKH 159
Query: 367 SHLKKVILLKRFVKALEKVRNINSRRPRQLPSDANFEAEKVLLNRQTSEERKKSEEWMLD 426
+ + + LE+ +++ + S + +K ++ EE K +E+
Sbjct: 160 EDVSQE------KEELEEEDGKKNKKKEKDESGTEEKKKKPKKEKKQKEESKSNED---- 209
Query: 427 YALQKVISKLAPAQRQRVTLLVEAFETIRPVQDAENGPQTSATVESHANLIQSLDASSNH 486
+KV K ++ + +D E + T + ++ D+ N
Sbjct: 210 ---KKVKGKKEKGEKGDL-----------EKEDEEKKKEHDETDQE----MKEKDSKKNK 251
Query: 487 SKEEINDRRDFEVTERARNDKNMDACKKNDESATVKSTATKAVKFPVCDTGIMEEEVTAE 546
KE +D E + + + K+ DES + K K G E+ +
Sbjct: 252 KKE-----KDESCAEEKKKKPDKEK-KEKDESTEKEDKKLKGKK------GKGEKPEKED 299
Query: 547 GEYKVQEKSIVKEDLKHGTSTTDVPYGVQERDQKYIKKWHLMYKQAVLSNTGKYDNKLPV 606
K +E ++++ D G +++++ KK K+ V+ + +
Sbjct: 300 EGKKTKEHDATEQEM--DDEAADHKEGKKKKNKDKAKK-----KETVIDEVCEKET---- 348
Query: 607 VGKDKEGREQGDAVFNGGNNSSCHNYNETDSDMDEEKKNVIELVQKAFD-EILLPETEDL 665
KDK+ E E D D++K+N +E + D ++ PE E
Sbjct: 349 --KDKDDDEGETKQKKNKKKEKKSEKGEKDVKEDKKKENPLETEVMSRDIKLEEPEAEKK 406
Query: 666 SSDD---RSKSRSYGSDELLEKSEGEREEMNATSFTETPKEAKKTENKPKSWSHLKKLIM 722
DD + KS+ G GE EE + K KK +PK
Sbjct: 407 EEDDTEEKKKSKVEG---------GESEEGKKKKKKDKKKNKKKDTKEPKMTE------- 450
Query: 723 LKRFVKALDKVRNINPRRPRELPSDANFEGEKVFLNRQTSEERKKSEEWMLDYALQKVIS 782
+ ++ D EG K ++ + +KK + K+ +
Sbjct: 451 --------------DEEEKKDDSKDVKIEGSKAKEEKKDKDVKKKKG----GNDIGKLKT 492
Query: 783 KLAPAQRQRVTLLIEAFETLRPIQDAE 809
KLA + L+ E E I+DAE
Sbjct: 493 KLAKIDEKIGALMEEKAEIENQIKDAE 519
Score = 40.4 bits (93), Expect = 0.006
Identities = 117/563 (20%), Positives = 204/563 (35%), Gaps = 137/563 (24%)
Query: 130 QLKSQKSTKKDGRSKQVGNARKGTQKTKTVHSE-DGNSQQNVKNVSMESSPFKPHDAPPS 188
++K++K + G+SK+ +KG V + D + +++ K VS K H+
Sbjct: 44 KVKAKKDEESSGKSKKDKEKKKGKNVDSEVKEDKDDDKKKDGKMVS------KKHEEGHG 97
Query: 189 TVNECDTSTKDKHMVTDYEVLQKSSTQEEPKPGSTTSVAYGVQERDQKYIKKWHLMYKHA 248
D EV + EE E++ K K+ KH
Sbjct: 98 ----------------DLEVKESDVKVEE-------------HEKEHKKGKE----KKHE 124
Query: 249 VLSNTGKCDNKVPLVEKEKEGGEEDNEGNNSYRNYSETDSDMDDEKKNVIELVQKAFDEI 308
L + K EK++ G EE N+ D EKK+ E
Sbjct: 125 ELEEEKEGKKKKNKKEKDESGPEEKNK-------------KADKEKKH----------ED 161
Query: 309 LLPEVEDLSSEGHSKSRGNETDEVLLEKSGGKIEERNTTTFTESPKEVPKMESKQKSWSH 368
+ E E+L E K++ E DE SG TE K+ PK E KQK S
Sbjct: 162 VSQEKEELEEEDGKKNKKKEKDE-----SG-----------TEEKKKKPKKEKKQKEESK 205
Query: 369 LKKVILLKRFVKALEKVRNINSRRPRQLPSDANFEAEKVLLNRQTSEERKKSEEWMLDYA 428
+ + VK ++ + EK L ++ E++K+ +E D
Sbjct: 206 SNE----DKKVKGKKE------------------KGEKGDLEKEDEEKKKEHDE--TDQE 241
Query: 429 LQKVISKLAPAQRQRVTLLVEAFETIRPVQDAENGPQTSATVESHANLIQSLDASSNHSK 488
+++ SK + + + E + D E + +T + L K
Sbjct: 242 MKEKDSKKNKKKEKDESCAEEK----KKKPDKEKKEKDESTEKEDKKLKGKKGKGEKPEK 297
Query: 489 EEINDR-RDFEVTERARNDKNMD----ACKKNDESATVKSTATKAVKFPVCDTGIMEEEV 543
E+ + ++ + TE+ +D+ D KKN + A K T V VC+ +++
Sbjct: 298 EDEGKKTKEHDATEQEMDDEAADHKEGKKKKNKDKAKKKET----VIDEVCEKETKDKD- 352
Query: 544 TAEGEYKVQEKSIVKEDLKHGTSTTDVPYGVQERDQKYIKKWHLMYKQAVLSNTGKYDNK 603
EGE K ++ ++ + G E+D K KK + V+S K +
Sbjct: 353 DDEGETKQKKNKKKEKKSEKG-----------EKDVKEDKKKENPLETEVMSRDIKLEEP 401
Query: 604 LPVVGKDKEGREQGDAVFNGGNNSSCHNYNETDSDMDEEKKNVIELVQKAFDEILLPETE 663
++ + E+ + GG E++ ++KK+ + +K E + E E
Sbjct: 402 EAEKKEEDDTEEKKKSKVEGG---------ESEEGKKKKKKDKKKNKKKDTKEPKMTEDE 452
Query: 664 DLSSDDRSKSRSYGSDELLEKSE 686
+ DD + GS EK +
Sbjct: 453 EEKKDDSKDVKIEGSKAKEEKKD 475
>At5g40450 unknown protein
Length = 2910
Score = 47.0 bits (110), Expect = 7e-05
Identities = 197/1071 (18%), Positives = 390/1071 (36%), Gaps = 126/1071 (11%)
Query: 116 DLPPLKRFVSMRRRQLKSQKSTKKDGRSKQVGNARKGTQKTKTVHSEDGNSQQNVKNVSM 175
D+ P + R + K+ K++ V + T++ + V D + +++ ++
Sbjct: 617 DIAPKTEEIQERPSESKASLEPKEE-----VDHISNETEEHEHVLERDVQQCETIESEAV 671
Query: 176 ESSPFKPHDAPPSTVNECDTSTKDKHM---VTDYEVLQKSSTQEE-------PKPGSTTS 225
E+ D PS + D T++ V + ++ S+ QEE P
Sbjct: 672 ETK----EDTQPSLDLKEDKETEEAETFKTVFSSDEVRSSAVQEEQFGEHTEPCSSEIKD 727
Query: 226 VAYGVQE----RDQKYIKKWHLMYKHAVLSNTGKCDNKVPLVEKEKEGGEEDN----EGN 277
++G +E + Q+ ++ + KH VL KVP E EK G E
Sbjct: 728 ESHGKEESVEVKSQETVQDENTEDKHDVL--------KVPSTESEKYQGNEPETVLVSNT 779
Query: 278 NSYRNYSETDSDM---------DDEKKNVIELV-QKAFDEILLPEVEDLSSEGHSKSRGN 327
SY ++ SD+ +DEK NV ++ + +E + + +E ++ N
Sbjct: 780 GSYEKSEKSPSDLVLNVDKEELNDEKINVDQVDGTQIMEEPIGLDSNGAEAEQIDQNITN 839
Query: 328 ETDEVLLEKSGGKIEERNTTTFTESPKEVPKMESKQKSWSHLKKVILLKRFVKALEKVRN 387
ET+E+L+ K ++ ++ + E P S++ S + +K+ L +
Sbjct: 840 ETEEILVAKPVSLLDVKSVEQMQKPKLESPSEVSEETSKTVDEKIEEKPEEEVTLYQEGQ 899
Query: 388 INSRRPRQLPSDANFEAEKVLLNRQTSEERKKSEEWMLDYALQKVISKLAPAQRQRVTLL 447
++ + + E + L Q EER + L + S++ + V
Sbjct: 900 VDGSYGLETKEETVSVPESIELEEQPQEERSVIDPTPLQKPTLESPSEVLEESSKTVDEK 959
Query: 448 VEAFETIRPVQDAENGPQTSATVESHANLIQSLDASSNHSKEEINDRRDFEVTERARNDK 507
+E E ++ E + + + +Q + N ++E + E ++D+
Sbjct: 960 IE--EKTDSIELGEIAQEERSVTD--LTPLQEESSQPNEQEKETKLEKHEPTNEEVKSDE 1015
Query: 508 NMDACKKNDESATVKSTATKAVKFPVCDTGIMEEEVTAEGEYKVQEKSIVKEDLKHGTST 567
++ + T +A EEE AE K+Q KS+ E ++ S
Sbjct: 1016 VIEVLSASPSKELEGETVVEAENIENIKEN--EEEQAAE---KIQ-KSL--ETVQTVESP 1067
Query: 568 TDVPYGVQERDQKYIKKWHLMYKQAVLSNTGKYDNKLPVVGKDKEGR-------EQGDAV 620
+ + + +E+D H+ + ++ K + + + + + EQ +
Sbjct: 1068 SSLLFSSEEQD-------HVTVAEEIVDEKAKEEVPMLQIKNEDDATKIHETRVEQARDI 1120
Query: 621 FNGGNNSSCHNYNETDSDMDEE-KKNVIELVQKAFDEILLPETEDLSSDDRSKSRSYGSD 679
N N+ + + E K E E ++ ET L S + ++ + +
Sbjct: 1121 GPSLTEICSINQNQPEEQVKEACSKEEQEKEISTNSENIVNETYALHSVEAAEEETATNG 1180
Query: 680 ELLEKSEGEREEMNATSFTETPKEAKKTENKPKSWSHL-----KKLIMLKRFVKALDKVR 734
E L+ E + + E KE ++ E K + L ++L +K V+ V
Sbjct: 1181 ESLDDVETTKSVL-----LEVRKEEEEAEMKTDAEPRLDAIEKEELETVKTVVQDAKIVN 1235
Query: 735 N--INPRRPRELPSDANFE--GEKVFLNRQTSEERKKSEEWMLDYALQKVISKL------ 784
N L D + E E V + + + S E +D + I++
Sbjct: 1236 NEETTAHESESLKGDNHQEKNAEPVEATQNLDDAEQISREVTVDTEREADITEKIEKVQE 1295
Query: 785 APAQRQRVTLLIEAFETLRPIQDAENGLRSSATVESLENPL------------QSLDASS 832
P + T+ E E+ ++ E +SS E E+ L +++D S+
Sbjct: 1296 GPTVIETPTIQGEDIESETSLELKEEVDQSSKDTEEHEHVLERDIPQCETLKAEAVDTST 1355
Query: 833 VLSAKTLLGKVSFSNDSTMEFSDKASDNPMPELCKPIKPVETISSCHEEAPTKRMVDEVP 892
V A L + ++ S+ + D + + K + V+T+ +E + +E
Sbjct: 1356 VEEAAILKTLETNISEPEAMHSETSLDLKVDKEQKEAETVKTVIFSNEVGTSDAQAEEFG 1415
Query: 893 EDLVSDLNTKTKDVIGGHGEQFSVTKSLILNGIVRSLR--SNLVVPEAPSNRLDEPTTDI 950
E +++ KD G E V + G + + L V S + E DI
Sbjct: 1416 EH-TEPCSSEIKDESQGSEESVEVKSKETVQGESSEEKDVNMLDVQSGESEKYQENEPDI 1474
Query: 951 KDVVEKDQLEKSEAPTSAVVESKNQLEKQGSTGLWFTVFKHMVSDMTENNSKTSTDVADE 1010
V + + +K E S V GL T + D+ E+ K S D E
Sbjct: 1475 SLVSKTENGDKFEEIPSVV----------EGAGLDETTHNQTLLDV-ESVVKQSLDTPSE 1523
Query: 1011 K------DSKYEDITTREISVSYENTPVVIQDMPFKDRAVVDAEVELRQIEAIKMVEDAI 1064
+ D K ED E+++ E + KD AV + +E R++ E+
Sbjct: 1524 EETSKTIDEKIEDKPKEEVTLHQEGREEGSYGLDTKDEAV--SVLESRELGEQPQQEELC 1581
Query: 1065 DSILPDTQPLPDNSTIDRTGGIYSEGLNQKEQKMESGNGIVEERKEESVSK 1115
+ + + +D+ E N ++ +E + V + S+S+
Sbjct: 1582 LANEQENETKLQEEQVDKHEPTKEEVSNDQQSPVEEISNEVIQVSSASLSE 1632
Score = 44.7 bits (104), Expect = 3e-04
Identities = 196/1092 (17%), Positives = 393/1092 (35%), Gaps = 185/1092 (16%)
Query: 154 QKTKTVHSEDGNSQQNVKNVSMESSPFKPHDAPPSTVNECDTSTKDKHMVTDYEVLQKSS 213
+K + +H E+ +V+ S + P + DA + ++ + TK++H + E+L
Sbjct: 1855 KKVEEIHEEEPKEAHDVEATSERNLPVETSDADNTLSSQLVSETKEEHKLQAGEIL---P 1911
Query: 214 TQEEPKPGSTTSVAYGVQERDQKYIKKWHLMYKHAVLSNTGKCDNKVPLVEKEKEGGE-E 272
T+ P+ S ++ + R+ + V + + VE E+ GE +
Sbjct: 1912 TEIIPRESSDEALVSMLASREDDKVALQEDNCADDVRETNDIQEERSISVETEESVGETK 1971
Query: 273 DNEGNNSYRN-YSETDSD----MDDEKKNVIELVQKAFDEILLPEVEDLSSEGHSKSRGN 327
E + R+ + ET + +++ + +I +K +EI E E + H + N
Sbjct: 1972 PKEHEDEIRDAHVETPTAPIILEENDSETLIAEAKKGNEEI--NETERTVALDHEEEFVN 2029
Query: 328 ETDEVLLEKSGGKIEE------RNTTTF-------TESPKEVPKMESKQKSWSHLKKVIL 374
L E K +E TT T + P + S + + + +
Sbjct: 2030 HEAPKLEETKDEKSQEIPETAKATETTIDQTLPIGTSQADQTPSLVSDKDDQTPKQVEEI 2089
Query: 375 LKRFVKALEKVRNINSRRPRQLPSDANFEAEKVLL---------------NRQTSEERKK 419
L+ K KV+ + +P ++ EA +L T EER
Sbjct: 2090 LEEETKETHKVQAEDIFSTETVPKESFIEAPVSMLASGEDEPVTPQEGDYAANTQEERHV 2149
Query: 420 SEEW-----------MLDYALQKVISKLAPAQRQRVTLLVEAFETIRPVQDAENGPQTSA 468
S E +K ++ ++ + V E P ++AE+G +T +
Sbjct: 2150 SAETEEKVGETKPKESQAEGAEKSDDQVEDESTKKTDVEVAGLENDYPTEEAEHGDETYS 2209
Query: 469 TVESHANLIQSLDASSNHSKEEINDRRDFEVT--------ERARNDKNMDACKKNDESAT 520
T+ L Q ++ ++ IND EV+ E + D +++ +K+ S
Sbjct: 2210 TLPVVGILTQL--QTTLETERAINDSASSEVSMIKEPADQEEKKGDDVVESNEKDFVSDI 2267
Query: 521 VKSTATKAVKFPVCDTGIMEEEVTAEGEYKVQEKSIV---KEDLKHGTSTTDVPYGVQER 577
+++ K + I EE A ++E ++ KE++K T ++
Sbjct: 2268 LEAKRLHGDKSGEAEK-IKEESGLAGKSLPIEEINLQEEHKEEVKVQEETREI------- 2319
Query: 578 DQKYIKKWHLMYKQAVLSNTGKYDNKLPVVGKDKEGREQGDAVFNGGNNSSCHNYNETDS 637
+ + + ++ + LS + + V+ +K+ + FNG + E
Sbjct: 2320 -AQVLPREEILISSSPLS----AEEQEHVISDEKQEEREPQQDFNGSTSEKISLQVEHLK 2374
Query: 638 DMDEEKK--------------NVIELVQKAFDEILLPETEDLSSD---DRSKSRSYGSDE 680
D + KK ++++ K D+ E E +SS+ D +R
Sbjct: 2375 DFETSKKEQKDETHETVKEEDQIVDIKDKKKDD---EEQEIVSSEVKKDNKDARELEVGN 2431
Query: 681 LLEKSEGEREEMNATSFTETPKEAKKTENKPKSWSHLKKLIMLKRFVKALDKVRNINPRR 740
+GE+EE+ + E +E + K S +I + D V +I
Sbjct: 2432 DFVSRDGEKEEVPHNAL-ENEEEMNEVVASEKQISDPVGVIKKASEAEHEDPVDDIKSND 2490
Query: 741 PRELPSDANFEGEKVFLNRQTSEERKKSEEWMLDYALQKVISKLAPAQRQRVTLLIEAFE 800
R+ P++ + +S+E D + P + L + + +
Sbjct: 2491 DRDFPTE--------------QAPKDQSDEVSAD--------ETVPKEAIGEELKVPSSK 2528
Query: 801 TLRPIQDAENGLRSSATVESLENPLQSLDASSVLSAKTLLGKVSFSNDSTMEFSDKASDN 860
L IQ+ N + A + L + S ++ + +V +++ EF+ D
Sbjct: 2529 VLDDIQENSN---TEAVTNFADRDLPVQNLSELIQSHQSPNQV---EETSFEFNKAQEDK 2582
Query: 861 PMPELCKPIKPVETISSCHEEAPTKRMVDEVPED--LVSDLNTKTKDVIGGHGEQFSVTK 918
+ I V+ E+ + E+ E ++DL +D+ G V
Sbjct: 2583 KEETVDALITNVQVQDQPKEDFEAAAIEKEISEQEHKLNDLTDVQEDI----GTYVKVQ- 2637
Query: 919 SLILNGIVRSLRSNLVVPEAPSNRLDEPTTDIKDVVEKDQLEKSEAPTSAVVESKNQLEK 978
VP+ DE D D V + E S +E K ++E
Sbjct: 2638 ----------------VPD------DEIKGDGHDSVAAQKEETSS------IEEKREVEH 2669
Query: 979 QGSTGLWFTVFKHMVSDMTENNSKTSTDVADEKDSKYEDITTREISVSYENTPVVIQDMP 1038
+ KH VS +NN+ + D K+ + E+ + T +V +++
Sbjct: 2670 VKAE--MEDAIKHEVSVEEKNNTSENIDHEAAKEIEQEE---------GKQTNIVKEEIR 2718
Query: 1039 FKDRAVVDAEVELRQIEAIKMVEDAIDSILPDTQPLPDNSTIDRTGGIYSEGLNQKEQKM 1098
+++ E+ +K +DAID P+ + + S++ +T ++ E +
Sbjct: 2719 EEEK-----EINQESFNNVKETDDAIDKTQPEIRDIESLSSVSKT-------QDKPEPEY 2766
Query: 1099 ESGNGIVEERKEESVSKEVNKPNQKLSRNWSNLKKVVLLRRFIKALEKVRKFNPRE--PR 1156
E N E E S E +K ++L + + L +K E K R+
Sbjct: 2767 EVPNQQKREITNEVPSLENSKIEEELQKKDEESENTKDLFSVVKETEPTLKEPARKSLSD 2826
Query: 1157 YLPLEPDSEDEK 1168
++ EP +E+++
Sbjct: 2827 HIQKEPKTEEDE 2838
Score = 41.6 bits (96), Expect = 0.003
Identities = 158/899 (17%), Positives = 343/899 (37%), Gaps = 100/899 (11%)
Query: 294 KKNVIELVQKAFDEILLPEVEDLSSEGHSKSRGNETDEVLLEKSGGKIEERNTTTFTESP 353
+K+ +E+ + +L E+ + G++ G ++ SG K E E+
Sbjct: 28 EKSSMEVQKVNLSTVLDDEITSGADRGNNIITGEADVAEVVNGSGDKETEELKRENGEAT 87
Query: 354 KEVPKMESKQKSWSHLKKVILLK----RFVKALEKVRNINSRRPRQLPSDANFEAEKVLL 409
K + ++ + + +++ + K EK+ + + D + E+V +
Sbjct: 88 KTISVLDDSKIVKDDQESIVVREPQSLNEKKEDEKIILSDVTLENKKEEDTTGKPEEVSV 147
Query: 410 NRQTSEERKKSEEWMLDYA-----LQKVISKLAPAQRQRVTL---LVEAFETIRPVQDAE 461
+ EE + + L+ + KV++ P + + L E I D
Sbjct: 148 EKPVIEEDQTEAKHSLEQEEDIGNISKVLTDTTPVKVDEYDIEKSLNSVCEEIPIKTDEV 207
Query: 462 NGPQTSATVESHANLIQSLDASSNHSKEEINDRRDFEVTERARNDKNMDACKKNDESATV 521
S TVE+ N ++ + ++ S EEI+ D V E D+ + + T+
Sbjct: 208 REETDSRTVETSVNGTEA-EHNATVSVEEISRNGDNTVNETVSEDQTATDGEPLHDVETI 266
Query: 522 KSTATKAVKFPVCDTGIMEEEVTAEGEYKVQEKSIVKEDLKHGTSTTDVPYGVQERDQKY 581
K A K V D I+ E T E K+ ++ +E+ T ++ ++
Sbjct: 267 KREAEPFYKTVVEDAKIVNTEETTAHESKILKEDNHQEEYAESVEATKNSDAAEQSSRE- 325
Query: 582 IKKWHLMYKQAVLSNTGKYDNKLPVVGKDKEGREQGDAVFNGGNNSSCHNYNETDSDMDE 641
V + K ++ + + + +E ++ G + + + +MD+
Sbjct: 326 -----------VTVDKEKEEDIIQNIEEVQESPSVMESPTIQGEDIESKASLDHEEEMDK 374
Query: 642 EKKNVIELVQKAFDEILLPETEDLSSDDR-SKSRSYGSDELLEKSEGEREEMNATSFTET 700
K+ E ++ +P++E L ++ + +++ S E+L+ + E E +A E
Sbjct: 375 ITKDTEEQEHVLVRDVPVPQSETLVTEAKTAETFSVQEAEILKTNINESEAHSAIGGEED 434
Query: 701 PKEAKKT--------ENKPKSWSHLKKLIMLKRFVKALDKVRNINPRRPR----ELPSDA 748
+E K+ ++K + S K I+ V++ D + E+ D
Sbjct: 435 GQETKENTEPSKDLKDDKEQEDSETVKTIISSDEVRSSDVQAEVFGEHTEPCSSEIKDDR 494
Query: 749 NFEGEKVFLN-RQTSEERKKSEEWMLDYALQKVISKLAPAQRQ--RVTLLIEAFETLRPI 805
+ E + + ++T E + + + + ++ L A+++ + + I+ +L I
Sbjct: 495 HGRDESIEVKAKETGPEIETAVDGESVHEIETTERVLLEAEKEEDKEEIKIDEEPSLNAI 554
Query: 806 QDAENGLRSSATVESLENPLQSLDASSVLSAKTLLGKVSFSNDSTMEFSDKASDNPMPEL 865
+ AE + + E + + + +SV +++L N +E + K SD
Sbjct: 555 EKAET---ENVKIVIEEPEIVNNEETSVHESESLK-----ENAEPVE-AVKNSDG----- 600
Query: 866 CKPIKPVETISSCHEE--APTKRMVDEVPEDLVSDLNTKTK-DVIGGHGEQFSVTKSLIL 922
+ I T+ EE AP + E P + + L K + D I E+ +L
Sbjct: 601 TEQISREVTVDRAKEEDIAPKTEEIQERPSESKASLEPKEEVDHISNETEE----HEHVL 656
Query: 923 NGIVRSLRSNLVVPEAPSNRLD-EPTTDIKDVVEKDQLE-------KSEAPTSAVVE--- 971
V+ + + EA + D +P+ D+K+ E ++ E E +SAV E
Sbjct: 657 ERDVQQCET--IESEAVETKEDTQPSLDLKEDKETEEAETFKTVFSSDEVRSSAVQEEQF 714
Query: 972 --------SKNQLEKQGSTGLWFTVFKHMVSDMTENNSKTSTDVADEKDSKYEDITTREI 1023
S+ + E G + V D + V + KY+ +
Sbjct: 715 GEHTEPCSSEIKDESHGKEESVEVKSQETVQDENTEDKHDVLKVPSTESEKYQGNEPETV 774
Query: 1024 SV----SYENTPVVIQDMPFK--DRAVVDAEVELRQIEAIKMVEDAI--DSILPDTQPLP 1075
V SYE + D+ + D ++ + Q++ +++E+ I DS + + +
Sbjct: 775 LVSNTGSYEKSEKSPSDLVLNVDKEELNDEKINVDQVDGTQIMEEPIGLDSNGAEAEQID 834
Query: 1076 DNSTIDRTGGIYSEGLN---------QKEQKMESGNGIVEERKEESVSKEVNKPNQKLS 1125
N T + + ++ ++ ++ K+ES + + EE + K KP ++++
Sbjct: 835 QNITNETEEILVAKPVSLLDVKSVEQMQKPKLESPSEVSEETSKTVDEKIEEKPEEEVT 893
>At4g37820 unknown protein
Length = 532
Score = 46.2 bits (108), Expect = 1e-04
Identities = 96/477 (20%), Positives = 189/477 (39%), Gaps = 79/477 (16%)
Query: 252 NTGKCDNKVPLVEKEKEGGEE--DNEGNNSYRNYSETDSDMDDEKKNVIELVQKAFDEIL 309
N G+ D + P +E+ K+ +E D EG+ + E D+ +K+N E+V++ +E
Sbjct: 63 NLGRKDLR-PRIEETKDVKDEVEDEEGSKN-----EGGGDVSTDKENGDEIVEREEEEKA 116
Query: 310 LPEVEDLSSEGHSKSRGNETDEVLLEKSGGKIEERNTTTFTESPKEVPKMESKQKSWS-- 367
+ E + +EG GNE + + G+ E+ +E E+ E+++ ++
Sbjct: 117 VEENNEKEAEGTGNEEGNE------DSNNGESEK--VVDESEGGNEISNEEAREINYKGD 168
Query: 368 HLKKVILLKRFVKALEKVR-----NINSRRPRQLPSDANFEAEKVLLNRQTSE-ERKKSE 421
++ K+ EKV NS + D + +VL E +E
Sbjct: 169 DASSEVMHGTEEKSNEKVEVEGESKSNSTENVSVHEDESGPKNEVLEGSVIKEVSLNTTE 228
Query: 422 EWMLDYALQKVISKLAPAQRQR--VTLLVEAFETIRPVQDAENGPQTSATVESHANL--- 476
D Q+ S+L ++ E ET +A ++S + ES ++
Sbjct: 229 NGSDDGEQQETKSELDSKTGEKGFSDSNGELPETNLSTSNATETTESSGSDESGSSGKST 288
Query: 477 -IQSLDASSNHSKEEINDRRDFEVTERARNDKNMDACKKNDESATVKSTATKAVKFPVCD 535
Q + ++ + + +V E +N+K DA DES K K
Sbjct: 289 GYQQTKNEEDEKEKVQSSEEESKVKESGKNEK--DASSSQDESKEEKPERKK-------- 338
Query: 536 TGIMEEEVTAEGEYKVQEKSIVKEDLKHGTSTTDVPYGVQERDQKYIKKWHLMYKQAVLS 595
+EE +++GE K +E K + + +S + E +K +
Sbjct: 339 ----KEESSSQGEGKEEEPE--KREKEDSSSQEESKEEEPENKEKE-------------A 379
Query: 596 NTGKYDNKLPVVGKDKEGREQGDAVFNGGNNSSCHNYNETDSDMDEEKKNVIELVQKAFD 655
++ + +N++ K+ E +E+ ++ GN + ET+ E ++ +K +
Sbjct: 380 SSSQEENEI----KETEIKEKEESSSQEGNENK-----ETEKKSSESQRKENTNSEKKIE 430
Query: 656 EILLPETEDLSSDDRSKSRSYGSDELLEKSEGEREEMNATSFTETPKEAKKTENKPK 712
++ ++ + D K+ E +RE N TS ET ++ KTE++ K
Sbjct: 431 QVESTDSSNTQKGDEQKT-----------DESKRESGNDTSNKETEDDSSKTESEKK 476
Score = 38.9 bits (89), Expect = 0.018
Identities = 52/251 (20%), Positives = 89/251 (34%), Gaps = 30/251 (11%)
Query: 114 HGDLPPLKRFVSMRRRQLKSQKSTKKDGRSKQVGNARKGTQKTKTVHSEDGNSQQNVKNV 173
+G+LP S +S S + K G Q+TK E Q + +
Sbjct: 256 NGELPETNLSTSNATETTESSGSDESGSSGKSTGY-----QQTKNEEDEKEKVQSSEEES 310
Query: 174 SMESSPFKPHDAPPSTVNECDTSTKDKHMVTDYEVLQKSSTQEEPKPGSTTSVAYGVQER 233
++ S DA S+ + K + + Q +EEP+ + + +
Sbjct: 311 KVKESGKNEKDA--SSSQDESKEEKPERKKKEESSSQGEGKEEEPEKREKEDSSSQEESK 368
Query: 234 DQKYIKKWHLMYKHAVLSNTGKCDNKVPLVEKEKEGGEEDNEGNNSYRNYSETDSDMDDE 293
+++ K K A S + + EKE+ +E NE + + SE+ +
Sbjct: 369 EEEPENK----EKEASSSQEENEIKETEIKEKEESSSQEGNENKETEKKSSESQRKENTN 424
Query: 294 KKNVIELVQKAFDEILLPEVEDLSSEGHSKSRGNETDEVLLEKSGGKIEERNTTTFTESP 353
+ IE +VE S K +TDE K E N T+ E+
Sbjct: 425 SEKKIE------------QVESTDSSNTQKGDEQKTDE-------SKRESGNDTSNKETE 465
Query: 354 KEVPKMESKQK 364
+ K ES++K
Sbjct: 466 DDSSKTESEKK 476
Score = 35.4 bits (80), Expect = 0.20
Identities = 54/314 (17%), Positives = 120/314 (38%), Gaps = 50/314 (15%)
Query: 477 IQSLDASSNHSKEEINDRRDF--EVTERARNDKNMDACKKNDESATVKSTATKAVKFPVC 534
++ + S N +++ ++ E+ ER +K ++ + + T +
Sbjct: 83 VEDEEGSKNEGGGDVSTDKENGDEIVEREEEEKAVEENNEKEAEGTGNEEGNED-----S 137
Query: 535 DTGIMEEEVT-AEGEYKVQEKSIVKEDLKHGTSTTDVPYGVQERDQKYIKKWHLMYKQAV 593
+ G E+ V +EG ++ + + + K ++++V +G +E+ + ++
Sbjct: 138 NNGESEKVVDESEGGNEISNEEAREINYKGDDASSEVMHGTEEKSNEKVE---------- 187
Query: 594 LSNTGKYDNKLPVVGKDKEGREQGDAVFNGGNNSSCHNYNETDSDMDEEKKNVIELVQKA 653
+ K ++ V + E + + + N E SD E+++ EL K
Sbjct: 188 VEGESKSNSTENVSVHEDESGPKNEVLEGSVIKEVSLNTTENGSDDGEQQETKSELDSKT 247
Query: 654 FDEIL------LPETEDLSSDDRSKSRSYGSDEL-----------LEKSEGEREEMNATS 696
++ LPET +S+ + S GSDE + E E+E++ ++
Sbjct: 248 GEKGFSDSNGELPETNLSTSNATETTESSGSDESGSSGKSTGYQQTKNEEDEKEKVQSSE 307
Query: 697 FTETPKEAKKTENKPKSWSHLKKLIMLKRFVKALDKVRNINPRRPRELPSDANFEGEKVF 756
KE+ K E S + D+ + P R ++ S + EG++
Sbjct: 308 EESKVKESGKNEKDASS---------------SQDESKEEKPERKKKEESSSQGEGKEEE 352
Query: 757 LNRQTSEERKKSEE 770
++ E+ EE
Sbjct: 353 PEKREKEDSSSQEE 366
Score = 33.5 bits (75), Expect = 0.76
Identities = 43/218 (19%), Positives = 87/218 (39%), Gaps = 8/218 (3%)
Query: 132 KSQKSTKKDGRSKQVGNARKGTQKTKTVHSEDGNSQQNV---KNVSMESSPFKPHDAPPS 188
+S K+ K S+ K +K K S G ++ + SS + + P
Sbjct: 314 ESGKNEKDASSSQDESKEEKPERKKKEESSSQGEGKEEEPEKREKEDSSSQEESKEEEPE 373
Query: 189 TVNECDTSTKDKHMVTDYEVLQK--SSTQEEPKPGSTTSVAYGVQERDQKYIKKWHLMYK 246
+ +S+++++ + + E+ +K SS+QE + T + Q ++ +K +
Sbjct: 374 NKEKEASSSQEENEIKETEIKEKEESSSQEGNENKETEKKSSESQRKENTNSEKKIEQVE 433
Query: 247 HAVLSNTGKCDNKVPLVEKEKEGGEEDNEGNNSYRNYSETDSDMDDEKKNVIELVQKAFD 306
SNT K D + K + G + N+ + S+T+S+ +E E + +
Sbjct: 434 STDSSNTQKGDEQKTDESKRESGNDTSNKETED--DSSKTESEKKEENNRNGETEETQNE 491
Query: 307 EILLPEVEDLSSEGHSKSRGNETDEVLLEKSGGKIEER 344
+ ++S K + E L E S G I ++
Sbjct: 492 QEQTKSALEISHTQDVKDARTDL-ETLPETSNGLISDK 528
Score = 33.1 bits (74), Expect = 1.00
Identities = 43/207 (20%), Positives = 79/207 (37%), Gaps = 32/207 (15%)
Query: 926 VRSLRSNLVVPEAPSNRLDEPTTDIKDVVEKDQLEKSEAPTS---AVVESKNQLEKQGST 982
V+S V E+ N D ++ + EK + +K E +S E + EK+ S+
Sbjct: 303 VQSSEEESKVKESGKNEKDASSSQDESKEEKPERKKKEESSSQGEGKEEEPEKREKEDSS 362
Query: 983 GLWFTVFKHMVSDMTENNSKTSTDVADEKDSKYEDITTREISVSYENTPVVIQDMPFKDR 1042
+ + EN K ++ +E + K +I +E S S E
Sbjct: 363 SQ-----EESKEEEPENKEKEASSSQEENEIKETEIKEKEESSSQEGNE----------- 406
Query: 1043 AVVDAEVELRQIEAIKMVEDAIDSILPDTQPLPDNSTIDRTGGIYSEGLNQK--EQKMES 1100
+ E E + E+ + + + + ++T +G QK E K ES
Sbjct: 407 ---NKETEKKSSESQRKENTNSEKKIEQVESTDSSNT--------QKGDEQKTDESKRES 455
Query: 1101 GNGIVEERKEESVSKEVNKPNQKLSRN 1127
GN + E+ SK ++ ++ +RN
Sbjct: 456 GNDTSNKETEDDSSKTESEKKEENNRN 482
Score = 33.1 bits (74), Expect = 1.00
Identities = 25/104 (24%), Positives = 47/104 (45%), Gaps = 5/104 (4%)
Query: 125 SMRRRQLKSQKSTKKDGRSKQVGNARKGTQK--TKTVHSEDGNSQQNVKNV-SMESSPFK 181
S ++K + +K+ S Q GN K T+K +++ E+ NS++ ++ V S +SS +
Sbjct: 382 SQEENEIKETEIKEKEESSSQEGNENKETEKKSSESQRKENTNSEKKIEQVESTDSSNTQ 441
Query: 182 PHDAPPS--TVNECDTSTKDKHMVTDYEVLQKSSTQEEPKPGST 223
D + + E T +K D + +E + G T
Sbjct: 442 KGDEQKTDESKRESGNDTSNKETEDDSSKTESEKKEENNRNGET 485
>At5g41140 putative protein
Length = 983
Score = 44.3 bits (103), Expect = 4e-04
Identities = 91/458 (19%), Positives = 192/458 (41%), Gaps = 38/458 (8%)
Query: 271 EEDNEGNNSYRNYSETDSDMDDEKKNVIELVQKAFD----EILLPEVEDLSSEGHSKSRG 326
E + E + SETD D D +K + ELV+ D +L + DL +E R
Sbjct: 437 ERNTEESRRMSCTSETDDDED--QKALDELVKGHMDAKEAHVLERRITDLYNEIEIYKRD 494
Query: 327 NETDEVLLEKSGG-----KIEERNTTTFTESPKEVPKMESKQKSWSHLKKVILLKRFVKA 381
E E+ +E+ K E + + E + +++ + + S L V L+ V++
Sbjct: 495 KEDLEIQVEQLSLDYEILKQENHDISYKLEQSQVQEQLKMQYECSSSLVNVNELENHVES 554
Query: 382 LEKVRNINSRRPRQLPSDANFEAEKVLLNRQTSEERKKSEEWMLDYALQKVISKLAPAQR 441
LE ++ + S++ + +++ + EE + + + + ++ V ++ Q
Sbjct: 555 LEA----KLKKQYKECSESLYRIKELETQIKGMEEELEKQAQIFEGDIEAV-TRAKVEQE 609
Query: 442 QRVTLLVEAFETIRPVQDAENGPQTSATVESHANLIQSLDASSNHSKEEINDRRDFEVTE 501
QR EA R + G + +L A+ + + + + R+ + +
Sbjct: 610 QRAIEAEEALRKTRWKNASVAGKIQDEFKRISEQMSSTLAANEKVTMKAMTETRELRMQK 669
Query: 502 RARNDKNMDACKKNDESATVKSTATKAVKFPVCDTGIMEEEV---TAEGEYKVQEKSIVK 558
R + M+A NDE + + T + +E+ +A+ EY+ ++K V
Sbjct: 670 RQLEELLMNA---NDELRVNRVEYEAKLNELSGKTDLKTKEMKRMSADLEYQKRQKEDVN 726
Query: 559 EDLKHGTSTTDVPYGVQERDQKYIKKWHLMYKQAVLSNTGK--YDNKLPVVGKDKEGREQ 616
DL H + + D + +K M +A LS + D K V+ K E
Sbjct: 727 ADLTHEITRRKDEIEILRLDLEETRK-SSMETEASLSEELQRIIDEKEAVITALKSQLET 785
Query: 617 GDAVFNGGNNSSCHNYNETDSDMDEEKKNVIEL---VQKAFDEILLPETEDLSSDDRSKS 673
A + +S +N +S+++ +K V+++ ++K +E+ E + S+D+ +K+
Sbjct: 786 AIAPCDNLKHSLSNN----ESEIENLRKQVVQVRSELEKKEEEMANLENREASADNITKT 841
Query: 674 RSYGSDELLEKSEGERE------EMNATSFTETPKEAK 705
+++ +++ EG+ + E ++ F E K+ K
Sbjct: 842 EQRSNEDRIKQLEGQIKLKENALEASSKIFIEKEKDLK 879
>At5g07820 putative protein
Length = 561
Score = 43.9 bits (102), Expect = 6e-04
Identities = 70/340 (20%), Positives = 135/340 (39%), Gaps = 38/340 (11%)
Query: 489 EEINDRRDFEVTERARNDKNMDACKKNDESATVKSTATKAVKFPV-CDTGIMEEEVTAEG 547
+++ ++ D EV + +R +N K+ + T+K+ + PV CD + + + A+
Sbjct: 216 DDVLEKTDLEVKKVSRISEN-----KSSKEDTLKNKEKAKIDEPVRCDDVLEKTSLDAQK 270
Query: 548 EYKVQEKSIVKED-LKHGTSTTDVPYGVQERDQKYIKKWHLMYKQAVLSNTGKYDNKLPV 606
++ E KE+ LK+ + R ++K + + +V K K
Sbjct: 271 VSRISENKNSKEERLKNLKNKEKTNIDEPVRPDDAVEKTLYVVESSVEKKKKKMSTKSVK 330
Query: 607 VGKDKEGREQGDAVFNGGNNSSCHNYNETDSDMDEEKKNVIELVQKAFDEILLPETEDLS 666
+ + ++ E+ G + S S+ V D + +T S
Sbjct: 331 ISETQQSSEKKIIRSTGKKSLSLLPSLPPPSE-----------VVTGSDPRPIRQTTSRS 379
Query: 667 SDDRSKSRSYGSDELLEKSEGE---REEMNATSFTETPKEAKKTEN-------KPKSWSH 716
+ + GS L+ + E R + T P K+ N +PK
Sbjct: 380 KTSLPEKKQSGSANLVTNPKPESKIRPKRIGLKVTPPPPPTKQQMNFKKGKVLEPKPEDS 439
Query: 717 LKKLIMLKRFVKALDKVR--NINPRRP-----RELPSDANFEG--EKVFLNRQTSEERKK 767
I K+ V K+R ++N ++ RE N EG EKV L + E +KK
Sbjct: 440 TTTSIKFKKIVVQEPKLRTSDVNKKKKSLKDKREGVGKINGEGKREKVVLRHRKVEVKKK 499
Query: 768 SEEWMLDYALQKVISKLAPAQRQRVTLLIEAFETLRPIQD 807
+ + + +++ ++KL ++ +V L+ AFET+ +QD
Sbjct: 500 LQT-LFNNVIEETVNKLEEVRKSKVKALVGAFETVISLQD 538
Score = 38.9 bits (89), Expect = 0.018
Identities = 21/55 (38%), Positives = 34/55 (61%), Gaps = 1/55 (1%)
Query: 1163 DSEDEKVQLRHQDMAERKGTEEWMLDYALRQVVSKLTPARKRKVELLVEAFETVV 1217
+ + EKV LRH+ + +K + + + + + V+KL RK KV+ LV AFETV+
Sbjct: 481 EGKREKVVLRHRKVEVKKKLQT-LFNNVIEETVNKLEEVRKSKVKALVGAFETVI 534
Score = 38.9 bits (89), Expect = 0.018
Identities = 25/106 (23%), Positives = 55/106 (51%), Gaps = 4/106 (3%)
Query: 357 PKMESKQKSWSHLKKVILLKRFVKALE---KVRNINSRRPRQLPSDANFEAEKVLLNRQT 413
PK E + KK+++ + ++ + K +++ +R + + EKV+L +
Sbjct: 434 PKPEDSTTTSIKFKKIVVQEPKLRTSDVNKKKKSLKDKREGVGKINGEGKREKVVLRHRK 493
Query: 414 SEERKKSEEWMLDYALQKVISKLAPAQRQRVTLLVEAFETIRPVQD 459
E +KK + + + +++ ++KL ++ +V LV AFET+ +QD
Sbjct: 494 VEVKKKLQT-LFNNVIEETVNKLEEVRKSKVKALVGAFETVISLQD 538
>At5g55660 putative protein
Length = 759
Score = 43.1 bits (100), Expect = 0.001
Identities = 70/303 (23%), Positives = 126/303 (41%), Gaps = 41/303 (13%)
Query: 455 RPVQDAENGPQTSATVESHANLIQSL---DASSNHSKEEINDRRDFEVTERARNDKNMDA 511
+ VQ+ +G + + + ++ + D H E ++ EVTE + D D
Sbjct: 30 KEVQENASGKEVQESKKEEDTGLEKMEIDDEGKQHEGESETGDKEVEVTEEEKKDVGED- 88
Query: 512 CKKNDESATV-KSTATKAVKFPVCDTGIMEEEVTAEGEYKVQEKSIVKEDLKHGTSTTDV 570
K+ E+ + + T K +K +G+ EE V ++S+ D K +
Sbjct: 89 -KEQPEADKMDEDTDDKNLKADDGVSGVATEEDA------VMKESVESADNKDAEN---- 137
Query: 571 PYGVQERDQKYIKKWHLMYKQAVLSNTGKYDNKLPVVGKDK----EGREQGDAVFNGGNN 626
P G QE++ K K L +A + G + KL VG DK + E+ + V
Sbjct: 138 PEGEQEKESKEEK---LEGGKANGNEEGDTEEKL--VGGDKGDDVDEAEKVENVDEDDKE 192
Query: 627 SSCHNYNETDSDMDEEKKNVIELVQKAFDE--------ILLPETEDLSSDDRSKSRSYGS 678
+ NE + +EE+ N E V++A E + PE ED ++ + ++
Sbjct: 193 EALKEKNEAEL-AEEEETNKGEEVKEANKEDDVEADTKVAEPEVEDKKTESKDENEDK-E 250
Query: 679 DELLEKSEGEREEMNATSFTETPKEAKKTENKPKSWSHLKKLIMLKRFVKALDKVRNINP 738
+E ++ E E+EE N KE KK + K + K + K+ ++ ++I P
Sbjct: 251 EEKEDEKEDEKEESN------DDKEDKKEDIKKSNKRGKGKTEKTRGKTKSDEEKKDIEP 304
Query: 739 RRP 741
+ P
Sbjct: 305 KTP 307
Score = 42.0 bits (97), Expect = 0.002
Identities = 137/703 (19%), Positives = 258/703 (36%), Gaps = 118/703 (16%)
Query: 133 SQKSTKKDGRSKQVGNARK----GTQKTKTVHS---EDGNSQQNVKNVSMESSPFKP--H 183
S K +++ K+V ++K G +K + +G S+ K V + K
Sbjct: 28 SGKEVQENASGKEVQESKKEEDTGLEKMEIDDEGKQHEGESETGDKEVEVTEEEKKDVGE 87
Query: 184 DAPPSTVNECDTSTKDKHMVTDYEV----LQKSSTQEEPKPGSTTSVAYGVQERDQKYIK 239
D ++ D T DK++ D V ++ + +E + A + +K K
Sbjct: 88 DKEQPEADKMDEDTDDKNLKADDGVSGVATEEDAVMKESVESADNKDAENPEGEQEKESK 147
Query: 240 KWHLMYKHAVLSNTGKCDNK---------VPLVEKEKEGGEEDNEGNNSYRNYSETDSDM 290
+ L A + G + K V EK + E+D E +N +E +
Sbjct: 148 EEKLEGGKANGNEEGDTEEKLVGGDKGDDVDEAEKVENVDEDDKEEALKEKNEAELAEEE 207
Query: 291 DDEKKNVIELVQKAFD-----EILLPEVEDLSSEGHSKSRGNETDEVLLEKSGGKIEERN 345
+ K ++ K D ++ PEVED +E SK + +E EK K +E+
Sbjct: 208 ETNKGEEVKEANKEDDVEADTKVAEPEVEDKKTE--SKDENEDKEE---EKEDEKEDEKE 262
Query: 346 TTTFTESPKEVPKMESKQKSWSHLKKVILLKRFVKALEKVRNINSRRPRQLPSDANFEAE 405
+ + K+ +S ++ +K + K+ E+ ++I + P SD
Sbjct: 263 ESNDDKEDKKEDIKKSNKRGKGKTEKT---RGKTKSDEEKKDIEPKTP--FFSDRPVRER 317
Query: 406 KVLLNRQTSEERKKSEEWMLDYA----LQKVISKLAPAQRQRVTLLVEAFETIRPVQDAE 461
K + ++ S E+ ++ L+ + + R++ + + TI
Sbjct: 318 KSVERLVAVVDKDSSREFHVEKGKGTPLKDIPNVAYKVSRKKSDEVFKQLHTI-----LF 372
Query: 462 NGPQTSAT-VESHANLIQSLDASSNHSKEEINDRRDFEVTERARNDKNMDACKKNDESAT 520
G + AT +++H + K ++ + FE + +K ++ C D S
Sbjct: 373 GGKRVKATQLKAHILRFSGYKWQGDEEKAKLKVKEKFE---KINKEKLLEFCDLFDISVA 429
Query: 521 VKST-----ATKAVKF---PVCDTGIMEEEVTAEGEYKVQEKSIVKEDLKHGTSTTDVPY 572
+T TK V+F P T ++ E E V+ K K+ S++
Sbjct: 430 KATTKKEDIVTKLVEFLEKPHATTDVLVNEK----EKGVKRKRTPKKSSPAAGSSSS--- 482
Query: 573 GVQERDQKYIKKWHLMYKQAVLSNTGKYDNKLPVVGKDKEGREQGDAVFNGGNNSSCHNY 632
+R K KK + NK V D E E
Sbjct: 483 ---KRSAKSQKK----------TEEATRTNKKSVAHSDDESEE----------------- 512
Query: 633 NETDSDMDEEKKNVIELVQKAFDEILLPETEDLSSDDRSKSRSYGSDELLEKSEGEREEM 692
E + D +EEK+ +E ++ +E +P+ S+D + S + + + E E E
Sbjct: 513 -EKEDDEEEEKEQEVE-EEEEENENGIPD----KSEDEAPQLSESEENVESEEESEEETK 566
Query: 693 NATSFTETPKEAKKTENKPKSWSHLKKLIMLKRF--VKALDKVRNINPRRPRELPSDANF 750
+ T + K++ K +S KK + + K + R+ R+ + SD
Sbjct: 567 KKKRGSRTSSDKKESAGKSRS----KKTAVPTKSSPPKKATQKRSAGKRKKSDDDSDT-- 620
Query: 751 EGEKVFLNRQTSEERKKSEEWMLDYALQ--KVISKLAPAQRQR 791
+ + S +RKK+E+ + A K +SK P +R
Sbjct: 621 -------SPKASSKRKKTEKPAKEQAAAPLKSVSKEKPVIGKR 656
>At5g16720 putative protein
Length = 600
Score = 42.0 bits (97), Expect = 0.002
Identities = 52/241 (21%), Positives = 99/241 (40%), Gaps = 26/241 (10%)
Query: 231 QERDQKYIKKWHLMYKHAVLSNTGKCDNKVPLVEKEKEGGEEDNEGNNSYR-NYSETDSD 289
+E++++ +++ +Y+ VL K NK+ +VE + E ++D E N N SE D D
Sbjct: 363 REKEKEQLQRELEVYRAKVLEYESKAKNKIIVVENDCEADDDDKEEENREEDNSSEMDVD 422
Query: 290 MDDEKKNVIELVQKAFDEILLPEVEDLSSEGHSKSRGNETDEVLLEKSGGKIEERNTTTF 349
+ ++I L V+ +S G S S E V+L++ +E+R T
Sbjct: 423 L---------------EKITLDCVQHMSMLGESLSEFEEERLVILDQL-KVLEDRLVTMQ 466
Query: 350 TESPKEVP-----KMESKQKSWSHLKKVILLKRFVKALEKVRNINSRRPRQLP--SDANF 402
+ E P E L + K + L+ N + + LP + NF
Sbjct: 467 DKESAEDPGEFSNSYEEASNGHGGLTMASMAKSLLPLLDAAENESEDGSQGLPESDEKNF 526
Query: 403 --EAEKVLLNRQTSEERKKSEEWMLDYALQKVISKLAPAQRQRVTLLVEAFETIRPVQDA 460
++EK+ + +Q ++ +E D K A + +L + + +R ++
Sbjct: 527 GSDSEKLEIIKQVDSVYERLQELETDGEFLKNCMSSAKKGDKGTDILKDILQHLRDLRTI 586
Query: 461 E 461
E
Sbjct: 587 E 587
>At2g03150 Unknown protein (T18E12.18; At2g03150)
Length = 1336
Score = 42.0 bits (97), Expect = 0.002
Identities = 98/495 (19%), Positives = 181/495 (35%), Gaps = 81/495 (16%)
Query: 503 ARNDKNMDACKKNDESATVKSTATKAVKFPVCDTGIMEEEVTAEGEYKVQEKSIVKE-DL 561
A D MD K D + K K D+G +E + TAE V++++ K D+
Sbjct: 839 AEIDNKMDGDSKKDGDSDEKKVMEVGKKSS--DSGSVEMKPTAESLEDVKDENASKTVDV 896
Query: 562 KHGTSTTDVPYGVQERDQKYIKKWHLMYKQAVLSNTGKYDNKLPVVGKDKEGREQGDA-V 620
K T + D K+ S++ K D K G+DK+ ++ ++
Sbjct: 897 KQETGSPDTK------------------KKEGASSSSKKDTK---TGEDKKAEKKNNSET 935
Query: 621 FNGGNNSSCHNYNETDSDMDEEKKNVIELVQKAFDEILLPETEDL--------------S 666
+ G +N +E + +K + E K I + + + +
Sbjct: 936 MSEGKKIDRNNTDEKEVKEKVTEKEIKERGGKDESRIQVKDRKKCEEPPRAGFILQTKRN 995
Query: 667 SDDRSKSRSYGSDELLEKSEGEREEMN--ATSFTETPKEAKKTENKPKSWSHLKKLIMLK 724
D + +S S D LL+ ++ + +E + + F E+ E + + + + LKKL +
Sbjct: 996 KDSKLRSLSASLDSLLDYTDKDLDESSFEISLFAESLYEMLQYQMGSRIFEFLKKLRV-- 1053
Query: 725 RFVKALDKVRNINPRRPRELPSDANFEGEKVFLNRQTSEERKKSEEWMLDYALQKVISKL 784
+ + RN R EL N E + ++T+E K VIS+
Sbjct: 1054 ----KIVRQRNQRKRHQEELSVKQN-EAKSQDKRQKTAEHEDKEAS---------VISES 1099
Query: 785 APAQRQRVTLLIEAFETLRPIQDAENGLRSSATVESLENPLQSLDASSVLSAKTLLGKVS 844
AP + + T E + R I D E ++ T+ S E T+ V+
Sbjct: 1100 APGKDDKETSGKETVDGSREIADKEAVAKTKETLGSKE--------------VTVGEAVN 1145
Query: 845 FSNDSTMEFSDKASDNPMPELCKPIKPVETISSCHEEAPTKRMVDEVPEDLVSDLNTKTK 904
++ E D D+P + P E EE P + + D+ + +
Sbjct: 1146 MEVENQDEEDDDGDDDPEED------PEEDPEEDPEEDPEEDPEECEEMDVANTEQEEPA 1199
Query: 905 DVIGGHGEQFSVTKSLILNGIVRSLRSNLVVPEAPSNRLDEPTTDIKDVVEKDQLEKSEA 964
+ E T + + I + N P ++ T+IK E ++ K +
Sbjct: 1200 EEPQKKEENLEKTSGTVADPITEAETDNRKEERGP----NDSKTEIKPKSETEKHGKQDG 1255
Query: 965 PTSAVVESKNQLEKQ 979
TS + + ++K+
Sbjct: 1256 GTSDAAKREETVDKE 1270
Score = 39.7 bits (91), Expect = 0.011
Identities = 132/630 (20%), Positives = 239/630 (36%), Gaps = 103/630 (16%)
Query: 8 KLLTVKGPKSTTKLYSESTDGIDGNNRNSTSDAGNKSQRVMTRRLSLKPVRISAKKPSLH 67
+LL+ + K T ++ ++ G+ + + K+ + + +R+ +PV
Sbjct: 709 RLLSQEVKKDTVEVTKDAEKKSPGDTSGTPTTGTKKTVKKIIKRVVKRPVNDG------- 761
Query: 68 KATCSSTIKDSHFPNHIDLPQE--------GSSSQGVSAVKVCTYAYCSLHGHHHGDLPP 119
KAT K S P H+ +P+ G+SS KV S + P
Sbjct: 762 KATGMKGEK-SDVPEHVAIPETTVPKEESTGTSSNKKIVKKVAETGDTSDPSAKANEQTP 820
Query: 120 LKRFVSMRRRQLKSQKSTKKDGRSK--QVGNARKGTQKTKTVHSEDGNS-QQNVKNVSME 176
K V +K K+ + K ++ N G K +DG+S ++ V V +
Sbjct: 821 AKTIVK--------KKIIKRVAKRKVAEIDNKMDGDSK------KDGDSDEKKVMEVGKK 866
Query: 177 SSPFKPHDAPPSTVNECDTSTKDKHMVTDYEVLQKSSTQE-EPKPGSTTSVAYGVQERDQ 235
SS + P+ E KD++ +V Q++ + + + K G+++S + +
Sbjct: 867 SSDSGSVEMKPTA--ESLEDVKDENASKTVDVKQETGSPDTKKKEGASSSSKKDTKTGED 924
Query: 236 KYIKKWHLMY-----KHAVLSNTGKCDNKVPLVEKE-KEGGEEDNEGNNSYRNYSETDSD 289
K +K + K +NT + + K + EKE KE G +D E+
Sbjct: 925 KKAEKKNNSETMSEGKKIDRNNTDEKEVKEKVTEKEIKERGGKD-----------ESRIQ 973
Query: 290 MDDEKKNVIELVQKAFDEILLPEVEDLSSEGHSKSRGNETDEVLLEKSGGKIEERN--TT 347
+ D KK E +A +L + S+ S S ++ LL+ + ++E + +
Sbjct: 974 VKDRKK--CEEPPRA--GFILQTKRNKDSKLRSLSASLDS---LLDYTDKDLDESSFEIS 1026
Query: 348 TFTESPKEVPKMESKQKSWSHLKKVILLKRFVKALEKVRNINSRRPRQLPSDANFEAEKV 407
F ES E+ + + + + LKK+ ++ VR N R+ Q K
Sbjct: 1027 LFAESLYEMLQYQMGSRIFEFLKKL--------RVKIVRQRNQRKRHQEELSVKQNEAKS 1078
Query: 408 LLNRQTSEERKKSEEWMLDYALQKVISKLAPAQRQRVTLLVEAFETIRPVQDAENGPQTS 467
RQ + E + E VIS+ AP + + T E + R + D E +T
Sbjct: 1079 QDKRQKTAEHEDKEA--------SVISESAPGKDDKETSGKETVDGSREIADKEAVAKTK 1130
Query: 468 ATVESHANLIQSLDASSNHSKEEINDRRDFEVTERARNDKNMDACKKNDESATVKSTATK 527
T+ S + +++E +D D + E D D + +E +
Sbjct: 1131 ETLGSKEVTVGEAVNMEVENQDEEDDDGDDDPEEDPEEDPEEDPEEDPEED------PEE 1184
Query: 528 AVKFPVCDTGIMEEEVTAEGEYKVQEKSIVKEDLKHGTSTTDVPYGVQERDQKYIKKWHL 587
+ V +T E+E AE K + E+L+ + T P E D +
Sbjct: 1185 CEEMDVANT---EQEEPAEEPQKKE------ENLEKTSGTVADPITEAETDNR------- 1228
Query: 588 MYKQAVLSNTGKYDNKLPVVGKDKEGREQG 617
K+ N K + K P +K G++ G
Sbjct: 1229 --KEERGPNDSKTEIK-PKSETEKHGKQDG 1255
Score = 31.2 bits (69), Expect = 3.8
Identities = 123/644 (19%), Positives = 239/644 (37%), Gaps = 105/644 (16%)
Query: 520 TVKSTATKAVKFPVCD---TGIMEEEVTAEGEYKVQEKSIVKEDLKHGTSTTDVPYGVQE 576
TVK + VK PV D TG+ E+ + E ++ KE+ GTS+
Sbjct: 745 TVKKIIKRVVKRPVNDGKATGMKGEKSDVPEHVAIPETTVPKEE-STGTSS--------- 794
Query: 577 RDQKYIKKWHLMYKQAVLSNTGKYDNKLP---VVGKDKEGREQGDAVFNGGNNSSCHNYN 633
++K +KK + + K + + P +V K R V N
Sbjct: 795 -NKKIVKK--VAETGDTSDPSAKANEQTPAKTIVKKKIIKRVAKRKVAEIDNKMD----G 847
Query: 634 ETDSDMDEEKKNVIELVQKAFDE--ILLPETEDLSSDDRSKSRSYGSDELLEKSEGEREE 691
++ D D ++K V+E+ +K+ D + + T + D + ++ S D E + ++
Sbjct: 848 DSKKDGDSDEKKVMEVGKKSSDSGSVEMKPTAESLEDVKDENASKTVDVKQETGSPDTKK 907
Query: 692 MNATSFTETPKEAKKTENKPKSWSHLKKLIMLKRFVKALDKVRNINPRRPRELPSDANFE 751
S + + K+ K E+K +K N S+ E
Sbjct: 908 KEGAS-SSSKKDTKTGEDKKA------------------EKKNN----------SETMSE 938
Query: 752 GEKVFLNRQTSEERKKSEEWMLDYALQKVISKLAPAQRQRVTLLIEAFETLRPIQDAENG 811
G+K+ +R ++E++ E+ +K I + R+ + + + E
Sbjct: 939 GKKI--DRNNTDEKEVKEK-----VTEKEIKERGGKDESRIQV--------KDRKKCEEP 983
Query: 812 LRSSATVESLENPLQSLDASSVLSAKTLLGKVSFSNDSTMEFSDKASDNPMPELCKPIKP 871
R+ +++ N + L +S S DS ++++DK D E+ +
Sbjct: 984 PRAGFILQTKRNK------------DSKLRSLSASLDSLLDYTDKDLDESSFEISLFAES 1031
Query: 872 VETISSCHEEAPTKRMVDEVPEDLVSDLNTKTKDVIGGHGEQFSVTKSLILNGIVRSLRS 931
+ + + + ++ +V N + + H E+ SV ++ + R +
Sbjct: 1032 LYEMLQYQMGSRIFEFLKKLRVKIVRQRNQRKR-----HQEELSVKQNEAKSQDKRQKTA 1086
Query: 932 NLVVPEAPSNRLDEPTTDIKDVVEKDQLEKSE--APTSAVVESKNQL-EKQGSTGLWFTV 988
EA P D K+ K+ ++ S A AV ++K L K+ + G
Sbjct: 1087 EHEDKEASVISESAPGKDDKETSGKETVDGSREIADKEAVAKTKETLGSKEVTVG----- 1141
Query: 989 FKHMVSDMTENNSKTSTDVADEKDSKYEDITTREISVSYENTPVVIQDMPFKDRAVVD-A 1047
V+ EN + D D+ + E+ + E P ++M + + A
Sbjct: 1142 --EAVNMEVENQDEEDDDGDDDPEEDPEEDPEEDPEEDPEEDPEECEEMDVANTEQEEPA 1199
Query: 1048 EVELRQIEAIKMVEDAIDSILPDTQPLPDNSTIDRTGG-----IYSEGLNQKEQKMESGN 1102
E ++ E ++ + P T+ DN +R I + +K K + G
Sbjct: 1200 EEPQKKEENLEKTSGTVAD--PITEAETDNRKEERGPNDSKTEIKPKSETEKHGKQDGGT 1257
Query: 1103 GIVEERKEESVSKEVNKPNQKLSRNWSNLKKVVLLRRFIKALEK 1146
+R EE+V KE+ + + RN + +V +R I +L K
Sbjct: 1258 SDAAKR-EETVDKELLQAFRFFDRNQAGYVRVEDMRVTIHSLGK 1300
>At5g24880 glutamic acid-rich protein
Length = 443
Score = 41.6 bits (96), Expect = 0.003
Identities = 93/464 (20%), Positives = 181/464 (38%), Gaps = 87/464 (18%)
Query: 132 KSQKSTKKDGRSKQVGNARKGTQKTKTVHSEDGNSQQNVKNVSMESSPFKPHDAPPSTVN 191
K++ + R+K +G RK Q T SEDG+S Q + P+ P+ +
Sbjct: 9 KAKPDNQNLSRTKSLG--RKPKQPLSTASSEDGSSDQKPEKP-------LPNYLKPTISS 59
Query: 192 ECDTS---TKDKHMVTDYEVLQKSSTQEEPKPGSTTSVAYGVQERDQKYIKKWHLMYKHA 248
D K+ + + D + L + + + P P S TS + R QK +
Sbjct: 60 RPDAGKLLKKNNNSLDDNQKLLRRRSFDRP-PSSLTSPSTSASPRIQKSLN--------- 109
Query: 249 VLSNTGKCDNKVPLVEKEK----------EGGEEDNEGNNSYR-----------NYSETD 287
+S + D P V +EK G +G N+ + N S T
Sbjct: 110 -VSPSRSRDR--PAVPREKPVTALRSSSFHGSRNVPKGGNTAKSPPVAPKKSGLNSSSTS 166
Query: 288 SDMDDEKKNVIELVQKAFDEILLPEVEDLSSEGHSKSRGNETDEVLLEKSGGKIEERNTT 347
S E + + + + EI L S S ++ + +E+L K+E +
Sbjct: 167 SKSKKEGSENVRIKKASDKEIA------LDSASMSSAQEDHQEEIL------KVESDHLQ 214
Query: 348 TFTESPKEVPKMESKQKSWSHLKKVILLKRFVKALEKV-RNINSRRPRQLPSDANFEAEK 406
++ E PK E ++K + ++ V+A E V S P + S +
Sbjct: 215 V-SDHDIEEPKYEKEEKE--------VQEKVVQANESVEEKAESSGPTPVASPVGKDCNA 265
Query: 407 VLLNRQTSEERKKSEEWMLDYALQKVISKLAPAQRQRVTLLVEAFETIRPVQDAENGPQT 466
V+ + E+ K+E+ +++ ++ + E + + + + E T
Sbjct: 266 VV--AELEEKLIKNED-----DIEEKTEEMKEQDNNQANKSEEEEDVKKKIDENE----T 314
Query: 467 SATVESHANLIQSLDASSNHSKEEINDRRDFEVTERARNDKNMDACKKNDESATVKSTAT 526
V++ + ++S++ ++ +EE+ + V E +K + K++D+ V+
Sbjct: 315 PEKVDTESKEVESVEETTQEKEEEVKEEGKERVEE---EEKEKEKVKEDDQKEKVEEEEK 371
Query: 527 KAVKFPVCDTGIMEEEVTAEGEYKVQEKSIVKEDLKHGTSTTDV 570
+ VK + EEE +AEG ++K +VK + ++ DV
Sbjct: 372 EKVKGDEEKEKVKEEE-SAEG----KKKEVVKGKKESPSAYNDV 410
Score = 37.7 bits (86), Expect = 0.041
Identities = 32/144 (22%), Positives = 69/144 (47%), Gaps = 11/144 (7%)
Query: 447 LVEAFETIRPVQDAENGPQTSATVESHANLI-----QSLDASSNHSKEEINDRRDFEVTE 501
+V+A E++ ++ ++ V N + + L + + +E+ + ++ + +
Sbjct: 236 VVQANESVEEKAESSGPTPVASPVGKDCNAVVAELEEKLIKNEDDIEEKTEEMKEQDNNQ 295
Query: 502 RARNDKNMDACKKNDESATVKS--TATKAVKFPVCDTGIMEEEVTAEGEYKVQEKSIVKE 559
++++ D KK DE+ T + T +K V+ T EEEV EG+ +V+E+ KE
Sbjct: 296 ANKSEEEEDVKKKIDENETPEKVDTESKEVESVEETTQEKEEEVKEEGKERVEEEEKEKE 355
Query: 560 DLKHGTSTTDVPYGVQERDQKYIK 583
+K D V+E +++ +K
Sbjct: 356 KVKE----DDQKEKVEEEEKEKVK 375
>At5g57120 unknown protein
Length = 330
Score = 41.2 bits (95), Expect = 0.004
Identities = 42/195 (21%), Positives = 81/195 (41%), Gaps = 6/195 (3%)
Query: 540 EEEVTAEGEYKVQEKSIVKEDLKHGTSTTDVPYGVQERDQKYIKKWHLMYKQAVLSNTGK 599
+ E A G + V+ K + V+ +++ +K+ + + V K
Sbjct: 68 DHEAAANGNTEANVVEAVENVKKDKKKKKNKETKVEVTEEEKVKETDAVIEDGVKEKKKK 127
Query: 600 YDNKLPVVGKDKEGREQGDAVFNGGNNSSCHNYNETDS-DMDEEKKNVIELVQKAFDEIL 658
+ K+ V ++K ++ DAV G +++ S + D++K+ V + +++ E
Sbjct: 128 KETKVKVTEEEKV--KETDAVIEDGVKEKKKKKSKSKSVEADDDKEKVSKKRKRSEPEET 185
Query: 659 LPETEDLSSDDRSKSRSYGSDELLEKSEGEREEMNATSFTETPKEAKKTENKPKSWSHLK 718
ETED DD R + ++E EG +E + T+ A+K+E K + K
Sbjct: 186 KEETED---DDEESKRRKKEENVVENDEGVQETPVKETETKENGNAEKSETKSTNQKSGK 242
Query: 719 KLIMLKRFVKALDKV 733
L K K +V
Sbjct: 243 GLSNSKEPKKPFQRV 257
Score = 32.3 bits (72), Expect = 1.7
Identities = 53/256 (20%), Positives = 96/256 (36%), Gaps = 57/256 (22%)
Query: 465 QTSATVESHANLIQSLDASSNHSKEEINDRRDFEVTERARNDKNMDACKKNDESATVKST 524
+ +A + AN++++++ K++ N EVTE + VK T
Sbjct: 70 EAAANGNTEANVVEAVENVKKDKKKKKNKETKVEVTEEEK----------------VKET 113
Query: 525 ATKAVKFPVCDTGIMEEEVTAEGEYKVQEKSIVKEDLKHGTSTTDVPYGVQERDQKYIKK 584
V + G+ E++ E + KV E+ VKE + + GV+E+ +K K
Sbjct: 114 DA------VIEDGVKEKKKKKETKVKVTEEEKVKE------TDAVIEDGVKEKKKKKSK- 160
Query: 585 WHLMYKQAVLSNTGKYDNKLPVVGKDKEGREQGDAVFNGGNNSSCHNYNETDSDMDEEKK 644
S + + D+ V K ++ E + ET+ D +E K+
Sbjct: 161 ----------SKSVEADDDKEKVSKKRKRSEPEE------------TKEETEDDDEESKR 198
Query: 645 -----NVIELVQKAFDEILLPETEDLSSDDRSKSRSYGSDELLEKSEGEREEMNATSFTE 699
NV+E + E + ETE + + KS + +++ K +E
Sbjct: 199 RKKEENVVE-NDEGVQETPVKETETKENGNAEKSETKSTNQKSGKGLSNSKEPKKPFQRV 257
Query: 700 TPKEAKKTENKPKSWS 715
E TEN +S
Sbjct: 258 NVDEIVYTENSNSYYS 273
Score = 32.0 bits (71), Expect = 2.2
Identities = 34/160 (21%), Positives = 65/160 (40%), Gaps = 8/160 (5%)
Query: 230 VQERDQKYIKKWHLMYKHAVLSNTGKCDNKVPLVEKEK----EGGEEDNEGNNSYRNYSE 285
V+ +++ +K+ + + V K + KV + E+EK + ED +
Sbjct: 102 VEVTEEEKVKETDAVIEDGVKEKKKKKETKVKVTEEEKVKETDAVIEDGVKEKKKKKSKS 161
Query: 286 TDSDMDDEKKNVIELVQKAFDEILLPEVEDLSSEGHSKSRGNETDEVLLEKSGGKIEERN 345
+ DD+K+ V + +++ E E ED E S+ + +E ++E G E
Sbjct: 162 KSVEADDDKEKVSKKRKRSEPEETKEETEDDDEE----SKRRKKEENVVENDEGVQETPV 217
Query: 346 TTTFTESPKEVPKMESKQKSWSHLKKVILLKRFVKALEKV 385
T T+ K E+K + K + K K ++V
Sbjct: 218 KETETKENGNAEKSETKSTNQKSGKGLSNSKEPKKPFQRV 257
>At5g65910 unknown protein
Length = 432
Score = 39.3 bits (90), Expect = 0.014
Identities = 42/156 (26%), Positives = 68/156 (42%), Gaps = 15/156 (9%)
Query: 154 QKTKTVHSEDGNSQQNVKNVSMESSPFKPHDAPPSTVNECDTST--KDKHMVTDYEV--L 209
QK + E G+S+ N V + P P A VN ++S DKH + E+ +
Sbjct: 252 QKRSKLPVEAGSSEANTVIVEPLTVPPSPETAAVKIVNPVESSDVETDKHPIESKEIQIV 311
Query: 210 QKSSTQEEPKPGSTTSVAYGVQ--ERDQKYIKKW-HLMYKHAVLSNTGKCDNKVPLVEKE 266
KS +E +++S VQ + D W + +V + G+ P E E
Sbjct: 312 DKSVIEERSTSTASSSRFINVQVDDEDDDDADDWLNDEETSSVSAIGGRSTTNHPFGEDE 371
Query: 267 KEGG------EEDNEGN--NSYRNYSETDSDMDDEK 294
++ E+DNEG+ SY+N + + SD D+K
Sbjct: 372 EDVSFSDLEEEDDNEGDVKVSYKNLTNSGSDSSDKK 407
>At5g45520 putative protein
Length = 1167
Score = 38.9 bits (89), Expect = 0.018
Identities = 50/225 (22%), Positives = 92/225 (40%), Gaps = 19/225 (8%)
Query: 481 DASSNHSKEEINDRRDFEVTERARNDKNMDACKKNDESATVKSTATKAVKFPVCDTGIME 540
D + E ++D E++++D+ ++ KK D+ + V+ + D G
Sbjct: 678 DGDKGKADLEEEKKQDEVEAEKSKSDEIVEGEKKPDDKSKVEKKGDGDKENADLDEGKKR 737
Query: 541 EEVTAEGEYKVQEKSIVKEDLKHGT---------STTDVPYGVQ---ERDQKYIKKWHLM 588
+EV A+ K + +V+ D K + TD V+ +RD+ +
Sbjct: 738 DEVEAK---KSESGKVVEGDGKESPPQESIDTIQNMTDDQTKVEKEGDRDKGKVDPEEGK 794
Query: 589 YKQAVLSNTGKYDNKLPVVGKDKEGREQGDAVFNGGNNSSCHNYNETDSDMDEEKKNVIE 648
V K DN + V K RE DA+ N ++ + E D ++EK N+ E
Sbjct: 795 KHDEVEGGIWKSDNGVEGVDKASPSRESTDAIENKPDDHQRGDKQEEKGDGEKEKVNLEE 854
Query: 649 LVQKAFDEIL--LPETEDLSSDDRSKSRSYGSDELLEKSEGEREE 691
K DEI + ++++ D KS S + +E +++E
Sbjct: 855 --WKKHDEIKEESSKQDNVTGGDVKKSPPKESKDTMESKRDDQKE 897
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.308 0.126 0.342
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 27,910,894
Number of Sequences: 26719
Number of extensions: 1304846
Number of successful extensions: 5350
Number of sequences better than 10.0: 183
Number of HSP's better than 10.0 without gapping: 19
Number of HSP's successfully gapped in prelim test: 165
Number of HSP's that attempted gapping in prelim test: 4852
Number of HSP's gapped (non-prelim): 525
length of query: 1222
length of database: 11,318,596
effective HSP length: 111
effective length of query: 1111
effective length of database: 8,352,787
effective search space: 9279946357
effective search space used: 9279946357
T: 11
A: 40
X1: 16 ( 7.1 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 42 (21.6 bits)
S2: 66 (30.0 bits)
Medicago: description of AC125478.8


