

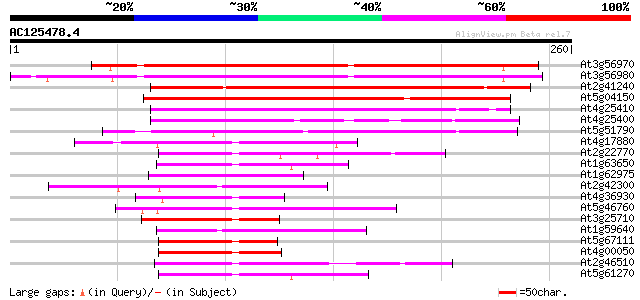
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC125478.4 + phase: 0
(260 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
At3g56970 putative bHLH transcription factor (bHLH038) 175 2e-44
At3g56980 putative bHLH transcription factor (bHLH039) 173 9e-44
At2g41240 putative bHLH transcription factor (bHLH100) 148 3e-36
At5g04150 myc - like protein 134 6e-32
At4g25410 putative protein 56 2e-08
At4g25400 putative bHLH transcription factor (bHLH118) 56 2e-08
At5g51790 putative protein 52 3e-07
At4g17880 putative transcription factor BHLH4 51 7e-07
At2g22770 putative bHLH transcription factor (bHLH020) 48 5e-06
At1g63650 putative transcription factor (BHLH2) 48 6e-06
At1g62975 putative bHLH transcription factor (bHLH125) 47 8e-06
At2g42300 putative bHLH transcription factor (bHLH048) 47 1e-05
At4g36930 putative bHLH transcription factor (AtbHLH024) / SPATU... 47 1e-05
At5g46760 putative transcription factor BHLH5 46 2e-05
At3g25710 putative bHLH transcription factor (bHLH032) 45 4e-05
At1g59640 putative bHLH transcription factor (bHLH031) 45 4e-05
At5g67111 putative bHLH transcription factor (bHLH073/ALCATRAZ) 44 7e-05
At4g00050 bHLH like transcription factor (bHLH016) 44 7e-05
At2g46510 putative bHLH transcription factor 44 7e-05
At5g61270 bHLH transcription factor like protein 44 9e-05
>At3g56970 putative bHLH transcription factor (bHLH038)
Length = 253
Score = 175 bits (443), Expect = 2e-44
Identities = 95/213 (44%), Positives = 145/213 (67%), Gaps = 11/213 (5%)
Query: 39 YSLPYHQ----FSSPKEHVEIERPPSPKLMAKKLNHNASERDRRKKINSLISSLRSLLPG 94
Y + +HQ S E EI+ P ++ KKLNHNASERDRRKKIN+L SSLRS LP
Sbjct: 44 YEVTHHQNSLGVSVSSEGNEIDNNP---VVVKKLNHNASERDRRKKINTLFSSLRSCLPA 100
Query: 95 EDQTKKMSIPVTISRVLKYIPDLQKQVQGLTKKKEELLSRISHRQEYAVNKESQRKKIPN 154
DQ+KK+SIP T+S+ LKYIP+LQ+QV+ L +KKEE+L R+S ++++ + + Q K + +
Sbjct: 101 SDQSKKLSIPETVSKSLKYIPELQQQVKRLIQKKEEILVRVSGQRDFELYDKQQPKAVAS 160
Query: 155 YNSAFVVSTSRLNDTELVIHISSYEANKIPLSEILMCLENNGLLLLNSSSSKTFGGRLFY 214
Y S VS +RL D E+++ +SS + + +S +L +E +G +L++ SSS++ G RLFY
Sbjct: 161 YLS--TVSATRLGDNEVMVQVSSSKIHNFSISNVLGGIEEDGFVLVDVSSSRSQGERLFY 218
Query: 215 NLHFQVDKTQRYE--CDDLIQKLSSIYEKQQNN 245
LH QV+ Y+ C++L +++ +YEK +N+
Sbjct: 219 TLHLQVENMDDYKINCEELSERMLYLYEKCENS 251
>At3g56980 putative bHLH transcription factor (bHLH039)
Length = 258
Score = 173 bits (438), Expect = 9e-44
Identities = 109/265 (41%), Positives = 155/265 (58%), Gaps = 25/265 (9%)
Query: 1 MVAFCPPQFSYSNMGW-LLEELEPESLISHKEKNYASLEYSLPYHQF------------- 46
M A PP F N GW E + L N L++ +P +
Sbjct: 1 MCALVPPLFP--NFGWPSTGEYDSYYLAGDILNNGGFLDFPVPEETYGAVTAVTQHQNSF 58
Query: 47 --SSPKEHVEIERPPSPKLMAKKLNHNASERDRRKKINSLISSLRSLLPGEDQTKKMSIP 104
S E EI+ P ++ KKLNHNASERDRR+KINSL SSLRS LP Q+KK+SIP
Sbjct: 59 GVSVSSEGNEIDNNP---VVVKKLNHNASERDRRRKINSLFSSLRSCLPASGQSKKLSIP 115
Query: 105 VTISRVLKYIPDLQKQVQGLTKKKEELLSRISHRQEYAVNKESQRKKIPNYNSAFVVSTS 164
T+SR LKYIP+LQ+QV+ L KKKEELL +IS ++ + K + NY S VS +
Sbjct: 116 ATVSRSLKYIPELQEQVKKLIKKKEELLVQISGQRNTECYVKQPPKAVANYIS--TVSAT 173
Query: 165 RLNDTELVIHISSYEANKIPLSEILMCLENNGLLLLNSSSSKTFGGRLFYNLHFQVDKTQ 224
RL D E+++ ISS + + +S +L LE + +L++ SSS++ G RLFY LH QV+K +
Sbjct: 174 RLGDNEVMVQISSSKIHNFSISNVLSGLEEDRFVLVDMSSSRSQGERLFYTLHLQVEKIE 233
Query: 225 RYE--CDDLIQKLSSIYEKQQNNHL 247
Y+ C++L Q++ +YE+ N+++
Sbjct: 234 NYKLNCEELSQRMLYLYEECGNSYI 258
>At2g41240 putative bHLH transcription factor (bHLH100)
Length = 181
Score = 148 bits (374), Expect = 3e-36
Identities = 79/176 (44%), Positives = 123/176 (69%), Gaps = 2/176 (1%)
Query: 66 KKLNHNASERDRRKKINSLISSLRSLLPGEDQTKKMSIPVTISRVLKYIPDLQKQVQGLT 125
KKLNHNASER+RRKKIN++ SSLRS LP +QTK +S+ T+S+ LKYIP+LQ+QV+ L
Sbjct: 2 KKLNHNASERERRKKINTMFSSLRSCLPPTNQTK-LSVSATVSQALKYIPELQEQVKKLM 60
Query: 126 KKKEELLSRISHRQEYAVNKESQRKKIPNYNSAFVVSTSRLNDTELVIHISSYEANKIPL 185
KKKEEL +IS +++ ++ + + + A VS++RL++TE+++ ISS + K
Sbjct: 61 KKKEELSFQISGQRDLVYTDQNSKSEEGVTSYASTVSSTRLSETEVMVQISSLQTEKCSF 120
Query: 186 SEILMCLENNGLLLLNSSSSKTFGGRLFYNLHFQVDKTQRYECDDLIQKLSSIYEK 241
+L +E +GL+L+ +SSS++ G RLFY++H Q+ K + ++L +L +YEK
Sbjct: 121 GNVLSGVEEDGLVLVGASSSRSHGERLFYSMHLQI-KNGQVNSEELGDRLLYLYEK 175
>At5g04150 myc - like protein
Length = 240
Score = 134 bits (336), Expect = 6e-32
Identities = 70/170 (41%), Positives = 116/170 (68%), Gaps = 2/170 (1%)
Query: 63 LMAKKLNHNASERDRRKKINSLISSLRSLLPGEDQTKKMSIPVTISRVLKYIPDLQKQVQ 122
++ KKLNHNASERDRR+K+N+L SSLR+LLP DQ +K+SIP+T++RV+KYIP+ ++++Q
Sbjct: 63 VLEKKLNHNASERDRRRKLNALYSSLRALLPLSDQKRKLSIPMTVARVVKYIPEQKQELQ 122
Query: 123 GLTKKKEELLSRISHRQEYAVNKESQRKKIPNYNSAFVVSTSRLNDTELVIHISSYEANK 182
L+++KEELL RIS + + + +S+ ++ + L DTE+ + I++ +
Sbjct: 123 RLSRRKEELLKRISRKTHQEQLRNKAMMDSIDSSSSQRIAANWLTDTEIAVQIATSKWTS 182
Query: 183 IPLSEILMCLENNGLLLLNSSSSKTFGGRLFYNLHFQVDKTQRYECDDLI 232
+ S++L+ LE NGL +++ SSS + R+FY LH Q+ + ++LI
Sbjct: 183 V--SDMLLRLEENGLNVISVSSSVSSTARIFYTLHLQMRGDCKVRLEELI 230
>At4g25410 putative protein
Length = 221
Score = 56.2 bits (134), Expect = 2e-08
Identities = 37/167 (22%), Positives = 85/167 (50%), Gaps = 4/167 (2%)
Query: 66 KKLNHNASERDRRKKINSLISSLRSLLPGEDQTKKMSIPVTISRVLKYIPDLQKQVQGLT 125
KKL H ER RR+++ +L ++LR+ LP + K ++ ++ + +I D + +++ L+
Sbjct: 43 KKLLHRDIERQRRQEMATLFATLRTHLPLKYIKGKRAVSDHVNGAVNFIKDTEARIKELS 102
Query: 126 KKKEELLSRISHRQEYAVNKESQRKKIPNYNSAFVVSTSRLNDTELVIHISSYEANKIPL 185
+++EL + + + A V+ ++ E+V+ +S +PL
Sbjct: 103 ARRDELSRETGQGYKSNPDPGKTGSDVGKSEPATVMVQPHVSGLEVVVSSNSSGPEALPL 162
Query: 186 SEILMCLENNGLLLLNSSSSKTFGGRLFYNLHFQVDKTQRYECDDLI 232
S++L ++ GL +++S +++ RL + + +V+ + C DL+
Sbjct: 163 SKVLETIQEKGLEVMSSFTTRV-NDRLMHTIQVEVNS---FGCIDLL 205
>At4g25400 putative bHLH transcription factor (bHLH118)
Length = 163
Score = 56.2 bits (134), Expect = 2e-08
Identities = 48/171 (28%), Positives = 83/171 (48%), Gaps = 12/171 (7%)
Query: 66 KKLNHNASERDRRKKINSLISSLRSLLPGEDQTKKMSIPVTISRVLKYIPDLQKQVQGLT 125
+KL H E+ RR+++ SL +SLRSLLP E K S + + YI LQ+ ++ +
Sbjct: 2 EKLVHKEIEKRRRQEMASLYASLRSLLPLEFIQGKRSTSDQVKGAVNYIDYLQRNIKDIN 61
Query: 126 KKKEELLSRISHRQEYAVNKESQRKKIPNYNSAFVVSTSRLNDTELVIHISSYEANKIPL 185
K+++L+ + + + + E + +I N+ VV L E+V+ I + P
Sbjct: 62 SKRDDLV--LLSGRSFRSSNEQEWNEISNH----VVIRPCLVGIEIVLSIL-----QTPF 110
Query: 186 SEILMCLENNGLLLLNSSSSKTFGGRLFYNLHFQVDKTQRYECDDLIQKLS 236
S +L L +GL +L S + RL + L +V+ + DL L+
Sbjct: 111 SSVLQVLREHGLYVLGYICS-SVNDRLIHTLQAEVNDLALIDLADLKDTLT 160
>At5g51790 putative protein
Length = 209
Score = 52.0 bits (123), Expect = 3e-07
Identities = 48/197 (24%), Positives = 88/197 (44%), Gaps = 15/197 (7%)
Query: 44 HQFSSPKEHVEIERPPSPKLMAKKLNHNASERDRRKKINSLISSLRSLLP-----GEDQT 98
HQ P+E E ++ KKL H ER RR+++ L +SLRS LP
Sbjct: 12 HQSHMPQERDETKKE-------KKLLHRNIERQRRQEMAILFASLRSQLPLKYIKALSSQ 64
Query: 99 KKMSIPVTISRVLKYIPDLQKQVQGLTKKKEELLSRISHRQEYAVNKESQRKKIPNYNSA 158
K ++ ++ + +I D Q +++ L+ +++EL I ++ A
Sbjct: 65 GKRAMSDHVNGAVSFIKDTQTRIKDLSARRDELKREIG--DPTSLTGSGSGSGSSRSEPA 122
Query: 159 FVVSTSRLNDTELVIHISSYEANKIPLSEILMCLENNGLLLLNSSSSKTFGGRLFYNLHF 218
V+ ++ E+V+ + PLS +L L GL +++S +++ RL Y +
Sbjct: 123 SVMVQPCVSGFEVVVSSLASGLEAWPLSRVLEVLHGQGLEVISSLTARV-NERLMYTIQV 181
Query: 219 QVDKTQRYECDDLIQKL 235
+V+ ++ L QKL
Sbjct: 182 EVNSFDCFDLAWLQQKL 198
>At4g17880 putative transcription factor BHLH4
Length = 589
Score = 50.8 bits (120), Expect = 7e-07
Identities = 40/141 (28%), Positives = 68/141 (47%), Gaps = 16/141 (11%)
Query: 31 EKNYASLEYSLPYHQFSSPKEHVEIERPPSPKLMAKK--------LNHNASERDRRKKIN 82
+ N++ LE S+ S+ V +E P+ +K LNH +ER RR+K+N
Sbjct: 373 DSNHSDLEASVAKEAESN---RVVVEPEKKPRKRGRKPANGREEPLNHVEAERQRREKLN 429
Query: 83 SLISSLRSLLPGEDQTKKMSIPVTISRVLKYIPDLQKQVQGLTKKKEELLSRISHRQEYA 142
SLR+++P + K S+ + + YI +L+ ++Q KEEL +I + A
Sbjct: 430 QRFYSLRAVVPNVSKMDKASL---LGDAISYISELKSKLQKAESDKEELQKQIDVMNKEA 486
Query: 143 VNKESQRK--KIPNYNSAFVV 161
N +S K K N S+ ++
Sbjct: 487 GNAKSSVKDRKCLNQESSVLI 507
>At2g22770 putative bHLH transcription factor (bHLH020)
Length = 320
Score = 48.1 bits (113), Expect = 5e-06
Identities = 40/153 (26%), Positives = 74/153 (48%), Gaps = 24/153 (15%)
Query: 70 HNASERDRRKKINSLISSLRSLLPGEDQTKKMSIPVTISRVLKYIPDLQKQVQGL----- 124
H +ER RR+K+N + +L +LLPG +T K ++ + +K++ LQ++V+ L
Sbjct: 133 HVLAERKRRQKLNERLIALSALLPGLKKTDKATV---LEDAIKHLKQLQERVKKLEEERV 189
Query: 125 -TKKKEELLSRISHRQEY--------------AVNKESQRKKIPNYNSAFVVSTSRLNDT 169
TKK ++ + + Q Y A S ++ + + +R++D
Sbjct: 190 VTKKMDQSIILVKRSQVYLDDDSSSYSSTCSAASPLSSSSDEVSIFKQTMPMIEARVSDR 249
Query: 170 ELVIHISSYEANKIPLSEILMCLENNGLLLLNS 202
+L+I + E NK + +IL LE L ++NS
Sbjct: 250 DLLIRVHC-EKNKGCMIKILSSLEKFRLEVVNS 281
>At1g63650 putative transcription factor (BHLH2)
Length = 596
Score = 47.8 bits (112), Expect = 6e-06
Identities = 30/91 (32%), Positives = 50/91 (53%), Gaps = 5/91 (5%)
Query: 69 NHNASERDRRKKINSLISSLRSLLPGEDQTKKMSIPVTISRVLKYIPDLQKQVQGLTKKK 128
NH SE+ RR+K+N +LRS++P + K+SI + ++Y+ DLQK+VQ L +
Sbjct: 405 NHALSEKKRREKLNERFMTLRSIIPSISKIDKVSI---LDDTIEYLQDLQKRVQELESCR 461
Query: 129 E--ELLSRISHRQEYAVNKESQRKKIPNYNS 157
E + +RI+ + + E +R NS
Sbjct: 462 ESADTETRITMMKRKKPDDEEERASANCMNS 492
>At1g62975 putative bHLH transcription factor (bHLH125)
Length = 259
Score = 47.4 bits (111), Expect = 8e-06
Identities = 26/72 (36%), Positives = 42/72 (58%)
Query: 65 AKKLNHNASERDRRKKINSLISSLRSLLPGEDQTKKMSIPVTISRVLKYIPDLQKQVQGL 124
+KK+ H ER RR++++SL LR+LLP + K S I + + YI DLQ +++ L
Sbjct: 73 SKKMKHRDIERQRRQEVSSLFKRLRTLLPFQYIQGKRSTSDHIVQAVNYIKDLQIKIKEL 132
Query: 125 TKKKEELLSRIS 136
+K+ + IS
Sbjct: 133 NEKRNRVKKVIS 144
>At2g42300 putative bHLH transcription factor (bHLH048)
Length = 327
Score = 47.0 bits (110), Expect = 1e-05
Identities = 34/137 (24%), Positives = 70/137 (50%), Gaps = 10/137 (7%)
Query: 19 EELEPESLISHKEKNYASLEYSLPYHQFSSP----KEHVEIERPPSPKLMAKKL----NH 70
E E +S++ ++ ++Y+S + + S K VE ++ P + A++ NH
Sbjct: 137 EPAETDSMVENQNQSYSSGKRKEREKKVKSSTKKNKSSVESDKLPYVHVRARRGQATDNH 196
Query: 71 NASERDRRKKINSLISSLRSLLPGEDQTKKMSIPVTISRVLKYIPDLQKQVQGLTKKKEE 130
+ +ER RR+KIN+ + L+ L+PG D K + + ++ ++ LQ+QV+ L+ +
Sbjct: 197 SLAERARREKINARMKLLQELVPGCD--KIQGTALVLDEIINHVQTLQRQVEMLSMRLAA 254
Query: 131 LLSRISHRQEYAVNKES 147
+ RI + + E+
Sbjct: 255 VNPRIDFNLDSILASEN 271
>At4g36930 putative bHLH transcription factor (AtbHLH024) / SPATULA
(SPT)
Length = 373
Score = 46.6 bits (109), Expect = 1e-05
Identities = 26/73 (35%), Positives = 43/73 (58%), Gaps = 7/73 (9%)
Query: 59 PSPKLMAKKLN----HNASERDRRKKINSLISSLRSLLPGEDQTKKMSIPVTISRVLKYI 114
PS + +K+ HN SE+ RR +IN + +L+SL+P ++T K S+ + ++Y+
Sbjct: 187 PSSRSSSKRCRAAEVHNLSEKRRRSRINEKMKALQSLIPNSNKTDKASM---LDEAIEYL 243
Query: 115 PDLQKQVQGLTKK 127
LQ QVQ LT +
Sbjct: 244 KQLQLQVQMLTMR 256
>At5g46760 putative transcription factor BHLH5
Length = 592
Score = 46.2 bits (108), Expect = 2e-05
Identities = 34/139 (24%), Positives = 65/139 (46%), Gaps = 12/139 (8%)
Query: 50 KEHVEIERPPS-PKLMAKK--------LNHNASERDRRKKINSLISSLRSLLPGEDQTKK 100
KE + +E P P+ +K LNH +ER RR+K+N SLR+++P + K
Sbjct: 387 KEAIVVEPPEKKPRKRGRKPANGREEPLNHVEAERQRREKLNQRFYSLRAVVPNVSKMDK 446
Query: 101 MSIPVTISRVLKYIPDLQKQVQGLTKKKEELLSRISHRQEYAVNKESQRKKIPNYNSAFV 160
S+ + + YI +L+ ++Q KEE+ ++ + N + + S+
Sbjct: 447 ASL---LGDAISYINELKSKLQQAESDKEEIQKKLDGMSKEGNNGKGCGSRAKERKSSNQ 503
Query: 161 VSTSRLNDTELVIHISSYE 179
ST+ + E+ + I ++
Sbjct: 504 DSTASSIEMEIDVKIIGWD 522
>At3g25710 putative bHLH transcription factor (bHLH032)
Length = 344
Score = 45.1 bits (105), Expect = 4e-05
Identities = 21/64 (32%), Positives = 43/64 (66%), Gaps = 3/64 (4%)
Query: 62 KLMAKKLNHNASERDRRKKINSLISSLRSLLPGEDQTKKMSIPVTISRVLKYIPDLQKQV 121
K +A +H+ +ER RR++IN+ ++ LRS+LP +T K S+ ++ V++++ +L++Q
Sbjct: 128 KALAASKSHSEAERRRRERINTHLAKLRSILPNTTKTDKASL---LAEVIQHMKELKRQT 184
Query: 122 QGLT 125
+T
Sbjct: 185 SQIT 188
>At1g59640 putative bHLH transcription factor (bHLH031)
Length = 264
Score = 45.1 bits (105), Expect = 4e-05
Identities = 29/97 (29%), Positives = 53/97 (53%), Gaps = 2/97 (2%)
Query: 69 NHNASERDRRKKINSLISSLRSLLPGEDQTKKMSIPVTISRVLKYIPDLQKQVQGLTKKK 128
+H+ +ER RR+KI+ + L+ L+PG + K + + + ++ YI LQ+QV+ L+ K
Sbjct: 146 SHSLAERARREKISERMKILQDLVPGCN--KVIGKALVLDEIINYIQSLQRQVEFLSMKL 203
Query: 129 EELLSRISHRQEYAVNKESQRKKIPNYNSAFVVSTSR 165
E + SR++ E KE ++ N F ++R
Sbjct: 204 EAVNSRMNPGIEVFPPKEFGQQAFENPEIQFGSQSTR 240
>At5g67111 putative bHLH transcription factor (bHLH073/ALCATRAZ)
Length = 210
Score = 44.3 bits (103), Expect = 7e-05
Identities = 22/55 (40%), Positives = 34/55 (61%), Gaps = 3/55 (5%)
Query: 70 HNASERDRRKKINSLISSLRSLLPGEDQTKKMSIPVTISRVLKYIPDLQKQVQGL 124
HN SE+ RR KIN + +L+ L+P ++T K S+ + ++Y+ LQ QVQ L
Sbjct: 98 HNLSEKKRRSKINEKMKALQKLIPNSNKTDKASM---LDEAIEYLKQLQLQVQTL 149
>At4g00050 bHLH like transcription factor (bHLH016)
Length = 399
Score = 44.3 bits (103), Expect = 7e-05
Identities = 22/57 (38%), Positives = 35/57 (60%), Gaps = 3/57 (5%)
Query: 70 HNASERDRRKKINSLISSLRSLLPGEDQTKKMSIPVTISRVLKYIPDLQKQVQGLTK 126
HN SER RR KIN + +L+ L+P +T K S+ + V++Y+ LQ QV +++
Sbjct: 218 HNQSERKRRDKINQRMKTLQKLVPNSSKTDKASM---LDEVIEYLKQLQAQVSMMSR 271
>At2g46510 putative bHLH transcription factor
Length = 662
Score = 44.3 bits (103), Expect = 7e-05
Identities = 33/138 (23%), Positives = 65/138 (46%), Gaps = 17/138 (12%)
Query: 68 LNHNASERDRRKKINSLISSLRSLLPGEDQTKKMSIPVTISRVLKYIPDLQKQVQGLTKK 127
LNH +ER RR+K+N +LRS++P + K S+ + + YI +LQ++V+ + +
Sbjct: 394 LNHVEAERQRREKLNQRFYALRSVVPNISKMDKASL---LGDAISYIKELQEKVKIMEDE 450
Query: 128 KEELLSRISHRQEYAVNKESQRKKIPNYNSAFVVSTSRLNDTELVIHISSYEANKIPLSE 187
+ +S V + + V +N+ +V IS +++ P S
Sbjct: 451 RVGTDKSLSESNTITVEESPE------------VDIQAMNEEVVVRVISPLDSH--PASR 496
Query: 188 ILMCLENNGLLLLNSSSS 205
I+ + N+ + L+ + S
Sbjct: 497 IIQAMRNSNVSLMEAKLS 514
>At5g61270 bHLH transcription factor like protein
Length = 366
Score = 43.9 bits (102), Expect = 9e-05
Identities = 27/110 (24%), Positives = 56/110 (50%), Gaps = 16/110 (14%)
Query: 70 HNASERDRRKKINSLISSLRSLLPGEDQTKKMSIPVTISRVLKYIPDLQKQVQGLTKKKE 129
HN SER RR +IN + +L+ LLP + K+SI + V++++ LQ QVQ ++ +
Sbjct: 171 HNESERRRRDRINQRMRTLQKLLPTASKADKVSI---LDDVIEHLKQLQAQVQFMSLRAN 227
Query: 130 -------------ELLSRISHRQEYAVNKESQRKKIPNYNSAFVVSTSRL 166
+ + I H+Q+ ++ Q+++ + + + + +R+
Sbjct: 228 LPQQMMIPQLPPPQSVLSIQHQQQQQQQQQQQQQQQQQFQMSLLATMARM 277
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.314 0.131 0.362
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 5,823,042
Number of Sequences: 26719
Number of extensions: 243170
Number of successful extensions: 1012
Number of sequences better than 10.0: 147
Number of HSP's better than 10.0 without gapping: 86
Number of HSP's successfully gapped in prelim test: 61
Number of HSP's that attempted gapping in prelim test: 899
Number of HSP's gapped (non-prelim): 153
length of query: 260
length of database: 11,318,596
effective HSP length: 97
effective length of query: 163
effective length of database: 8,726,853
effective search space: 1422477039
effective search space used: 1422477039
T: 11
A: 40
X1: 16 ( 7.2 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 42 (22.0 bits)
S2: 59 (27.3 bits)
Medicago: description of AC125478.4


